Abstract
Background
Prompt identification of patients suspected to have COVID-19 is crucial for disease control. We aimed to develop a deep learning algorithm on the basis of chest CT for rapid triaging in fever clinics.
Methods
We trained a U-Net-based model on unenhanced chest CT scans obtained from 2447 patients admitted to Tongji Hospital (Wuhan, China) between Feb 1, 2020, and March 3, 2020 (1647 patients with RT-PCR-confirmed COVID-19 and 800 patients without COVID-19) to segment lung opacities and alert cases with COVID-19 imaging manifestations. The ability of artificial intelligence (AI) to triage patients suspected to have COVID-19 was assessed in a large external validation set, which included 2120 retrospectively collected consecutive cases from three fever clinics inside and outside the epidemic centre of Wuhan (Tianyou Hospital [Wuhan, China; area of high COVID-19 prevalence], Xianning Central Hospital [Xianning, China; area of medium COVID-19 prevalence], and The Second Xiangya Hospital [Changsha, China; area of low COVID-19 prevalence]) between Jan 22, 2020, and Feb 14, 2020. To validate the sensitivity of the algorithm in a larger sample of patients with COVID-19, we also included 761 chest CT scans from 722 patients with RT-PCR-confirmed COVID-19 treated in a makeshift hospital (Guanggu Fangcang Hospital, Wuhan, China) between Feb 21, 2020, and March 6, 2020. Additionally, the accuracy of AI was compared with a radiologist panel for the identification of lesion burden increase on pairs of CT scans obtained from 100 patients with COVID-19.
Findings
In the external validation set, using radiological reports as the reference standard, AI-aided triage achieved an area under the curve of 0·953 (95% CI 0·949–0·959), with a sensitivity of 0·923 (95% CI 0·914–0·932), specificity of 0·851 (0·842–0·860), a positive predictive value of 0·790 (0·777–0·803), and a negative predictive value of 0·948 (0·941–0·954). AI took a median of 0·55 min (IQR: 0·43–0·63) to flag a positive case, whereas radiologists took a median of 16·21 min (11·67–25·71) to draft a report and 23·06 min (15·67–39·20) to release a report. With regard to the identification of increases in lesion burden, AI achieved a sensitivity of 0·962 (95% CI 0·947–1·000) and a specificity of 0·875 (95 %CI 0·833–0·923). The agreement between AI and the radiologist panel was high (Cohen's kappa coefficient 0·839, 95% CI 0·718–0·940).
Interpretation
A deep learning algorithm for triaging patients with suspected COVID-19 at fever clinics was developed and externally validated. Given its high accuracy across populations with varied COVID-19 prevalence, integration of this system into the standard clinical workflow could expedite identification of chest CT scans with imaging indications of COVID-19.
Funding
Special Project for Emergency of the Science and Technology Department of Hubei Province, China.
Introduction
As of July 19, 2020, 14 043 176 confirmed COVID-19 cases and 597 583 deaths had been reported globally.1 Early identification of patients with COVID-19 has been recommended by WHO to control transmission and to prevent depletion of hospital resources.2, 3
RT-PCR testing is the gold standard for confirming severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection.4, 5 However, not all countries have sufficient RT-PCR testing capacity. Additionally, due to technological constraints, even in developed countries, RT-PCR can take up to 3 days to provide a result.6 Furthermore, studies have found that RT-PCR testing can produce false negative results,7 which could result in patients with COVID-19 remaining unidentified in the community, enabling the epidemic to continue to spread despite aggressive interventions such as regional lockdowns.8 Chest CT has been used to supplement RT-PCR testing of patients with suspected COVID-19.9, 10, 11, 12
In China, patients with symptoms such as cough and fever are required to visit fever clinics. Patients with lung opacities indicating viral infection (eg, ground-glass opacities and consolidations) on chest CT with COVID-19 related epidemiological history, are defined as suspected cases, and are isolated and tested with RT-PCR (appendix p 5).13 In the guidance issued by the British Society of Thoracic Imaging, chest CT is used as a radiological decision tool, but is limited to seriously ill patients with suspected COVID-19, for whom chest x-ray results are uncertain or normal.14, 15
Research in context.
Evidence before this study
We searched Google Scholar for deep learning studies on the triage of patients with suspected COVID-19 on the basis of chest CT published between Dec 1, 2019, and March 22, 2020, using the search terms “COVID-19” OR “2019-nCoV” OR “Coronavirus Disease 2019” OR “Novel Coronavirus” AND “chest CT” AND “Triage” AND “Deep learning”. Our search yielded no studies that developed and validated deep learning algorithms to triage patients with suspected COVID-19. After removal of the search term “Triage”, we identified seven studies (one peer-reviewed publication and six preprint articles) that developed and validated deep learning algorithms for differential diagnosis associated with COVID-19 on the basis of assembled CT datasets that contained a small number of real-time PCR confirmed COVID-19 cases. Assembled datasets that combined COVID-19 cases with other types of pneumonia might not represent the distribution of COVID-19 in real-world settings. Few studies applied external validation to test the performance of algorithms and therefore could not rule out the possibility of model overfitting (ie, whereby an algorithm can perform well on patients from the same data source used for algorithm training, but poorly on data obtained from different sources).
Added value of this study
We developed a deep learning algorithm for triaging patients with suspected COVID-19 and analysing lesion burden of patients with confirmed COVID-19 on the basis of chest CT. We trained the algorithm on the largest available set of confirmed COVID-19 cases and validated the algorithm on multiple datasets, which indicated the algorithm was robust with high clinical efficacy. We compared two proposed AI triage pathways (scan-to-second-reader triage and scan-to-fever-clinician triage) with standard of care in a fever clinic, and showed that both workflows increased the efficiency of suspected case identification. We also considered the potential value of deep learning in COVID-19 clinical management across different health-care systems, which showed that the developed AI system might also assist radiologists to precisely assess how lesion burden changed over time on CT imaging since AI's performance was satisfactory with 96·2% sensitivity and 87·5% specificity.
Implications of all the available evidence
The robust and satisfactory performance of our deep learning algorithm indicates its potential clinical use for screening patients with suspected COVID-19 in fever clinics and monitoring disease progression among patients with confirmed COVID-19. Shortening the time to diagnosis would enable earlier isolation and treatment of affected patients, which is crucial to curb the pandemic.
In addition to supplementing COVID-19 diagnosis when RT-PCR testing is not available or returns false negative results,16 CT imaging is important for monitoring changes in disease burden.17, 18 According to the Chinese COVID-19 clinical guidance, patients with lung opacities on CT that increase by 50% within 24–48 h require immediate clinical intervention.5
During the COVID-19 pandemic, heavy reliance on CT imaging in Chinese hospitals has increased the burden on radiologists.18 At fever clinics, CT reports are usually required within 1 h of the chest scan. At treatment hospitals, radiologists need to carefully compare opacities on CT scans across time to alert cases suspected of deterioration.19 Time pressure, heavy workload, and a shortage of experienced radiologists resulted in challenges for imaging-based management of COVID-19.
In previous studies, deep learning-based artificial intelligence (AI) algorithms have been applied in multiple imaging tasks such as the diagnosis of skin cancer malignancy,20 breast cancer detection,21 and cerebral haemorrhage triage.22 To expedite chest CT-based triage in fever clinics, we aimed to develop a fully automated deep learning algorithm to flag suspected COVID-19 cases and analyse lesion burdens. By validating the algorithm on fever clinic cases across regions with variable COVID-19 prevalence, we aimed to assess the clinical value of the developed algorithm in real-world scenarios.
Methods
Study design
We did a retrospective diagnostic study, using CT images obtained from Tongji Hospital (Wuhan, China), and CT images and radiological reports obtained from three fever clinics (Tianyou Hospital [Wuhan, China], Xianning Central Hospital [Xianning, China], and The Second Xiangya Hospital [Changsha, China]). We also obtained unenhanced chest CT scans from patients with RT-PCR-confirmed COVID-19 treated in a single makeshift hospital (Guanggu Fangcang Hospital, Wuhan China) to validate the sensitivity of the algorithm.
We trained a U-Net-based deep learning model23 to segment lung opacities on chest CT. Opacity segmentation could automatically analyse lung lesion volumes and alert positive CT scans to expedite patient triage. Full details of algorithm development are in the appendix (pp 1–3). The triage cutoff threshold was determined on the basis of the receiver operating characteristic curve of an internal validation set. The accuracy and efficiency of AI triage was then assessed on an external validation dataset. Figure 1A shows the research pipeline of the study.
Figure 1.
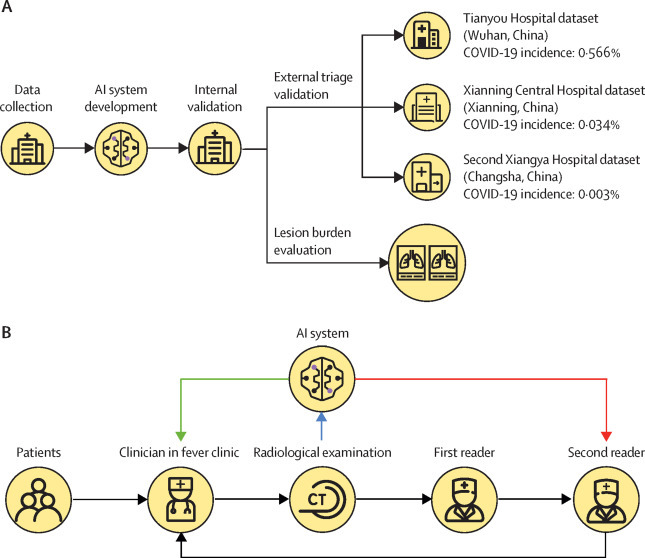
Development and validation of a deep learning algorithm to provide rapid triage in fever clinics and to automatically analyse lung opacities on the basis of chest CT scans
(A) Overview of the development and validation of the algorithm. (B) Evaluation of triage efficiency; black lines show the standard workflow in Chinese fever clinics; after a patient's CT examination is completed, a first reader drafts a radiology report in a first-in-first-out order and then a second radiologist revises and approves the first reader's report before sending it to a fever clinician; after receiving the radiological report the fever clinician decides whether the patient qualifies as a suspected case and should receive RT-PCR testing; we proposed that through directly notifying either the second radiologist (ie, scan-to-second-reader triage; red line) or the fever clinician (scan-to-fever-clinician triage; green line) of suspected cases triaged by AI, the workflow in fever clinics could be expedited. AI=artificial intelligence.
This study was approved by the Institutional Review Board of each participating hospital (appendix p 13). Written informed consent was waived due to the retrospective nature of the study. All data were fully anonymised.
CT image datasets and algorithm development
Patients with RT-PCR-confirmed COVID-19 who were admitted to Tongji Hospital (Wuhan, China) between Feb 1, 2020, and March 3, 2020, were identified and their unenhanced chest CT scans (appendix p 8) were retrieved from the Picture Archiving and Communication System of Tongi Hospital (video). The scans were obtained using a variety of scanner models and manufacturers. We also collected patient demographic information and RT-PCR test results from electronic medical records. Unenhanced CT chest scans for 2191 adult patients (aged >14 years) with COVID-19 and 1000 adult patients without COVID-19 who were admitted to Tongji Hospital during the same time period and had double negative RT-PCR test results were selected for algorithm development. The patients in the non-COVID-19 group might or might not have had positive CT findings. For patients who had undergone multiple CT scans, we used the first scan that had COVID-19 imaging manifestation for algorithm development.
The dataset was randomly split into a development set (1674 patients with COVID-19; 800 patients without COVID-19) and an internal validation set (439 patients with COVID-19; 200 patients without COVID-19) in a ratio of 8:2.23 Positive cases in the development set were annotated by radiologists (appendix p 4). 105 cases were excluded due to difficulty with annotation. After data annotation, the development set was randomly split into a training set (1318 patients with COVID-19; 640 patients without COVID-19) and a testing set (329 patients with COVID-19; 160 patients without COVID-19) with a ratio of 8:2.23 Full details of data inclusion and exclusion criteria and data partition are shown in figure 2A .
Figure 2.
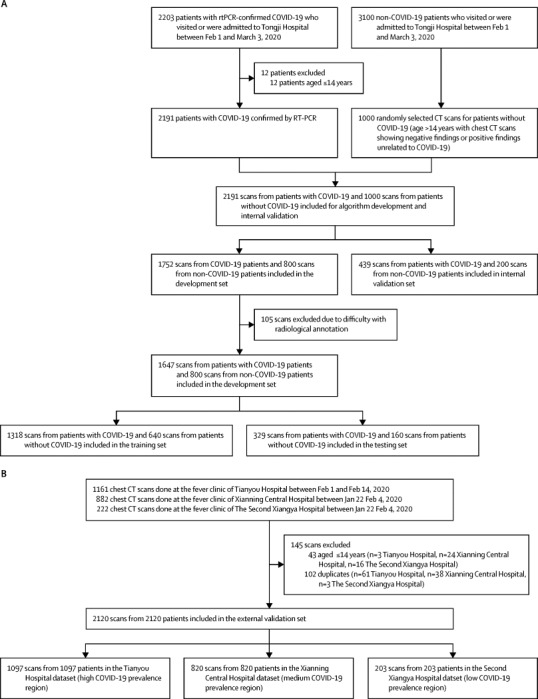
Data collection
(A) Dataset for algorithm development and internal validation. (B) Dataset for external validation.
Data collection for external validation of triage performance
To assess the accuracy and efficiency of the algorithm for patient triage across populations with different COVID-19 prevalences, we retrospectively collected consecutive unenhanced chest CT scans done over 2 weeks between Jan 22, 2020, and Feb 14, 2020, at three fever clinics: Tianyou Hospital (Wuhan, China; area of high COVID-19 prevalence), Xianning Central Hospital (Xianning, China; area of medium COVID-19 prevalence), and The Second Xiangya Hospital (Changsha, China; area of low COVID-19 prevalence). Corresponding radiological reports were retrieved from the Radiology Information System of each hospital. We obtained RT-PCR results if available. Patients with positive CT findings might not have had a COVID-19 related epidemiological history or blood test results, thus, such patients were not suspected to have COVID-19, and were not tested with RT-PCR.5
The external validation set included 2120 scans from 2120 patients (1097 scans from Tianyou Hospital; 820 scans from Xianning Central Hospital; 203 scans from The Second Xiangya Hospital; figure 2B; appendix p 8). Of the 2120 patients included in the external validation set, 411 patients at Tianyou Hospital, 369 patients at Xianning Central Hospital, and 130 patients at The Second Xiangya Hospital had an RT-PCR test, of whom 118, 87, and 12 were COVID-19 positive, respectively.
Triage performance was assessed for accuracy and efficiency; the original radiological reports were used as the reference standard. Two radiologists who were independent of those who wrote or approved the original reports, classified cases into four categories: 1, clear mention of suspected COVID-19 in the radiological impression section; 2, ambiguous radiological impression description, but presence of COVID-19 imaging features in the radiological findings section; 3, ambiguous radiological impression description but absence of COVID-19 imaging features in the radiological findings section; 4, negative radiological findings. The first radiologist (HQ) rated all reports and marked cases for which categorisation was unclear. The second radiologist (ZD) subsequently reviewed the unclear cases and made final decisions. The two radiologists were masked to the results of AI-aided triage. Cases with scores of 1 or 2 were categorised as COVID-19-positive, and cases with scores of 3 or 4 were defined as COVID-19-negative.
Figure 1B shows a typical workflow in a Chinese fever clinic and scan-to-second-reader triage and scan-to-fever-clinician triage were proposed to expedite clinical workflow. To measure AI triage time, we recorded the time AI took to flag each true positive case. To measure the triage time of standard of care, we calculated the time intervals between CT exam completion and initial draft report and between CT exam completion and senior radiologists' report approval based on timestamps recorded by the Radiology Information System of each hospital or fever clinic.
Only a small proportion of patients who were included in the external validation set were diagnosed with COVID-19, thus to validate the sensitivity of the algorithm in a larger sample of patients with COVID-19, we included 761 unenhanced chest CT scans from 722 patients with RT-PCR-confirmed COVID-19 treated in a makeshift hospital (Guanggu Fangcang Hospital) in Wuhan, China, between Feb 21, 2020, and March 6, 2020. Since only asymptomatic individuals or those with mild COVID-19 were admitted to Fangcang hospitals in China, this patient population represented more challenging cases for both chest CT and AI since disease manifestation on images could be subtle or even absent. Considering the issue of false negatives with RT-PCR testing,10 to test the specificity of the algorithm on non-COVID-19 cases, we included 686 scans from 651 patients who visited Tianyou Hospital or The Third People's Hospital of Shenzhen for respiratory diseases between Oct 1, 2019, and Oct 31, 2019, before the COVID-19 outbreak (405 scans from 385 patients from Tianyou Hospital; 281 scans from 266 patients from The Third People's Hospital of Shenzhen). 652 (95%) of 686 chest CT scans had positive findings such as ground glass opacities, pulmonary fibrosis, consolidations, inter_stitial thickening, pleural effusion, emphysema, and nodules or masses.
Data collection for assessment of change in lesion burden
The developed model could automatically calculate lung lesion burden volumes, thus, we collected pairs of CT scans from the same patients with COVID-19 to assess the accuracy of AI for the identification of lesion burden increase in comparison to radiologists. Since radiologists are at a disadvantage compared with AI when assessing lung lesion burden volumes (in cm3 or percentages) using the naked eye, we developed a qualitative task in which radiologists judged whether a new scan showed an increase in lung lesion burden volume when compared with a previous scan. This task was also more clinically relevant than estimating the lung lesion burden volume in cm3 or percentages because in real-world settings, radiologists are responsible for reporting disease progression, such as increases in lesion volume or size. We consecutively included 100 patients with RT-PCR-confirmed COVID-19 who were admitted to Tongji Hospital between Dec 26, 2019, and Jan 31, 2020 (no cases overlapped with algorithm development or internal validation) and had undergone at least two CT scans. For patients with more than two scans, the first two scans were selected. A panel of three radiologists (CW and others) served as the reference standard. Each radiologist independently classified each second scan as either increase (increase in lesion burden volume) or no increase (no change or decrease in lesion burden volume) and marked cases that were difficult to categorise. Consensus was reached through majority vote among the three radiologists. For the AI algorithm, cases with lesion burden volumes segmented in the second scan that were larger than that in the first scan were categorised as increase, and cases with no changes were categorised as no increase.
Statistical analysis
Sample size was calculated before collection of the external validation dataset, and was estimated on the basis of performance targets of 90% for sensitivity and 80% for specificity (appendix p 4). To assess the accuracy of triage, receiver operating characteristic curves were plotted and the area under the curve was calculated. Sensitivity, specificity, positive predictive value, and negative predictive value at a fixed threshold (a specificity above the target threshold of 80%) were estimated and 95% CIs were estimated with bootstrapping (10 000 replicates). To assess the efficiency of AI-aided triage, we used log transformation since time intervals might not have a normal distribution. To assess the efficiency of AI-aided triage, reduction in time to triage was assessed using Student's t tests. The full statistical analysis plan is described in the appendix (p 4).
We used Cohen's kappa coefficient to assess the agreement between AI and the radiologist panel with regard to increase in lesion burden. The interrater agreement of the radiologist panel was calculated using Fleiss' kappa.
Continuous variables were reported as median and IQR. Categorical variables were reported as frequencies and percentages. A two-sided p value of less than 0·05 was considered statistically significant. All statistical analyses were done using R (version 3·6.2).
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
The clinical characteristics of the patients in the Tongji hospital dataset used for development and internal validation of the model are summarised in table 1 . The dataset included 3086 CT images (2086 patients with COVID-19 and 1000 patients without COVID-19). For the internal validation set, a specificity of 85% was selected as a cutoff for flagging positive cases in the external validation datasets. AI-aided triage achieved an area under the curve of 0·985 (95% CI 0·982–0·989) with a sensitivity of 0·973 (0·966–0·980), specificity of 0·850 (0·827–0·875), positive predictive value of 0·934 (0·924–0·946), and a negative predictive value of 0·934 (0·917–0·952).
Table 1.
Characteristics of the Tongji dataset used for algorithm development and internal validation
| Tongji dataset (n=3086) | Development set (n=2447) | Internal validation set (n=639) | ||
|---|---|---|---|---|
| Sex | ||||
| Male | 1544 (50%) | 1235 (50%) | 309 (48%) | |
| Female | 1542 (50%) | 1212 (50%) | 330 (52%) | |
| Age, years | 55 (39–67) | 55 (39–67) | 55 (38–66) | |
| COVID-19 positive CT scans | 2086 (68%) | 1647 (67%) | 439 (69%) | |
| COVID-19 negative CT scans | 1000 (32%) | 800 (33%) | 200 (31%) | |
| CT manufacturers | ||||
| GE Medical System (Chicago, IL, USA) | 140 (5%) | 122 (5%) | 18 (3%) | |
| MinFound Medical Systems (Shaoxing, China) | 6 (<1%) | 6 (<1%) | 0 | |
| Siemens (Munich, Germany) | 2180 (71%) | 1723 (70%) | 457 (72%) | |
| Toshiba (Tokyo, Japan) | 74 (2%) | 51 (2%) | 23 (4%) | |
| United Imaging Healthcare (Shanghai, China) | 686 (22%) | 545 (22%) | 141 (22%) | |
Data are n (%), or median (IQR).
The external validation dataset included 2120 CT scans (median age 43 years [IQR: 31–56]; 1041 [49%] women, 1079 [51%] men). Regarding accuracy, using radiological reports as the reference standard, AI-aided triage achieved an area under the curve of 0·953 (95% CI 0·949–0·959), with a sensitivity of 0·923 (95% CI 0·914–0·932), specificity of 0·851 (0·842–0·860), a positive predictive value of 0·790 (0·777–0·803), and a negative predictive value of 0·948 (0·941–0·954). The sensitivity and specificity of AI-aided triage were significantly higher than the performance targets of 90% and 80% (p=0·0299 for sensitivity; p<0·0001 for specificity). The performance of AI-aided triage for each hospital cohort is shown in table 2 and receiver operating characteristics are shown in figure 3 . AI-aided triage achieved an area under the curve of 0·966 (95% CI 0·961–0·971) for the Tianyou Hopsital dataset, 0·931 (0·921–0·945) for the Xianning Central Hospital dataset, and 0·908 (0·888–0·929) for The Second Xiangya Hospital dataset.
Table 2.
Characteristics of the external validation dataset and accuracy of AI-aided triage
| External validation set (n=2120) | Tianyou Hospital dataset*(n=1097) | Xianning Central Hospital dataset†(n=820) | The Second Xiangya Hospital dataset‡(n=203) | ||
|---|---|---|---|---|---|
| Sex | |||||
| Male | 1079 (51%) | 506 (46%) | 463 (56%) | 110 (54%) | |
| Female | 1041 (49%) | 591 (54%) | 357 (44%) | 93 (46%) | |
| Age, years | 43 (31–56) | 48 (36–58) | 34 (27–49) | 45 (34–63) | |
| CT scans, n | 2120 | 1097 | 820 | 203 | |
| CT manufacturers | |||||
| GE Medical System (Chicago, IL, USA) | 1730 (82%) | 978 (90%) | 752 (92%) | 0 | |
| Siemens (Munich, Germany) | 271 (13%) | 0 | 68 (8%) | 203 (100%) | |
| United Imaging Healthcare (Shanghai, China) | 119 (6%) | 119 (11%) | 0 | 0 | |
| CT findings | |||||
| Positive | 802 (38%)§ | 547 (50%) | 180 (22%) | 75 (37%) | |
| Negative | 1318 (62%)¶ | 550 (50%) | 640 (78%) | 128 (63%) | |
| AI-aided triage performance‖ | |||||
| Sensitivity (95% CI) | 0·923 (0·914–0·932) | 0·934 (0·925–0·944) | 0·900 (0·880–0·924) | 0·893 (0·862–0·932) | |
| Specificity (95% CI) | 0·851 (0·842–0·860) | 0·855 (0·840–0·868) | 0·859 (0·846–0·874) | 0·789 (0·752–0·828) | |
| Positive predictive value (95% CI) | 0·790 (0·777–0·803) | 0·865 (0·851–0·878) | 0·643 (0·613–0·673) | 0·713 (0·662–0·764) | |
| Negative predictive value (95% CI) | 0·948 (0·941–0·954) | 0·929 (0·919–0·940) | 0·968 (0·962–0·976) | 0·927 (0·905–0·953) | |
| AUC (95% CI) | 0·953 (0·949–0·959) | 0·966 (0·961–0·971) | 0·931 (0·921–0·945) | 0·908 (0·888–0·929) | |
| RT-PCR testing | |||||
| Positive | 217/910 (24%) | 118/411 (29%) | 87/369 (24%) | 12/130 (9%) | |
| Negative | 693/910 (76%) | 293/411 (71%) | 282/369 (76%) | 118/130 (91%) | |
| AI-aided triage performance** | |||||
| Sensitivity (95% CI) | 0·876 (0·854–0·898) | 0·907 (0·883–0·935) | 0·839 (0·803–0·880) | 0·833 (0·750–1·000) | |
| Specificity (95% CI) | 0·519 (0·501–0·539) | 0·386 (0·358–0·401) | 0·660 (0·633–0·686) | 0·517 (0·473–0·562) | |
| Positive predictive value (95% CI) | 0·363 (0·343–0·384) | 0·373 (0·345–0·401) | 0·432 (0·392–0·469) | 0·149 (0·109–0·189) | |
| Negative predictive value (95% CI) | 0·930 (0·918–0·944) | 0·911 (0·889–0·939) | 0·930 (0·913–0·949) | 0·968 (0·957–1·000) | |
| AUC (95% CI) | 0·774 (0·757–0·791) | 0·725 (0·699–0·751) | 0·837 (0·810–0·866) | 0·679 (0·608–0·781) | |
| RT-PCR positive and CT positive cases | 191/217 (88%) | 109/118 (92%) | 72/87 (83%) | 10/12 (83%) | |
| Sensitivity of AI-aided triage on RT-PCR positive and CT positive cases (95% CI) | 0·974 (0·966–0·987) | 0·972 (0·964–0·989) | 0·972 (0·963–1·000) | 1·000 (1·000–1·000) | |
Data are n (%), median (IQR), or n/N (n%), unless otherwise stated. AUC=area under the receiver operating curve.
Dataset from Wuhan (Hubei province): 2018 population, 8 837 300 (according to National Bureau of Statistics of China), 50 006 confirmed COVID-19 cases (calculated up to March 25, 2020, according to the National Health Commission of the People's Republic of China), and a disease prevalence of 0·566% (ie, number of confirmed cases in the total population).
Dataset from Xianning (Hubei province): 2018 population, 2 485 000 (according to National Bureau of Statistics of China), 836 confirmed COVID-19 cases (calculated up to March 25, 2020, according to the National Health Commission of the People's Republic of China), and and a disease prevalence of 0·034% disease prevalence.
Dataset from Changsha (Hunan province): 2018 population, 7 288 600 (according to 2018 National Bureau of Statistics of China data), 242 confirmed COVID-19 cases (calculated up to March 25, 2020, according to the National Health Commission of the People's Republic of China), and a disease prevalence of 0·003% disease prevalence.
Of 802 positive CT scans, 772 clearly mentioned COVID-19 signs in the radiological impression section and 30 had an ambiguous radiological impression description but described COVID-19 signs in the radiological findings section (11 in the Tianyou Hopsital dataset, nine in the Xianning Central hospital dataset, and ten in the Second Xiangya Hospital dataset).
Of 1318 negative CT studies, 593 had negative CT findings and 725 had positive findings not associated with COVID-19 (324 in the Tianyou Hopsital dataset, 291 in the Xianning Central Hospital dataset, and 110 in the Second Xiangya Hopsital dataset).
Radiological CT findings were used as the reference standard.
RT-PCR was used as the reference standard.
Figure 3.
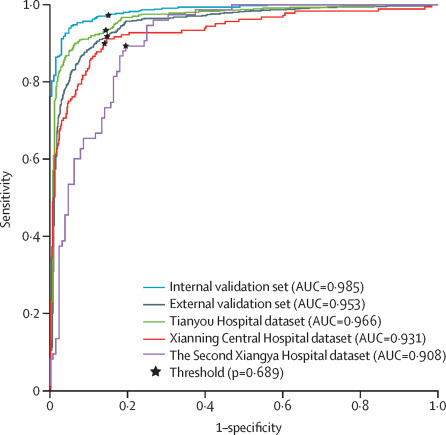
AI triage accuracy for the internal validation set, external validation set overall, and three individual hospital datasets
The black points indicate sensitivity and specificity thresholds used. AUC=area under the receiver operating curve.
Since RT-PCR is the gold standard for COVID-19 confirmation we also assessed AI's performance with RT-PCR as the reference standard. For patients with positive RT-PCR results from Tianyou Hospital, Xianning Central Hospital, and The Second Xiangya Hospital, using RT-PCR as the reference standard, AI-aided triage achieved sensitivities of 0·907 (95% CI 0·883–0·935), 0·839 (0·803–0·880), and 0·833 (0·750–1·000), and specificities of 0·386 (95% CI 0·358–0·401), 0·660 (0·633–0·686), and 0·517 (0·473–0·562), respectively. In the Guanggu Fangcang Hospital dataset (761 unenhanced chest CT scans from 722 patients [including duplicate scans]; median age 49 years [IQR 37–57]; 355 [49%] women, 367 [51%] men) which only included RT-PCR-positive patients, AI-aided triage had a sensitivity of 0·736 (95% CI 0·720–0·752 (appendix p 6). Of the 761 CT scans from the Guanggu Fangcang Hospital dataset, 618 (81%) patients had radiological features associated with COVID-19. The sensitivity of AI for the detection of abnormal opacities on these CT scans was 0·886 (95% CI 0·873–0·898; appendix p 6). For the 686 scans of confirmed non-COVID-19 patients done at Tianyou Hospital and The Third People's Hospital of Shenzhen before the COVID-19 outbreak (median age 57 years [IQR 43–68]; 246 [38%] women, 405 [62%] men), the specificity of AI for patients without COVID-19 was 0·822 (95% CI 0·808–0·836; appendix p 6).
AI took a median of 0·55 min (IQR 0·43–0·63) to flag positive cases compared with the standard-of-care workflow, which took radiologists 16·21 min (11·67–25·71) to draft a report and 23·06 min (15·67–39·20) to approve a report and send it to fever clinicians. In the scan-to-second-reader triage workflow, the time taken to flag positive cases was reduced by a median of 15·73 min (IQR 11·05–25·25); and in the scan-to-fever-clinician triage workflow whereby AI directly informed fever clinicians, the time taken to flag positive cases was reduced by a median of 22·62 min (IQR 15·12–38·63; table 3 ). AI significantly improved the efficiency of scan-to-second reader triage at the three fever clinics studied (14·03 min [IQR 10·13–20·55] for Tianyou Hospital; 24·31 min (15·13–43·40) for Xianning Central Hospital; 25·24 min (19·65–38·48) for The Second Xiangya Hospital; p<0·0001) and also improved the efficiency of scan-to-fever-clinician triage at each hospital (18·77 min [13·88–26·73] Tianyou Hospital; 47·03 min (26·53–95·83) for Xianning Central Hospital; 198·28 min [45·31–675·26] for The Second Xiangya Hospital; p<0·0001).
Table 3.
Triage efficiency for the external validation set
| External validation set | Tianyou Hospital dataset | Xianning Central Hospital dataset | The Second Xiangya Hospital dataset | ||
|---|---|---|---|---|---|
| True positive scans, n | 698 | 511 | 129 | 58 | |
| Median draft report time (IQR), min* | 16·21 (11·67–25·71) | 14·50 (10·75–21·11) | 24·75 (15·67–43·72) | 25·73 (20·10–38·84) | |
| Median report approval time (IQR), min* | 23·06 (15·67–39·20) | 19·23 (14·33–27·33) | 47·37 (27·12–96·35) | 198·77 (45·85–675·83) | |
| Median AI triage time (IQR), min* | 0·55 (0·43–0·63) | 0·58 (0·45–0·64) | 0·50 (0·42–0·58) | 0·48 (0·44–0·53) | |
| Median reduction in triage time under scan-to-second-reader triage workflow (IQR), min | 15·73 (11·05–25·25) | 14·03 (10·13–20·55) | 24·31 (15·13–43·40) | 25·24 (19·65–38·48) | |
| p value† | <0·0001 | <0·0001 | <0·0001 | <0·0001 | |
| t | 257·42 | 243·09 | 114·59 | 69·32 | |
| Median reduction in triage time under scan-to-fever-clinician triage workflow (IQR), min | 22·62 (15·12–38·63) | 18·77 (13·88–26·73) | 47·03 (26·53–95·83) | 198·28 (45·31–675·26) | |
| p value† | <0·0001 | <0·0001 | <0·0001 | <0·0001 | |
| t | 188·08 | 253·61 | 87·37 | 50·78 | |
Raw data did not follow a normal distribution.
Calculated by comparing the results with zero.
Of the 100 patients with RT-PCR-confirmed COVID-19 (median age 53 years (IQR 41–65); 50 [50%] women, 50 [50%] men) who had two CT scans at Tongji Hospital, based on consensus the radiologist panel rated 52 cases as showing an increase in lesion burden volume and 48 cases as showing no increase. The median time interval between two scans was 5 days (IQR 4–8). For 35 (35%) of 100 cases, at least one radiologist indicated difficulty in categorising changes in lesion burden. Agreement between the three radiologists was high (Fleiss' kappa 0·786). AI had a sensitivity of 0·962 (0·947–1·000) for the identification of cases with increases in lesion burden volume and a specificity of 0·875 (0·833–0·923) for cases with no increases in lesion burden volume (table 4 ). Agreement between AI and the radiologist panel was high (Cohen's kappa coefficient 0·839, 95% CI 0·718–0·940).
Table 4.
Performance of AI and radiologists for the identification of changes in lesion burden between two CT scans
| AI | Radiologist 1 | Radiologist 2 | Radiologist 3 | |
|---|---|---|---|---|
| True positive* | 50 | 47 | 52 | 51 |
| True negative† | 42 | 47 | 42 | 45 |
| False positive‡ | 6 | 1 | 6 | 3 |
| False negative§ | 2 | 5 | 0 | 1 |
| Accuracy (95% CI) | 0·920 (0·900–0·950) | 0·940 (0·925–0·962) | 0·940 (0·925–0·962) | 0·960 (0·950–0·988) |
| Sensitivity (95% CI) | 0·962 (0·947–1·000) | 0·904 (0·872–0·951) | 1·000 (1·000–1·000) | 0·981 (0·974–1·000) |
| Specificity (95% CI) | 0·875 (0·833–0·923) | 0·979 (0·971–1·000) | 0·875 (0·833–0·923) | 0·938 (0·917–0·974) |
Data are n, unless stated otherwise. 52 patients had an increase in lesion burden volume and were defined as positive. 48 patients did not have any increase in lesion burden volume and were defined as negative. We presented the complete information to show interrater variability. AI=artificial intelligence.
Correct prediction of lesion burden volume increase.
Correct prediction of no increase in lesion burden volume.
Incorrect prediction of lesion burden volume increase.
Incorrect prediction of no increase in lesion burden volume.
Discussion
To the best of our knowledge, this study was the first to develop and validate an AI algorithm for triaging suspected COVID-19 cases on the basis of chest CT in fever clinics. A large sample of chest CT scans from RT-PCR-confirmed COVID-19 cases were obtained to develop the deep learning algorithm and consecutive cases were collected from regions of varying COVID-19 prevalence to assess the accuracy and efficiency of AI triage, using radiological reports as the reference standard.
The 2-week imaging workload was higher in high-prevalence regions (Tianyou Hospital and Xianning Central Hospital) than in low-prevalence regions (The Second Xiangya Hospital). Additionally, the requirement for radiological responsiveness inside the epidemic centre was higher than that outside the epidemic centre. Regarding the accuracy of AI-aided triage, the overall sensitivity and specificity were above the performance targets of 90% and 80%. The general pattern across patient populations showed that COVID-19 prevalence could influence AI's performance, with highest performance in the Tianyou Hospital dataset (high prevalence) and the lowest performance in The Second Xiangya Hospital dataset (low prevalence). One explanation could be that the algorithm was trained entirely on a dataset collected in Wuhan (a high-prevalence region) and testing the algorithm on an external population with low disease prevalence could decrease its performance. Considering the efficiency of AI-guided triage, the proposed scan-to-second-reader triage and scan-to-fever-clinician triage workflows reduced time to triage compared with standard clinical workflow across different fever clinics. Although the accuracy of AI-aided triage was lower in the Second Xiangya Hospital dataset than the Tianyou Hospital and Xianning Central Hospital datasets, of the ten patients with RT-PCR-confirmed COVID-19 who had positive CT scans, AI successfully flagged all ten cases and shortened the time from scan-to-fever-clinician. Moreover, considering that AI-aided triage will be of greater importance in medical contexts where workload is high and medical resources are scarce, the guarantee of reliable performance in populations with high disease prevalence is important.
Since radiological reference standard is not the gold standard for diagnosis of COVID-19, the accuracy of AI-aided triage was also assessed using an external validation dataset, which included patients with RT-PCR results. The sensitivity of AI-aided triage for patients with RT-PCR-confirmed COVID-19 was similar to that based on the radiological reference standard; however, the specificity of AI-aided triage was markedly lower than that of the radiological reference standard. At fever clinics, only patients with an epidemiological history or radiological features of COVID-19 had RT-PCR testing.5 Since RT-PCR can produce false negatives,7 negative results could not rule out virus infection. Therefore, selection bias and potential false negatives among these patients with negative RT-PCR test results could have compromised the specificity of AI-aided triage.11 To further validate the performance of AI-aided triage in patients with and without COVID-19, we collected CT scans from patients with RT-PCR-confirmed COVID-19 who were asymptomatic or had mild symptoms who were admitted to a fangcang hospital in Wuhan and CT scans from patients with various respiratory diseases, who were admitted to Tianyou Hopsital or The Third People's Hospital of Shenzhen before the COVID-19 outbreak. AI-aided triage was found to be reliable in these patients, which substantiates its efficacy in assisting COVID-19 identification.
Although the current study was done in China, the proposed AI-aided triage has potential uses in other geographical settings. The WHO suggested that for symptomatic patients with suspected COVID-19, chest imaging was recommended for the diagnostic workup of COVID-19 if RT-PCR testing was unavailable or delayed, or when initial RT-PCR testing was negative but patients had high clinical suspicion of COVID-19.24 In these scenarios, when chest CT is used as a surrogate tool to identify suspected COVID-19 cases, AI-aided triage could facilitate timely isolation of patients with suspected COVID-19 and alleviate pressure on medical staff, especially in regions with high disease prevalence. Additionally, in countries where RT-PCR testing is available with timely results, AI-aided triage might help to notify incidental findings. According to the report of a US doctor on March 25, 2020, patients who visited the emergency department for reasons other than COVID-19, such as a traffic accident, were found to have SARS-CoV-2 infection.25 In this scenario, AI could notify incidental COVID-19 findings on CT and alert medical staff of timely nosocomial infection prevention. Lung lesion burden assessment could also potentially be automated by AI to inform therapeutic management.
Several studies have applied deep learning to the clinical evaluation of COVID-19 using chest CT scans. Previous studies18, 26, 27 have applied deep learning to differentiate COVID-19 from other chest diseases including influenza A and community-acquired pneumonia, and in one previous study28 an algorithm was developed to segment and quantify COVID-19 opacities on chest CT. However, some of these studies were not validated externally or were tested on non-consecutively collected clinical cases,27, 28 whereas others did not specify the clinical context in which their algorithms could be applied, or focused on a narrow set of differential diagnoses, which would not cover the full disease spectrum in real-world clinical contexts.18, 26
The current study has several limitations. First, the algorithm was trained on data from Tongji Hospital only, which could compromise the robustness of the algorithm, as indicated by the results for the Second Xiangya Hospital dataset. Future studies could use multiple data sources to train models to improve generalisability. Second, we adopted the U-Net model structure to assess the feasibility of AI-aided triage for COVID-19. More methodologically rigorous algorithms could be developed to improve case classification. Third, the comparison of efficiency between AI-aided triage and standard of care with regard to time taken to triage was estimated in an ideal scenario where clinicians would respond instantly to AI notifications. However, in real-world clinical settings, this might not be realistic. Therefore, a prospective randomised control trial is needed to more accurately estimate the reduction in time to triage gained from AI. Fourth, since the main purpose of the study was to develop an AI algorithm for chest CT triage, clinical and laboratory information, with the exception of radiological findings and RT-PCR results, were not collected. Fifth, we did not directly compare the accuracy of AI for lesion burden analysis with individual radiologists. Future studies could systematically compare the accuracy of quantitative and qualitative lesion burden analysis of AI and radiologists.
The current study showed the efficacy of a deep learning algorithm for the triage of patients with suspected COVID-19 in fever clinics. The integration of AI into the standard clinical workflow has the potential to relieve burden on clinicians and expedite the isolation of suspected cases and disease control.
Data sharing
Access to datasets from Tongji Hospital, Second Xiangya Hospital, Tianyou Hospital, Xianning Central Hospital, The Third People's Hospital of Shenzhen, and Guanggu Fangcang Hospital used with permission for this study, should be requested directly from these institutions via their data access request systems. Subject to the institutional review boards' ethical approval, the corresponding author agrees to share de-identified individual participant data with academic researchers following completion of a data use agreement. The coding used to train the artificial intelligence (AI) model are dependent on annotation, infrastructure, and hardware, and thus cannot be released. However, all experimental and implementation details that can be shared are described in detail in this Article and the appendix. Major components of our algorithm are available in the Pytorch open source repository. The corresponding author agrees to apply the AI algorithm to data provided by other academic researchers on their behalf for research purposes only, following completion of a data use agreement form. Proposals for de-identified individual participant data or the algorithm should be directed to wwang@vip.126.com.
Acknowledgments
Acknowledgments
We thank Xiaoxiang Zhang and Jin Li (Department of Computer Centre, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China) for providing support with the data collection. We thank Qiufeng Huang, Dayong Zhang, Tian Tian, and Wei Cai for their assistance with validation dataset annotation for lesion burden analysis. We thank Ben Yarbrough for proofreading and providing language suggestions for this manuscript.
Contributors
WW, SW, ZL, and LX conceived and designed the study. WW, ZL, and LX managed the study. MW, CX, LH, SX, CQ, JW, GD, JL, XC, and CZ collected the data. KC, PY, and RZ developed and validated the algorithm. MW, CQ, SX, LH, HZ, CW, and TZ collected, anonymised, and prepared CT scans and reports from LX, JW, and GD, respectively. LX arranged for radiologists to rate the CT scans and reports. YC and RZ did the statistical analysis. CX, WW, MW, and ZL wrote the initial draft. All authors critically reviewed the report, and all read and approved the final version.
Declaration of interests
SW, RZ, CX, YC, and PY, and KC are employees of Beijing Infervision Technology. All other authors declare no competing interests.
Supplementary Materials
Lung lesion distribution of a patient with RT-PCR-confirmed COVID-19
References
- 1.WHO Coronavirus disease (COVID-19) situation report–181. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200719-covid-19-sitrep-181.pdf?sfvrsn=82352496_2
- 2.WHO WHO Director-General's opening remarks at the media briefing on COVID-19 −16 March 2020. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19—16-march-2020
- 3.WHO Operational considerations for case management of COVID-19 in health facility and community. March 19, 2020. https://apps.who.int/iris/bitstream/handle/10665/331492/WHO-2019-nCoV-HCF_operations-2020.1-eng pdf?sequence=1&isAllowed=y
- 4.WHO Laboratory testing for 2-19 novel coronavirus (2019-nCoV) in suspected human cases. Interim guidance. https://www.who.int/publications-detail/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117
- 5.National Health Commission of the People's Republic of China Chinese clinical guidance for COVID-19 pneumonia diagnosis and treatment (7th edition) http://www.nhc.gov.cn/yzygj/s7653p/202003/46c9294a7dfe4cef80dc7f5912eb1989/files/ce3e6945832a438eaae415350a8ce964.pdf (in Chinese).
- 6.Cable News Network South Korea pioneers coronavirus drive-through testing station. March 3, 2020. https://www.cnn.com/2020/03/02/asia/coronavirus-drive-through-south-korea-hnk-intl/index.html
- 7.Kucirka LM, Lauer SA, Laeyendecker O, Boon D, Lessler J. Variation in false-negative rate of reverse transcriptase polymerase chain reaction-based SARS-CoV-2 tests by time since exposure. Ann Intern Med. 2020 doi: 10.7326/M20-1495. published online May 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.WHO Report of the WHO–China Joint Mission on Coronavirus Disease 2019 (COVID-19) https://www.who.int/docs/default-source/coronaviruse/who-china-joint-mission-on-covid-19-final-report.pdf
- 9.Kanne JP, Little BP, Chung JH, Elicker BM, Ketai LH. Essentials for radiologists on COVID-19: an update—radiology scientific expert panel. Radiology. 2020;296:e113–e114. doi: 10.1148/radiol.2020200527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi H, Han X, Jiang N. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 2020;20:425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ai T, Yang Z, Hou H. Correlation of chest CT and RT-PCR testing in Coronavirus Disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296:e32–e40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zu ZY, Jiang MD, Xu PP. Coronavirus Disease 2019 (COVID-19): a perspective from China. Radiology. 2020;296:e15–e25. doi: 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mao B, Liu Y, Chai YH. Assessing risk factors for SARS-CoV-2 infection in patients presenting with symptoms in Shanghai, China: a multicentre, observational cohort study. Lancet Digital Health. 2020;2:e323–e330. doi: 10.1016/S2589-7500(20)30109-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.British Society of Thoracic Imaging Radiology decision tool for suspected COVID-19. https://www.bsti.org.uk/media/resources/files/NHSE_BSTI_APPROVED_Radiology_on_CoVid19_v6_ucQ1tNv.pdf
- 15.Chua F, Armstrong-James D, Desai SR. The role of CT in case ascertainment and management of COVID-19 pneumonia in the UK: insights from high-incidence regions. Lancet Respir Med. 2020;8:438–440. doi: 10.1016/S2213-2600(20)30132-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.National Health Commission of the People's Republic of China Chinese clinical guidance for COVID-19 pneumonia diagnosis and treatment (5th edition) http://www.nhc.gov.cn/yzygj/s7653p/202002/3b09b894ac9b4204a79db5b8912d4440/files/7260301a393845fc87fcf6dd52965ecb.pdf (in Chinese).
- 17.Wang Y, Dong C, Hu Y. Temporal changes of CT findings in 90 patients with COVID-19 pneumonia: a longitudinal study. Radiology. 2020;296:e55–e64. doi: 10.1148/radiol.2020200843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gozes O, Frid-Adar M, Greenspan H. Rapid AI development cycle for the coronavirus (COVID-19) pandemic: initial results for automated detection & patient monitoring using deep learning CT image analysis. arXiv. 2020 https://arxiv.org/abs/2003.05037 published online March 10. (preprint) [Google Scholar]
- 19.Forsberg D, Rosipko B, Sunshine JL. Radiologists' variation of time to read across different procedure types. J Digit Imaging. 2017;30:86–94. doi: 10.1007/s10278-016-9911-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esteva A, Kuprel B, Novoa RA. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKinney SM, Sieniek M, Godbole V. International evaluation of an AI system for breast cancer screening. Nature. 2020;577:89–94. doi: 10.1038/s41586-019-1799-6. [DOI] [PubMed] [Google Scholar]
- 22.Chilamkurthy S, Ghosh R, Tanamala S. Deep learning algorithms for detection of critical findings in head CT scans: a retrospective study. Lancet. 2018;392:2388–2396. doi: 10.1016/S0140-6736(18)31645-3. [DOI] [PubMed] [Google Scholar]
- 23.Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. MICCAI. 2015;9351:234–241. [Google Scholar]
- 24.WHO Use of chest imaging in COVID-19. June 11, 2020. https://www.who.int/publications/i/item/use-of-chest-imaging-in-covid-19
- 25.Rothfeld M, Sengupta S, Goldstein J, Rosenthal BM. 13 deaths in a day: an ‘apocalyptic’ coronavirus surge at an NYC hospital. New York Times. https://www.nytimes.com/2020/03/25/nyregion/nyc-coronavirus-hospitals.html
- 26.Li L, Qin L, Xu Z. Artificial Intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020 doi: 10.1148/radiol.2020200905. published online March 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang K, Liu X, Shen J. Clinically applicable AI system for accurate diagnosis, quantitative measurements and prognosis of COVID-19 pneumonia using computed tomography. Cell. 2020;181:1423–1433. doi: 10.1016/j.cell.2020.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shan F, Gao Y, Wang J. Lung infection quantification of COVID-19 in CT images with deep learning. arXiv. 2020 https://arxiv.org/abs/2003.04655 published online March 10. (preprint) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lung lesion distribution of a patient with RT-PCR-confirmed COVID-19
Data Availability Statement
Access to datasets from Tongji Hospital, Second Xiangya Hospital, Tianyou Hospital, Xianning Central Hospital, The Third People's Hospital of Shenzhen, and Guanggu Fangcang Hospital used with permission for this study, should be requested directly from these institutions via their data access request systems. Subject to the institutional review boards' ethical approval, the corresponding author agrees to share de-identified individual participant data with academic researchers following completion of a data use agreement. The coding used to train the artificial intelligence (AI) model are dependent on annotation, infrastructure, and hardware, and thus cannot be released. However, all experimental and implementation details that can be shared are described in detail in this Article and the appendix. Major components of our algorithm are available in the Pytorch open source repository. The corresponding author agrees to apply the AI algorithm to data provided by other academic researchers on their behalf for research purposes only, following completion of a data use agreement form. Proposals for de-identified individual participant data or the algorithm should be directed to wwang@vip.126.com.


