Abstract
The development of an effective vaccine against SARS-CoV-2 is urgently needed. We generated SARS-CoV-2 RBD-Fc fusion protein and evaluated its potency to elicit neutralizing antibody response in mice. RBD-Fc elicited a higher neutralizing antibodies titer than RBD as evaluated by a pseudovirus neutralization assay and a live virus based microneutralization assay. Furthermore, RBD-Fc immunized sera better inhibited cell–cell fusion, as evaluated by a quantitative cell–cell fusion assay. The cell–cell fusion assay results correlated well with the virus neutralization potency and could be used for high-throughput screening of large panels of anti-SARS-CoV-2 antibodies and vaccines without the requirement of live virus infection in BSL3 containment. Moreover, the anti-RBD sera did not enhance the pseudotyped SARS-CoV-2 infection of K562 cells. These results demonstrate that Fc fusion can significantly improve the humoral immune response to recombinant RBD immunogen, and suggest that RBD-Fc could serve as a useful component of effective vaccines against SARS-CoV-2.
Keywords: SARS-CoV-2, Subunit vaccine, Receptor-binding domain (RBD), Cell–cell fusion assay
1. Introduction
The coronavirus disease 2019 (COVID-19) outbreak has become a global pandemic responsible for over 20 million confirmed cases and over 0.7 million deaths worldwide as of Aug 11, 2020. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the novel betacoronavirus, is the etiological agent of COVID-19. Vaccines are a highly effective strategy in preventing the spread of infectious diseases with advantages of low production and distribution cost; stark reduction in morbidity; and minimal negative long-term effects on overall health. Currently, no approved vaccines are available for COVID19 patients although tremendous scientific and industrial efforts have been dedicated. Several approaches to the development of COVID-19 vaccines have emerged including DNA vaccines, RNA vaccines, viral vector vaccines, recombinant subunit vaccines, inactivated and attenuated virus vaccines [1]. As of Aug 10, 2020, 28 candidate vaccines are undergoing clinical evaluations with >139 vaccines in preclinical development stages (DRAFT landscape of COVID-19 candidate vaccines, WHO). Among those, Cansino’s recombinant adenovirus type-5 vectored vaccine has achieved the fastest clinical progress as the first-in-human trial vaccine, reporting tolerability and high immunogenicity after 28 days post-vaccination [2].
The spike (S) glycoprotein of coronaviruses including SARS-CoV and MERS-CoV has been demonstrated to be an appropriate immunogen capable of eliciting neutralizing antibodies [3]. Both viruses are phylogenetically close to SARS-CoV-2 [4], thus supporting the rationale of using the SARS-CoV-2 S protein as a subunit vaccine. Subunit vaccines contain recombinant antigen proteins with strong immunogenicity capable of efficiently stimulating the host immune system. Advantages of subunit vaccines include easy manufacture, low cost, and overall safety, since they do not contain genetic material and have a low probability to induce severe adverse reactions. For example, subunit vaccines are reported to be safer than other vaccines such as virus like particles, inactivated whole viruses and an rDNA expressed S protein, which have been shown to induce the cytokine Th2-type immunopathology in SARS-CoV [5]. Several subunit vaccines based on the full-length SARS-CoV-2 S proteins are under-development, with five candidates in clinical trials. However, the full-length S protein immunogen contains many non-neutralizing epitopes that could result in enhanced infection through the antibody-dependent enhancement (ADE) effect, which may occur through the FcγR-mediated internalization of antibody-bound virions in FcγR-expressing cells [6], [7]. The risk of ADE, observed for SARS-CoV, MERS-CoV, HIV-1, Zika and dengue virus vaccination, has become a major concern in vaccine development [8], [9], [10].
The SARS-CoV-2 receptor-binding domain (RBD) is the protruding site of the S protein that mediates viral cell fusion during the initial infection event through binding of the human receptor angiotensin-converting enzyme 2 (hACE2) [11], [12]. Based on its high homology to SARS-CoV, SARS-CoV-2 RBD is corroborated to contain immune dominant epitopes capable of eliciting antibodies that can neutralize viral infection and block viral entry by competing hACE2 binding. The antigenic regions on SARS-CoV-2 were also confirmed by computational studies exploring T cell and B cell epitopes [13]. Compared to the full-length S protein based subunit vaccine, reducing the immunogen to only the neutralizing epitope containing RBD can not only specifically mount neutralizing antibody titers, but could also mitigate the risk of ADE, which in most cases is mediated by the non-neutralizing antibodies [14]. Therefore, SARS-CoV-2 RBD has been selected as a primary target for vaccine design [15], [16]. Ravichandran et al. recently found that compared to full-length S, SARS-CoV-2 RBD immunogen elicited a higher titer of neutralizing antibodies with 5-fold higher affinity [17]. Yang et al. also showed that RBD can induce a potent antibody response in the immunized mice, rabbits and non-human primates [18]. Zang et al. have showed that the anti-RBD sera from mice did not promote (both pseudotyped and authentic) SARS-CoV-2 infection of Fcγ receptor-bearing cells [19].
Given those advantages, we chose the SARS-CoV-2 RBD as a subunit vaccine. To boost the vaccination-induced antibody response, we fused RBD to the IgG1 Fc. The Fc fragment can serve as a vaccine adjuvant by promoting cellular and humoral immune responses, probably by facilitating antigen delivery and presentation through interacting with Fcγ receptors on antigen-presenting cells. In addition, the Fc-fusion can also improve recombinant immunogen solubility and stability and extend their in vivo half-life after injection by interacting with human neonatal Fcγ-receptor (hFcRn) [20]. To stimulate anti-RBD antibody titers, we also utilized a well-accepted, highly effective adjuvant, MF59™. MF59 has been proven to increase neutralizing antibody production and boost the Th2 and Th1 immune responses. Several vaccines containing MF59 as adjuvants are undergoing clinical trials (Phases I-III). Notably, MF59-adjuvanted seasonal influenza vaccine (Fluad™) has already been licensed, showing acceptable safety and tolerability, and improved immunogenicity [21].
2. Materials and methods
2.1. Expression of SARS-CoV-2 RBD and RBD-Fc
The gene of SARS-CoV-2 RBD domain (residues 330–532) was synthesized by IDT (Coralville, Iowa), then cloned in frame to human IgG1 Fc or 6 × His tag in the mammalian cell expression plasmid. These proteins were expressed in the Expi293™ expression system (Thermo Fisher Scientific) and purified by protein A resin (GenScript) or Ni-NTA resin (Thermo Fisher Scientific).
2.2. SDS-PAGE and Western blot
The purified SARS-CoV-2 RBD and RBD-Fc were analyzed by SDS-PAGE and Western blot (WB). Protein concentration was measured spectrophotometrically (NanoVue, GE Healthcare), 2 μg of proteins were separated by NuPAGE® 4–12% Bis-Tris Gels (Life Technologies), protein purity was estimated as > 95%. For Western blot, the separated proteins were transferred to nitrocellulose membranes. After blocking at room temperature using 5% non-fat milk in PBST for 1 h, the blots were incubated for 2 h at room temperature with 100 nM hACE2-mFc (mouse Fc, Sino Biological, Beijing, China). After three washes, the blots were incubated with anti-mouse IgG-horseradish peroxidase (HRP) conjugated secondary antibody (1:1000, Sigma-Aldrich) for 1 h at room temperature. The membranes were reacted with enhanced ECL chemiluminescence reagent (Millipore, Billerica, USA) and exposed by Bio-Rad detection system.
2.3. Deglycosylation
Protein Deglycosylation Mix II (NEB) was used for deglycosylation reaction following manufacturer's instructions. Briefly, 2 μg proteins were mixed with Deglycosylation Mix Buffer 1 (non-denaturing reaction) or Buffer 2 (denaturing reaction, with an additional incubation at 75 °C for 10 min), then incubated with Protein Deglycosylation Mix II for reaction (25 °C for 30 min, and then 37 °C for 16 h). The deglycosylated proteins were analyzed by SDS-PAGE. The MW of enzymes in Protein Deglycosylation Mix II is approximately 36, 100, 147 and 231 kDa.
2.4. Construction of cell line
Genes of hACE2 (OriGene, Rockville, MD) and the full length S protein of SARS-CoV-2 (codon optimized and synthesized by IDT) were subcloned into our in-house mammalian cell expression plasmid and used to construct stable cell line 293T-ACE2 and 293T-S, which were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco) with 10% fetal bovine serum (FBS), 1% Penicillin/Streptomycin (P/S) and 200 μg/ml zeocin (Thermo Fisher).
2.5. Flow cytometry analysis
For the determination of 293T-S cell line, 500 nM hACE2-Fc (Sino Biological, Beijing, China) were incubated with cells for 30 min at 4 °C. Cells were washed and then incubated with PE conjugated anti-human Fc antibody (Sigma-Aldrich) for 30 min at 4 °C. Bound antibodies were detected by flow cytometry using BD LSR II (San Jose, CA). For the 293T-ACE2 cell line, 500 nM RBD-His followed by PE conjugated anti-His antibody (Sigma-Aldrich) were used for analysis.
2.6. Mouse immunization
Four groups of 8–10 week old female BALB/c mice (n = 5) were immunized twice (day 0 and day 14) subcutaneously with RBD proteins (10 μg/mouse) with or without adjuvant MF59. Group 1 was immunized with RBD-Fc fusion, group 2 was immunized with RBD-Fc fusion in emulsion with MF59, group 3 was immunized with RBD in emulsion with MF59, group 4 served as a control and was immunized subcutaneously with Dulbecco's phosphate-buffered saline DPBS (Gibco™). Sera were collected before (pre-vaccination), after 13 days, and after 27 days vaccination.
2.7. ELISA
For evaluation of affinity of RBD and RBD-Fc to hACE2, both proteins were coated on a 96-well plate (Costar) at 200 ng/well in PBS overnight at 4 °C. The plate was blocked using 3% skim milk for 1 h at room temperature (RT). We then added serially diluted hACE2-mFc (mouse Fc, Sino Biological, Beijing, China) and incubated for 2 h at RT. The plates were washed 4 times with 0.05% tween in phosphate buffered saline (PBST). Anti-mouse IgG–horseradish peroxidase (HRP) conjugated secondary antibody (Sigma-Aldrich) was added to the plate followed by incubation for 1 h at RT. After another 4 washes with PBST, the plate was incubated with a 3,3′,5,5′-tetramethylbenzidine substrate solution (TMB, Sigma-Aldrich) for 3 min. The reaction was stopped using 1 M H2SO4 followed by reading absorbance of each well at 450 nm. For detection of anti-RBD or anti-(S1 + S2) antibodies in mouse serum, the SARS-CoV-2 RBD proteins or S1 + S2 (Sino Biological, Beijing, China) were coated at 200 ng/well in PBS overnight at 4 °C. After blocking, serially diluted mouse serum were added and incubated for 2 h at RT. For antibody isotyping, the bound RBD-specific antibodies were detected by anti-mouse IgG, IgM, IgA HRP conjugated secondary antibody (Sigma-Aldrich), respectively. For competitive ELISA, ~20 nM (4 μg/ml) biotinylated hACE2 (Sino Biological, Beijing, China) was incubated with serially diluted mouse serum, and the mixtures were added to RBD coated wells. After washing, bound hACE2 was detected by Streptavidin-HRP secondary antibody (Sigma-Aldrich).
2.8. Pseudovirus neutralization assay
The pseudovirus neutralization assay was performed based on previous protocols. Briefly, HIV-1 backbone based pseudovirus was packaged in 293T cells by co-transfecting with plasmid encoding SARS-CoV-2 S protein and plasmid encoding luciferase expressing HIV-1 genome (pNL4-3.luc.RE) using polyethylenimine (PEI). Pseudovirus-containing supernatants were collected 48 h later and concentrated using Lenti-X™ concentrator kit (Takara, CA). Pseudovirus neutralization assay was then performed by incubation of SARS-CoV-2 pseudovirus with serially diluted mouse serum for 1 h at 37 °C, followed by addition of the mixture into pre-seeded 293T-ACE2 cells. The mixture was then centrifuged at 1000 × g for 1 h at RT. The medium was replaced 4 hrs later. After 24 h, luciferase expression was determined by Bright-Glo kits (Promega, Madison, WI) and read using BioTek synergy multi-mode reader (Winooski, VT). The 50% pseudovirus neutralizing antibody titer (NT50) was calculated using the Graphpad Prism 7.
2.9. Microneutralization assay
The standard live virus-based microneutralization (MN) assay was used. Briefly, serially five-fold (start from 1:5) and duplicate dilutions of mouse serum were incubated with 100 pfu of SARS-CoV-2 at room temperature for 2 h before transferring into designated wells of confluent Vero E6 cells (ATCC, CRL-1586) grown in 96-well microtiter plates. Vero E6 cells cultured with medium with or without virus were included as positive and negative controls, respectively. After incubation at 37 °C for 4 days, individual wells were observed under the microscopy for the status of virus-induced formation of cytopathic effect (CPE). The titer of mouse serum (NT100) was expressed as the lowest dilution folds capable of completely preventing virus-induced CPE in 100% of the wells.
2.10. Cell-Cell fusion inhibition assay
To test mouse serum mediated inhibition of cell fusions, the β-gal reporter gene based quantitative cell fusion assay was used. Briefly, 293T-S cells were infected with T7 polymerase-expressing vaccinia virus (vTF7-3), while 293T-ACE2 cells were infected with vaccinia virus (vCB21R Lac-Z) encoding T7 promotor controlled β-gal. Two hours after infection, cells were incubated with fresh medium and transferred to 37 °C for overnight incubation. The next day, 293T-S cells were pre-mixed with serially diluted mouse serum at 37 °C for 1 h followed by incubation with 293T-ACE2 cells at a 1:1 ratio for 3 h at 37 °C. Then cells were then lysed, and the β-gal activity was measured using β-galactosidase assay kit (substrate CPRG, G-Biosciences, St. Louis, MO) following the manufacturer’s protocols. Fusion inhibition percentage (sample reading, F) was normalized by maximal fusion (reading, Fmax) of 293T-S and 293T-ACE2 cells in the absence of inhibitors using this formula: Fusion inhibition % = [(Fmax-F)/(Fmax − Fblank)] × 100%, in which Fblank refers to the OD reading of 293T-S and 293T incubation wells. Fusion inhibition percentage was plotted against serum dilution folds from which IC50 was calculated in Graphpad Prism 7.
2.11. ADE assay
FcγRII expressing cell lines K562 (ATCC, CCL-243) were used to perform ADE assays. Briefly, the mouse serum was serially diluted, mixed with SARS-CoV-2 pseudovirus, and incubated at 37 °C for 1 h. Then, the mixtures were added to the pre-seeded plates with K562 cells. The following infection and culturing steps were carried out as described above in the pseudovirus neutralization assay. Pseudovirus infected K562 or 293T-ACE2 cells were set as the negative and positive controls, respectively.
2.12. Statistical analysis
All experiments were conducted in duplicate, and data were averaged and presented as the mean ± standard deviation (SD). Significant differences were determined by one-way analysis of variance followed by Tukey’s test, using the Graphpad Prism (version 7) package. Statistical significance was defined as P < 0.05.
3. Results and discussion
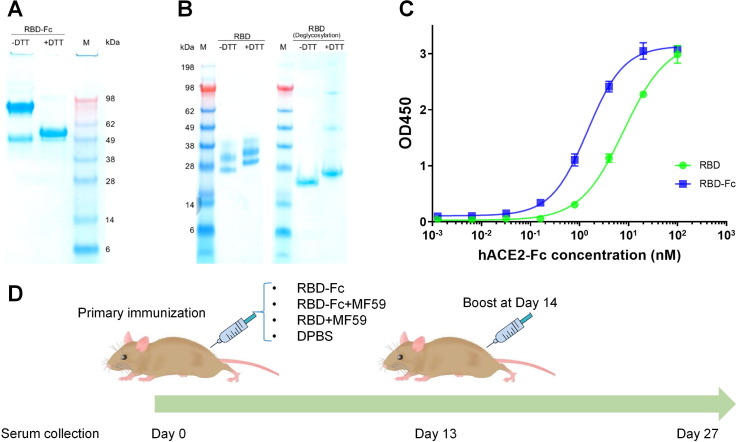
Recombinant SARS-CoV-2 RBD (without Fc) and RBD-Fc proteins were produced in Expi293™ mammalian cells, and then verified by SDS-PAGE and Western blot. RBD-Fc showed a homogenous band (Fig. 1 A) while the RBD exhibited relatively heterogeneous bands due to varying extents of glycosylation, which was also found by Yang et al. [18]. The RBD protein showed a single band upon deglycosylation, confirming the heterogeneous bands resulting from glycosylation (Fig. 1B). Besides, Western blot results showed that RBD with different glycoforms can react with hACE2 (Fig. S1). ELISA binding to hACE2 further validated the qualities of both RBD proteins (Fig. 1C). RBD antigens produced in this study were also used for panning against our in-house phage antibody libraries to retrieve high affinity binders [29], [22]. Four groups of 8–10 week old female BALB/c mice (n = 5) were immunized subcutaneously at day 0 and boosted at day 14 with RBD-Fc, RBD-Fc in emulsion with MF59 (RBD-Fc + MF59), and RBD in emulsion with MF59 (RBD + MF59) for each group at a dose of 10 μg of protein per mouse. The fourth group received injection of DPBS, which served as the negative control (Fig. 1D). On day 0 (pre-immunization), day 13 and day 27, mouse sera were collected and analyzed for RBD binding, pseudovirus and live virus neutralization, and cell–cell fusion inhibition.
Fig. 1.
Characterization of RBD and RBD-Fc, mouse immunization with recombinant RBD proteins. (A) SDS-PAGE of RBD-Fc (2 μg, ~100 kDa without DTT and ~50 kDa with DTT) are consistent with their theoretically calculated MWs. (B) SDS-PAGE of RBD (with 6 × His tag, 2 μg) in the presence or absence of DTT. The apparent molecular weight (MW) of RBD (heterogeneity ranging from 25 to 38 kDa) due to glycosylation was verified by deglycosylation. (C) ELISA measurement of binding of the recombinant RBD and RBD-Fc to hACE2-mFc (mouse Fc, Sino Biological). 200 ng RBD or RBD-Fc was coated on plate with incubation of serially diluted hACE2-mFc. Binding was detected by using HRP conjugated anti-mouse Fc antibody. Experiments were performed in duplicate and the error bars denote ± SD, n = 2. (D) Mouse immunization and sera sampling schedule. Four groups of BALB/c mice (n = 5) received same doses of the RBD vaccine or the control DPBS on day 0 and boost again on day 14. Sera were collected on day 0 (pre-vaccination), day 13 and 27 (post-vaccination).
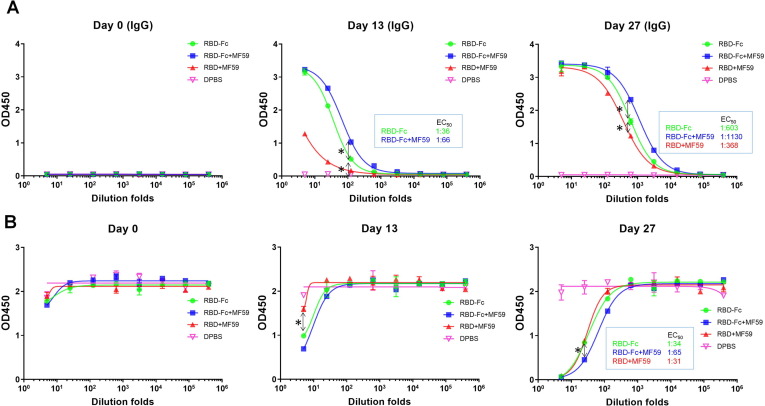
Anti-RBD sera from each trial group were firstly evaluated for RBD binding as measured by ELISA (Fig. 2 A). The anti-RBD antibody in post immune mouse sera were also isotyped by anti-mouse IgG, IgM and IgA antibodies, respectively (Fig. S2). The RBD antibody titers were calculated as the dilution folds that retained 50% of maximal binding signal (EC50). The recombinant RBD (His tag) was used as the detection antigen to avoid the interference of anti-human Fc antibody titers in mice. The impact of His tag on the detection of RBD binding titer is marginal (Fig. S2C). Results showed that for the sera collected at 13 and 27 days post immunization, the anti-RBD antibodies were mostly composed of the IgG isotype with only marginally detectable IgM (Fig. S2A) and no detectable IgA isotype (Fig. S2B). The low IgM titer detected at day 13 and 27 may correlate to the fact that IgM is typically rapidly mounted post infection (within one week) followed by isotype switching into IgG isotype [23]. The lack of IgA titer may result from IgA usually deriving from mucosa immunity, leading to low titers in sera [24]. For the IgG isotype antibodies, the pre-immunization sera showed no binding to RBD, while the day 13 sera from all three RBD immunized groups exhibited varying extents of binding to the RBD. Interestingly, the RBD binding titer elicited by the RBD + M59 group on day 13 was much less than titers of the RBD-Fc (titer 1:36) and RBD-Fc + M59 (titer 1:66) groups, indicating the immune stimulation roles of Fc fusion. However, for post-boosted sera on day 27, the RBD binding titers of RBD + MF59 group was significantly increased to 1:368. In contrast, on day 27 the titers of RBD-Fc (titer 1:603) and RBD-Fc + MF59 (titer 1:1130) groups were only improved by 17-fold compared to those of day 13, indicating distinct humoral response kinetics against RBD and RBD-Fc immunogens. Interestingly, although to a lesser extent than before receiving the booster, the RBD-Fc and RBD-Fc + MF59 groups exhibited 1.6 and 3 folds higher titers, over the RBD + MF59 group (P < 0.05) respectively, assuring the enhancing role of Fc in elicitation antibody response by the RBD immunogen. It is also intriguing that the RBD-Fc + MF59 group exhibited slightly higher titers than the RBD-Fc group, probably due to the adjuvant role of MF59. We also correlated the RBD binding titer to the full-length S ectodomain binding titer for the day 27 sera. The ELISA showed that the RBD binding sera also bound to S1 + S2 with similar titers (Fig. S2D), which suggest that the RBD recognition antibody in the sera can also bind to full length S.
Fig. 2.
Evaluation of binding (A) and competition with hACE2 (B) of mouse sera to the SARS-CoV-2 RBD as measured by ELISA. (A) 200 ng of RBD was coated and 5-fold serially diluted serum was added after blocking. After washing, the binding was detected by HRP conjugated anti-mouse IgG antibody. (B) 200 ng of RBD were coated and 5-fold serially diluted mouse serum was added in the presence of ~20 nM biotinylated hACE2 followed by PBST washing. For detection, streptavidin-HRP secondary antibody was used. Experiments were performed in duplicate and the error bars denote ± SD, n = 2. Statistical significance was defined as *: P < 0.05.
hACE2 blocking is a surrogate indicator for anti-SARS-CoV-2 antibody neutralizing activity. To preliminarily evaluate the neutralizing titers of post-immunization mouse serum, we performed the hACE2 competitive ELISA, in which serially diluted mouse sera in the presence of the biotinylated hACE2 were added into RBD coated plates. Bound hACE2 was detected by the streptavidin-HRP secondary antibody. Results showed the three RBD immunogen groups developed discernable hACE2 competitive titers on day 13 compared to the PBS control group; further significantly boosted to 1:34, 1:65, 1:31 for the RBD-Fc, RBD-Fc + MF59, RBD + MF59 groups respectively on day 27 (Fig. 2B). Consistent with the above RBD binding titer, the RBD-Fc and RBD-Fc + MF59 groups sera showed 1.1 folds and 2.1 folds higher competitive titers respectively than the RBD + MF59 group (P < 0.05), supporting the role of Fc in mounting neutralization titers. The competitive ELISA results gave the specific hACE2 blocking titers elicited by RBD immunogens, which presumably predict their neutralization activity [25].
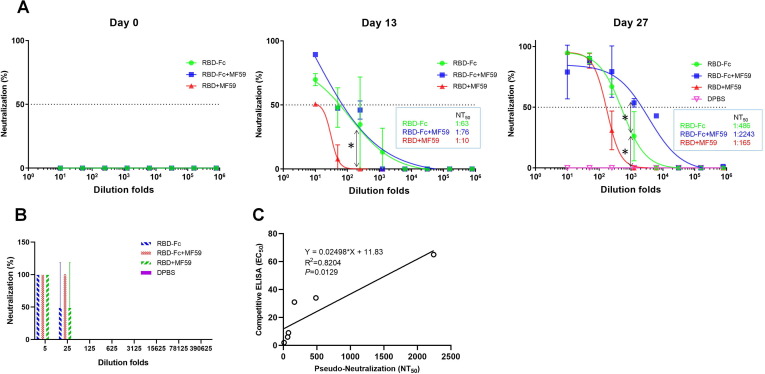
Next we exploited the SARS-CoV-2 S pseudotyped HIV-1 to evaluate the neutralization activity of those anti-RBD sera. The SARS-CoV-2 pseudovirus was packaged by co-transfecting HEK 293T cells with pCDNA3.1-S plasmid encoding codon-optimized full-length SARS-CoV-2 S protein and pNL4-3.luc.RE plasmid containing the luciferase expressing HIV-1 genome. Serially diluted mouse sera were pre-incubated with pseudovirus followed by infection of 293T cells stably expressing hACE2 (293T-ACE2). As shown in Fig. 3 A, on day 13 all of the RBD immunized groups sera showed substantial 50% neutralizing antibody titers (NT50, 1:63, 1:76, 1:10) compared to the pre-immune sera on day 0, which were largely boosted to 1:486, 1:2243, 1:165 on day 27 for the RBD-Fc, RBD-Fc + MF59 and RBD + MF59 groups respectively. Intriguingly, unlike the marginal differences for the RBD binding and hACE2 competitive titers across the sera of the three RBD immunized groups on day 27, the pseudovirus neutralization titers were significantly distinct with the RBD-Fc + MF59 group showing highest titers, 4.6-fold higher than the RBD-Fc group and 13.6-fold than the RBD + MF59 group (P < 0.05). In addition to the pseudovirus neutralization, we also evaluated the live virus neutralization potency of those mouse anti-RBD sera (day 27) by using a microneutralization (MN) assay. In this assay, the cytopathic effect (CPE) of Vero E6 was observed after 4 days incubation with live virus, which was pre-mixed with the anti-RBD sera. The neutralization titer of mouse serum (NT100) was expressed as the lowest dilution folds capable of completely preventing virus-induced CPE in 100% of the wells. Consistently, the NT100 of serum in RBD-Fc + MF59 group (1:25) was higher than those of the RBD-Fc and RBD + MF59 groups (1:5) (Fig. 3B). These results clearly demonstrated that Fc fusion could significantly augment the elicitation of neutralizing antibody titers by RBD immunogen, and the adjuvant MF59 can further stimulate the antigenicity of the RBD-Fc fusion proteins. Interestingly, we found that the pseudovirus neutralization titer positively correlated with the hACE2 competition titer (Fig. 3C), demonstrating the utility of our hACE2 competition ELISA in predicting neutralization titers in convalescent plasma therapy and in detecting of the presence of neutralizing antibodies in serological tests during the COVID19 pandemic.
Fig. 3.
Potent neutralization of SARS-CoV-2 pseudovirus (A) and live virus (B) by mouse serum, correlation analysis for pseudo-neutralization and competitive ELISA (C). (A) Pseudoviruses were pre-incubated with serially diluted serum and then used to infect 293T-ACE2 cells. 24 hrs later, luciferase activities in cell lysates were recorded. 50% neutralizing antibody titers (NT50) was obtained by non-linear fitting of plots of neutralization against serum dilution folds in Graphpad Prism 7. (B) Neutralization of live virus by a microneutralization assay. Virus-induced cytopathic effects (CPE) were observed under the microscopy. The neutralization capacity (NT100) was expressed as the lowest dilution folds capable of completely preventing virus induced CPE in 100% of the wells. Experiments were performed in duplicate and the error bars denote ± SD, n = 2. Statistical significance was defined as *: P < 0.05. (C) Correlation analysis between pseudo-neutralization antibody titers (NT50) and competitive ELISA (EC50) for sera of day 13 and day 27. Correlation and linear regression analyses were performed in GraphPad Prism using Pearson’s correlation coefficients. Statistical significance was calculated using the two-tailed test. The dashed lines indicate the standard deviations of the linear regression plots.
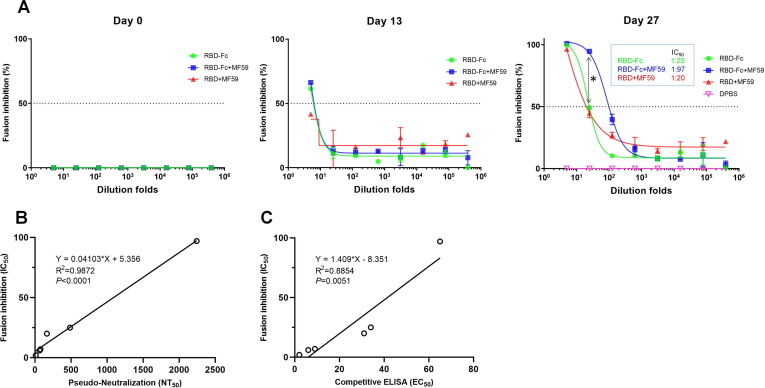
To further evaluate whether the anti-RBD sera could prevent SARS-CoV-2 S-mediated cell–cell fusion, we established a quantitative cell fusion assay using β-galactosidase (β-gal) as a reporter gene. We constructed 293T cell lines stably overexpressing SARS-CoV-2 S (293T-S) and hACE2 (293T-ACE2) respectively (Fig. S3). In this assay, the 293T-S cells were infected with T7 polymerase-expressing vTF7-3 vaccinia virus and the 293T-ACE2 cells were infected with T7 promotor controlled β-gal expressing vCB21R vaccinia virus. Therefore, β-gal expression is only allowed after cell–cell fusion, which can be quantified by monitoring β-gal activity. Serially diluted mouse sera were pre-mixed with infected 293T-S cells followed by incubation with infected 293T-ACE2 cells. As shown in Fig. 4 A, the pre-vaccination sera at day 0 and the PBS control mouse sera did not inhibit the cell–cell fusion. However, the day 27 RBD-Fc + MF59 sera showed obvious cell–cell fusion inhibition with a 50% fusion inhibition antibody titers (IC50) of 1:97, which was 3.9-fold higher than the inhibition titers of the RBD-Fc (1:25) and 4.9-fold higher than the RBD + MF59 (1:20) group (P < 0.05). Interestingly, in this assay, the RBD-Fc and the RBD + MF59 groups did not show significant differences (P > 0.05).
Fig. 4.
Inhibition of cell-cell fusion (A) by mouse serum, correlation analysis for fusion inhibition assay with pseudo-nertralization (B) and competitive ELISA (C). (A) A β-galactosidase (β-Gal) reporter gene-based quantitative cell–cell fusion assay was used, in which T7 polymerase expressing 293T-S pre-incubated with mouse serum followed by mixing with T7 promotor controlled β-Gal expressing 293T-ACE2 cells. After 3 hrs incubation, the β-Gal activity was detected by a chromogenic reaction using the β-Gal substrate CPRG. Fusion inhibition percentage was plotted against serum dilution folds from which 50% fusion inhibition antibody titers (IC50) was calculated in Graphpad Prism 7. Experiments were performed in duplicate and the error bars denote ± SD, n = 2. Statistical significance was defined as *: P < 0.05, n.s.: P > 0.05. (B) Correlation analysis between cell–cell fusion inhibition (IC50) and pseudo-neutralization antibody titers (NT50) for sera of day 13 and day 27. (C) Correlation analysis between cell–cell fusion inhibition (IC50) and competitive ELISA (EC50) for sera of day 13 and day 27. Correlation and linear regression analyses were performed in GraphPad Prism using Pearson’s correlation coefficients. Statistical significance was calculated using the two-tailed test. The dashed lines indicate the standard deviations of the linear regression plots.
We also observed a nearly perfect correlation of the cell–cell fusion inhibition titer with the pseudovirus neutralization titer (R2 = 0.9872, P < 0.0001, Fig. 4B), which may be attributed to their mechanism of action. Anti-RBD antibodies typically neutralize virus by blocking viral entry. For SARS-CoV-2, virus entry and cell–cell fusion share similar mechanisms. Both viral entry and cell–cell fusion are initiated by the S protein binding to the receptor ACE2, followed by S1 subunit triggered susceptibility to protease cleavage, which causes S1 dissociation and conformational change of the S2 subunit. Then, the fusion peptide (FP) in S2 is exposed for anchoring into host cell membrane and heptad repeats (HR1 and HR2) establishes the six helical-bundle resulting in the membrane fusion between viral and host cells [26]. Molecules, including antibodies, interfering with any of the above processes can block viral entry as well as cell–cell fusion [11]. Based on their similar mechanisms, the inhibitory activity of antibodies for cell–cell fusion can be a highly relevant predictor of the antibody neutralizing activity. This is further supported by our and others’ SARS-CoV-2 neutralizing antibodies, which shows both potent inhibition of S mediated cell–cell fusion and neutralization of SARS-CoV-2 [27]. Although highly correlated, there are differences between these two assays in terms of different environments on the cell and viral surface such as S protein conformation/ density, accessibility of proteases. Due to the high correlation to the virus neutralization, this method allows for high-throughput screening and is therefore well suited for the characterization of cell–cell fusion mediated by SARS-CoV-2 or other viruses. In addition, this assay is highly effective in screening potential neutralizing antibodies with fast speed since this assay can be finished within one day without the requirement of in biosafety level 3 facilities while the virus neutralization assays typically require several days which is of particular relevance in the context of the global pandemic, which urgently needs vaccines and candidate antibody drugs.
From the mechanism, one can also envision that RBD antibodies showing ACE2 competition is sufficient, but not necessary for virus neutralization and cell–cell fusion, since antibodies disturbing other entry steps, rather than ACE2/RBD binding, can also neutralize virus and inhibit cell–cell fusion, as exampled by the antibody 47D11[27]. In this regard, one pertinent result from this study is that we found the extent of the correlation of ACE2 competition ELISA titer with the neutralizing titer (R2 = 0.8204, P = 0.0129, Fig. 3C) and competition ELISA titer with cell–cell fusion inhibition titer (R2 = 0.8854, P = 0.0051, Fig. 4C) were lower than that of neutralizing titer to cell–cell fusion inhibition titer (R2 = 0.9872, P < 0.0001, Fig. 4B).
Finally, we evaluated whether the anti-RBD mouse sera can enhance SARS-CoV-2 infection of FcγRII expressing K562 cells [28]. The results showed that SARS-CoV-2 pseudovirus alone cannot infect the K562 cells (Fig. S4). In addition, treatment with serially diluted (ranging from 1:250 to 1:107) anti-RBD sera did not enhance SARS-CoV-2 pseudovirus infection, indicating that the anti-RBD sera may not promote ADE.
4. Conclusion
SARS-CoV-2 has presented a global health crisis requiring a rapid response from the scientific community in the form of a treatment or vaccine. Our study has identified the potency of the recombinant SARS-CoV-2 RBD as an immunogen in the context of the Fc fusion and MF59 adjuvant. We conclude that the RBD-Fc fusion vaccine with MF59 adjuvant is the most potent RBD based immunogen to elicit neutralizing antibodies that can potently inhibit SARS-CoV-2 pseudovirus and live virus infection. In addition, we established a high throughput quantitative cell–cell fusion assay based on β-gal as a reporter gene and confirmed the anti-RBD sera could prevent S-mediated cell–cell fusion. This assay showed a strong correlation with the neutralization assay, suggesting it can be used for high-throughput screening of large panels of anti-SARS-CoV-2 antibodies and vaccines. These results demonstrated that human IgG1 Fc fragment can significantly improve the humoral immune response to the recombinant SARS-CoV-2 RBD, and provided important information for further development of RBD-based SARS-CoV-2 subunit vaccines.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgement
We would like to thank the members of the Center for Antibody Therapeutics Doncho Zhelev, Du-San Baek, Liyong Zhang, and Xiaojie Chu for their helpful discussions. This work was supported by the University of Pittsburgh Medical Center.
Author contributions
DSD, XL, WL, CTT and JWM conceived and designed the research; XL performed research; AD conducted the microneutralization assay. CC, ZS and MS made the RBD and characterized them; DSD, XL, WL and CA wrote the first draft of the article, and all authors discussed the results and contributed to the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.vaccine.2020.09.058.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Amanat F., Krammer F. SARS-CoV-2 vaccines: status report. Immunity. 2020;52:583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu F.-C., Li Y.-H., Guan X.-H., Hou L.-H., Wang W.-J., Li J.-X., et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet. 2020;395:1845–1854. doi: 10.1016/S0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Du L., He Y., Zhou Y., Liu S., Zheng B.J., Jiang S. The spike protein of SARS-CoV–a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tseng C.T., Sbrana E., Iwata-Yoshikawa N., Newman P.C., Garron T., Atmar R.L., et al. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS One. 2012;7:e35421. doi: 10.1371/journal.pone.0035421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu L., Wei Q., Lin Q., Fang J., Wang H., Kwok H., et al. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI insight. 2019;4:e123158. doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor A., Foo S.-S., Bruzzone R., Vu Dinh L., King N.J.C., Mahalingam S. Fc receptors in antibody-dependent enhancement of viral infections. Immunol Rev. 2015;268:340–364. doi: 10.1111/imr.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J., Zand M.S. The potential for antibody-dependent enhancement of SARS-CoV-2 infection: Translational implications for vaccine development. J Clin Transl Sci. 2020:1–4. [Google Scholar]
- 9.Wan Y., Shang J., Sun S., Tai W., Chen J., Geng Q., et al. Molecular mechanism for antibody-dependent enhancement of coronavirus entry. J Virol. 2020;94:e02015–e2019. doi: 10.1128/JVI.02015-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poland G.A. Tortoises, hares, and vaccines: A cautionary note for SARS-CoV-2 vaccine development. Vaccine. 2020;38:4219–4220. doi: 10.1016/j.vaccine.2020.04.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimitrov D.S. The secret life of ACE2 as a receptor for the SARS virus. Cell. 2003;115:652–653. doi: 10.1016/S0092-8674(03)00976-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baruah V., Bose S. Immunoinformatics-aided identification of T cell and B cell epitopes in the surface glycoprotein of 2019-nCoV. J Med Virol. 2020;92:495–500. doi: 10.1002/jmv.25698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eroshenko N., Gill T., Keaveney M.K., Church G.M., Trevejo J.M., Rajaniemi H. Implications of antibody-dependent enhancement of infection for SARS-CoV-2 countermeasures. Nat Biotechnol. 2020;38:789–791. doi: 10.1038/s41587-020-0577-1. [DOI] [PubMed] [Google Scholar]
- 15.Zhu X., Liu Q., Du L., Lu L., Jiang S. Receptor-binding domain as a target for developing SARS vaccines. J Thoracic Dis. 2013;5:S142–S148. doi: 10.3978/j.issn.2072-1439.2013.06.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tai W., He L., Zhang X., Pu J., Voronin D., Jiang S., et al. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. 2020;17:613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ravichandran S., Coyle E.M., Klenow L., Tang J., Grubbs G., Liu S., et al. Antibody signature induced by SARS-CoV-2 spike protein immunogens in rabbits. Sci Transl Med. 2020;12:eabc3539. doi: 10.1126/scitranslmed.abc3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J., Wang W., Chen Z., Lu S., Yang F., Bi Z., et al. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature. 2020 doi: 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- 19.Zang J, Gu C, Zhou B, Zhang C, Yang Y, Xu S, et al. Immunization with the receptor-binding domain of SARS-CoV-2 elicits antibodies cross-neutralizing SARS-CoV-2 and SARS-CoV without antibody-dependent enhancement. Cell Discov. 2020;6:61. doi: 10.1038/s41421-020-00199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y., Li R., Wang M., Liu Y., Yin Y., Zai X., et al. Fc-Based recombinant henipavirus vaccines elicit broad neutralizing antibody responses in mice. Viruses. 2020;12 doi: 10.3390/v12040480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khurana S., Chearwae W., Castellino F., Manischewitz J., King L.R., Honorkiewicz A., et al. Vaccines with MF59 adjuvant expand the antibody repertoire to target protective sites of pandemic avian H5N1 influenza virus. Sci Transl Med. 2010;2 doi: 10.1126/scitranslmed.3000624. 15ra5-ra5. [DOI] [PubMed] [Google Scholar]
- 22.Sun Z., Chen C., Li W., Martinez D.R., Drelich A., Baek D.S., et al. Potent neutralization of SARS-CoV-2 by human antibody heavy-chain variable domains isolated from a large library with a new stable scaffold. mAbs. 2020;12:1778435. doi: 10.1080/19420862.2020.1778435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cafruny W.A., Chan S.P.K., Harty J.T., Yousefi S., Kowalchyk K., McDonald D., et al. Antibody response of mice to lactate dehydrogenase-elevating virus during infection and immunization with inactivated virus. Virus Res. 1986;5:357–375. doi: 10.1016/0168-1702(86)90029-8. [DOI] [PubMed] [Google Scholar]
- 24.Li Y., Jin L., Chen T. The effects of secretory IgA in the mucosal immune system. Biomed Res Int. 2020;2020:2032057. doi: 10.1155/2020/2032057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan C.W., Chia W.N., Qin X., Liu P., Chen M.I.C., Tiu C., et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2–spike protein–protein interaction. Nat Biotechnol. 2020 doi: 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- 26.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., et al. Cell entry mechanisms of SARS-CoV-2. PNAS. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A., et al. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chawla T., Chan K.R., Zhang S.L., Tan H.C., Lim A.P., Hanson B.J., et al. Dengue virus neutralization in cells expressing Fc gamma receptors. PLoS One. 2013;8:e65231. doi: 10.1371/journal.pone.0065231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W, Schäfer A, Kulkarni S.S, Liu X, Martinez D.R, Chen C, et al. High potency of a bivalent human VH domain in SARS-CoV-2 animal models. Cell. 2020 doi: 10.1016/j.cell.2020.09.007. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.