Bloodstream infections (BSIs) have emerged as a worldwide concern because of their increasing incidence, mortality, and associated healthcare cost. Strains in ESKAPE including Enterococcus faecium (E. faecium), Staphylococcus aureus (S. aureus), Klebsiella pneumonia (K. pneumonia), Acinetobacter baumannii (A. baumannii), Pseudomonas aeruginosa (P. aeruginosa), and Enterobacter spp., as well as Escherichia coli (E. coli), account for more than half of the causative pathogens of BSI.[1] Moreover, the incidences and resistance rates of ESKAPE and E. coli have changed in recent years. A study in Rome found that E. coli, followed by S. aureus, was the most frequently identified microorganism; further, the study revealed that, regardless of hospital or community-onset, the number of extended-spectrum β-lactamases-producing E. coli was increasing, and carbapenemase-producing K. pneumoniae had become more prevalent during the study period.[1] A recent study suggested upward resistance trends of ESKAPE and E. coli organisms isolated from blood cultures from 2014 to 2018 in a hospital in Italy.[2] The frequencies and resistance rates of ESKAPE and E. coli are most likely interrelated with the spectrum of disease, geographical situation, antimicrobial management, and hand hygiene. Since 2012, some requirements have been gradually executed according to the Measures for the Administration of Clinical Application of Antibiotics (Decree No. 84 of the Ministry of Health), the Guidelines for Clinical Application of Antibiotics (2015 edition), and the Notice on Continuous Management of Clinical Application of Antibiotics (General Office of the National Health Commission, 2018) at our hospital. However, it is unclear whether these measures are effective to minimize the antibiotic resistance rate. This study investigated the resistance rates and antimicrobial resistance indexes (ARIs) of ESKAPE and E. coli isolated from blood samples at our hospital, and aimed to contribute to the research on ESKAPE and E. coli in China as well as the availability of methods for antibiotic administration.
A single-center retrospective study was conducted in the Huaihe Hospital of Henan University, a Class A tertiary general hospital. The Bioethics and Medical Ethics Committee of Henan University has approved this study (No. 4102010117436). Information on the resistance rates of ESKAPE and E. coli isolated from blood from January 2015 to December 2019 was collected from the database of the Microbiology Laboratory, using the WHONET 5.6 software (World Health Organization Collaborating Centre for Surveillance of Antimicrobial Resistance); no personal patient information was extracted in the study. All isolates were from the first specimens collected and considered to be indicative of community-acquired BSIs as the sample collection occurred within 48 h of hospitalization, and repeat specimens were excluded. Clinical and Laboratory Standards Institute M100-S20 was used as the reference standard for determining antimicrobial susceptibility. The ARI was estimated as previously reported by Ewing et al.[3] Briefly, scores of 0, 0.5, and 1 were assigned for susceptibility, intermediate resistance, and resistance, respectively, for each patient, and the total score of each antibiotic for each microorganism of ESKAPE and E. coli was divided by the number of the patients tested, then the sum of scores for all antibiotics was divided by the number of the antibiotics tested for each microorganism. Consequently, an ARI of 0 and 1 indicated pan-susceptible and pan-resistant microorganisms, respectively.
The antibiotic resistance rates for each microorganism of ESKAPE and E. coli from 2015 to 2019 were assessed with Pearson Chi-square or Fisher's exact test, and statistical significance was set at a two-tailed P value < 0.05.
There were 1343 isolates from blood culture specimens from 2015 to 2019, of which 982 indicated ESKAPE strains and E. coli; there were 528, 164, 92, 73, 40, 39, 39, and 7 cases of E. coli, K. pneumoniae, S. aureus, E. faecium, A. baumannii, P. aeruginosa, E. cloacae, and E. aerogenes, respectively. The analysis indicated that there was no statistically significant difference in the antibiotic resistance rates of E. coli, except for cefepime, for which the resistance rates significantly differed among five years (χ2 = 10.314, P = 0.035) and showed an annual declining trend using linear by linear association test (Z = 9.882, P = 0.002) [Figure 1A]. E. coli maintained stable resistance tendencies in recent years and was frequently susceptible to carbapenems. However, a recent study reported that the susceptibility rates of E. coli to cephalosporins decreased significantly in 2017 compared with 1998 in a large teaching hospital in China.[4]
Figure 1.
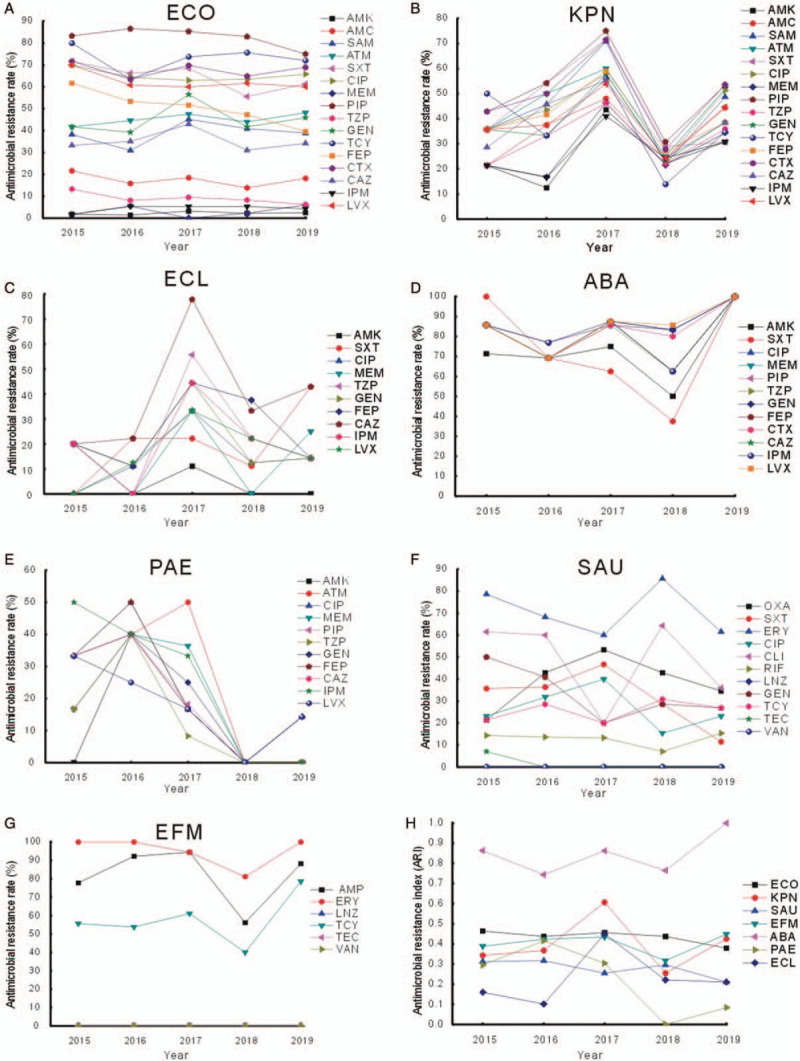
Resistance profiles of (A) Escherichia coli (ECO), (B) Klebsiella pneumoniae (KPN), (C) Enterobacter cloacae (ECL), (D) Acinetobacter baumannii (ABA), (E) Pseudomonas aeruginosa (PAE), (F) Staphylococcus aureus (SAU), (G) Enterococcus faecium (EFM), and (H) antimicrobial resistance index of ESKAPE and E. coli from 2015 to 2019. AMC: Amoxicillin; AMK: Amikacin; AMP: Ampicillin; ATM: Aztreonam; CAZ: Ceftazidime; CIP: Ciprofloxacin; CLI: Clindamycin; CTX: Cefotaxime; ERY: Erythromycin; ESKAPE: Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumonia, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.; FEP: Cefepime; GEN: Gentamicin; IPM: Imipenem; LNZ: Linezolid; LVX: Levofloxacin; MEM: Meropenem; OXA: Oxacillin; PIP: Piperacillin; RIF: Rifampicin; SAM: Ampicillin/sulbactam; SXT: Trimethoprim–sulfamethoxazole; TCY: Tetracycline; TZP: Piperacillin/tazobactam; TEC: Teicoplanin; VAN: Vancomycin.
K. pneumoniae showed general resistance (20–70%) to β-lactamases, aminoglycosides, and quinolones at our hospital, and the resistance rate increased significantly from 2015 to 2017 and subsequently decreased in 2018 and 2019, showing statistically significant differences in the resistance to ampicillin/sulbactam (χ2 = 15.903, P = 0.003), piperacillin (χ2 = 13.348, P = 0.009), cefotaxime (χ2 = 12.571, P = 0.013), sulfamethoxazole–trimethoprim (χ2 = 20.555, P = 0.000), ciprofloxacin (χ2 = 11.691, P = 0.019), gentamicin (χ2 = 10.611, P = 0.030), and tetracycline (χ2 = 13.403, P = 0.009). The other antibiotics included in this figure revealed the same tendency, despite no statistical significance [Figure 1B]. The resistance profile of E. cloacae presented a similar tendency—increasing in 2017 and subsequently decreasing in 2018 and 2019 [Figure 1C]. This tendency was possibly associated with the following factors: more rigorous antibiotic administration, including a classification system for antibiotic management by the usage grade (unrestricted, restricted, and special use); antibiotic use management in pediatric, elderly, and pregnant and postpartum patients; and hand hygiene, which has been emphasized since 2017 at our hospital.
Since 2015, A. baumannii showed a remarkably high susceptibility to all antibiotics tested, despite a slight decline of sensitivity to several antibiotics in 2018 [Figure 1D]. Notably, P. aeruginosa, a non-fermentative bacterium, presented continuously declining resistance rates to the antibiotics tested since 2016, with statistically significant differences in resistance to piperacillin (χ2 = 4.595, P = 0.040), cefepime (χ2 = 5.525, P = 0.025), ceftazidime (χ2 = 5.246, P = 0.025), and imipenem (χ2 = 7.062, P = 0.008) [Figure 1E]. P. aeruginosa can develop antibiotic resistance during therapy, which makes it difficult to eliminate; however, in this study, P. aeruginosa was susceptible to almost all of the tested antibiotics since 2018, which was mostly attributable to stringent supervision of antibiotic use.
There was no significant difference in the antibiotic resistance rates of S. aureus, except for clindamycin (χ2 = 9.740, P = 0.044), and vancomycin-and linezolid-resistant S. aureus strains were not reported from 2015 to 2019 [Figure 1F]. E. faecium, a gram-positive facultative anaerobe, steadily showed high resistance rates to ampicillin, erythromycin, and tetracycline and susceptibility to linezolid, teicoplanin, and vancomycin [Figure 1G]. However, proportions of vancomycin-resistant E. faecium isolated from patients with BSIs have increased since 2012 in Europe,[5] indicating a geographic difference for vancomycin-resistant Enterococcus (VRE). Therefore, E. faecium resistance should be evaluated regularly, although VRE was not detected at our hospital during the study period.
This study demonstrated that the ARI tendencies consistently correlated with the alterations in the ESKAPE and E. coli resistance rates during the study period [Figure 1H]. Therefore, ARI could be a significant indicator of the antibiotic resistance.
In summary, this study identified the resistance rates and ARI of ESKAPE and E. coli at our hospital, with a focus on not only the efficacy of empirical therapy for eliminating these pathogens but also predicting the tendency of antibiotic resistance, which was instrumental for infection control and antibiotic administration.
Conflicts of interest
None.
Footnotes
How to cite this article: Wei DD, Gao J, Yang RL, Bai CY, Lin XH. Antimicrobial resistance profiles of ESKAPE and Escherichia coli isolated from blood at a tertiary hospital in China. Chin Med J 2020;133:2250–2252. doi: 10.1097/CM9.0000000000000987
References
- 1.De Angelis G, Fiori B, Menchinelli G, D’Inzeo T, Liotti FM, Morandotti GA, et al. Incidence and antimicrobial resistance trends in bloodstream infections caused by ESKAPE and Escherichia coli at a large teaching hospital in Rome, a 9-year analysis (2007–2015). Eur J Clin Microbiol Infect Dis 2018; 37:1627–1636. doi: 10.1007/s10096-018-3292-9. [DOI] [PubMed] [Google Scholar]
- 2.De Socio GV, Rubbioni P, Botta D, Cenci E, Belati A, Paggi R, et al. Measurement and prediction of antimicrobial resistance in bloodstream infections by ESKAPE pathogens and Escherichia coli. Glob Antimicrob Resist 2019; 19:154–160. doi: 10.1016/j.jgar.2019.05.013. [DOI] [PubMed] [Google Scholar]
- 3.Ewing J, McCaughan J, Moore J, Fairley D, Sutherland B, Reid A, et al. Relative resistance index (RRI)—a scoring system for antibiotic resistance in Pseudomonas aeruginosa. Br J Biomed Sci 2017; 74:198–202. doi: 10.1080/09674845.2017.1338500. [DOI] [PubMed] [Google Scholar]
- 4.Tian L, Zhang Z, Sun Z. Antimicrobial resistance trends in bloodstream infections at a large teaching hospital in China: a 20-year surveillance study (1998-2017). Antimicrob Resist Infect Control 2019; 8:86.doi: 10.1186/s13756-019-0545-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ayobami O, Willrich N, Reuss A, Eckmanns T, Markwart R. The ongoing challenge of vancomycin-resistant Enterococcus faecium and Enterococcus faecalis in Europe: an epidemiological analysis of bloodstream infections. Emerg Microbes Infect 2020; 9:1180–1193. doi: 10.1080/22221751.2020.1769500. [DOI] [PMC free article] [PubMed] [Google Scholar]


