Abstract
Early clinical results of chimeric antigen receptor (CAR) T cell therapy targeting B cell maturation antigen (BCMA) for multiple myeloma (MM) appear promising, but relapses associated with residual low-to-negative BCMA-expressing MM cells have been reported, necessitating identification of additional targets. The orphan G protein–coupled receptor, class C group 5 member D (GPRC5D), normally expressed only in the hair follicle, was previously identified as expressed by mRNA in marrow aspirates from patients with MM, but confirmation of protein expression remained elusive. Using quantitative immunofluorescence, we determined that GPRC5D protein is expressed on CD138+ MM cells from primary marrow samples with a distribution that was similar to, but independent of, BCMA. Panning a human B cell–derived phage display library identified seven GPRC5D-specific single-chain variable fragments (scFvs). Incorporation of these into multiple CAR formats yielded 42 different constructs, which were screened for antigen-specific and antigen-independent (tonic) signaling using a Nur77-based reporter system. Nur77 reporter screen results were confirmed in vivo using a marrow-tropic MM xenograft in mice. CAR T cells incorporating GPRC5D-targeted scFv clone 109 eradicated MM and enabled long-term survival, including in a BCMA antigen escape model. GPRC5D(109) is specific for GPRC5D and resulted in MM cell line and primary MM cytotoxicity, cytokine release, and in vivo activity comparable to anti-BCMA CAR T cells. Murine and cynomolgus cross-reactive CAR T cells did not cause alopecia or other signs of GPRC5D-mediated toxicity in these species. Thus, GPRC5D(109) CAR T cell therapy shows potential for the treatment of advanced MM irrespective of previous BCMA-targeted therapy.
INTRODUCTION
Antibody-based therapies, including recent advances such as bispecific antibodies (1) and chimeric antigen receptor (CAR) T cell therapies (2–6), are revolutionizing the treatment of B cell malignancies. Although the management of multiple myeloma (MM) has advanced recently, it is still considered incurable, and the prognosis for patients with multiply relapsed and refractory MM remains grim.
Early clinical results with CAR T cell therapies targeting B cell maturation antigen (BCMA) are promising (7). However, although BCMA is expressed on most malignant plasma cells, expression is heterogeneous, potentially leading to variable responses (8). In addition, expression of BCMA on the cell surface varies over time because of γ secretase–mediated shedding of the extracellular domain (9); this and potentially other mechanisms may cause therapeutic selection of BCMA-low or BCMA-negative MM plasma cells. BCMA down-regulation has been reported in patients with MM who relapsed after BCMA-targeted CAR T cell therapy (8, 10), similar to relapses after CD19-targeted (11, 12) and CD22-targeted (13) CAR T cell therapy for B cell malignancies. Developing immunotherapies for additional targets may mitigate antigen loss and effectively treat patients with low or variable BCMA expression.
One potential alternative CAR T cell target for MM is the orphan G protein–coupled receptor, class C group 5 member D (GPRC5D). Earlier work discovered GPRC5D expression in two anatomic locations: the hair follicle (14–16), considered an immune-privileged site (17–19), and the bone marrow from patients with MM (20, 21). These two latter studies identified GPRC5D mRNA in the unsorted bone marrow of patients with MM; however, the only report to evaluate protein expression on MM samples did not detect it on the surface of MM cells (22). Until now, evidence of GPRC5D protein expression on MM cells and an extensive evaluation of potential “on-target/off-tumor” toxicity remain lacking.
Through immunohistochemical analyses, we demonstrate that GPRC5D is expressed on malignant bone marrow plasma cells, whereas normal tissue expression is limited to the hair follicle. We developed and evaluated an optimized, human-derived, GPRC5D-targeted CAR T cell therapy. Using a reporter line that provides a specific readout of signaling from the CAR, we identified CAR designs optimized for spacer length (23) and low antigen-independent (tonic) signaling (24–26). Last, we provide preclinical evidence that a GPRC5D-targeted CAR T cell therapy candidate is safe and effective. Despite GPRC5D expression in the hair follicle, we show that anti-cynomolgus and anti-murine cross-reactive GPRC5D CAR T cells do not induce alopecia or cause other clinical signs of damage to the skin in these species. On the basis of these results, we anticipate that GPRC5D will become an important clinical target for MM immunotherapy.
RESULTS
Expression of GPRC5D by MM cells
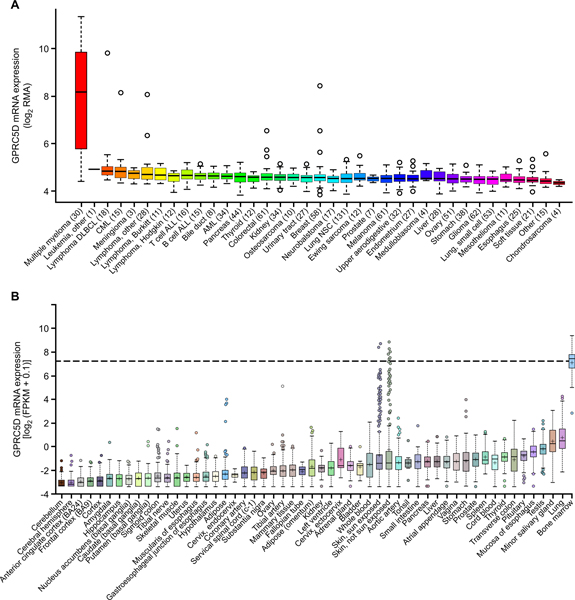
In evaluating potential cell surface targets for immunotherapy of MM, we sought to identify antigens with near ubiquitous expression on MM plasma cells and limited expression on essential normal tissue cells. Using the Cancer Cell Line Encyclopedia (CCLE), we evaluated mRNA expression of GPRC5D in silico across >1000 different malignant cell lines, including 30 MM cell lines. As a control, we evaluated SDC1 (CD138), a common surface marker of normal and malignant plasma cells. Although SDC1 is highly expressed in MM cell lines, it is also highly expressed in cell lines from the majority of tumor types, with upper aerodigestive tract tumors having the highest expression (fig. S1A). GPRC5D mRNA was highly expressed in MM cell lines (n = 30), but in contrast to SDC1, no other tumor types exhibited substantial expression (Fig. 1A). Similarly, analysis of data from the Genotype-Tissue Expression (GTEx) database of primary normal (nonmalignant) tissue types demonstrated high expression of SDC1 mRNA in the esophagus, skin, lung, and liver, among other tissues (fig. S1B), whereas GPRC5D mRNA was not highly expressed in any normal tissues aside from the skin, in which it was variably expressed, in agreement with previous reports (14–16). Furthermore, analysis of RNA expression data on human bone marrow samples showed that primary malignant and normal plasma cells expressed 1000- and 500-fold more GPRC5D mRNA than B cells from peripheral blood, respectively (Fig. 1B and fig. S1C).
Fig. 1. High expression of GPRC5D mRNA in MM cells and variable expression in skin.
(A) mRNA expression of GPRC5D in malignant cell lines (n = 1036; CCLE, accessed in September 2013, Affymetrix). RMA, robust multiarray average; DLBCL, diffuse large B cell lymphoma; CML, chronic myeloid leukemia; ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; NSC, non–small cell. (B) mRNA expression of GPRC5D in normal tissues according to GTEx RNASeq data (GTEx ENSG00000111291.4). The dashed line represents the expression of GPRC5D in CD138-sorted primary MM cells (BLUEPRINT RNA-seq, n = 9). FPKM, fragments per kilobase of transcript per million mapped reads.
To evaluate potential correlations between GPRC5D expression and clinical outcomes, we analyzed the Multiple Myeloma Research Foundation (MMRF) CoMMpass trial (NCT0145429), a publicly available longitudinal study with accompanying CD138-sorted RNA-seq expression data from 765 patients (research.themmrf.org/; version IA13). A previous investigation of 48 patients independent of the CoMMpass cohort (20 ) reported that GPRC5D expression above the median correlated with a worse prognosis. Our analysis of the CoMMpass cohort confirms this finding, as GPRC5D expression above the median in this large dataset correlated with shorter progression-free survival (P = 0.0031; fig. S2A). GPRC5D expression did not correlate with International Staging System score or any evaluated common cytogenetic abnormality (fig. S2, B and C).
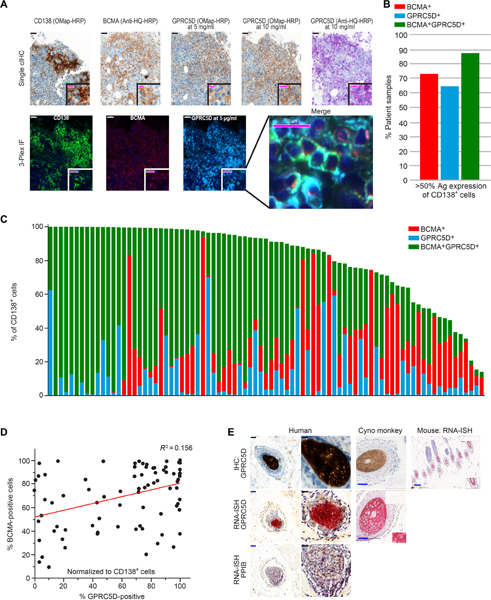
Similar to an earlier report (22), we did not identify GPRC5D on MM cells using any commercially available or internally developed flow cytometric reagents. These reagents were incompatible with quantitation of cellular antigen density. We used protein immunohistochemistry (IHC) to evaluate protein expression by primary malignant plasma cells. The specificity of anti-GPRC5D IHC was validated using K562 cells engineered to express GPRC5D and human MM cell lines endogenously expressing GPRC5D (fig. S3). We also performed multiplex quantitative immunofluorescence (Q-IF) for CD138, BCMA, and GPRC5D on primary bone marrow samples; representative images are presented in Fig. 2A. Using a cutoff of ≥50% antigen expression on CD138+ cells, which has been used in some trials of BCMA-targeted CAR T cell therapy (NCT02215967 and NCT02658929), we observed that 65% (54 of 83) of samples have GPRC5D expression above this level, 73% (61 of 83) of samples meet this threshold for BCMA, and 88% (73 of 83) meet this cutoff when expression of either BCMA or GPRC5D is considered (Fig. 2, B and C). GPRC5D expression on CD138 cells was independent of BCMA expression (R2 = 0.156; Fig. 2D).
Fig. 2. Expression of GPRC5D protein on primary MM cells and in the hair follicle.
(A) Representative immunostaining of primary myeloma bone marrow for CD138, BCMA, and GPRC5D. Scale bars, 50 μm (black and white) and 20 μm (magenta). (B) Percentage of patient bone marrow samples with >50% of CD138 + cells expressing the indicated antigen(s) (Ag; n = 83). (C) Automated Q-IF in 83 bone marrow samples from patients with MM stained as in (A). Each column represents an individual patient sample. (D) Correlation of BCMA and GPRC5D expression on CD138+ cells; R2 = 0.156. (E) GPRC5D staining of the hair follicle, the only tissue type in which positive IHC staining was confirmed by RNA in situ hybridization (RNA-ISH; RNAscope; results are summarized in table S1). Scale bars, 50 μm (blue) and 20 μm (black).
We examined expression of GPRC5D on normal tissue by immunostaining core biopsies of 30 primary tissues, each from three human donors. Of these, 24 did not express GPRC5D protein (table S1); IHC of the tissue types that showed any sign of positive staining was repeated using samples from non-human primates (NHPs) (cynomolgus monkey; 96% amino acid homology to human; antibody cross-reactive), yielding similar results. Among non–plasma cell normal tissues, IHC was positive in cells from the hair follicle bulb and the peribronchial glands; the hair follicle bulb was the only tissue in which expression was confirmed by RNA-ISH (RNAscope) and quantitative polymerase chain reaction (PCR) (table S1 and Fig. 2E). Results from quantitative PCR assessment of expression in skin indicated a weakly positive signal (table S1), consistent with expression being limited to a rare cell type in the skin. These results are in agreement with the GTEx data and previous reports of GPRC5D expression in the hair follicle (14–16).
Development of a GPRC5D-targeted CAR
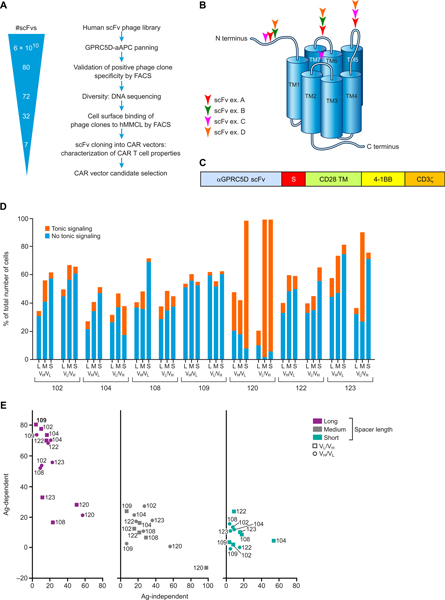
To select and identify GPRC5D-specific single-chain variable fragments (scFvs), NIH-3T3 fibroblasts were stably transduced with human GPRC5D cDNA via a retrovirus to generate stable artificial antigen-presenting cells (hGPRC5D-aAPCs). Expression of GPRC5D by these cells was confirmed by flow cytometry, and a highly antigen-expressing subclone was expanded. hGPRC5D-aAPCs were used to screen scFvs from a human B cell–derived scFv phage display library (screening and validation strategy; Fig. 3A). Ultimately, 32 distinct clones were identified, including light- and heavy-chain complementarity-determining regions (CDRs) covering five and three subfamilies, respectively, and with HCDR3 length ranging from 6 to 23 amino acids. The top seven clones that exhibited the highest specific binding to human MM cell lines MM.1S and NCI-H929 but did not bind to GPRC5D-negative cell lines (derived from other hematologic malignancies) were selected for development into CAR constructs. Epitope mapping of a subset of these scFv clones demonstrated diverse epitope binding; all four extracellular domains of GPRC5D were bound by at least one of the identified scFvs (Fig. 3B).
Fig. 3. Development of GPRC5D-targeted CARs.
(A) Human B cell–derived scFv phage display library screening strategy. Positive “hits” were confirmed by scFv binding to NIH-3T3 fibroblasts expressing GPRC5D, but not those expressing an irrelevant protein. Sequencing identified individual clones, which underwent a second validation step of binding to human MM cell lines MM.1S and NCI-H929, but not to the acute myeloid leukemia cell line SET2. scFv clones with the strongest specific binding to the MM cell lines were selected for cloning into a CAR vector. (B) Linear, conformational, and discontinuous epitope binding of a subset of GPRC5D-targeted scFvs assessed by enzyme-linked immunosorbent assay (ELISA)–based technology. (C) GPRC5D-targeted scFvs were cloned into CAR constructs including one of three spacers of varying lengths (S), a CD28 transmembrane (TM) domain, and 4–1BB and CD3ζ signaling domains. (D) Antigen-independent (tonic) signaling of CARs containing the indicated scFvs and spacers. Jurkat Nur77-RFP reporter cells were transduced with 1 of 42 CAR/green fluorescent protein (GFP) bicistronic constructs. Viable GFP+ Jurkat cells (5 × 105) were plated and monitored for RFP expression 11 days after transduction in the absence of target antigen. Expression of both RFP and GFP indicated tonic signaling; expression of GFP alone indicated CAR transduced without tonic signaling. (E) Antigen-dependent versus antigen-independent signaling of candidate CARs. Antigen-dependent signalling was measured after culturing Jurkat Nur77-RFP reporter cells 2:1 with MM.1S cells (expressing endogenous GPRC5D) for 20 hours. Percent CAR T cell signaling represents the proportion of GFP+ (CAR-transduced) cells that are also RFP+ (activated). Data are representative of two experiments.
To select an scFv for clinical development as a CAR, we engineered CARs incorporating all top seven GPRC5D-targeted human scFvs identified above in various structural formats: the variable heavy chain/variable light chain (VH/VL) or VL/VH orientation of each scFv, each with one of three IgG4/IgG2–derived spacer domains of varying lengths [short, hinge only, 12 amino acids; medium, hinge-CH3, 119 amino acids; or long, hinge-CH2-CH3, 228 amino acids, with CH2 modifications to limit Fc receptor binding as previously described (23)], for a total of 42 CAR constructs. All CARs designed for this initial evaluation contained a CD28 transmembrane domain and 4–1BB and CD3ζ signaling domains (Fig. 3C).
Because antigen-independent tonic signaling can impede the overall efficacy of CAR T cell therapy (24–26), we first screened for CARs that conveyed limited tonic signaling. We generated a reporter Jurkat T cell line that expressed red fluorescent protein (RFP) specifically downstream of CD3ζ signaling. The RFP gene was inserted in-frame downstream of the endogenous NR4A1 (Nur77) gene after a “self-cleaving” T2A element via homologous recombination; RFP expression thus indicates transcription of the immediate early gene NR4A1 (fig. S4), which is not influenced by cytokine-mediated or Toll-like receptor–mediated signals (27). The Nur77-RFP Jurkat T cell line was stably transduced with a bicistronic construct containing GFP and 1 of the 42 CAR constructs described above, and tonic signaling was determined as the percentage of GFP-expressing (CAR-transduced) cells that were also positive for RFP. Results of the assay varied substantially among CAR constructs, and the constructs incorporating human GPRC5D–targeted scFv 109 [GPRC5D(109)] consistently displayed the least tonic signaling as indicated by this assay (Fig. 3D). Certain CAR constructs that were associated with the most tonic signaling in cells expressing such CARs also inhibited growth of the Jurkat reporter cell line and were excluded from further evaluation (120 VH/VL and VL/VH with the short spacer and 123 VL/VH with the medium spacer).
Using the remaining cell lines, we compared the various CARs’ antigen-independent signaling with their antigen-dependent signaling via CAR binding to GPRC5D as measured using this reporter system. To assess antigen-specific signaling, the CAR/GFP-modified Jurkat Nur77-RFP reporter cells were cocultured 1:2 with MM.1S myeloma cells, which endogenously express GPRC5D, and RFP was measured after 20 hours. Results of this assay demonstrated that incorporation of a long spacer increased antigen-mediated signaling through the CAR but did not increase antigen-independent (tonic) signaling (Fig. 3E). The GPRC5D(109)-containing CAR in the VL/VH orientation with a long spacer was the most responsive to antigen exposure and displayed the lowest tonic signaling (Fig. 3E; representative flow plots, fig. S4B).
In vitro activity of GPRC5D-targeted CAR T cell therapy
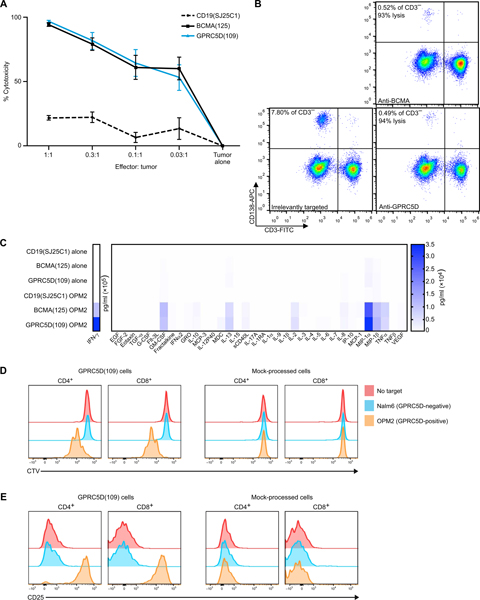
We tested the in vitro activity of primary human T cells modified to express 4-1BB–containing GPRC5D(109) CAR T cells after coculture with human MM cell lines with a range of GPRC5D mRNA expression (fig. S5A) and primary MM cells. After a 24-hour coculture, these cells efficiently induced cytotoxicity in OPM2 human MM cells (which express endogenous BCMA and GPRC5D) across a broad range of effector–to–tumor cell (E:T) ratios, from 80% at 0.03:1 E:T ratio to 98% at 1:1 E:T ratio, comparable to coculture with BCMA-targeted CAR T cells (28) (Fig. 4A), with cytotoxicity from both MM antigen-targeted cocultures significantly above background cytotoxicity seen with irrelevantly CD19-targeted (SJ25C1) CAR T cells (P < 0.001; Fig. 4A) (3). Similar cytotoxicity after coculture was seen against other GPRC5D mRNA high (MM.1S) and low (RPMI-8226) human MM cell lines (fig. S5B). GPRC5D(109)-containing CAR T cells also eradicated primary MM cells obtained via bone marrow aspirate. As shown in Fig. 4B, coculture with either GPRC5D- or BCMA-targeted CAR T cells from the same donor reduced the CD138+ MM cell fraction of CD3− viable bone marrow mononuclear cells (BMMCs) by >90% relative to coculture with irrelevantly targeted CAR T cells (representative of coculture with n = 5 primary samples; additional cocultures of primary samples are presented in fig. S6).
Fig. 4. Cytotoxicity, cytokine secretion, proliferation, and activation of GPRC5D-targeted CAR T cells in the presence of MM cell targets.
(A) Cell killing of OPM2–firefly luciferase (ffLuc) MM cells induced by CAR T cells incorporating the indicated scFv after 24 hours of coculture, as indicated by adenosine triphosphate–dependent bioluminescence after addition of luciferin; normalized to tumor cell–alone control (pooled data from two experiments each performed in triplicate, mean ± SEM; P < 0.001). Effectors counted as CAR+ viable cells. (B) Flow cytometry of primary BMMCs from a patient with multiply relapsed MM after overnight coculture with CAR T cells incorporating the indicated scFv at a 1:1 ratio of CAR+ viable T cells to BMMCs. To avoid contribution by T cell expansion or transduction efficiency, percentage of CD3− cells is reported (representative of primary samples from five patients with MM). (C) Cytokines produced by CAR+ viable T cells incorporating the indicated scFv after 1:1 coculture with OPM2 MM cells or alone for 24 hours, measured in the supernatant by multiplex Luminex assay. (D) Proliferation and (E) activation of mock-transduced or GPRC5D(109)-expressing CAR T cells cultured alone, with B-ALL (Nalm6; GPRC5D−), or with MM (OPM2; endogenous GPRC5D+) cells at a 1:1 ratio of CAR+ viable T cells to tumor cells. T cells were stained with CellTrace Violet (CTV) before coculture and stained for CD4, CD8, and CD25 after 72 hours. (D) Proliferation indicated by dilution of CTV fluorescence. (E) Activation indicated by increased CD25 fluorescence. Representative data are from two or more experiments, unless otherwise stated.
Cytokine secretion profiles after coculture with OPM2 MM cells were similar between CAR T cells targeting GPRC5D and those targeting BCMA. GPRC5D- or BCMA-targeted CAR T cells cocultured with OPM2 cells had polyfunctional cytokine secretion profiles when compared with either irrelevantly targeted CD19-targeted (SJ25C1) control or cells cultured in the absence of target cells. The largest increases were seen in secretion of interferon-γ (IFN-γ), macrophage inflammatory protein 1-α (MIP-1α), and tumor necrosis factor–α (TNFα) (effector); granulocyte-macrophage colony-stimulating factor (GM-CSF) and interleukin-2 (IL-2) (stimulatory); MIP-1β and IL-8 (chemo-attractive); and sCD40L and IL-13 (regulatory) (Fig. 4C and fig. S7).
We also measured the proliferation and activation of GPRC5D(109) CAR T cells (Fig. 4, D and E). These responses were similarly specific; GPRC5D(109) T cells proliferated (as indicated by dilution of CellTrace Violet) and up-regulated the activation marker CD25 in the presence of OPM2 cells but not upon coculture with B cell acute lymphoblastic leukemia (B-ALL) Nalm6 cells; mock-transduced T cells did not respond to MM cells.
Specificity of scFv clone 109 for GPRC5D
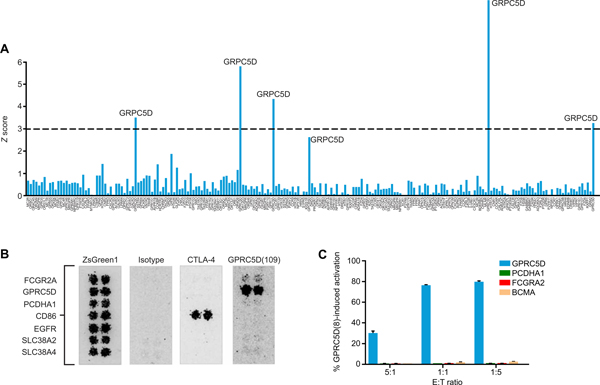
To evaluate the potential for off-target binding of anti-GPRC5D clone 109, we measured its specificity among G protein–coupled receptors (GPCRs). We transiently expressed anti-GPRC5D scFv clone 109, including the long spacer, in human embryonic kidney (HEK) 293 cells using a cell surface expression vector that included cytoplasmic mCherry. In parallel, we transiently expressed the cDNA for each human GPCR in a vector with cytoplasmic GFP; of these, 202 passed quality control of >25% transduction and were screened for off-target binding. Using an automated flow cytometric assay that detects cell-cell interaction, we determined that scFv clone 109, as a cell surface chimeric receptor, interacted exclusively with GPRC5D (Fig. 5A).
Fig. 5. Specific binding of GPRC5D by scFv clone 109.
(A) Binding of HEK293 cells transiently expressing a library of human GPCRs with cytoplasmic GFP to cocultured HEK293 cells transiently expressing anti-GPRC5D scFv clone 109, a long spacer, and cytoplasmic mCherry (both cell types in suspension), quantified by automated flow cytometric analysis. Prespecified threshold for significance (dashed line): Z score = 3, P < 0.0027. (B) Binding of anti-GPRC5D scFv clone 109-mIgG2a Fc chimeric antibody to HEK293 cells expressing the indicated cell surface proteins. Confirmation of binding to potential off-target proteins and nonspecific binders identified in a microarray screen of >4400 transmembrane proteins is shown. ZsGreen1, transfection control; Isotype, irrelevant scFv-mIgG2a Fc, negative control; CTLA-4/CD86 interaction, positive control. (C) Evaluation of GPRC5D(109) CAR activation by potential off-target proteins PCDH1A and FCGR2A. Jurkat Nur77-RFP activation reporter cells expressing a bicistronic plasmid containing a GPRC5D(109) CAR and GFP were cocultured with K562 cells expressing the indicated antigens, GPRC5D (positive control), or BCMA (negative control). Activation is determined as %RFP+GFP+/total GFP+ cells.
The specificity of clone 109 for GPRC5D among cell surface proteins generally was confirmed using an scFv-Fc IHC assay in which individual HEK293 cell populations, each expressing 1 of 4417 human plasma membrane proteins, were grown in microarray spots and treated with an anti-GPRC5D clone 109 scFv-mIgG2a Fc antibody or an mIgG2a Fc isotype control. Cell microarrays were assessed for binding by automated fluorescent microscopy after treatment with a fluorescently labeled secondary antibody. The clone 109–containing antibody bound strongly to GPRC5D and initially indicated potential weak-to-medium binding of two additional proteins, protocadherin α1 (PCDHA1) and Fcγ receptor 2A (CD32a; FCGR2A), a protein with known potential for Fc interaction (29). A small-scale second assay including only these proteins indicated potential for binding (Fig. 5B). Nonetheless, after coculture of K562 cells individually expressing these proteins with GPRC5D(109) CAR-expressing Jurkat Nur77-RFP reporter cells, neither coculture resulted in activation, as measured by RFP signal (Fig. 5C), confirming that these potential off-target interactions are nonspecific and did not stimulate GPRC5D(109) CAR signaling in the presence of these proteins. Further, activation of Jurkat Nur77-RFP reporter cells mediated by the GPRC5D(109) CAR after coculture with OPM2 MM cells was abolished when they were instead cocultured with OPM2 cells in which GPRC5D was knocked out using CRISPR-Cas9 (fig. S8). Together, these results show that scFv clone 109 specifically recognizes GPRC5D.
In vivo activity of GPRC5D-targeted CAR T cell therapy
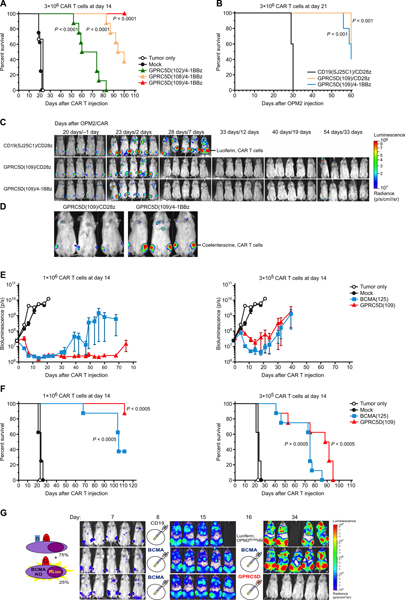
To evaluate the in vivo activity of GPRC5D-targeted CAR T cell therapy, we used the OPM2 human myeloma cell xenograft model that causes bone marrow–predominant disease (28, 30). Nonobese diabetic scid gamma (NSG) mice were injected via tail vein with OPM2-ffLuc cells, which were allowed to engraft and expand for 14 days before a single tail vein injection of CAR T cells (with comparable transduction efficiency; representative example, fig. S9), and tumor burden was monitored by bioluminescence imaging (BLI). In vivo activity was compared among CAR T cells incorporating one of three highly active anti-GPRC5D scFv clones, which also had low antigen-independent tonic signaling (102, 108, and 109); all were made with the same long spacer, CD28 transmembrane domain, and 4–1BB and CD3ζ signaling domains. Treatment with each of the GPRC5D-targeted CAR T cells increased survival (Fig. 6A). At 100 days after CAR T cell injection, only mice treated with GPRC5D(109)-containing CAR T cells maintained 100% survival.
Fig. 6. Eradication of MM cells and improvement of survival by GPRC5D-targeted CAR T cells in a murine xenograft model.
NSG mice were injected intravenously with OPM2-ffLuc cells to establish a bone marrow–tropic MM xenograft and then treated with a single intravenous injection of CAR T cells at the indicated time and dose. (A) Survival of mice treated 14 days after OPM2 injection with 3 × 106 4-1BB–containing CAR+ viable T cells incorporating the indicated anti-GPRC5D scFv clones (n = 8 per arm). (B) Survival of mice treated at 21 days after OPM2 injection with 3 × 106 CAR+ viable T cells gene-modified to express a bicistronic construct encoding extGLuc and a CAR incorporating scFv CD19(SJ25C1) or GPRC5D(109) and either a 4-1BB or CD28 costimulatory domain (n = 5 per arm). (C) Tumor burden (d-luciferin BLI of OPM-ffLuc) of mice from (B). (D) CAR T cell homing (coelenterazine BLI of extGLuc CAR T cells) of mice from (B) performed on day 7 after CAR T cell treatment. (E and F) Dose response of GPRC5Dand BCMA-targeted CAR+ viable T cells, administered 14 days after OPM2 injection (n = 8 mice per arm). (E) Tumor burden as assessed by BLI of OPM-ffLuc and (F) survival. (G) NSG mice were injected with a mixed population of unmanipulated OPM2WT (75%) + GFP/ffLuc+ CRISPR-mediated OPM2BCMA-KO (25%). On day 8 and day 16, mice were injected with the indicated CAR+ viable T cells. BLI monitors only OPM2BCMA-KO, which are the only cells to express GFP/ffLuc (n = 5 mice per arm, representative of two experiments). Note that, in the two BLI images of mice with the highest tumor burden, a higher minimum color scale threshold was used compared to the shown scale to remove BLI scatter. All P values shown are relative to mock-transduced (A, E, and F) or irrelevantly targeted (B) CAR T cells. Average myeloma distribution and percent weight change were assessed by two-way analysis of variance (ANOVA). P ≤ 0.05 is considered significant. The log-rank (Mantel-Cox) test was used to calculate statistical significance of survival experiments, with P values adjusted for multiple comparisons via Benjamini-Hochberg correction.
Evaluation of GPRC5D(109) CAR T cells in a large burden of disease model (treatment 3 weeks after tumor engraftment) demonstrated CAR T cells homing to and rapidly eradicating MM by day 7 after treatment with either a 4–1BB or a CD28 costimulatory signaling domain. All aspects of the CARs other than the costimulatory domain were kept constant, including the long spacer and CD28 transmembrane domain. In this case, we used a bicistronic construct including a membrane-tethered exterior Gaussia luciferase, separated by a P2A element from the CAR, to allow in vivo BLI of CAR T cells after injection of coelenterazine, a distinct substrate from the luciferin required for ffLuc BLI (31). A large burden OPM2-ffLuc MM model was used to distinguish between these two similar CARs; the ample amount of antigen was intended to drive CAR T cell expansion and ease monitoring of their homing and accumulation. Mice were treated with 3 × 106 CAR+ viable T cells 21 days after OPM2 engraftment. Regardless of costimulatory domain, GPRC5D(109) CAR T cell therapy comparably extended survival (Fig. 6B). GPRC5D(109) CAR constructs with either costimulatory domain eradicated OPM2 cells between days 2 and 7 after CAR T cell injection, and eradication was predominately durable (Fig. 6C). The deaths of treated mice around day 60, in the absence of OPM2 BLI signal, were secondary to xenogeneic graft-versus-host disease, a known, donor-dependent limitation of injecting human T cells into NSG mice (32, 33).
To assess whether GPRC5D-targeted CAR T cells accumulate at the site of the xenograft, we conducted BLI after coelenterazine injection at 1 week after treatment, the time we previously found to be the peak of in vivo expansion of BCMA-targeted CAR T cell therapy in this model (28). This assay demonstrated that CAR T cells with either costimulatory domain localized to the site of the MM xenograft (Fig. 6D).
In addition to the OPM2 model, in vivo analysis of mice bearing xenografts of the human MM cell line, RPMI-8226, with low expression of GPRC5D mRNA (fig. S5) similarly showed that T cells gene-modified with this GPRC5D-targeted CAR mediated antitumor activity and in vivo CAR T cell expansion (fig. S10).
Given the promising clinical results reported with BCMA-targeted CAR T cell therapy to treat MM (7), we directly compared GPRC5D(109)/4-1BBz with BCMA-targeted CAR T cell therapy, both including an identical CAR backbone (Fig. 6, E and F). We treated mice 14 days after OPM2 injection as in Fig. 6A; however, we used lower doses of CAR T cells than in previous experiments. When treating with 1 × 106 or 3.3 × 105 GPRC5D or BCMA-targeted CAR+ T cells, a dose response was noted in the kinetics of tumor regression (Fig. 6E). GPRC5D(109) CAR T cells were comparable to BCMA-targeted CAR T cells in inducing tumor regression (Fig. 6E) and in extending survival (Fig. 6F) across both doses.
Because loss or down-regulation of BCMA is implicated in relapse after BCMA-targeted CAR T cell therapy (8, 10), we evaluated GPRC5D-targeted CAR T cell therapy in a model of BCMA loss-mediated relapse. We injected a mixture of OPM2WT cells spiked with a subpopulation of GFP/ffLuc+ CRISPR-mediated OPM2BCMA-KO cells so that the BCMA-KO cells could be specifically imaged. BCMA(125) CAR T cells eradicated OPM2WT cells (fig. S11), whereas the OPM2BCMA-KO subpopulation progressed. Antigen escape-mediated tumor progression could be rescued by GPRC5D(109) CAR T cells (Fig. 6G).
Lack of on-target/off-tumor toxicity induced by GPRC5D-targeted CAR T cells
To evaluate potential activation of GPRC5D-targeted CAR T cells by essential normal cells, primary human T cells were genetically modified to express the GPRC5D(109) CAR and cocultured with a panel of isolated primary human cell types, after which cytokine release was measured. Whereas coculture of GPRC5D(109) CAR T cells with positive control OPM2 MM cells caused substantial IFN-γ, IL-2, and TNFα release, quantities of cytokines in the media after coculture with any of the 20 normal tissue types investigated were minimal; for example, IFN-γ was 2600-fold higher after OPM2 coculture when compared to the highest value after coculture with cells isolated from normal tissue (fig. S12).
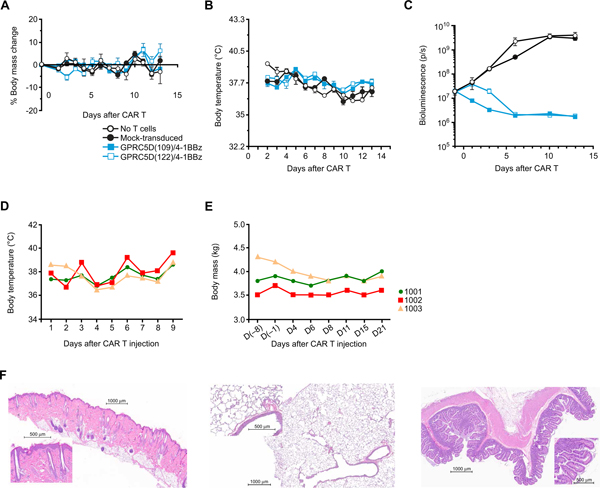
Given the expression of GPRC5D in cells from the hair follicle (Fig. 2E and table S1), we sought to evaluate on-target/off-tumor binding in a relevant in vivo model. To find a murine or cynomolgus cross-reactive anti-GPRC5D scFv, we used the Nur77 Jurkat reporter cell line. Nur77 Jurkat cell populations were stably transduced to express one of six anti-GPRC5D scFvs and cocultured 1:1 with K562 cells engineered to express human, murine, or cynomolgus GPRC5D (the latter two forms have 82 and 96% amino acid homology to human, respectively). scFv clone 109 was not cross-reactive to either mGPRC5D or cGPRC5D, but multiple clones were cross-reactive to mGPRC5D and cGPRC5D (fig. S13). Of these, clones 122 and 108 were selected for further experimentation in murine and cynomolgus models, respectively, given their high antigen-dependent signaling-to-tonic signaling ratios (fig. S13). Both GPRC5D(109)- and GPRC5D(122)-containing CAR T cells were well tolerated by mice; neither affected body mass or temperature (Fig. 7, A and B). Both CAR T cell therapies eradicated OPM2 cells injected 14 days earlier (Fig. 7C). There was no fur loss or other clinical sign of toxicity.
Fig. 7. Lack of overt toxicity caused by GPRC5D-targeted CAR T cells in murine and NHP models.
(A to C) Mice were injected with 3 × 106 human CAR+ viable T cells expressing a CAR containing a human/murine cross-reactive anti-GPRC5D scFv (clone 122) 14 days after injection of OPM2; nontreated mice and mice injected with mock-transduced T cells or T cells expressing a CAR containing scFv clone 109 (which recognizes only human GPRC5D) served as controls. (A) Body mass, (B) body temperature, and (C) BLI of OPM2-ffLuc cells. (D to E) Three cynomolgus monkeys were injected with autologous cynomolgus T cells modified to express a CAR containing a human/cynomolgus cross-reactive anti-GPRC5D scFv clone 108 (1 × 107 CAR+caspase− T cells/kg) and 1 × 107 cGPRC5D+caspase− autologous aAPC T cells/kg on day 4 (D4) and then euthanized for pathologic evaluation at 21 days (full protocol in fig. S14). (D) Body temperature, (E) body mass, and (F) pathologic investigation of skin, lung, and small intestine (representative images). All abnormal pathologic findings are summarized in Table 1.
In the NHP model, autologous cynomolgus T cells were genetically modified with the cGPRC5D cross-reactive CAR GPRC5D(108) (protocol schematic, fig. S14). After blood was taken for CAR T cell production, NHPs were treated with lymphodepleting conditioning cyclophosphamide (40 mg/kg on days −4 and −2). On day 1, 10 × 106 CAR+caspase3− autologous cynomolgus T cells/kg were injected into three NHPs. Because of the concern for hair follicle expression of GPRC5D, to increase the sensitivity of detecting toxicity, NHPs were also treated with the topical skin irritant imiquimod on a small region of the back 4 days before CAR T cell injection. To boost CAR T cell expansion, each NHP received 10 × 106 cGPRC5D+caspase3− autologous artificial antigen-presenting T cells/kg 4 days after CAR T cell injection. The transduction rate of the CAR into NHP T cells was 36 to 49% of CD3+ cells. After gene transfer, preinfusion NHP T cells remained viable (80 to 92%) and were functional, with IFN-γ release approaching 10,000 pg/ml of supernatant after coculture with cells expressing GPRC5D (1:1 E:T ratio) and antigen-specific target cell lysis consistent between CAR T cells from the three NHPs (fig. S15, A and D). Although there was insufficient material to assess post-infusion CAR T cell expansion, persistence was detected in the peripheral blood in all three NHPs and in the bone marrow in two at the time of sacrifice for gross and histologic examination (day 21 after infusion; fig. S15E).
After GPRC5D(108)-containing CAR T cell therapy, there was no acute infusion-related toxicity. After treatment, clinical observation, body temperature, and weight curves remained stable (Fig. 7, D and E). No adverse events, increases in proinflammatory cytokines, or relevant changes in clinical chemistry were observed in any of the subjects. There was no fur loss or other clinical or pathological signs of damage to the skin, lungs, or other tissues (Fig. 7F and Table 1).
Table 1.
Summary of pathologic findings from cynomolgus injected with human/cynomolgus cross-reactive anti-GPRC5D(scFv clone 108) CAR T cells.
| Tissue | Finding | Number of individuals (n = 3) | Degree |
|---|---|---|---|
| Spleen | Increased cellularity in lymphoid tissue and white pulp | 2 | Minimal (1) Mild (1) |
| Lymph node, mesenteric | Increased number and size in lymphoid follicle and germinal center | 3 | Minimal (1) Mild (2) |
| Liver | Focal fibrosis with pigmented macrophages | 2 | Minimal |
| Focal hepatocellular necrosis | 1 | Minimal | |
| Lung, left diaphragmatic lobe | Embolus containing nuclear material in the capillary | 1 | Minimal |
DISCUSSION
These studies demonstrate that GPRC5D is an attractive target for the immunotherapy of MM. This receptor is expressed on MM cells and absent from nearly all healthy tissues, with the exception of the hair follicle, which may be immune-privileged. Further, GPRC5D-targeted CAR T cells eradicate MM cells from xenograft models and do not cause overt toxicity or pathology in mice or NHPs.
We confirmed that GPRC5D protein is consistently expressed on MM cells with a membranous pattern by IHC staining. The single prior reported study to evaluate protein expression did not identify GPRC5D on the cell surface using flow cytometric analyses (22), likely due to the unavailability of reliable, high-quality flow cytometry reagents. Our study was limited by a lack of reagents to quantify antigen density; the development of such reagents should allow investigation of whether the GPRC5D− MM cells seen in our IHC analysis are a distinct subpopulation or part of the normal distribution of expression. Nonetheless, the activity of GRPC5D(109) CAR T cells against MM cell lines with relatively high and low GPRC5D mRNA expression is encouraging.
The distribution of GPRC5D expression on CD138+ cells across patient samples is similar to that of BCMA. However, the intrapatient expression patterns are independent of each other; therefore, the percentage of CD138+ cells expressing BCMA does not correlate with an expected percent GPRC5D expression for any individual patient. Consistent with this, although in most samples, the largest population of CD138+ cells expresses both BCMA and GPRC5D, in several patient samples, single-antigen BCMA expression was more prevalent, whereas in others, GPRC5D expression was dominant. One limitation of these results is that the effects of processing primary bone marrow samples may result in artefactual antigen loss in some cases, which would cause underestimation of BCMA and GPRC5D expression.
We explored the potential for on-target/off-tumor toxicity by targeting GPRC5D. GPRC5D is strongly expressed in the hair follicles and variably in lung tissue; the latter was apparent by IHC but not confirmed by RNA-ISH. Coculture of GPRC5D-targeted CAR T cells with 20 different primary cell types from essential normal tissues induced only minimal IFN-γ release, at least 2600-fold lower than coculture with MM cells. To further ensure the specificity of GPRC5D-targeted CAR T cells for MM cells, it will also be important to evaluate GPRC5D expression in additional primary cell targets from multiple donors when improved flow reagents become available. NHPs and mice treated with species cross-reactive CAR T cells showed no clinical signs of toxicity. Potential hair loss may be a risk acceptable for patients; however, the hair follicle is considered an immune-privileged site (17–19), which may explain the apparent lack of skin or hair toxicity in NHPs and mice treated with species cross-reactive CAR T cells. However, although we did not observe any acute toxicities targeting GPRC5D in murine and NHP studies, the conclusions are limited by the use of surrogate species cross-reactive CAR binders and, in the NHP study, the small number of animals treated (n = 3) and the inability to evaluate CAR T cell expansion. The potential for on-target/off-tumor toxicity against normal plasma cells is similar to toxicity with CD19-targeted CAR T cell therapy, where hypogammaglobulinemia may occur. If hypogammaglobulinemia or cytopenias result, treatment with pooled donor intravenous immunoglobulin is available. We concluded that GPRC5D is an attractive target for immunotherapy of MM given its preferential expression on MM cells.
Our GPRC5D-targeted CAR development strategy was designed to maximize its clinical activity. To avoid host anti-murine CAR immune responses upon translation into patients, which have, in some cases, been found to correlate with limited clinical responses in patients retreated with murine scFv-based CAR T cells targeting CD19 (6), we screened a human B cell–derived phage display library and identified seven human anti-GPRC5D scFvs. As the orientation of the scFv can affect the affinity of a CAR, we evaluated these scFvs in both the VH/VL and VL/VH orientations. The distance between gene-modified T cells and the specific binding domain on the target antigen also influences optimal CAR T cell function (34); we therefore evaluated each of these scFvs with three different length spacer domains (23). To minimize antigen-independent (tonic) signaling, another factor found to adversely affect CAR T cell activity (24–26), scFvs were evaluated using a Jurkat T cell line designed to express RFP in-frame immediately downstream of endogenous Nur77 via homologous recombination; this Nur77-RFP reporter construct serves as a surrogate for T cell activation (27). A flow cytometric assay of these reporter cells transduced with each of the 42 potential CAR constructs (seven scFvs in both VH/VL and VL/VH orientations, each with three spacers) for antigen-independent (tonic) signaling and antigen-mediated activation clearly and rapidly differentiated between the CAR designs. Confirming the utility of this reporter assay, primary T cells modified with CARs incorporating the GPRC5D(109) scFv showed superior in vivo activity compared with GPRC5D-targeted CARs containing other scFvs. This approach is useful in screening not only CARs but also synthetic T cell receptor–based or tumor-infiltrating lymphocyte-based cell therapies for activation.
We have shown that GPRC5D-targeted CAR T cells had antitumor activity in an in vivo BCMA antigen escape relapse model. It is not yet firmly established whether or how the degree of BCMA expression may affect the efficacy of BCMA-targeted CAR T cell therapy, but it is possible that targeting a second antigen with an independent expression pattern, such as GPRC5D, may increase the frequency, depth, and/or duration of responses in patients harboring BCMA-low or BCMA-negative MM plasma cell reservoirs. Clinical investigation of GPRC5D-targeted CAR T cell therapy should be pursued for patients with advanced MM, regardless of previous BCMA-targeted therapy.
MATERIALS AND METHODS
Study design
The objectives of this study were (i) to determine whether GPRC5D is an attractive target for the immunotherapy of MM and (ii) to develop an optimized CAR targeting GPRC5D to treat MM, including post-BCMA antigen escape-mediated relapse. We screened primary MM samples and normal tissue by multiplex Q-IF, scoring by HALO image analysis using the same settings across samples. Toward the second objective, we screened a human scFv phage display library to develop GPRC5D-targeted CARs. Candidate CARs were screened for tonic signaling using a Jurkat Nur77 reporter line. The efficacy and safety of one CAR with low tonic signaling, GPRC5D(109), were extensively evaluated via in vitro cytotoxicity, cytokine, proliferation, and activation assays, as well as murine xenograft safety and efficacy studies. Safety was further evaluated with species cross-reactive GPRC5D-targeted CARs. Studies were planned with the minimum number of animals per treatment group to reproducibly observe statistically significant differences (n = 5 to 8 per arm per experiment). All murine experiments were replicated at least twice, using T cells from different donors in each replicate. Tumor engraftment was defined by baseline BLI before cellular therapy. Outlier mice with baseline tumor burden discordant from others in the experiment were excluded before randomization and CAR T cell infusion. No data were excluded at any later point. Researchers imaging and collecting data from mice were unaware of treatment group allocation, but data were not analyzed in a blinded fashion.
Human scFv phage display library screen
A human B cell–derived scFv phage display library (E-ALPHA, Eureka Therapeutics) was panned using NIH-3T3 aAPCs stably expressing GPRC5D. Positive phage clones were first confirmed by their capacity to specifically bind 3T3-GPRC5D aAPCs via flow cytometry. Confirmed clones were then sequenced to assess diversity. Unique clones were further validated by flow cytometry to assess specific binding to human MM cell lines and not human cell lines from other hematologic malignancies.
Epitope mapping
A library of overlapping 15-nucleotide oligomer peptides covering the extracellular domains of GPRC5D was synthesized and chemically linked to flexible scaffolds to assess linear, conformational, and discontinuous epitope binding of GPRC5D-targeted scFvs using Pepscan’s proprietary ELISA-based technology (Pepscan) (35).
Transduction of human T cells
Primary human T cells were isolated from whole blood obtained from healthy donors or the New York Blood Center, Memorial Sloan Kettering (MSK) Institutional Review Board (IRB) no. 95–054, Blood-Works IRB no. 20140680, or Key Biologics IRB no. 25042/1. T cells were stimulated with phytohemagglutinin (2 mg/ml; Sigma) or CD3/CD28 Dynabeads (Thermo Fisher) at a 1:1 ratio for 24 hours and grown in the presence of IL-2 ± IL-7 and IL-15. T cells were spinoculated with gamma retrovirus or lentivirus on days 2 to 3 after activation. Gamma retrovirus was gibbon ape leukemia virus pseudotyped and used in vitro for tests of cytotoxicity, primary MM cell coculture, and cytokine profiling in mice to investigate antitumor activity and T cell proliferation and homing and in NHPs to assess tolerability. Lentivirus was vesicular stomatitis virus glycoprotein (VSV-G) pseudotyped and used in vitro for cytotoxicity, proliferation, and activation assays and in vivo to assess antitumor activity and toxicity. Transduction efficiency was determined by flow cytometric analysis on days 4 to 10. All experiments were normalized for CAR+ viable cells.
T cell signaling
A Jurkat T cell leukemia Nur77-RFP reporter cell line was generated by inserting a 2A-RFP sequence in-frame with the endogenous Nur77 gene by homologous recombination. This Jurkat Nur77-RFP line was further engineered to express various anti-GPRC5D CAR-2A-GFP bicistronic constructs. Cells were plated either alone or 2:1 with MM.1S cells for 20 hours. Antigen-independent (tonic signaling) and antigen-dependent activation were assessed by measuring changes in RFP expression by flow cytometry. Signaling of transduced cells was calculated as the ratio of GFP+RFP+ cells to total GFP+ cells.
Generation of GPCR transient expression library
A cDNA library of all human GPCRs was generated, and each cDNA was cloned into a transient expression construct including a C-terminus fused GFP. Constructs were transfected into HEK293 cells cultured in suspension. Transfection was confirmed by flow cytometric analysis for GFP, and membrane localization was confirmed manually by fluorescent microscopy. Two hundred two GPCR-transfected HEK293 cell populations with >25% GFP+ and correct localization were used for screening.
High-throughput scFv-GPCR cell-cell binding screen
A construct containing GPRC5D scFv clone 109, connected by a long IgG4 spacer to the murine programmed death-ligand 1 (PD-L1) transmembrane domain with RFP fused to the C terminus, was generated and transfected into HEK293 cells cultured in suspension. On day 2 after transfection, GPCR library and GPRC5D scFv 109–expressing cells were cocultured 1:1 in 96-well plates. Cell-cell conjugates were detected as GFP+RFP+ doublets via automated flow cytometry.
Cell surface protein binding screen
Expression vectors for 4417 cDNAs (representing >3500 distinct genes) encoding native human plasma membrane proteins were reverse-transfected into HEK293 cells for screening in duplicate. Cells were grown in microarray spots on glass slides. Transduction efficiency was checked by transducing four spots per slide with pIRES-hEGFR-IRES-ZsGreen1 control vector to confirm a mean ZsGreen signal of at least 1.5-fold greater than background, a previously defined threshold. Cells were fixed, and slides were stained with anti-GPRC5D clone 109 scFv-Fc (6.7 mg/ml) for 1 hour. Binding was detected with Alexa Fluor 647 (A647)–congugated anti-mouse IgG. A647 signal was recorded by automated fluorescence microscopy. Images were analyzed with ImageQuant software. Positive hits were classified as strong, medium, weak, or very weak via visual inspection. The identity of all binding antigens was confirmed by Sanger sequencing of the vector.
Murine experiments
All in vivo studies were conducted in compliance with Institutional Animal Care and Use Committee (IACUC)–approved protocols [Memorial Sloan Kettering Cancer Center (MSKCC) 00-05-065 or Juno 15–06]. Six- to 12-week-old NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice (The Jackson Laboratory) were injected subcutaneously with RPMI-8226 cells or systemically via tail vein with OPM2 cells (30) stably transduced with ffLuc. Injection of D-luciferin substrate (Millipore-Sigma) allowed for longitudinal in vivo BLI. A single dose of human genetically modified CAR T cells was administered at the indicated time points. In some cases, T cells were modified with a bicistronic construct including a CAR and membrane-tethered external Gaussia luciferase (31), which could be imaged after injection of coelenterazine substrate (NanoLight Technology). BLI was conducted using an IVIS Spectrum, and images were analyzed using Living Image software (PerkinElmer). Survival was graphically represented as Kaplan-Meier curves. The log-rank (Mantel-Cox) test was used to test statistical significance, with P values adjusted for multiple comparisons via Benjamini-Hochberg correction.
NHP experiments
This study was conducted in compliance with protocol 20127278, approved by the Charles River Laboratories IACUC. Whole blood was drawn from seven cynomolgus monkeys (Charles River Laboratories), and peripheral blood mononuclear cells (PBMCs) were isolated, viably frozen, and shipped. After 13 days, PBMCs were thawed, and T cells were isolated, activated, and transduced (day−9 to day −5). In parallel, baseline blood and bone marrow were obtained (at 14 days after the initial blood draw). CAR T cell viability, transduction efficiency, and functionality (IFN-γ release and lysis of target cells) were assessed in vitro, and three animals with highly viable, functional CAR T cells on these in vitro assays were selected for further experimentation. Lymphodepleting chemotherapy [cyclophosphamide (40 mg/kg); Cardinal Health] was administered 4 and 2 days before CAR T cell injection; the first dose was accompanied by imiquimod (skin irritant) application. After lymphodepletion, baseline blood and bone marrow samples were collected 1 day before administration of 1 × 107 cGPRC5D-targeted CAR+caspase− autologous T cells/kg body weight [in which the CAR consisted of anti-GPRC5D(108), a long spacer, 4-1BB endodomain, and CD3ζ ] as a single dose. On day 4, 1 × 107 autologous aAPC-cGPRC5D+caspase− T cells/kg were injected. Clinical observations, temperatures, weights, and serum and bone marrow evaluations were conducted over the following 21 days, after which animals were humanely euthanized with sodium pentobarbital (Vortech Pharmaceuticals) and necropsy and pathology were performed. Genomic DNA was extracted from cynomolgus PBMC and bone marrow samples on day 21, with cynomolgus anti-GPRC5D CAR T cells serving as a positive CAR-expressing control. A nested two-step PCR was performed using CAR-specific primers. PCR products were visualized on an agarose gel, with an expected CAR band size of 1267 base pairs. Bands were extracted and sequenced to confirm the presence of CAR DNA in the samples.
Statistical analysis
Statistical analysis was performed using GraphPad Prism (GraphPad Software). All statistical tests are two-tailed. Unless otherwise indicated, log-rank Mantel-Cox test was used for survival curves, and unpaired t test was used for comparison of experimental groups to controls. Original data are in data file S1.
Supplementary Material
Acknowledgments:
We would like to thank S. Yoo, P. Carlson, T. Cox, J. Freeth, C. Hauskins, C. Herr, Y. Ho, C. de Imus, A. Lickteig, S. Morkowski, M. Myers, and L. Torrey for technical assistance; J. Moore (MSKCC Editorial and Grant Services) for editorial support; S. Weil (MSKCC Medical Graphics) for assistance with presentation of data; and M. Blake, C. Davis, C. Hordo, R. Guzman, B. Moyer, R. Ponce, R. Salmon, S. Moulis, J. Schwartz, S. Sequeira, E. Levine, A. Jungbluth, M. Roshal, Q. Gao, A. Dogan, S. Mailankody, S. Monette, S. Giralt, and O. Landgren for scientific and strategic discussions.
Funding: E.L.S. is a Special Fellow of The Leukemia & Lymphoma Society and an American Society of Hematology Scholar. Additional support was provided by an MSKCC Technology Development Grant, the Multiple Myeloma Research Foundation, the Lymphoma Research Foundation, and the Society of Immunotherapy for Cancer. S.C.A. reports support from the NIH (R01 HG008325 and R01 CA198095) and the Albert Einstein Cancer Center (P30 CA013330). R.J.B. reports support from the NIH (R01 CA138738-05, P01 CA059350, and PO1 CA190174-01), the annual Terry Fox Run for Cancer Research organized by the Canada Club of New York, Kate’s Team, the Carson Family Charitable Trust, the William Lawrence and Blanche Hughes Foundation, the Emerald Foundation, and the Experimental Therapeutics Center of MSKCC. All MSK investigators acknowledge the MSKCC Core Facilities Grants (P30 CA008748 and U54 OD020355-01).
Footnotes
Competing interests: E.L.S., R.J.B., and C.L. have licensed intellectual property to and collect royalties from Juno Therapeutics, A Celgene Company. E.L.S., R.J.B., and I.R. receive research funding from Juno Therapeutics, A Celgene Company. E.L.S. and R.J.B. are consultants for Juno Therapeutics, A Celgene Company. E.L.S. is a consultant for Fate Therapeutics. C.F.D.L. has received research funding and is a consultant for Celgene, Janssen, Takeda, and Amgen. P.W., H.L., Y.X., and C.L. are employed by and hold equity in Eureka Therapeutics. I.R. receives research funding from Fate Therapeutics. K.H., J.J., T.J.L., M.T.P., and E.O. are employed by and hold equity in Juno Therapeutics, A Celgene Company. B.S. and M.G. were employed at Juno Therapeutics, A Celgene Company during their involvement in all work within this manuscript. B.S. is currently an employee and equity shareholder of Lyell Immunopharma. M.G. is currently an employee of Poseida Therapeutics and is an equity shareholder of Celgene and Poseida Therapeutics. R.M. and E.P. were employed by MSKCC during their involvement in all work within this manuscript. R.M. is currently employed by AgenTus Therapeutics Inc. E.P. is currently employed by Regeneron. E.L.S., C.L., and R.J.B. acknowledge filed patent application WO2016/090312 “Chimeric antigen receptors targeting g-protein coupled receptor and uses thereof” related to the work disclosed in this paper. All other authors declare that they have no competing interests.
Data and materials availability: All data associated with this study are present in the paper or the Supplementary Materials. OPM2, RPMI-8226, and MM1.S human myeloma cell lines expressing ffLuc and guide RNAs for CRISPR KO of GPRC5D and BCMA are available from R.J.B. under a material transfer agreement with MSKCC.
REFERENCES AND NOTES
- 1.Kantarjian H, Stein A, Gökbuget N, Fielding AK, Schuh AC, Ribera J-M, Wei A, Dombret H, Foà R, Bassan R, Arslan Ö, Sanz MA, Bergeron J, Demirkan F, Lech-Maranda E, Rambaldi A, Thomas X, Horst H-A, Brüggemann M, Klapper W, Wood BL, Fleishman A, Nagorsen D, Holland C, Zimmerman Z, Topp MS, Blinatumomab versus chemotherapy for advanced acute lymphoblastic leukemia. N. Engl. J. Med 376, 836–847 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davila ML, Riviere I, Wang X, Bartido S, Park J, Curran K, Chung SS, Stefanski J, Borquez-Ojeda O, Olszewska M, Qu J, Wasielewska T, He Q, Fink M, Shinglot H, Youssif M, Satter M, Wang Y, Hosey J, Quintanilla H, Halton E, Bernal Y, Bouhassira DCG, Arcila ME, Gonen M, Roboz GJ, Maslak P, Douer D, Frattini MG, Giralt S, Sadelain M, Brentjens R, Efficacy and toxicity management of 19–28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci. Transl. Med 6, 224ra25 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park JH, Rivière I, Gonen M, Wang X, Sénéchal B, Curran KJ, Sauter C, Wang Y, Santomasso B, Mead E, Roshal M, Maslak P, Davila M, Brentjens RJ, Sadelain M, Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. N. Engl. J. Med 378, 449–459 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maude SL, Laetsch TW, Buechner J, Rives S, Boyer M, Bittencourt H, Bader P, Verneris MR, Stefanski HE, Myers GD, Qayed M, De Moerloose B, Hiramatsu H, Schlis K, Davis KL, Martin PL, Nemecek ER, Yanik GA, Peters C, Baruchel A, Boissel N, Mechinaud F, Balduzzi A, Krueger J, June CH, Levine BL, Wood P, Taran T, Leung M, Mueller KT, Zhang Y, Sen K, Lebwohl D, Pulsipher MA, Grupp SA, Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med 378, 439–448 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee DW, Kochenderfer JN, Stetler-Stevenson M, Cui YK, Delbrook C, Feldman SA, Fry TJ, Orentas R, Sabatino M, Shah NN, Steinberg SM, Stroncek D, Tschernia N, Yuan C, Zhang H, Zhang L, Rosenberg SA, Wayne AS, Mackall CL, T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: A phase 1 dose-escalation trial. Lancet 385, 517–528 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turtle CJ, Hanafi L-A, Berger C, Hudecek M, Pender B, Robinson E, Hawkins R, Chaney C, Cherian S, Chen X, Soma L, Wood B, Li D, Heimfeld S, Riddell SR, Maloney DG, Immunotherapy of non-Hodgkins lymphoma with a defined ratio of CD8+ and CD4+ CD19-specific chimeric antigen receptor-modified T cells. Sci. Transl. Med 8, 355ra116–355ra116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ali SA, Shi V, Maric I, Wang M, Stroncek DF, Rose JJ, Brudno JN, Stetler-Stevenson M, Feldman SA, Hansen BG, Fellowes VS, Hakim FT, Gress RE, Kochenderfer JN, T cells expressing an anti–B-cell maturation antigen chimeric antigen receptor cause remissions of multiple myeloma. Blood 128, 1688–1700 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brudno JN, Maric I, Hartman SD, Rose JJ, Wang M, Lam N, Stetler-Stevenson M, Salem D, Yuan C, Pavletic S, Kanakry JA, Ali SA, Mikkilineni L, Feldman SA, Stroncek DF, Hansen BG, Lawrence J, Patel R, Hakim F, Gress RE, Kochenderfer JN, T cells genetically modified to express an anti-B-cell maturation antigen chimeric antigen receptor cause remissions of poor-prognosis relapsed multiple myeloma. J. Clin. Oncol 36, 2267–2280 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laurent SA, Hoffmann FS, Kuhn P-H, Cheng Q, Chu Y, Schmidt-Supprian M, Hauck SM, Schuh E, Krumbholz M, Rübsamen H, Wanngren J, Khademi M, Olsson T, Alexander T, Hiepe F, Pfister H-W, Weber F, Jenne D, Wekerle H, Hohlfeld R, Lichtenthaler SF, Meinl E, γ-Secretase directly sheds the survival receptor BCMA from plasma cells. Nat. Commun 6, 7333 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen AD, Garfall AL, Stadtmauer EA, Lacey SF, Lancaster E, Vogl DT, Weiss BM, Ambrose DE, Nelson AM, Chen F, Plesa G, Kulikovskaya I, Gonzalez V, Gupta M, Young RM, Dengel K, O’keefe L, Le S, Richardson C, Isaacs RE, Melenhorst JJ, Levine BL, June CH, Milone MC, Safety and efficacy of B-cell maturation antigen (BCMA)-specific chimeric antigen receptor T cells (CART-BCMA) with cyclophosphamide conditioning for refractory multiple myeloma (MM). Blood 130, 505 (2017). [Google Scholar]
- 11.Gardner R, Wu D, Cherian S, Fang M, Hanafi L-A, Finney O, Smithers H, Jensen MC, Riddell SR, Maloney DG, Turtle CJ, Acquisition of a CD19-negative myeloid phenotype allows immune escape of MLL-rearranged B-ALL from CD19 CAR-T-cell therapy. Blood 127, 2406–2410 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sotillo E, Barrett DM, Black KL, Bagashev A, Oldridge D, Wu G, Sussman R, Lanauze C, Ruella M, Gazzara MR, Martinez NM, Harrington CT, Chung EY, Perazzelli J, Hofmann TJ, Maude SL, Raman P, Barrera A, Gill S, Lacey SF, Melenhorst JJ, Allman D, Jacoby E, Fry T, Mackall C, Barash Y, Lynch KW, Maris JM, Grupp SA, Thomas-Tikhonenko A, Convergence of acquired mutations and alternative splicing of CD19 enables resistance to CART-19 immunotherapy. Cancer Discov. 5, 1282–1295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fry TJ, Shah NN, Orentas RJ, Stetler-Stevenson M, Yuan CM, Ramakrishna S, Wolters P, Martin S, Delbrook C, Yates B, Shalabi H, Fountaine TJ, Shern JF, Majzner RG, Stroncek DF, Sabatino M, Feng Y, Dimitrov DS, Zhang L, Nguyen S, Qin H, Dropulic B, Lee DW, Mackall CL, CD22-targeted CAR T cells induce remission in B-ALL that is naive or resistant to CD19-targeted CAR immunotherapy. Nat. Med 24, 20–28 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao Y, Wang X, Yan H, Zeng J, Ma S, Niu Y, Zhou G, Jiang Y, Chen Y, Zhang M, Comparative transcriptome analysis of fetal skin reveals key genes related to hair follicle morphogenesis in cashmere goats. PLOS ONE 11, e0151118 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoue S, Nambu T, Shimomura T, The RAIG family member, GPRC5D, is associated with hard-keratinized structures. J. Invest. Dermatol 122, 565–573 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Kim Y-J, Yoon B, Han K, Park BC, Comprehensive transcriptome profiling of balding and non-balding scalps in trichorhinophalangeal syndrome type I patient. Ann. Dermatol 29, 597–601 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paus R, Nickoloff BJ, Ito T, A ‘hairy’ privilege. Trends Immunol. 26, 32–40 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Marr AK, Breitkopf T, Leung G, Hao J, Wang E, Kwong N, Akhoundsadegh N, Chen L, Mui A, Carr N, Warnock GL, Shapiro J, McElwee KJ, Hair follicle mesenchyme-associated PD-L1 regulates T-cell activation induced apoptosis: A potential mechanism of immune privilege. J. Invest. Dermatol 134, 736–745 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Westgate GE, Craggs RI, Gibson WT, Immune privilege in hair growth. J. Invest. Dermatol 97, 417–420 (1991). [DOI] [PubMed] [Google Scholar]
- 20.Atamaniuk J, Gleiss A, Porpaczy E, Kainz B, Grunt TW, Raderer M, Hilgarth B, Drach J, Ludwig H, Gisslinger H, Jaeger U, Gaiger A, Overexpression of G protein-coupled receptor 5D in the bone marrow is associated with poor prognosis in patients with multiple myeloma. Eur. J. Clin. Investig 42, 953–960 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Cohen Y, Gutwein O, Garach-Jehoshua O, Bar-Haim A, Kornberg A, GPRC5D is a promising marker for monitoring the tumor load and to target multiple myeloma cells. Hematology 18, 348–351 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Frigyesi I, Adolfsson J, Ali M, Christophersen MK, Johnsson E, Turesson I, Gullberg U, Hansson M, Nilsson B, Robust isolation of malignant plasma cells in multiple myeloma. Blood 123, 1336–1340 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Hudecek M, Sommermeyer D, Kosasih PL, Silva-Benedict A, Liu L, Rader C, Jensen MC, Riddell SR, The nonsignaling extracellular spacer domain of chimeric antigen receptors is decisive for in vivo antitumor activity. Cancer Immunol. Res 3, 125–135 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gomes-Silva D, Mukherjee M, Srinivasan M, Krenciute G, Dakhova O, Zheng Y, Cabral JMS, Rooney CM, Orange JS, Brenner MK, Mamonkin M, Tonic 4–1BB costimulation in chimeric antigen receptors impedes T cell survival and is vector-dependent. Cell Rep. 21, 17–26 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Long AH, Haso WM, Shern JF, Wanhainen KM, Murgai M, Ingaramo M, Smith JP, Walker AJ, Kohler ME, Venkateshwara VR, Kaplan RN, Patterson GH, Fry TJ, Orentas RJ, Mackall CL, 4–1BB costimulation ameliorates T cell exhaustion induced by tonic signaling of chimeric antigen receptors. Nat. Med 21, 581–590 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe N, Bajgain P, Sukumaran S, Ansari S, Heslop HE, Rooney CM, Brenner MK, Leen AM, Vera JF, Fine-tuning the CAR spacer improves T-cell potency. Oncoimmunology 5, e1253656 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ashouri JF, Weiss A, Endogenous Nur77 is a specific indicator of antigen receptor signaling in human T and B cells. J. Immunol 198, 657–668 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith EL, Staehr M, Masakayan R, Tatake IJ, Purdon TJ, Wang X, Wang P, Liu H, Xu Y, Garrett-Thomson SC, Almo SC, Riviere I, Liu C, Brentjens RJ, Development and evaluation of an optimal human single-chain variable fragment-derived BCMA-targeted CAR T cell vector. Mol. Ther 26, 1447–1456 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams TE, Nagarajan S, Selvaraj P, Zhu C, Concurrent and independent binding of Fc receptors IIa and IIIb to surface-bound IgG. Biophys. J 79, 1867–1875 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawson MA, Paton-Hough JM, Evans HR, Walker RE, Harris W, Ratnabalan D, Snowden JA, Chantry AD, Cui R, NOD/SCID-GAMMA mice are an ideal strain to assess the efficacy of therapeutic agents used in the treatment of myeloma bone disease. PLOS ONE 10, e0119546 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santos EB, Yeh R, Lee J, Nikhamin Y, Punzalan B, Punzalan B, La Perle K, Larson SM, Sadelain M, Brentjens RJ, Sensitive in vivo imaging of T cells using a membrane-bound Gaussia princeps luciferase. Nat. Med 15, 338–344 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.King MA, Covassin L, Brehm MA, Racki W, Pearson T, Leif J, Laning J, Fodor W, Foreman O, Burzenski L, Chase TH, Gott B, Rossini AA, Bortell R, Shultz LD, Greiner DL, Human peripheral blood leucocyte non-obese diabetic-severe combined immunodeficiency interleukin-2 receptor gamma chain gene mouse model of xenogeneic graft-versus-host-like disease and the role of host major histocompatibility complex. Clin. Exp. Immunol 157, 104–118 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Covassin L, Laning J, Abdi R, Langevin DL, Phillips NE, Shultz LD, Brehm MA, Human peripheral blood CD4 T cell-engrafted non-obese diabetic-scid IL2rγnull H2-Ab1tm1Gru Tg (human leucocyte antigen D-related 4) mice: A mouse model of human allogeneic graft-versus-host disease. Clin. Exp. Immunol 166, 269–280 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haso W, Lee DW, Shah NN, Stetler-Stevenson M, Yuan CM, Pastan IH, Dimitrov DS, Morgan RA, FitzGerald DJ, Barrett DM, Wayne AS, Mackall CL, Orentas RJ, Anti-CD22-chimeric antigen receptors targeting B-cell precursor acute lymphoblastic leukemia. Blood 121, 1165–1174 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Timmerman P, Puijk WC, Meloen RH, Functional reconstruction and synthetic mimicry of a conformational epitope using CLIPS™ technology. J. Mol. Recognit 20, 283–299 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.