Abstract
We demonstrate sequential optical activation of two types of mRNAs in the same mammalian cell through the sequential photocleavage of small molecule caging groups (‘photo-cages’) tethered to the 5’ untranslated region (5’-UTR) of an mRNA. Synthetic ‘photo-cages’ were conjugated onto target mRNA using RNA-TAG, an enzymatic site-specific RNA modification technique. Translation of mRNA was severely reduced upon conjugation of the ‘photo-cages’ onto the 5’-UTR. However, subsequent photo-release of the ‘cages’ from the mRNA transcript triggered activation of translation with single-cell spatiotemporal resolution. To achieve sequential photo-activation of two mRNAs in the same cell, we synthesized a pair of ‘photo-cages’ which can be selectively cleaved from mRNA upon photo-irradiation with different wavelengths of light. Sequential photo-activation of two mRNAs enabled precise optical control of translation of two unique transcripts. We believe that this modular approach to precisely and rapidly control gene expression will serve as a powerful tool in future biological studies that require controlling translation of multiple transcripts with high spatiotemporal resolution.
Keywords: gene regulation, optogenetics, translation, RNA modification, multiplexing
Graphical Abstract
INTRODUCTION
The ability to precisely control gene expression is important for a wide range of applications in basic biological research, genetics, as well as gene therapies. [1–2] Stimuli-responsive control in confined time and space is enabled by inducible gene expression systems. For instance, small-molecule inducers, such as doxycycline, have been widely used in regulating synthetic gene circuits. [3] However, chemical inducers have slow diffusion rates, potential toxicity, and off-target effects on living systems, limiting their utility when high spatiotemporal resolution is required. [4] In contrast, optogenetic approaches utilize light as an external stimulus to regulate cellular processes. [5] Light irradiation is convenient to apply to biological samples such as live cells and organisms. The adverse effects of light-irradiation on living systems can be minimized by optimizing the wavelength and irradiation period of the light source. Perhaps most importantly, light can be applied with high spatial-temporal resolution, offering fast activation and deactivation dynamics. [6–7]
There has been an extensive body of research aimed at developing optogenetic tools to regulate gene expression. In a common strategy, a light-sensitive protecting group (‘photo-cage’) is chemically installed on a biologically relevant target (e.g. metabolites, oligonucleotides, or proteins) rendering the substrate inactive. Subsequently, light irradiation triggers the release of the ‘photo-cage’ from the target biological molecule, restoring its biological activity. [8–10] For instance, optogenetic approaches often involve the installation of light-responsive protein domains or amino acids to achieve photo-chemical manipulation of proteins such as nucleases, proteases and transcription factors. [11–16] Caged oligonucleotides have also been used to control gene expression at the level of transcription or translation. [17–24] By conjugating ‘photo-cages’ onto nucleotide bases, hybridization between the antisense oligonucleotide and the target DNA/RNA can be controlled precisely using light. While most studies have focused on optogenetic control of transcription, there are advantages to methods that optically control gene expression at the level of translation. [25–29, 41] Since mRNA can be processed by cellular translation machinery immediately upon cytoplasm entry, controlling gene expression through direct manipulation of mRNA provides more rapid changes in cellular protein concentration compared to the regulation of transcription. [30] Moreover, in-vitro transcribed mRNA (IVT-mRNA) is only transiently active in cytosol and is completely degraded via cellular metabolism, typically within 24 hours. Thus, unlike the use of plasmid DNA or viral vectors which may be integrated into the cellular genome, mRNA-based gene expression regulation systems do not pose the risk of insertional mutagenesis. [31] Thus, a technique that enables optogenetic manipulation of translation would benefit from the high spatial-temporal resolution inherent to optical control as well as fast dynamics of mRNA processing.
Previously, we demonstrated a technique which enabled precise optical control of mRNA translation through the conjugation of light-sensitive ‘cages’ onto IVT-mRNA. [32] Laser irradiation (405 nm) on live cells removed the ‘photo-cages’ from cytoplasmic mRNA, subsequently activating translation with single cell precision. We speculated we could significantly expand this tool by enabling sequential photo-activation of two mRNAs within the same cell. Here, leveraging the multiplex capability of our mRNA caging/uncaging platform, we describe a sequentially light-activated translation regulatory system that utilizes two ‘photo-cages’ to cage two types of mRNAs. Irradiation with longer wavelength light (456–488 nm) activates one mRNA, while subsequent irradiation with shorter wavelength light (365–405 nm) activates the other mRNA. This multiplexed gene expression regulatory system provides a high degree of flexibility and shows potential for enabling the study of multiple regulatory genes with high spatial-temporal resolution.
RESULTS AND DISCUSSION
To allow for site-specific and covalent conjugation of small molecule effectors onto target mRNA, we previously developed a technology named RNA transglycosylation at guanosine (RNA-TAG), which utilizes a bacterial tRNA guanine transglycosylase (TGT) to exchange a guanine nucleobase within a specific 17-nucleotide RNA stem-loop structure (‘Tag’) with synthetic enzyme substrate analogs (Figure 1A, Figure S2). [33–34] In E. Coli, tRNAs carrying Asn, Asp, His and Tyr are post-transcriptionally modified by TGT, which exchanges the guanine at the wobble position of the anticodon loop of the tRNA with the enzyme’s natural substrate pre-queuosine1 (preQ1). [35–36] Recognition of the RNA substrate by TGT does not require the full sequence of tRNA. Instead, a minimal 17-nucleotide RNA stem-loop from the anticodon loop of the tRNA (the 17-nucleotide ‘Tag’ sequence) is sufficient to promote TGT recognition and labeling. [37–38] TGT also accepts a wide range of synthetic preQ1 derivatives as small molecule substrates. By genetically inserting the ‘Tag’ sequence into an RNA of interest, we previously demonstrated TGT labeling is able to covalently conjugate a variety of functional small molecules, such as fluorophores and affinity tags, site-specifically onto the target RNA. [33–34] The versatility of this RNA modifying platform enabled us to adapt this technique to regulate translation, through conjugation of small-molecule effectors directly onto an mRNA transcript.
Figure 1. mRNA photo-caging using the RNA-TAG technique.
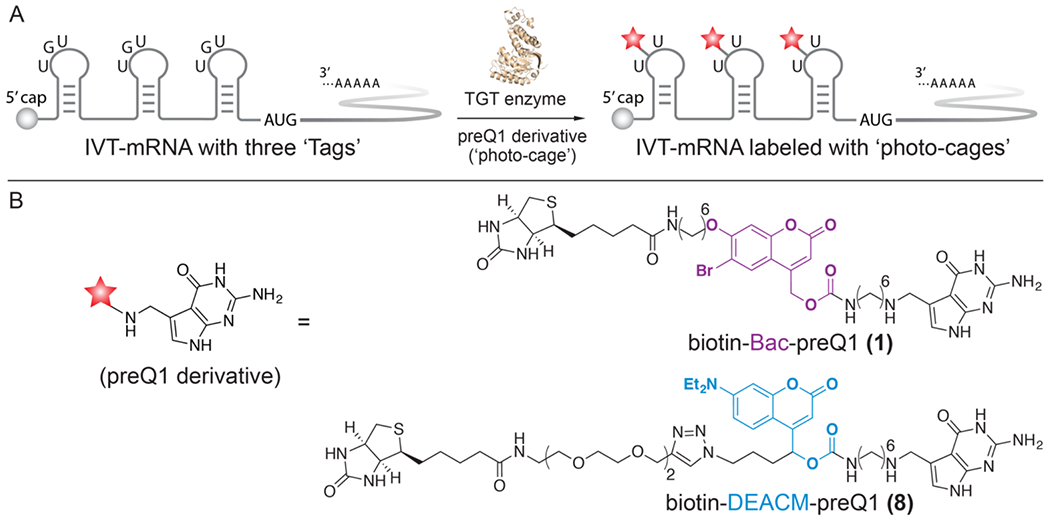
A) To facilitate TGT enzymatic labeling, three enzyme recognition sequences, ‘Tags’, are genetically inserted along the 5’-UTR of an IVT-mRNA. Subsequent conjugation of the ‘photo-cages’ severely reduces mRNA translation activity. B) Chemical structures of two sequentially activable preQ1 derivatives (‘photo-cages’), biotin-Bac-preQ1 and biotin-DEACM-preQ1.
To achieve optical-control of mRNA translation, we covalently conjugated the synthetic ‘photo-cage’, biotin-Bac-preQ1 (1), at three different locations along the 5’-UTR of a mature IVT-mRNA (Figure 1). The first conjugation site was located adjacent to the 5’-cap of the mRNA, specifically, at the 11th base after the 5’-cap. The second conjugation site was located in the middle of the 5’-UTR. The third conjugation was located at the 11th base upstream of the AUG start codon (Figure 1A). To determine the effect of such conjugation on translation, both the labeled and unlabeled IVT-mRNAs were delivered into mammalian cells through transient transfection. As a result, translation efficiency of the labeled IVT-mRNA was severely reduced to approximately 10% of the activity relative to the unlabeled IVT-mRNA. [32] We hypothesized that this phenomenon is due to the steric hindrance of these bulky ‘photo-cages’ conjugated at the 5’-UTR, which is where translation initiation takes place. Upon irradiation with 405 nm laser light, the Bac linker was photo-cleaved, leaving a minimal residue on the mRNA. As a result, translation activity of the IVT-mRNA was recovered after photo-uncaging, demonstrating successful photo-activation of gene expression. Importantly, mRNA translation was only observed in laser irradiated cells, and not in adjacent cells, demonstrating single-cell resolution of activation.
Previously, we chose the photo-cleavable linker 6-bromo-7-aminoethoxycoumarin-4-ylmethoxycarbonyl (Bac) to synthesize the ‘photo-cage’, biotin-Bac-preQ1 (1). [32] To allow for sequential photo-activation of mRNA translation, an additional photo-sensitive linker that is responsive to a longer wavelength of light was desired. Inspired by previously reported work, [39] we chose to explore [7-(diethylamino)coumarin-4-yl]-methyl (DEACM) as an additional photo-activable linker that can be sequentially activated. The DEACM linker has a wide absorbance spectrum and was previously reported to be cleaved in cellular conditions by irradiation with 470 nm light. [40] Thus, the DEACM linker should form a sequentially photo-activable linker pair with our previously reported Bac linker, which is uncaged by irradiation with 365–405 nm light. To synthesize the new ‘photo-cage’ biotin-DEACM-preQ1 (8) (Scheme 1), the DEACM-based building block (2) was subjected to sequential mesylation and azidation to generate azide compound (3). Next, the DEACM NHS-ester (4) was obtained through deprotection of the silica protecting group and N-succinimidyl carbonate (DSC) treatment. Subsequently, the preQ1 derivative (5) was coupled to the DEACM NHS-ester (4) to yield the preQ1-DEACM conjugate (6). To introduce a biotin affinity handle to facilitate purification of the labeled mRNAs, the preQ1-DEACM conjugate (6) was further coupled with commercially available biotin-PEG4-alkyne (7) via click chemistry to obtain the final TGT enzymatic substrate biotin-DEACM-preQ1 (8).
Scheme 1. Synthesis of the biotin-DEACM-preQ1 (8).
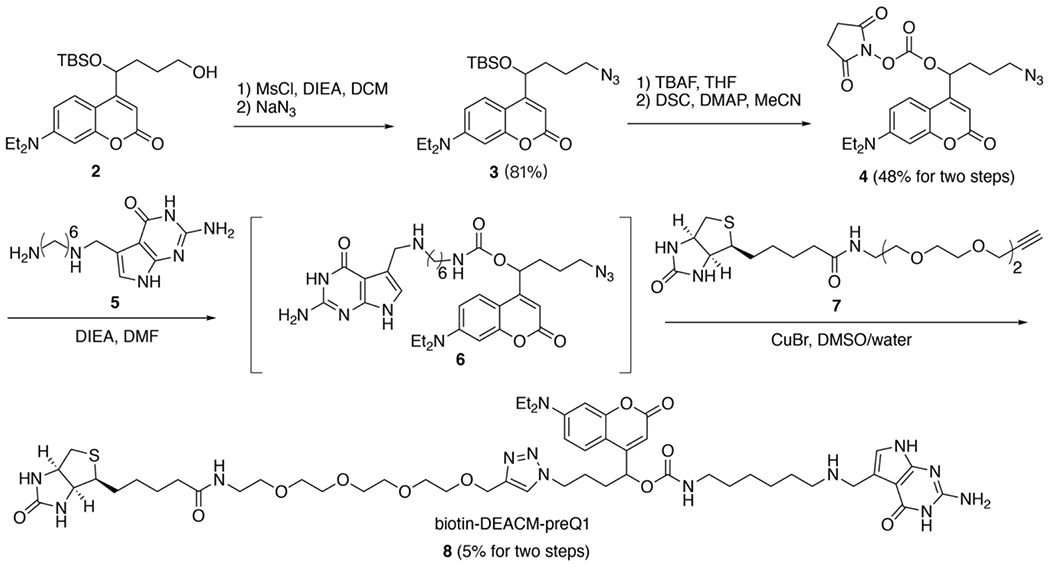
Boc = tert-butyloxycarbonyl protecting group, MsCl = methanesulfonyl chloride, DCM = dichloromethane, TBS = tert-butyldimethylsilyl protective group, TBAF = tetra-«-butylammonium fluoride, THF = tetrahydrofuran, DSC = N-succinimidyl carbonate, DMAP = 4-dimethylaminopyridine, DIEA = N,N-diisopropylethylamine, DMF = dimethylformamide, DMSO = dimethyl sulfoxide.
To demonstrate that biotin-Bac-preQ1 (1) and biotin-DEACM-preQ1 (8) can be sequentially released from RNA upon photo-irradiation with two wavelengths of light, in-vitro uncaging of labeled RNA oligos was performed using either 365 nm or 456 nm light followed by 18% denaturing polyacrylamide gel electrophoresis (PAGE) analysis. The ‘Tag’ oligo was used as RNA substrate for TGT labeling (Figure 2A). To covalently conjugate the photo-cage onto the ‘Tag’ oligo, TGT labeling using either biotin-Bac-preQ1 or biotin-DEACM-preQ1 as small molecule substrate was carried out in a dark room with minimal red ambient light to prevent undesired degradation of the light-sensitive preQ1 derivatives (Figure S1). Labeled ‘Tag’ oligo was further purified by streptavidin-biotin pull-down followed by denaturing-PAGE analysis. Covalent conjugation of the ‘photo-cage’ increased the molecular weight of the ‘Tag’ oligo, resulting in significant RNA band shifts shown in denaturing-PAGE (Figure 2B). [32–33] To trigger the cleavage of the ‘photo-cage’, the ‘Tag’ oligo labeled with biotin-DEACM-preQ1 was irradiated with a 456 nm lamp, while the ‘Tag’ oligo labeled with biotin-Bac-preQ1 was irradiated with a 365 nm lamp. We observed that upon irradiation with 456 nm light, only biotin-DEACM-preQ1 was released from the ‘Tag’ oligo, not the biotin-Bac-preQ1. Biotin-Bac-preQ1 was released from the ‘Tag’ oligo upon irradiation with 365 nm light. Thus, by using 456 nm and 365 nm light sources, sequential release of the ‘photo-cages’ from RNA was achieved.
Figure 2. TGT labeling of the RNA ‘Tag’ oligo and in-vitro sequential photo-uncaging.
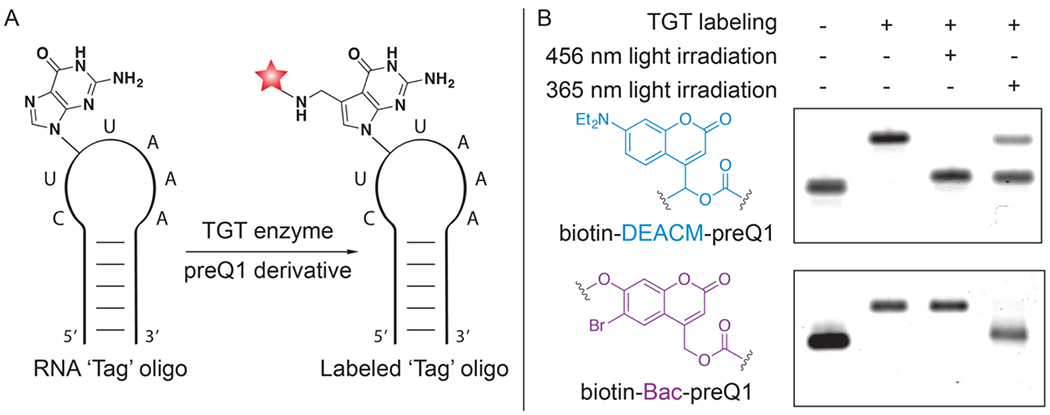
A) In-vitro conjugation of RNA ‘Tag’ oligo with preQ1 derivatives through RNA-TAG. B) Denaturing 18% PAGE analysis of enzymatic labeling and subsequent photo-uncaging of the RNA ‘Tag’ oligos. Compared to the unlabeled RNA oligo, an upper gel shift in the second column demonstrates covalent attachment of the ‘photo-cage’ onto the RNA oligo. Lower gel shift in the third or fourth column demonstrates photo-release of the ‘photo-cage’ from the RNA oligo. After photo-cleavage, the resulting RNA band was only slightly up-shifted compared to the starting material, supporting that only a minimal residue was left on the RNA after photo-cleavage.
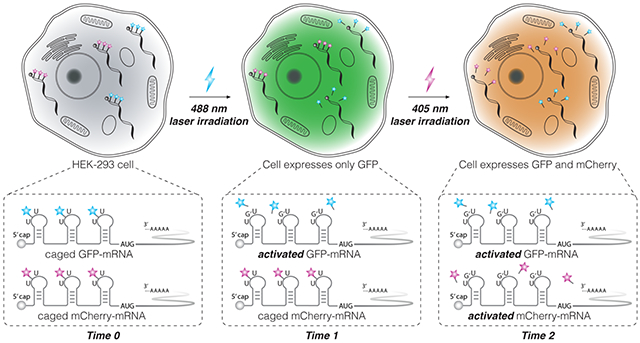
Having demonstrated that biotin-DEACM-preQ1 and biotin-Bac-preQ1 can be sequentially released from RNA oligos in-vitro, we examined whether these ‘photo-cages’ can be sequentially photo-released from mRNA in live mammalian cells. Mature IVT-mRNAs coding for either the green fluorescent protein (GFP) or the red fluorescent protein (RFP) mCherry, with three TGT labeling sites located along the 5’-UTR, were synthesized following a previously reported mRNA transcription and maturation protocol. [32] Using TGT enzymatic labeling, the mature IVT-mRNA coding for GFP was conjugated with biotin-DEACM-preQ1 while the IVT-mRNA coding for RFP was conjugated with biotin-Bac-preQ1. To get rid of unlabeled IVT-mRNAs, biotin-streptavidin affinity purification was performed (60% recovery). Equal amounts of photocaged IVT-mRNAs coding for GFP or RFP were co-transfected into HEK-293 cells using lipofectamine reagent (Figure 3A). mRNA translation activity was quantified by fluorescence imaging. Minimal translation activity was observed for the photocaged mRNAs, shown as dark cells (Figure 3B, Figure S7). Two hours post transfection, selected cells were irradiated with either 488 nm for 30 seconds or 405 nm wavelength of laser for 10 seconds to trigger the release of the ‘photo-cages’ from mRNA. [32] To allow time for sufficient mRNA translation and maturation, cells were imaged 8 hours after laser irradiation. As expected from our in-vitro studies, the 488 nm irradiation only triggered the release of the biotin-DEACM-preQ1 from mRNA. As a result, recovered protein expression of GFP was observed only in cells that were irradiated with the 488 nm laser (Figure 3B). Importantly, expression of RFP was not observed in these cells, which was expected because the biotin-Bac-preQ1 used to cage the RFP-mRNA is not responsive to 488 nm irradiation. In contrast, expression of both GFP and RFP was observed in cells that were irradiated with 405 nm wavelength of laser, because both biotin-Bac-preQ1 and biotin-DEACM-preQ1 have absorbance at 405 nm wavelength. These observations were consistent with our in-vitro uncaging experiments, that the longer wavelength of light only triggered the release of biotin-DEACM-preQ1, whereas the shorter wavelength of light triggered the release of both ‘photocages’ from mRNA. Moreover, expression of fluorescent protein was only observed in laser irradiated cells, not in adjacent cells, demonstrating photo-activation of gene expression with high cellular resolution.
Figure 3. Live cell photo-activation of mRNA translation.
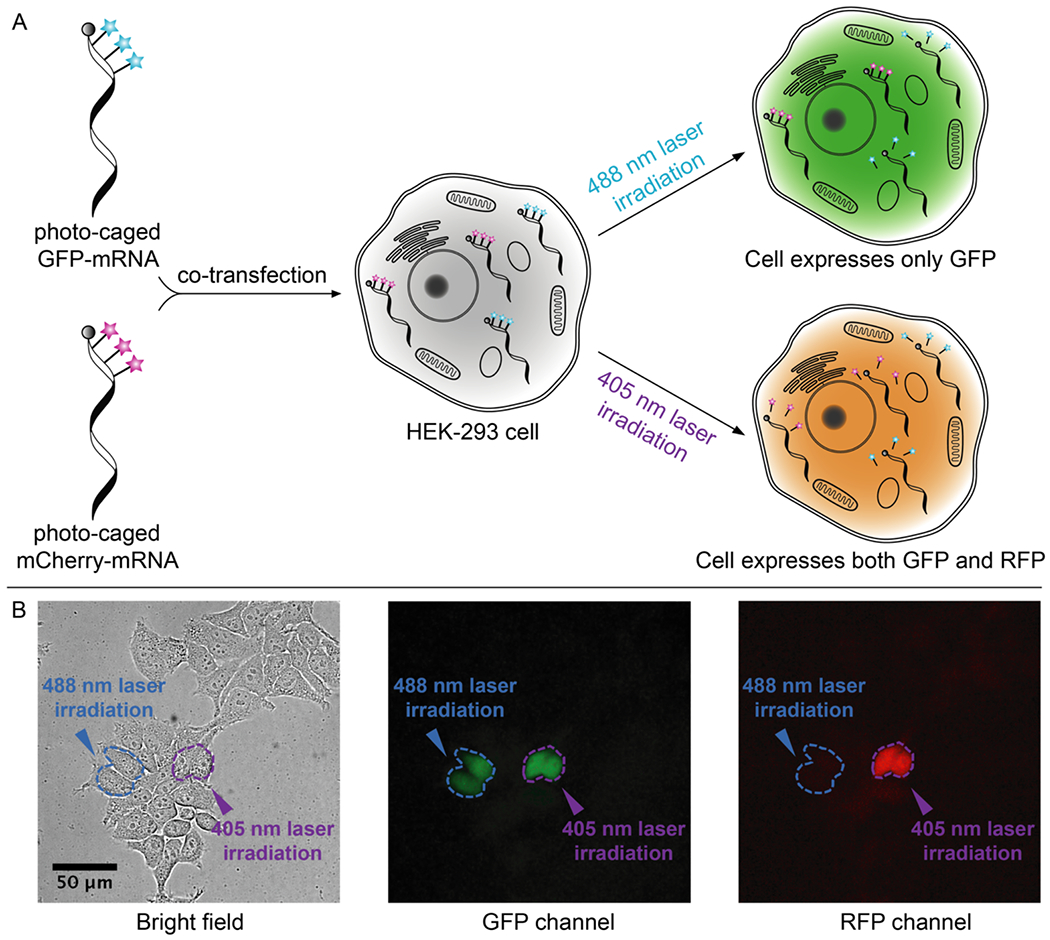
A) HEK-293 cells are co-transfected with caged GFP-mRNA and caged mCherry-mRNA, followed by photo-uncaging with either 488 nm (30 seconds) or 405 nm laser (10 seconds) irradiation. B) Live cell fluorescence imaging. Selected cells (circled in blue) irradiated with 488 nm laser only express GFP, shown as green cells in the GFP channel and dark cells in the RFP channel. Selected cells (circled in purple) irradiated with 405 nm light express both GFP and RFP. Scale bar = 50 μm.
Next, we demonstrated sequential photo-activation of two mRNAs in the same living cell. Caged IVT-mRNAs coding for GFP and RFP were co-transfected into HEK-293 cells. Two hours post transfection, the selected cell was first irradiated with 488 nm laser light to trigger the release of biotin-DEACM-preQ1 from GFP-mRNA. Five hours post transfection, the same cell was then irradiated with 405 nm laser light to trigger the release of the biotin-Bac-preQ1 from RFP-mRNA. Cells were continuously imaged to quantify protein expression level (Figure 4A, Figure S9). As shown in the fluorescence images, the first irradiation with 488 nm laser light triggered the expression of GFP, while the expression of RFP remained silenced. As expected, the subsequent irradiation with 405 nm laser light activated the expression of RFP. GFP and RFP expression levels in laser irradiated cells were quantified by measuring relative fluorescence intensity (Figure 4B). Using this model, we demonstrated sequential photo-activation of two mRNAs in the same cell by using two wavelengths of lights.
Figure 4. Live cell sequential photo-activation of two mRNAs within the same cell.
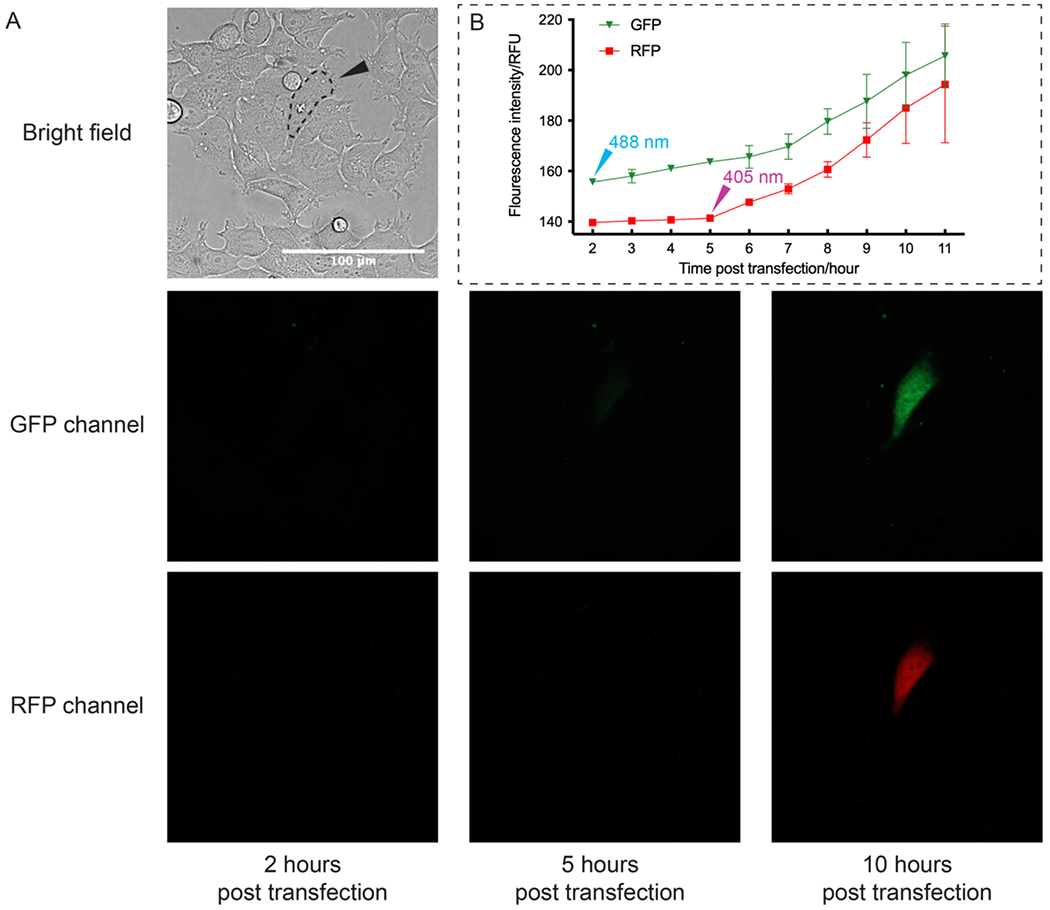
A) 2 hours post transfection, the selected cell (circled in dash line) was first irradiated with 488 nm laser light to activate GFP-mRNA. 5 hours post transfection, the same cell was irradiated with 405 nm laser light to activate RFP-mRNA (Scale bar = 100 μm). 3 more replicates were shown in Figure S9. B) Relative fluorescence unit (RFU) of GFP and RFP from the selected cells was measured using Fiji software as mean gray value. Average fluorescence intensities from 3 replicates were plotted against time. Error bars represent standard deviation calculated from 3 replicates.
CONCLUSION
In conclusion, we have developed a technique that allows sequential photo-activation of two mRNAs with single-cell resolution. We demonstrated the synthesis and photochemical properties of two ‘photo-cages’, biotin-Bac-preQ1 (1) and biotin-DEACM-preQ1 (8). These ‘photocages’ were covalently and site-specifically conjugated onto the 5’-UTR of mRNA through TGT enzymatic labeling. As a result, translation efficiency of the labeled mRNA was severely diminished compared to the unlabeled mRNA. This pair of ‘photo-cages’ can be released from mRNA transcripts sequentially upon irradiation with 365 nm/405 nm light (lamp excitation) or 405 nm/488 nm light (laser excitation), leading to translational activation of the corresponding mRNA. Irradiation with a longer wavelength of light only cleaves one ‘photo-cage’ (biotin-DEACM-preQ1) from RNA, while a shorter wavelength of light cleaves both ‘photo-cages’. By using the appropriate order of photo-irradiation, sequential photo-activation of two mRNAs within the same cell was demonstrated by live cell fluorescence imaging. We believe that the ability to sequentially photo-activate two genes with high spatial-temporal resolution provides a powerful and versatile optogenetic tool to build robust, complex, and scalable synthetic gene networks. Such a tool may improve capabilities to precisely manipulate biological networks, which can aid studies of gene regulatory mechanisms, promote the engineering of artificial biological systems, and facilitate the development of novel therapeutic applications.
Supplementary Material
ACKNOWLEDGMENTS
The project or effort depicted is sponsored by the Defense Advanced Research Projects Agency Biological Technologies Office (BTO) Safe Genes Program under Contract Number HR0011-18-2-0039. The content of the information does not necessarily reflect the position or the policy of the government, and no official endorsement should be inferred.
Footnotes
Supporting Information
Experimental methods, supplemental figures and schemes, and associated references are listed in the Supporting Information. The supporting information is available free of charge online at http://pubs.acs.org.
Supplemental methods and general materials; chemical synthesis of compound (3), compound (4) and compound (8); NMR and HRMS spectrums; HPLC trace of purified compound (8); in vitro transcription and maturation of mRNA; in vitro TGT labeling and purification of 17-nt RNA oligo and mRNA transcripts; live cell fluorescence imaging and photo-uncaging experiments.
The authors declare no competing financial interest.
REFERENCES
- [1].Lanctôt C, Cheutin T, Cremer M, Cavalli G, & Cremer T (2007). Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat. Rev. Genet, 8(2), 104–115. [DOI] [PubMed] [Google Scholar]
- [2].Jaenisch R, & Bird A (2003). Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet, 33(3), 245–254. [DOI] [PubMed] [Google Scholar]
- [3].Kistner A, Gossen M, Zimmermann F, Jerecic J, Ullmer C, Lübbert H, & Bujard H (1996). Doxycycline-mediated quantitative and tissue-specific control of gene expression in transgenic mice. Proc. Natl. Acad. Sci. U. S. A, 93(20), 10933–10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Motta-Mena LB, Reade A, Mallory MJ, Glantz S, Weiner OD, Lynch KW, & Gardner KH (2014). An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat. Chem. Biol, 10(3), 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Fenno L, Yizhar O, & Deisseroth K (2011). The development and application of optogenetics. Annu. Rev. Neurosci, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Deiters A (2009). Light activation as a method of regulating and studying gene expression. Curr. Opin. Chem. Biol, 13(5–6), 678–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gardner L, & Deiters A (2012). Light-controlled synthetic gene circuits. Curr. Opin. Chem. Biol, 16(3–4), 292–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Adams SR, & Tsien RY (1993). Controlling cell chemistry with caged compounds. Annu. Rev. Physiol, 55(1), 755–784. [DOI] [PubMed] [Google Scholar]
- [9].Mayer G, & Heckel A (2006). Biologically active molecules with a “light switch”. Angew. Chem., Int. Ed, 45(30), 4900–4921. [DOI] [PubMed] [Google Scholar]
- [10].Tang X, & Dmochowski IJ (2007). Regulating gene expression with light-activated oligonucleotides. Mol. BioSyst, 3(2), 100–110. [DOI] [PubMed] [Google Scholar]
- [11].Courtney T, & Deiters A (2018). Recent advances in the optical control of protein function through genetic code expansion. Curr. Opin. Chem. Biol, 46, 99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Liu Q, & Tucker CL (2017). Engineering genetically-encoded tools for optogenetic control of protein activity. Curr. Opin. Chem. Biol, 40, 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kaberniuk AA, Shemetov AA, & Verkhusha VV (2016). A bacterial phytochrome-based optogenetic system controllable with near-infrared light. Nat. Methods, 13(7), 591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zetsche B, Volz SE, & Zhang F (2015). A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol, 33(2), 139–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Taslimi A, Zoltowski B, Miranda JG, Pathak GP, Hughes RM, & Tucker CL (2016). Optimized second-generation CRY2-CIB dimerizers and photoactivatable Cre recombinase. Nat. Chem. Biol, 12(6), 425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Nihongaki Y, Kawano F, Nakajima T, & Sato M (2015). Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat. Biotechnol, 33(7), 755–760. [DOI] [PubMed] [Google Scholar]
- [17].Mikat V, & Heckel A (2007). Light-dependent RNA interference with nucleobase-caged siRNAs. RNA, 13(12), 2341–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Tang X, Maegawa S, Weinberg ES, & Dmochowski IJ (2007). Regulating gene expression in zebrafish embryos using light-activated, negatively charged peptide nucleic acids. J. Am. Chem. Soc, 129(36), 11000–11001. [DOI] [PubMed] [Google Scholar]
- [19].Blidner RA, Svoboda KR, Hammer RP, & Monroe WT (2008). Photoinduced RNA interference using DMNPE-caged 2’-deoxy-2’-fluoro substituted nucleic acids in vitro and in vivo. Mol. Bio Syst, 4(5), 431–440. [DOI] [PubMed] [Google Scholar]
- [20].Tang X, Swaminathan J, Gewirtz AM, & Dmochowski IJ (2008). Regulating gene expression in human leukemia cells using light-activated oligodeoxynucleotides. Nucleic Acids Res, 36(2), 559–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Young DD, Lusic H, Lively MO, Yoder JA, & Deiters A (2008). Gene silencing in mammalian cells with light-activated antisense agents. Chem Bio Chem, 9(18), 2937–2940. [DOI] [PubMed] [Google Scholar]
- [22].Ouyang X, Shestopalov IA, Sinha S, Zheng G, Pitt CL, Li WH, ... & Chen JK (2009). Versatile synthesis and rational design of caged morpholinos. J. Am. Chem. Soc, 131(37), 13255–13269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jain PK, Shah S, & Friedman SH (2011). Patterning of gene expression using new photolabile groups applied to light activated RNAi. J. Am. Chem. Soc, 133(3), 440–446. [DOI] [PubMed] [Google Scholar]
- [24].Young DD, Lively MO, & Deiters A (2010). Activation and deactivation of DNAzyme and antisense function with light for the photochemical regulation of gene expression in mammalian cells. J. Am. Chem. Soc, 132(17), 6183–6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ogasawara S (2014). Control of cellular function by reversible photoregulation of translation. ChemBioChem, 15(18), 2652–2655. [DOI] [PubMed] [Google Scholar]
- [26].Shestopalov IA, Sinha S, & Chen JK (2007). Light-controlled gene silencing in zebrafish embryos. Nat. Chem. Biol, 3(10), 650–651. [DOI] [PubMed] [Google Scholar]
- [27].Wang Y, Wu L, Wang P, Lv C, Yang Z, & Tang X (2012). Manipulation of gene expression in zebrafish using caged circular morpholino oligomers. Nucleic Acids Res, 40(21), 11155–11162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Govan JM, Young DD, Lusic H, Liu Q, Lively MO, & Deiters A (2013). Optochemical control of RNA interference in mammalian cells. Nucleic Acids Res, 41(22), 10518–10528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ando H, Furuta T, Tsien RY, & Okamoto H (2001). Photo-mediated gene activation using caged RNA/DNA in zebrafish embryos. Nat. Genet, 28(4), 317–325. [DOI] [PubMed] [Google Scholar]
- [30].Sonenberg N, & Hinnebusch AG (2009). Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell, 136(4), 731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sahin U, Karikó K, & Türeci Ö (2014). mRNA-based therapeutics—developing a new class of drugs. Nat. Rev. Drug Discovery, 13(10), 759. [DOI] [PubMed] [Google Scholar]
- [32].Zhang D, Zhou CY, Busby KN, Alexander SC, & Devaraj NK (2018). Light-activated control of translation by enzymatic covalent mRNA Labeling. Angew. Chem., Int. Ed, 57(11), 2822–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Alexander SC, Busby KN, Cole CM, Zhou CY, & Devaraj NK (2015). Site-Specific covalent labeling of RNA by enzymatic transglycosylation. J. Am. Chem. Soc, 137(40), 12756–12759. [DOI] [PubMed] [Google Scholar]
- [34].Ehret F, Zhou CY, Alexander SC, Zhang D, & Devaraj NK (2017). Site-specific covalent conjugation of modified mRNA by tRNA guanine transglycosylase. Mol. Pharmaceutics, 15(3), 737–742. [DOI] [PubMed] [Google Scholar]
- [35].Chen YC, Brooks AF, Goodenough-Lashua DM, Kittendorf JD, Showalter HD, & Garcia GA (2011). Evolution of eukaryal tRNA-guanine transglycosylase: insight gained from the heterocyclic substrate recognition by the wild-type and mutant human and Escherichia coli tRNA-guanine transglycosylases. Nucleic Acids Res, 39(7), 2834–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Stengl B, Reuter K, & Klebe G (2005). Mechanism and Substrate Specificity of tRNA-Guanine Transglycosylases (TGTs): tRNA-Modifying Enzymes from the Three Different Kingdoms of Life Share a Common Catalytic Mechanism. ChemBioChem, 6(11), 1926–1939. [DOI] [PubMed] [Google Scholar]
- [37].Hurt JK, Olgen S, & Garcia GA (2007). Site-specific modification of Shigella flexneri virF mRNA by tRNA-guanine transglycosylase in vitro. Nucleic Acids Res, 35(14), 4905–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Curnow AW, Kung FL, Koch KA, & Garcia GA (1993). tRNA-guanine transglycosylase from Escherichia coli: gross tRNA structural requirements for recognition. Biochemistry, 32(19), 5239–5246. [DOI] [PubMed] [Google Scholar]
- [39].Goguen BN, Aemissegger A, & Imperiali B (2011). Sequential activation and deactivation of protein function using spectrally differentiated caged phosphoamino acids. J. Am. Chem. Soc, 133(29), 11038–11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yamazoe S, Liu Q, McQuade LE, Deiters A, & Chen JK (2014). Sequential gene silencing using wavelength-selective caged morpholino oligonucleotides. Angew. Chem., Int. Ed, 53(38), 10114–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Holstein JM, Anhauser L, & Rentmeister A (2016). Modifying the 5’-cap for click reactions of eukaryotic mRNA and to tune translation efficiency in living cells. Angew. Chem., Int. Ed, 55(36), 10899–10903. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


