A multicenter collection of bacteremic isolates of Escherichia coli (n = 423), Klebsiella pneumoniae (n = 372), Pseudomonas aeruginosa (n = 300), and Acinetobacter baumannii complex (n = 199) was analyzed for susceptibility. Xpert Carba-R assay and sequencing for mcr genes were performed for carbapenem- or colistin-resistant isolates. Nineteen (67.
KEYWORDS: Enterobacteriaceae, KPC, carbapenemases, colistin, mcr-1
ABSTRACT
A multicenter collection of bacteremic isolates of Escherichia coli (n = 423), Klebsiella pneumoniae (n = 372), Pseudomonas aeruginosa (n = 300), and Acinetobacter baumannii complex (n = 199) was analyzed for susceptibility. Xpert Carba-R assay and sequencing for mcr genes were performed for carbapenem- or colistin-resistant isolates. Nineteen (67.8%) carbapenem-resistant K. pneumoniae (n = 28) and one (20%) carbapenem-resistant E. coli (n = 5) isolate harbored blaKPC (n = 17), blaOXA-48 (n = 2), and blaVIM (n = 1) genes.
INTRODUCTION
The increase in antimicrobial-resistant infections is a concern worldwide (1). The World Health Organization has published a list of antibiotic-resistant bacteria that pose a substantial threat to human health (2). Acinetobacter baumannii complex, Pseudomonas aeruginosa, and Enterobacteriaceae isolates are at the top of the list.
In Taiwan, the increase in antimicrobial-resistant Gram-negative bacteria has caused a significant increase in infections, and an association with poorer patient outcomes has been observed (3). Generally, antimicrobial resistance focuses on nosocomial infections; however, there are increasing concerns regarding community-acquired antimicrobial-resistant microorganisms (4). A community may act as a reservoir and breeding ground for the transmission of microorganisms (5). The growing numbers of senior and long-term-care facilities further complicate the transmission dynamics of antimicrobial-resistant bacteria between the community and hospital (6, 7). The increasing interchange between patients and microorganisms as they move back and forth between hospitals and communities may blur the distinction between community-acquired and hospital-acquired pathogens (8).
The Surveillance of Multicenter Antimicrobial Resistance in Taiwan (SMART) is an ongoing surveillance study conducted by the Taiwan Centers for Disease Control. SMART has been monitoring the in vitro resistance of clinically important bacteria obtained from hospitals throughout Taiwan since 2017 (9, 10). In this study, we analyzed data on antimicrobial susceptibility and major resistance mechanisms, especially for carbapenem and colistin resistance, of clinically important Gram-negative bacteria from 18 hospitals in Taiwan in 2019, with emphasis on isolates from community-acquired infections.
Escherichia coli, Klebsiella pneumoniae, P. aeruginosa, and A. baumannii complex isolates obtained from patients with bloodstream infections were included in this study. When multiple isolates of the same species were obtained from the same patient, only the first isolate was included. The identification of isolates was confirmed using the Phoenix PMIC/ID-30 identification system (Becton, Dickinson, Sparks, MD). Community-acquired isolates were defined as isolates obtained within 48 h after admission to a hospital with symptoms and signs of infection on admission, in the absence of recent hospitalization or residence in a skilled-nursing facility, and with no history of antibiotic therapy within the last 3 months. Hospital-acquired isolates were defined as those obtained >48 h after admission from patients who initially did not have symptoms or signs of infection. The study was approved by the research ethics committees or institutional review boards of the participating hospitals.
For all antibiotics tested, except for colistin, MICs were determined using the Vitek 2 antimicrobial susceptibility system (AST-NB card; bioMérieux, Marcy-l’Étoile, France) (10). The MICs for colistin were determined using the broth microdilution method as recommended by CLSI (11). E. coli ATCC 25922 and P. aeruginosa ATCC 27853 were used as quality control strains. According to the MIC breakpoints recommended by CLSI for colistin, an MIC of ≤2 μg/ml was considered intermediate and an MIC of ≥4 μg/ml was considered resistant for Enterobacteriaceae, P. aeruginosa, and A. baumannii complex isolates (11).
Carbapenem-nonsusceptible Enterobacteriaceae, P. aeruginosa, and A. baumannii complex isolates were tested for genes encoding blaKPC, blaNDM, blaIMP, blaVIM, and blaOXA-48 using the Xpert Carba-R assay (Cepheid, Sunnyvale, CA). The sequence types (STs) were determined using multilocus sequence typing (MLST) for isolates harboring the carbapenemase gene. Screening for mcr-1 to mcr-5 genes was performed for colistin-resistant Enterobacteriaceae, P. aeruginosa, and A. baumannii complex isolates (12).
During the study period, 1,294 bloodstream isolates (only 1 per patient was included), including E. coli (n = 423), K. pneumoniae (n = 372), P. aeruginosa (n = 300), and A. baumannii complex (n = 199), were collected consecutively. Of the 1,294 isolates studied, 772 (59.6%) were community acquired and 522 (40.3%) were hospital acquired. The in vitro activities of the antimicrobial agents tested are shown in Table 1. There were no significant differences (P > 0.05) in the percentage of resistance to ampicillin-sulbactam, ciprofloxacin, levofloxacin, and trimethoprim-sulfamethoxazole between hospital-acquired and community-acquired E. coli isolates. Carbapenems, amikacin, and colistin were the most-active agents tested against E. coli. The community-acquired isolates showed significantly higher (P < 0.001) rates of susceptibility to cefazolin (65.3% versus 41.0%), cefmetazole (92.1% versus 72.3%), cefotaxime (72.4% versus 42.2%), ceftazidime (83.8% versus 57.8%), and cefepime (92.1% versus 92.1%).
TABLE 1.
In vitro susceptibility to tested antimicrobial agents among bloodstream isolates of Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii complex obtained from patients at 18 participating hospitals in Taiwan in 2019 and their in vitro susceptibility among community-acquired and hospital-acquired isolates
| Bacterial species (na) and antimicrobial agent | MIC (μg/ml) |
No. (%) of isolates with indicated susceptibilityb |
No. (%) of isolates with susceptibility category: |
P value | |||||
|---|---|---|---|---|---|---|---|---|---|
| Range | 50% | 90% | S | I | R | Community acquired | Hospital acquired | ||
| E. coli (423/340/83) | |||||||||
| Ampicillin-sulbactam | ≤2 to ≥32 | 16 | ≥32 | 156 (36.9) | 76 (18.0) | 191 (45.2) | 133 (39.1) | 23 (27.7) | 0.058 |
| Cefazolin | ≤4 to ≥64 | ≤4 | ≥64 | 256 (60.5) | 167 (39.5) | 222 (65.3) | 34 (41.0) | <0.001 | |
| Cefmetazole | ≤1 to ≥64 | ≤1 | 8 | 373 (88.2) | 26 (6.1) | 24 (5.7) | 313 (92.1) | 60 (72.3) | <0.001 |
| Cefotaxime | ≤1 to ≥64 | ≤1 | ≥64 | 281 (66.4) | 3 (0.7) | 139 (32.9) | 246 (72.4) | 35 (42.2) | <0.001 |
| Ceftazidime | ≤1 to ≥64 | ≤1 | 16 | 333 (78.7) | 3 (0.7) | 87 (20.6) | 285 (83.8) | 48 (57.8) | <0.001 |
| Cefepime | ≤1 to ≥64 | ≤1 | 2 | 377 (89.1) | 19 (4.5) | 27 (6.4) | 313 (92.1) | 64 (77.1) | <0.001 |
| Piperacillin-tazobactam | ≤4 to ≥128 | ≤4 | 8 | 387 (91.5) | 21 (5.0) | 15 (3.5) | 316 (92.9) | 71 (85.5) | 0.046 |
| Ertapenem | ≤0.5 to 4 | ≤0.5 | ≤0.5 | 417 (98.6) | 1 (0.2) | 5 (1.2) | 337 (99.1) | 80 (96.4) | 0.093 |
| Imipenem | ≤0.25 to 1 | ≤0.25 | ≤0.25 | 422 (99.8) | 0 (0) | 1 (0.2) | 340 (100) | 81 (97.6) | 0.038 |
| Meropenem | ≤0.25 | ≤0.25 | ≤0.25 | 422 (99.8) | 0 (0) | 1 (0.2) | 340 (100) | 82 (98.8) | 0.196 |
| Ciprofloxacin | ≤0.25 to ≥4 | ≤0.25 | ≥4 | 232 (54.8) | 36 (8.5) | 155 (36.6) | 192 (56.5) | 40 (48.2) | 0.179 |
| Levofloxacin | ≤0.12 to ≥8 | 1 | ≥8 | 201 (47.5) | 80 (18.9) | 142 (33.6) | 165 (48.5) | 36 (43.4) | 0.462 |
| Gentamicin | ≤1 to ≥16 | ≤1 | ≥16 | 334 (79.0) | 0 (0) | 89 (21.0) | 266 (78.2) | 68 (81.9) | 0.549 |
| Amikacin | ≤2 to 16 | ≤2 | 4 | 422 (99.8) | 0 (0) | 1 (0.2) | 340 (100) | 82 (98.8) | 0.196 |
| TMP-SMXc | ≤1 to ≥16 | ≤1 | ≥16 | 242 (57.2) | 181 (42.8) | 201 (59.1) | 41 (49.4) | 0.137 | |
| Tigecycline | ≤0.5 to 4 | ≤0.5 | ≤0.5 | NA | NA | NA | NA | NA | |
| Colistin | ≤0.5 to ≥16 | ≤0.5 | ≤0.5 | 414 (97.9) | 9 (2.1) | 333 (97.9)d | 81 (97.6)d | 0.691 | |
| K. pneumoniae (372/239/133) | |||||||||
| Ampicillin-sulbactam | ≤2 to ≥32 | 8 | ≥32 | 246 (66.1) | 8 (2.2) | 118 (31.7) | 185 (77.4) | 61 (45.9) | <0.001 |
| Cefazolin | ≤4 to ≥64 | ≤4 | ≥64 | 262 (70.4) | 110 (29.6) | 198 (82.8) | 64 (48.1) | <0.001 | |
| Cefmetazole | ≤1 to ≥64 | ≤1 | ≥64 | 299 (80.4) | 22 (5.9) | 51 (13.7) | 210 (87.9) | 89 (66.9) | <0.001 |
| Cefotaxime | ≤1 to ≥64 | ≤1 | ≥64 | 279 (75) | 17 (4.6) | 76 (20.4) | 203 (84.9) | 76 (57.1) | <0.001 |
| Ceftazidime | ≤1 to ≥64 | ≤1 | ≥64 | 285 (76.6) | 15 (4.0) | 72 (19.4) | 207 (86.6) | 78 (58.6) | <0.001 |
| Cefepime | ≤1 to ≥64 | ≤1 | 32 | 325 (87.4) | 7 (1.9) | 40 (10.8) | 225 (94.1) | 100 (75.2) | <0.001 |
| Piperacillin-tazobactam | ≤4 to ≥128 | ≤4 | ≥128 | 298 (80.1) | 14 (3.8) | 60 (16.1) | 216 (90.4) | 82 (61.7) | <0.001 |
| Ertapenem | ≤0.5 to ≥8 | ≤0.5 | ≤0.5 | 335 (90.1) | 9 (2.4) | 28 (7.5) | 227 (95.0) | 108 (81.2) | <0.001 |
| Imipenem | ≤0.25–≥16 | ≤0.25 | 1 | 343 (92.2) | 12 (3.2) | 17 (4.6) | 228 (95.4) | 115 (86.5) | 0.004 |
| Meropenem | ≤0.25–≥16 | ≤0.25 | ≤0.25 | 349 (93.8) | 1 (0.3) | 22 (5.9) | 233 (97.5) | 116 (87.2) | <0.001 |
| Ciprofloxacin | ≤0.25 to ≥4 | ≤0.25 | ≥4 | 263 (70.7) | 17 (4.6) | 92 (24.7) | 190 (79.5) | 73 (54.9) | <0.001 |
| Levofloxacin | ≤0.12 to ≥8 | ≤0.12 | ≥8 | 244 (65.6) | 55 (14.8) | 73 (19.6) | 182 (76.2) | 62 (46.6) | <0.001 |
| Gentamicin | ≤1 to ≥16 | ≤1 | ≥16 | 300 (80.6) | 11 (3.0) | 61 (16.4) | 214 (89.5) | 86 (64.7) | <0.001 |
| Amikacin | ≤2 to ≥64 | ≤2 | ≤2 | 359 (96.5) | 0 (0) | 13 (3.5) | 234 (97.9) | 125 (94.0) | 0.073 |
| TMP-SMX | ≤1 to ≥16 | ≤1 | ≥16 | 267 (71.8) | 105 (28.2) | 195 (81.6) | 72 (54.1) | <0.001 | |
| Tigecycline | ≤0.5 to ≥8 | ≤0.5 | 2 | NA | NA | NA | NA | NA | |
| Colistin | ≤0.5 to ≥16 | ≤0.5 | ≤0.5 | NA | 356 (95.7) | 16 (4.3) | 231 (96.6)d | 125 (93.9)d | 0.286 |
| P. aeruginosa (300/146/154) | |||||||||
| Ceftazidime | ≤1 to ≥64 | 4 | 16 | 257 (85.7) | 19 (6.3) | 24 (8) | 136 (93.2) | 121 (78.6) | <0.001 |
| Cefepime | ≤1 to ≥64 | 2 | 8 | 272 (90.7) | 13 (4.3) | 15 (5) | 141 (96.6) | 131 (85.1) | 0.001 |
| Piperacillin-tazobactam | ≤4 to ≥128 | 8 | ≥128 | 238 (79.3) | 24 (8) | 38 (12.7) | 128 (87.7) | 110 (71.4) | 0.001 |
| Imipenem | ≤0.25 to ≥16 | 2 | ≥16 | 257 (85.7) | 0 (0) | 43 (14.3) | 135 (92.5) | 122 (79.2) | 0.002 |
| Meropenem | ≤0.25 to ≥16 | ≤0.25 | 4 | 260 (86.7) | 12 (4) | 28 (9.3) | 139 (95.2) | 121 (78.6) | <0.001 |
| Ciprofloxacin | ≤0.25 to ≥4 | ≤0.25 | 1 | 257 (85.7) | 13 (4.3) | 30 (10) | 127 (87.0) | 130 (84.4) | 0.622 |
| Levofloxacin | ≤0.12 to ≥8 | 0.5 | 4 | 252 (84) | 7 (2.3) | 41 (13.7) | 127 (87.0) | 125 (81.2) | 0.208 |
| Gentamicin | ≤1 to ≥16 | ≤1 | 2 | 282 (94) | 2 (0.7) | 16 (5.3) | 136 (93.2) | 146 (94.8) | 0.63 |
| Amikacin | ≤2 to ≥64 | ≤2 | 4 | 296 (98.7) | 1 (0.3) | 3 (1) | 146 (100) | 150 (97.4) | 0.123 |
| Colistin | ≤0.5 to ≥16 | ≤0.5 | ≤0.5 | NA | 295 (98.3) | 5 (1.7) | 144 (98.6)d | 151 (98.4)d | 0.999 |
| A. baumannii complex (199/47/152) | |||||||||
| Ampicillin-sulbactam | ≤2 to ≥32 | ≤2 | ≥32 | 122 (61.3) | 16 (8.0) | 61 (30.7) | 32 (68.1) | 90 (59.2) | 0.307 |
| Ceftazidime | ≤1 to ≥64 | 16 | ≥64 | 96 (48.2) | 25 (12.6) | 78 (39.2) | 24 (51.1) | 72 (47.4) | 0.739 |
| Cefepime | ≤1 to ≥64 | 8 | ≥64 | 105 (52.8) | 8 (4.0) | 86 (43.2) | 26 (55.3) | 79 (52.0) | 0.74 |
| Piperacillin-tazobactam | ≤4 to ≥128 | 32 | ≥128 | 98 (49.2) | 7 (3.5) | 94 (47.2) | 25 (53.2) | 73 (48.0) | 0.617 |
| Imipenem | ≤0.25 to ≥16 | ≤0.25 | ≥16 | 115 (57.8) | 0 (0) | 84 (42.2) | 32 (68.1) | 83 (54.6) | 0.128 |
| Meropenem | ≤0.25 to ≥16 | 0.5 | ≥16 | 112 (56.3) | 2 (1.0) | 85 (42.7) | 32 (68.1) | 80 (52.6) | 0.067 |
| Ciprofloxacin | ≤0.25 to ≥4 | 0.5 | ≥4 | 110 (55.3) | 1 (0.5) | 88 (44.2) | 27 (57.4) | 83 (54.6) | 0.867 |
| Levofloxacin | ≤0.12 to ≥8 | ≤0.12 | ≥8 | 111 (55.8) | 30 (15.1) | 58 (29.1) | 27 (57.4) | 84 (55.3) | 0.867 |
| Gentamicin | ≤1 to ≥16 | ≤1 | ≥16 | 119 (59.8) | 7 (3.5) | 73 (36.7) | 31 (66.0) | 88 (57.9) | 0.395 |
| Amikacin | ≤2 to ≥64 | ≤2 | ≥64 | 161 (80.9) | 5 (2.5) | 33 (16.6) | 39 (83.0) | 122 (80.3) | 0.832 |
| TMP-SMX | ≤1 to ≥16 | ≤1 | ≥16 | 113 (56.8) | 86 (43.2) | 32 (68.1) | 81 (53.3) | 0.092 | |
| Tigecycline | ≤0.5 to ≥8 | ≤0.5 | 4 | NA | NA | NA | NA | NA | |
| Colistin | ≤0.5 to 2 | ≤0.5 | ≤0.5 | 182 (91.5) | 17 (8.5) | 45 (95.7)d | 137 (90.1)d | 0.370 | |
Number of total tested/community-acquired/hospital-acquired isolates.
S, susceptible; I, intermediate; R, resistant; NA, not available.
TMP-SMX, trimethoprim-sulfamethoxazole.
Isolates with intermediate resistance to colistin.
Hospital-acquired K. pneumoniae isolates were less susceptible than community-acquired K. pneumoniae isolates (Table 1). Both community-acquired and hospital-acquired isolates demonstrated the highest rates of susceptibility to amikacin (97.9% and 96.6%, respectively) and colistin (94.0% and 93.9%, respectively). The rates of susceptibility to third-generation cephalosporins (cefotaxime and ceftazidime) were >84% for community-acquired K. pneumoniae isolates but <60% for hospital-acquired isolates. Compared with community-acquired isolates, hospital-acquired K. pneumoniae isolates exhibited a significantly lower susceptibility to ciprofloxacin (54.9% versus 79.5%) and levofloxacin (46.6% versus 76.2%).
The rates of susceptibility of P. aeruginosa isolates to ciprofloxacin and levofloxacin were <80%, with nonsignificant differences between community-acquired and hospital-acquired isolates. The rates of susceptibility to agents against A. baumannii isolates were <65% except for amikacin (80.9%) and colistin (91.5%); the rates were similar in community-acquired and hospital-acquired isolates.
The carbapenem resistance rates were 1.2% (5/423) in E. coli, 7.5% (28/372) in K. pneumoniae, 14.3% (43/300) in P. aeruginosa, and 42.7% (85/199) in A. baumannii complex isolates. The rate of community-acquired isolates in colistin-resistant isolates was 40% (2/5) for P. aeruginosa and 13.3% (2/17) for A. baumannii complex. Approximately 77.7% (7/9) of E. coli and 50% (8/16) of K. pneumoniae that were resistant to colistin were community-acquired isolates.
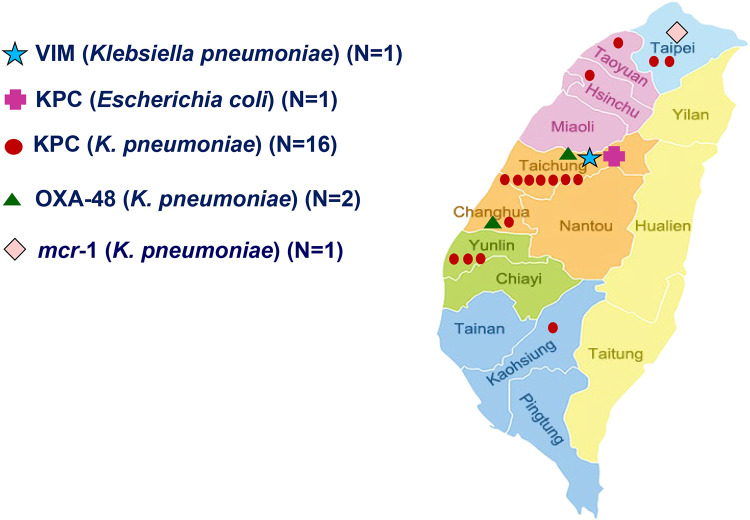
Carbapenem-resistant E. coli (n = 5), K. pneumoniae (n = 28), P. aeruginosa (n = 43), and A. baumannii complex (n = 85) isolates were screened for carbapenemase genes (Table 2). Carbapenemase genes were detected mostly in K. pneumoniae isolates (67.8%, 19/28). Among the carbapenem-resistant K. pneumoniae isolates, 57.1% (16/28) harbored blaKPC. Twenty-one percent (4/19) of carbapenemase-producing K. pneumoniae isolates were community acquired. All of the community-acquired carbapenemase-producing K. pneumoniae strains belonged to ST11. Of the carbapenemase-producing Enterobacteriaceae, 75% (15/20) were isolated from samples collected in central Taiwan, including all community-acquired carbapenemase-producing K. pneumoniae isolates (Fig. 1).
TABLE 2.
Characteristics of 20 Enterobacteriaceae isolates with carbapenemase-mediated genes
| Carbapenemase gene | Species | STa | MIC (μg/ml) |
Site of acquisitionb | ||||
|---|---|---|---|---|---|---|---|---|
| Ertapenem | Imipenem | Meropenem | Ciprofloxacin | Colistin | ||||
| blaKPC | K. pneumoniae | 11 | ≥8 | ≥16 | ≥16 | ≥4 | >32 | HA |
| blaKPC | K. pneumoniae | 11 | ≥8 | ≥16 | ≥16 | ≥4 | 1 | HA |
| blaOXA-48 | K. pneumoniae | 307 | ≥8 | 1 | 1 | ≥4 | 2 | HA |
| blaOXA-48 | K. pneumoniae | 11 | ≥8 | 8 | 4 | ≥4 | 2 | CA |
| blaKPC | K. pneumoniae | 11 | ≥8 | ≥16 | ≥16 | ≥4 | 1 | HA |
| blaKPC | K. pneumoniae | 11 | ≥8 | ≥16 | ≥16 | ≥4 | 16 | CA |
| blaKPC | K. pneumoniae | 11 | ≥8 | ≥16 | ≥16 | ≥4 | 16 | CA |
| blaKPC | K. pneumoniae | 11 | ≥8 | ≥16 | ≥16 | 1 | 1 | HA |
| blaKPC | K. pneumoniae | 11 | ≥8 | ≥16 | ≥16 | ≥4 | 1 | CA |
| blaVIM | K. pneumoniae | NA | ≤0.5 | 2 | ≥16 | 0.5 | 1 | HA |
| blaKPC | K. pneumoniae | 11 | ≥8 | ≥16 | ≥16 | ≥4 | 1 | HA |
| blaKPC | K. pneumoniae | 11 | ≥8 | ≥16 | ≥16 | ≥4 | 1 | HA |
| blaKPC | K. pneumoniae | 2640 | 4 | ≥16 | ≥16 | ≥4 | 1 | HA |
| blaKPC | K. pneumoniae | 11 | ≥8 | ≥16 | ≥16 | ≥4 | 16 | HA |
| blaKPC | K. pneumoniae | 11 | ≥8 | ≥16 | ≥16 | ≥4 | 1 | HA |
| blaKPC | K. pneumoniae | 11 | ≥8 | ≥16 | ≥16 | ≥4 | 1 | HA |
| blaKPC | K. pneumoniae | 11 | ≥8 | ≥16 | ≥16 | ≥4 | 1 | HA |
| blaKPC | K. pneumoniae | 11 | ≥8 | ≥16 | ≥16 | ≥4 | 1 | HA |
| blaKPC | K. pneumoniae | 11 | ≥8 | ≥16 | ≥16 | ≥4 | 1 | HA |
| blaKPC | E. coli | 3492 | ≥8 | ≥16 | ≥16 | ≥4 | 1 | HA |
ST, type was not present in the MLST database. NA, not available.
CA, community acquired; HA, hospital acquired.
FIG 1.
Geographical distribution of E. coli and K. pneumoniae isolates carrying blaKPC, blaOXA-48, blaVIM, and mcr-1, identified by 2019 Surveillance of Multicenter Antimicrobial Resistance in Taiwan (SMART) program. KPC, K. pneumoniae carbapenemase; OXA-48, oxacillinase-48 carbapenem-hydrolyzing class D β-lactamase; mcr-1, mobilized colistin resistance-1; VIM, Verona integron-encoded metallo-β-lactamase.
In this study, we demonstrated that the susceptibility rates of Gram-negative clinically important pathogens to several medically important antibiotics were similarly low in community-acquired and hospital-acquired isolates. Previously published surveillance studies in Taiwan reported that Gram-negative bacilli generally demonstrated higher rates of antimicrobial resistance in Taiwan than in Western countries (10, 13).
The extensive spread of resistance in E. coli isolates seen in this study was consistent with a previous study (14). We found a higher prevalence of third-generation cephalosporin resistance in E. coli isolates in this study than reported in Western countries, i.e., ∼10% (15, 16). Prevalence rates similar to those from our findings have been reported in Africa, where third-generation cephalosporin resistance was found to be 10% to 30% (17). Subgroup analysis in our study revealed that 27.1% of community-acquired E. coli isolates were resistant to cefotaxime. In Taiwan, the proportion of third-generation cephalosporin-resistant E. coli isolates causing community-onset bacteremia was 0.5% from 2001 to 2002 (18) and 19.7% in 2015 (4). Moreover, the activity of antibiotics in an oral formulation, such as ampicillin-sulbactam, fluoroquinolones, and trimethoprim-sulfamethoxazole (TMP-SMX), in community-acquired E. coli infection was low. These findings have been observed in other studies (19–21). Controlling the spread of drug-resistant E. coli isolates in the community may be a challenge because of their broad distribution in the ecosystem (22).
In our study, 67.8% of carbapenem-nonsusceptible K. pneumoniae and 20% of carbapenem-nonsusceptible E. coli isolates carried the carbapenemase genes. ST11 KPC-2-producing K. pneumoniae isolates are endemic in Taiwan and China (23, 24), as demonstrated in our study. Moreover, the incidence of fluoroquinolone-resistant community-acquired carbapenemase-producing K. pneumoniae infection exceeded that of hospital-acquired K. pneumoniae infection. The incidence of fluoroquinolone-resistant Enterobacteriaceae correlated with fluoroquinolone usage (25, 26). Continued surveillance of carbapenem-resistant Enterobacteriaceae in the community is needed to reveal its nature (27).
The resistance pattern of P. aeruginosa isolates found in our study is consistent with other reports in Asia (28). Our data reveal that the rates of susceptibility to fluoroquinolones are similar in community-acquired and hospital-acquired P. aeruginosa isolates. Community-acquired P. aeruginosa infections had markedly high mortality rates in other studies (29). It has been demonstrated that the avoidance of fluoroquinolone-based empirical regimens for P. aeruginosa infections in settings with high rates of fluoroquinolone resistance improves patient outcomes and future susceptibility (30). Further studies are needed to assess the clinical impact of antimicrobial stewardships aimed at curbing the inappropriate use of fluoroquinolones.
Our results showed high resistance in A. baumannii complex isolates, which is consistent with previous studies (31). We further demonstrated that susceptibility to carbapenem is equally low in community-acquired and hospital-acquired A. baumannii complex isolates. A study in Taiwan revealed that the mortality rates were comparable between community-acquired and hospital-acquired A. baumannii bacteremia, and unfavorable outcomes were associated with carbapenem resistance (32). The spread of A. baumannii infection may be the result of patient migration between homes, hospitals, and long-term-care facilities (33).
This study has a number of limitations. The SMART project was an observational study. The prevalence of resistance among key pathogens may be influenced by several clinical parameters, but these data were not reported, and thus subgroup analysis based on these factors was not possible. Moreover, the role of environmental contamination is underreported in current studies. Several studies have demonstrated the importance of environmental surveillance in the investigation of antimicrobial resistance (34, 35).
In conclusion, we demonstrated the extent of antimicrobial resistance in clinically important Gram-negative bacteria in Taiwan. Because a community may act as a breeding ground for emerging resistance, the importance of antimicrobial resistance surveillance in communities cannot be overemphasized.
ACKNOWLEDGMENTS
This work was supported by grants from the Taiwan Centers for Disease Control and Prevention, Minister of Health and Welfare, Executive Yuan, Taiwan (MOHW108-CDC-C-114-134504).
Investigators from the SMART Program 2019 include Shio-Shin Jean (Wan Fang Hospital, Taipei), Wen-Sen Lee (Wan Fang Hospital, Taipei), Min-Chi Lu (China Medical University Hospital, Taichung), Zhi-Yuan Shi (Taichung Veterans General Hospital, Taichung), Yao-Shen Chen (Kaohsiung Veterans General Hospital, Kaohsiung), Lih-Shinn Wang (Buddhist Tzu Chi General Hospital, Hualien), Shu-Hui Tseng (Ministry of Health and Welfare, Taipei), Chao-Nan Lin (National Pingtung University of Science and Technology, Pingtung), Hung-Jen Tang (Chi Mei Hospital, Tainan), Yu-Hui Chen (Chi Mei Hospital, Tainan), Wang-Huei Sheng (National Taiwan University Hospital, Taipei), Chang-Pan Liu (MacKay Memorial Hospital, Taipei), Ting-Shu Wu (Chang Gung Memorial Hospital, Taoyuan), Chun-Ming Lee (St. Joseph’s Hospital, Yunlin), Po-Liang Lu (Kaohsiung Medical University Hospital, Kaohsiung), Muh-Yong Yen (Taipei City Hospital, Taipei), Pei-Lan Shao (National Taiwan University Hospital, Hsin-Chu), Shu-Hsing Cheng (Taoyuan General Hospital, Taoyuan), Chi-Ying Lin (National Taiwan University Hospital, Yun-Lin), Ming-Huei Liao (National Pingtung University of Science and Technology, Pingtung), Yen-Hsu Chen (Kaohsiung Medical University, Kaohsiung), Wen-Chien Ko (National Cheng Kung University Hospital, Tainan), Fu-Der Wang (Taipei Veterans General Hospital, Taipei), and Po-Ren Hsueh (National Taiwan University Hospital, Taipei).
REFERENCES
- 1.Hawkey PM, Warren RE, Livermore DM, McNulty CAM, Enoch DA, Otter JA, Wilson A. 2018. Treatment of infections caused by multidrug-resistant Gram-negative bacteria: report of the British Society for Antimicrobial Chemotherapy/Healthcare Infection Society/British Infection Association Joint Working Party. J Antimicrob Chemother 73:iii2–iii78. doi: 10.1093/jac/dky027. [DOI] [PubMed] [Google Scholar]
- 2.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, WHO Pathogens Priority List Working Group. 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 3.Lai CC, Chen YS, Lee NY, Tang HJ, Lee SS, Lin CF, Lu PL, Wu JJ, Ko WC, Lee WS, Hsueh PR. 2019. Susceptibility rates of clinically important bacteria collected from intensive care units against colistin, carbapenems, and other comparative agents: results from Surveillance of Multicenter Antimicrobial Resistance in Taiwan (SMART). Infect Drug Resist 12:627–640. doi: 10.2147/IDR.S194482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin WP, Huang YS, Wang JT, Chen YC, Chang SC. 2019. Prevalence of and risk factor for community-onset third-generation cephalosporin-resistant Escherichia coli bacteremia at a medical center in Taiwan. BMC Infect Dis 19:245. doi: 10.1186/s12879-019-3880-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsai CS, Hung YP, Lee JC, Lee NY, Chen PL, Syue LS, Li MC, Li CW, Ko WC. 2018. Community-onset Clostridium difficile infection at a tertiary medical center in southern Taiwan, 2007–2015. J Microbiol Immunol Infect 51:243–250. doi: 10.1016/j.jmii.2016.08.013. [DOI] [PubMed] [Google Scholar]
- 6.Kim YA, Kim JJ, Kim H, Lee K. 2017. Community-onset extended-spectrum-beta-lactamase-producing Escherichia coli sequence type 131 at two Korean community hospitals: the spread of multidrug-resistant E. coli to the community via healthcare facilities. Int J Infect Dis 54:39–42. doi: 10.1016/j.ijid.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 7.Mao YC, Chang CL, Huang YC, Su LH, Lee CT. 2018. Laboratory investigation of a suspected outbreak caused by Providencia stuartii with intermediate resistance to imipenem at a long-term care facility. J Microbiol Immunol Infect 51:214–219. doi: 10.1016/j.jmii.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 8.McGowan JE Jr, Hall EC, Parrott PL. 1989. Antimicrobial susceptibility in Gram-negative bacteremia: are nosocomial isolates really more resistant? Antimicrob Agents Chemother 33:1855–1859. doi: 10.1128/aac.33.11.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jean SS, Lu MC, Shi ZY, Tseng SH, Wu TS, Lu PL, Shao PL, Ko WC, Wang FD, Hsueh PR. 2018. In vitro activity of ceftazidime-avibactam, ceftolozane-tazobactam, and other comparable agents against clinically important Gram-negative bacilli: results from the 2017 Surveillance of Multicenter Antimicrobial Resistance in Taiwan (SMART). Infect Drug Resist 11:1983–1992. doi: 10.2147/IDR.S175679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee YL, Lu MC, Shao PL, Lu PL, Chen YH, Cheng SH, Ko WC, Lin CY, Wu TS, Yen MY, Wang LS, Liu CP, Lee WS, Shi ZY, Chen YS, Wang FD, Tseng SH, Lin CN, Chen YH, Sheng WH, Lee CM, Liao MH, Hsueh PR. 2019. Nationwide surveillance of antimicrobial resistance among clinically important Gram-negative bacteria, with an emphasis on carbapenems and colistin: results from the Surveillance of Multicenter Antimicrobial Resistance in Taiwan (SMART) in 2018. Int J Antimicrob Agents 54:318–328. doi: 10.1016/j.ijantimicag.2019.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Clinical and Laboratory Standards Institute. 2020. Performance standards for antimicrobial susceptibility testing–30th ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 12.Rebelo AR, Bortolaia V, Kjeldgaard JS, Pedersen SK, Leekitcharoenphon P, Hansen IM, Guerra B, Malorny B, Borowiak M, Hammerl JA, Battisti A, Franco A, Alba P, Perrin-Guyomard A, Granier SA, De Frutos Escobar C, Malhotra-Kumar S, Villa L, Carattoli A, Hendriksen RS. 2018. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill 23 https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2018.23.6.17-00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jean SS, Lee WS, Yu KW, Liao CH, Hsu CW, Chang FY, Ko WC, Chen RJ, Wu JJ, Chen YH, Chen YS, Liu JW, Lu MC, Lam C, Liu CY, Hsueh PR. 2016. Rates of susceptibility of carbapenems, ceftobiprole, and colistin against clinically important bacteria collected from intensive care units in 2007: results from the Surveillance of Multicenter Antimicrobial Resistance in Taiwan (SMART). J Microbiol Immunol Infect 49:969–976. doi: 10.1016/j.jmii.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 14.Tsai WL, Hung CH, Chen HA, Wang JL, Huang IF, Chiou YH, Chen YS, Lee SS, Hung WY, Cheng MF. 2018. Extended-spectrum beta-lactamase-producing Escherichia coli bacteremia: comparison of pediatric and adult populations. J Microbiol Immunol Infect 51:723–731. doi: 10.1016/j.jmii.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Bou-Antoun S, Davies J, Guy R, Johnson AP, Sheridan EA, Hope RJ. 2016. Descriptive epidemiology of Escherichia coli bacteraemia in England, April 2012 to March 2014. Euro Surveill 21 https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2016.21.35.30329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Mee-Marquet NL, Blanc DS, Gbaguidi-Haore H, Dos Santos Borges S, Viboud Q, Bertrand X, Quentin R. 2015. Marked increase in incidence for bloodstream infections due to Escherichia coli, a side effect of previous antibiotic therapy in the elderly. Front Microbiol 6:646. doi: 10.3389/fmicb.2015.00646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lester R, Musicha P, van Ginneken N, Dramowski A, Hamer DH, Garner P, Feasey NA. 2019. Prevalence and outcome of bloodstream infections due to third-generation cephalosporin-resistant Enterobacteriaceae in sub-Saharan Africa: a systematic review. J Antimicrob Chemother 75:492–507. doi: 10.1093/jac/dkz464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun HY, Chen SY, Chang SC, Pan SC, Su CP, Chen YC. 2006. Community-onset Escherichia coli and Klebsiella pneumoniae bacteremia: influence of health care exposure on antimicrobial susceptibility. Diagn Microbiol Infect Dis 55:135–141. doi: 10.1016/j.diagmicrobio.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 19.Garau J, Xercavins M, Rodríguez-Carballeira M, Gómez-Vera JR, Coll I, Vidal D, Llovet T, Ruíz-Bremón A, 1999. Emergence and dissemination of quinolone-resistant Escherichia coli in the community. Antimicrob Agents Chemother 43:2736–2741. doi: 10.1128/AAC.43.11.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacDougall C, Powell JP, Johnson CK, Edmond MB, Polk RE. 2005. Hospital and community fluoroquinolone use and resistance in Staphylococcus aureus and Escherichia coli in 17 US hospitals. Clin Infect Dis 41:435–440. doi: 10.1086/432056. [DOI] [PubMed] [Google Scholar]
- 21.Kahlmeter G, Poulsen HO. 2012. Antimicrobial susceptibility of Escherichia coli from community-acquired urinary tract infections in Europe: the ECO.SENS study revisited. Int J Antimicrob Agents 39:45–51. doi: 10.1016/j.ijantimicag.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 22.Rogers BA, Sidjabat HE, Paterson DL. 2011. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother 66:1–14. doi: 10.1093/jac/dkq415. [DOI] [PubMed] [Google Scholar]
- 23.Chen CM, Guo MK, Ke SC, Lin YP, Li CR, Vy Nguyen HT, Wu LT. 2018. Emergence and nosocomial spread of ST11 carbapenem-resistant Klebsiella pneumoniae co-producing OXA-48 and KPC-2 in a regional hospital in Taiwan. J Med Microbiol 67:957–964. doi: 10.1099/jmm.0.000771. [DOI] [PubMed] [Google Scholar]
- 24.Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y. 2011. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother 66:307–312. doi: 10.1093/jac/dkq431. [DOI] [PubMed] [Google Scholar]
- 25.Kuo SC, Shih SM, Lauderdale TY, Chang IS, Chen YC, Hsiung CA, Chang SC. 2020. Policy-driven revolution of prescription record in outpatient use of fluoroquinolones. J Microbiol Immunol Infect 53:133–140. doi: 10.1016/j.jmii.2018.05.002. [DOI] [PubMed] [Google Scholar]
- 26.Wu HH, Liu HY, Lin YC, Hsueh PR, Lee YJ. 2016. Correlation between levofloxacin consumption and the incidence of nosocomial infections due to fluoroquinolone-resistant Escherichia coli. J Microbiol Immunol Infect 49:424–429. doi: 10.1016/j.jmii.2011.12.019. [DOI] [PubMed] [Google Scholar]
- 27.Jean S-S, Lee W-S, Ko W-C, Hsueh P-R. 2020. In vitro susceptibility of ceftaroline against clinically important Gram-positive cocci, Haemophilus species and Klebsiella pneumoniae in Taiwan: results from the Antimicrobial Testing Leadership and Surveillance (ATLAS) in 2012–2018. J Microbiol Immunol Infect doi: 10.1016/j.jmii.2020.04.017. [DOI] [PubMed] [Google Scholar]
- 28.Feng W, Huang Q, Wang Y, Yuan Q, Li X, Xia P, Sun F. 2019. Changes in the resistance and epidemiological characteristics of Pseudomonas aeruginosa during a ten-year period. J Microbiol Immunol Infect doi: 10.1016/j.jmii.2019.08.017. [DOI] [PubMed] [Google Scholar]
- 29.Rojas A, Palacios-Baena ZR, López-Cortés LE, Rodríguez-Baño J. 2019. Rates, predictors and mortality of community-onset bloodstream infections due to Pseudomonas aeruginosa: systematic review and meta-analysis. Clin Microbiol Infect 25:964–970. doi: 10.1016/j.cmi.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen LH, Hsu DI, Ganapathy V, Shriner K, Wong-Beringer A. 2008. Reducing empirical use of fluoroquinolones for Pseudomonas aeruginosa infections improves outcome. J Antimicrob Chemother 61:714–720. doi: 10.1093/jac/dkm510. [DOI] [PubMed] [Google Scholar]
- 31.Diekema DJ, Hsueh PR, Mendes RE, Pfaller MA, Rolston KV, Sader HS, Jones RN. 2019. The microbiology of bloodstream infection: 20-year trends from the SENTRY Antimicrobial Surveillance Program. Antimicrob Agents Chemother 63:e00355-19. doi: 10.1128/AAC.00355-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chusri S, Chongsuvivatwong V, Silpapojakul K, Singkhamanan K, Hortiwakul T, Charernmak B, Doi Y. 2019. Clinical characteristics and outcomes of community and hospital-acquired Acinetobacter baumannii bacteremia. J Microbiol Immunol Infect 52:796–806. doi: 10.1016/j.jmii.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 33.Chen CH, Kuo HY, Hsu PJ, Chang CM, Chen JY, Lu HH, Chen HY, Liou ML. 2018. Clonal spread of carbapenem-resistant Acinetobacter baumannii across a community hospital and its affiliated long-term care facilities: a cross sectional study. J Microbiol Immunol Infect 51:377–384. doi: 10.1016/j.jmii.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 34.Henriksen T-H, Abebe W, Amogne W, Getachew Y, Weedon-Fekjær H, Klein J, Woldeamanuel Y. 2019. Association between antimicrobial resistance among Enterobacteriaceae and burden of environmental bacteria in hospital acquired infections: analysis of clinical studies and national reports. Heliyon 5:e02054. doi: 10.1016/j.heliyon.2019.e02054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng W, Faheem A, McGeer A, Simor AE, Gelosia A, Willey BM, Watt C, Richardson DC, Wong H, Ostrowska K, Vernich L, Muller MP, Gnanasuntharam P, Porter V, Katz K. 2017. Community- and healthcare-associated methicillin-resistant Staphylococcus aureus strains: an investigation into household transmission, risk factors, and environmental contamination. Infect Control Hosp Epidemiol 38:61–67. doi: 10.1017/ice.2016.245. [DOI] [PubMed] [Google Scholar]