Class A β-lactamases are a major cause of β-lactam resistance in Gram-negative bacteria. The recently FDA-approved cyclic boronate vaborbactam is a reversible covalent inhibitor of class A β-lactamases, including CTX-M extended-spectrum β-lactamase and KPC carbapenemase, both frequently observed in the clinic. Intriguingly, vaborbactam displayed different binding kinetics and cell-based activity for these two enzymes, despite their similarity. A 1.0-Å crystal structure of CTX-M-14 demonstrated that two catalytic residues, K73 and E166, are positively charged and neutral, respectively.
KEYWORDS: beta-lactamases, beta-lactams, carbapenemase, cyclic boronic acid, vaborbactam
ABSTRACT
Class A β-lactamases are a major cause of β-lactam resistance in Gram-negative bacteria. The recently FDA-approved cyclic boronate vaborbactam is a reversible covalent inhibitor of class A β-lactamases, including CTX-M extended-spectrum β-lactamase and KPC carbapenemase, both frequently observed in the clinic. Intriguingly, vaborbactam displayed different binding kinetics and cell-based activity for these two enzymes, despite their similarity. A 1.0-Å crystal structure of CTX-M-14 demonstrated that two catalytic residues, K73 and E166, are positively charged and neutral, respectively. Meanwhile, a 1.25-Å crystal structure of KPC-2 revealed a more compact binding mode of vaborbactam versus CTX-M-14, as well as alternative conformations of W105. Together with kinetic analysis of W105 mutants, the structures demonstrate the influence of this residue and the unusual conformation of the β3 strand on the inactivation rate, as well as the stability of the reversible covalent bond with S70. Furthermore, studies of KPC-2 S130G mutant shed light on the different impacts of S130 in the binding of vaborbactam versus avibactam, another recently approved β-lactamase inhibitor. Taken together, these new data provide valuable insights into the inhibition mechanism of vaborbactam and future development of cyclic boronate inhibitors.
INTRODUCTION
The β-lactam antibiotics are among the most commonly prescribed antibacterial agents used to treat a variety of infections due to their broad spectrum of activity and favorable pharmacological properties (1–3). The most frequent mechanism of β-lactam resistance involves the production of β-lactam-degrading enzymes known as β-lactamases, which hydrolyze the amide bond of the four-membered β-lactam ring essential to the activity of these antibiotics (4–7). β-Lactamases are divided into four classes (Ambler classes A, B, C, and D) based on their amino acid sequence homology. Classes A, C, and D are serine β-lactamases (SBLs), which use a serine residue for hydrolysis, whereas class B enzymes are metalloenzymes that instead use zinc ions for the hydrolysis reaction (8).
Class A β-lactamases are the most prevalent class of β-lactamases and are usually found in Gram-negative pathogens (9–11). Many class A β-lactamases display hydrolytic activity against a wide range of β-lactam compounds (12–14). Extended-spectrum β-lactamases (ESBLs) can hydrolyze penicillins, monobactams, and cephalosporins belonging to the first, second, and third generations. Currently, the class A CTX-M family of β-lactamases are the most prevalent ESBLs worldwide (15, 16). Carbapenemases have an even broader spectrum of activity than ESBLs and can inactivate all classes of β-lactam antibiotics, including the classical β-lactamase inhibitors (i.e., clavulanate, sulbactam, and tazobactam). The class A Klebsiella pneumoniae carbapenemase (KPC) is the most common cause of carbapenem resistance seen in the United States (13, 14, 17).
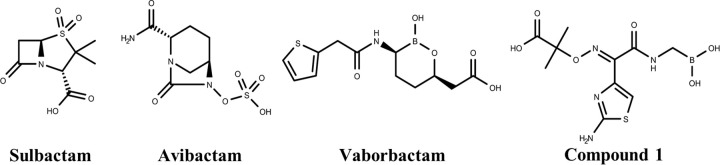
The growing problem of antibiotic resistance mediated by the production of ESBLs and carbapenemases has prompted the search for novel β-lactamase inhibitors (Fig. 1) (18, 19). In 2015, the FDA approved a novel non-β-lactam β-lactamase inhibitor, avibactam, in combination with the third-generation cephalosporin ceftazidime to combat bacteria producing class A and C β-lactamases such as CTX-M, KPC, and AmpC (20). Unfortunately, even this inhibitor has demonstrated cases of resistance due to porin mutations and increased expression of KPC-3 (21), as well as various mutations in KPC-2 and KPC-3 (22), suggesting that additional inhibitor discovery is necessary. An emerging class of novel β-lactamase inhibitors is boronic acids, which have undergone extensive investigation due to their ability to form a stable reversible covalent bond with the active-site serine residue of SBLs (23–25). Boronic acids potently inhibit SBLs because of their effective mimicking of the tetrahedral transition state that is part of the acylation or deacylation steps of SBL catalysis (26, 27). Currently, boronic acid transition state inhibitors (BATSIs) have exhibited inhibition against a wide range of SBLs (28, 29). Most recently, an FDA-approved cyclic boronate inhibitor, vaborbactam (formerly RPX7009), in combination with the carbapenem meropenem, demonstrated potent activity against Gram-negative bacteria, including KPC-producing carbapenem-resistant Enterobacteriaceae (CRE). Specifically, vaborbactam can inhibit class A β-lactamases such as CTX-M, KPC, SHV, and TEM and class C β-lactamases such as P99, MIR, and FOX (30, 31).
FIG 1.
Chemical structures of serine β-lactamase inhibitors.
A recent study showed different binding kinetics of vaborbactam during its inhibition of KPC-2 versus CTX-M-15 despite the highly similar active sites of these two class A enzymes (31). Compared with CTX-M-15, the inactivation and koff rates of KPC-2 are ∼3 and 50 times lower, respectively, leading to an ∼15-fold difference in the binding affinity. The importance of binding kinetics for drug efficacy has received increasing attention, with many studies demonstrating that the residence time (i.e., the duration of drug occupation of the target binding site) corresponds more to the drug’s in vivo efficacy than the equilibrium dissociation constant (Kd) (32–34). However, most of these studies focused on how the residence time can influence the pharmacokinetics of the drug inside animals or humans (35, 36). It is largely unknown how binding kinetics can impact the activity of a drug on the cellular level, especially relative to their bactericidal effects (37). While the study comparing KPC-2 and CTX-M-15 was informative, it also highlighted the need to elucidate the unique binding kinetics of vaborbactam for KPC-2 and CTX-M, as well as its influence on vaborbactam’s cell-based activity.
In this study, we examined the structural basis for vaborbactam’s inhibition of two clinically relevant class A enzymes, CTX-M-14 and KPC-2. Crystals of CTX-M-14 routinely diffract to sub-angstrom resolution, allowing us to visualize hydrogen atoms on catalytically important residues and gain insights into vaborbactam’s mode of inhibition of class A β-lactamases (19, 38). Most importantly, the crystal structure of vaborbactam with KPC-2, combined with binding kinetics and mutagenesis studies, helps shed light on the origin and manifestation of vaborbactam’s long residence time in KPC-2 as well as the importance of the inactivation rate and Kd for vaborbactam’s cell-based activity.
RESULTS AND DISCUSSION
Ultrahigh-resolution X-ray crystal structure of CTX-M-14 with vaborbactam.
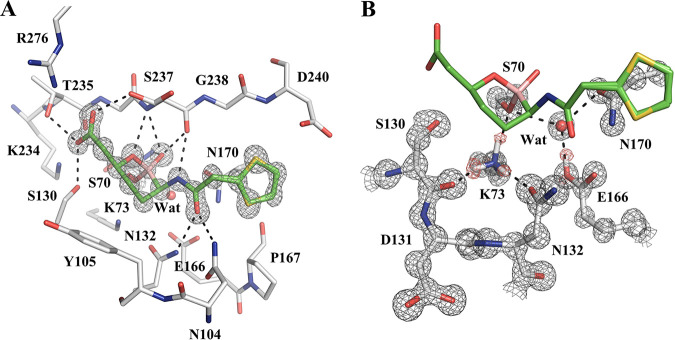
The structure of the class A ESBL CTX-M-14 complexed with vaborbactam was solved to a 1.0-Å resolution (Fig. 2; Table S1). CTX-M-14 was crystallized in the P21 space group, with two molecules in the asymmetric unit (ASU). The structure was refined to an Rwork and Rfree of 0.1046 and 0.1209, respectively. Two copies of vaborbactam were observed in each monomer, with one covalently linked with S70 in the active site and the other one binding noncovalently outside the active site, near residues N104, Y105, and T227 and likely a crystal artifact. The interactions that vaborbactam forms with the CTX-M-14 active site mirror those seen in the previous CTX-M-15–vaborbactam complex (PDB code 4XUZ) (Fig. 2A) (30). However, the CTX-M-15–vaborbactam complex was determined to a resolution of 1.5 Å, whereas solving our complex at a 1.0-Å resolution allowed us to see additional detailed and important structural features, such as the protonation state of catalytic residues. We were particularly interested in the protonation state of K73 and E166, two residues that participate in the acylation and deacylation reaction central to the catalysis of class A β-lactamases (39–41).
FIG 2.
Vaborbactam complex crystal structure with CTX-M-14 class A β-lactamase. (A) Vaborbactam covalently bound in the active site of CTX-M-14 (1.0-Å resolution; PDB code 6V7H). The unbiased Fo-Fc omit map of vaborbactam is contoured at 3σ and represented with gray mesh. Hydrogen bonds are shown as black dashed lines. (B) Protonation state of K73 and E166 in the CTX-M-14–vaborbactam complex. The 2Fo-Fc omit map (gray) is contoured at 3σ around the protein residues, and the unbiased Fo-Fc omit map (red) is contoured at 2σ around K73 and E166, corresponding to hydrogen atoms located on the terminal groups of K73 and E166.
The vaborbactam carboxylate group forms hydrogen bonds (HBs) with S130, T235, and S237, three residues that usually interact with the C-3/4-carboxylate group of β-lactam substrates. The two boronic acid oxygens (i.e., endocyclic ring oxygen and exocyclic hydroxyl) are placed inside the “oxyanion hole” of CTX-M-14 formed by the backbone amides of S70 and S237. Like most serine hydrolases, this oxyanion hole, which is a small subpocket surrounded by several HB donor atoms, helps coordinate the carbonyl group oxygen of the substrate next to the scissile bond (42, 43). Whereas the exocyclic oxygen interacts with both backbone NH groups of the oxyanion hole, the endocyclic oxygen forms only one HB with the NH group of S237. The NH of the amide group of vaborbactam donates a hydrogen bond to the S237 backbone carbonyl, while the carbonyl of the same amide group of vaborbactam accepts a hydrogen bond from the N104 and N132 side chain amide groups. Meanwhile, nonpolar contacts are observed between the six-membered ring and Y105 and between the five-membered thiophene ring and the Cβ atoms of P167 and N170.
Examination of the Fo-Fc electron density map shows hydrogen atoms on several residues, including the catalytically important residues K73 and E166 (Fig. 2B). There are three positive peaks on the Nζ of K73, corresponding to a positively charged lysine, which serves as an HB donor to the side chains of S70 and N132 and the main-chain carbonyl group of S130. Additionally, a single positive peak on the E166 carboxyl group indicates that it is neutral and in its glutamic acid form. For the first time, these results shed light on the protonation states of these two residues in the vaborbactam complex.
Crystal structure of KPC-2 with vaborbactam.
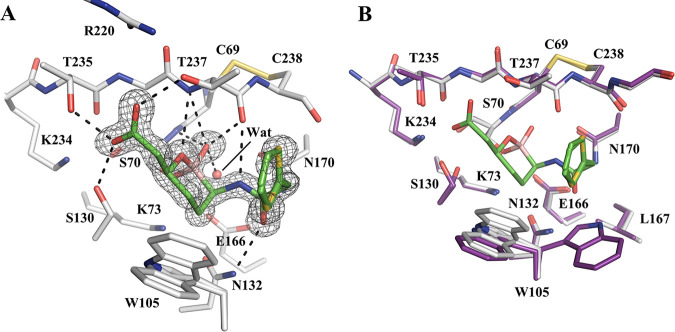
The KPC-2 complex with vaborbactam was solved to a 1.25-Å resolution (Fig. 3; Table S1). The KPC-2–vaborbactam complex crystal belongs to the space group P22121, with one protein molecule in the ASU. The structure was refined to an Rwork and Rfree of 0.1566 and 0.1789, respectively. The Fo-Fc map showed an unambiguous density in the KPC-2 active site corresponding to two closely related conformations of vaborbactam, differing from one another by a flip of the thiophene ring. Similar to the CTX-M-14 complex, vaborbactam forms several HBs with the enzyme (Fig. 3A), including the hydroxyl side chains of S130, T235, and T237, the oxyanion hole, and N132. The patterns of HB with vaborbactam are nearly identical for the two enzymes. One main difference in side chain polar groups interacting with vaborbactam is T237, which, like S237 in CTX-M-14, forms an HB with the vaborbactam carboxylate group. In addition, compared with N104 in CTX-M-14, P104 in KPC-2 abolishes the HB with the vaborbactam side chain carbonyl group. However, the HB between this carbonyl group and N132 is maintained in both KPC-2 and CTX-M-14. A key difference in the binding mode of vaborbactam between KPC-2 and CTX-M-14 is the conformation of the thiophene moiety of the inhibitor, which reorients inward to make hydrophobic interactions with W105, and the hydrocarbon atoms of the T237 side chain and C238 backbone. These contacts between vaborbactam and KPC-2 largely mimic the interactions that the β-lactam antibiotics cefotaxime and faropenem form with KPC-2 (44).
FIG 3.
Vaborbactam complex crystal structure with class A carbapenemase KPC-2. (A) Vaborbactam covalently bound in the active site of KPC-2 (1.25-Å resolution; PDB code 6V7I). The unbiased Fo-Fc omit map of vaborbactam is contoured at 3σ and represented with gray mesh. Hydrogen bonds are shown as black dashed lines. (B) Superimposition of KPC-2–vaborbactam (gray) and KPC-2 apo (PDB code 5UL8; purple) structures.
Vaborbactam induces some conformational changes in the KPC-2 active site upon binding. In the apoenzyme (PDB code 5UL8) (44), W105 alternates between two conformations that flank opposite sides of the active site, whereas in the complex structure, W105 adopts two conformations that are positioned to make hydrophobic contacts with the six-membered ring and thiophene moiety of vaborbactam (Fig. 3B). One conformation of W105 is similar to that in the apoenzyme, whereas the other conformation is unique to the particular complex, allowing the protein to establish more nonpolar contacts with vaborbactam. In the apoenzyme, S130 also has two conformations. Conformation 1 is located closer to K73, with a distance of 3.5 Å between S130 Oγ and K73 Nζ, and is the conformation normally observed in class A β-lactamases (44). Conformation 2 moves closer to K234, forming a stronger HB (2.7 Å) than conformation 1 (3.0 Å). In the complex structure, S130 adopts a single conformation, conformation 2, forming an HB with the vaborbactam carboxylate group and K234.
Effects of W105 mutations on vaborbactam binding kinetics.
Substitution of the W105 residue in KPC-2 has been previously shown to affect substrate binding and catalysis as well inhibition by some β-lactamase inhibitors (45). W105 appears to establish more nonpolar interactions with vaborbactam in KPC-2 than the corresponding Y105 in CTX-M-14. To probe the contribution of W105 to the vaborbactam activity and binding kinetics, various W105 substitutions were introduced in KPC-2 by site-directed mutagenesis, and mutant proteins were purified for biochemical studies. As expected, W105 substitutions significantly affected the kinetics of vaborbactam binding and KPC-2 hydrolysis of β-lactam substrates.
An enzymatic competition assay using the chromogenic β-lactamase substrate nitrocefin revealed that vaborbactam behaves as a two-step tight-binding inhibitor that initially binds to SBLs in a noncovalent fashion (characterized by the affinity constant [K]). Subsequently, covalent bond formation occurs between the active-site serine and boron of vaborbactam (characterized by the rate constant [k2]) (31). Vaborbactam exhibited various degrees of acylation efficiency (or inactivation rate, characterized by k2/K) with the W105 mutants (Table 1). Wild-type KPC-2 had a k2/K of 5.8 × 103 M−1 s−1. Mutants that had aromatic residue substitutions at residue 105, such as W105F and W105Y, had comparable acylation efficiencies at 4.1 × 103 M−1 s−1 and 3.8 × 103 M−1 s−1, respectively. Generally, residue 105 mutants that replaced tryptophan with a nonaromatic residue had much lower acylation efficiencies with vaborbactam (e.g., 20-fold decrease for the W105D mutant compared to the wild-type KPC-2), except the W105N mutant, which had a 2-fold decrease compared with the wild-type KPC-2. In contrast to acylation efficiency, W105 substitutions had minimal impacts on the off rate of dissociation (characterized by koff) of vaborbactam. For example, the largest difference in koff was roughly 2-fold when wild-type KPC-2 was compared to the W105L mutant.
TABLE 1.
Impact of KPC-2 W105 mutations on reaction with vaborbactam
| Enzyme mutation | k2/K (M−1 s−1) | koff (s−1) | Residence time (min) | Kd (nM) |
|---|---|---|---|---|
| WT | 5.8 × 103 ± 0.3 × 103 | 4.3 × 10−5 ± 0.6 × 10−5 | 394 ± 50 | 7.4 ± 0.9 |
| W105F | 4.1 × 103 ± 0.1 × 103 | 3.9 × 10−5 ± 0.5 × 10−5 | 437 ± 59 | 9.3 ± 1.1 |
| W105N | 2.9 × 103 ± 0.1 × 103 | 5.0 × 10−5 ± 0.6 × 10−5 | 337 ± 46 | 17 ± 2 |
| W105L | 0.63 × 103 ± 0.01 × 103 | 2.3 × 10−5 ± 0.3 × 10−5 | 721 ± 101 | 37 ± 4 |
| W105V | 0.94 × 103 ± 0.1 × 103 | 3.5 × 10−5 ± 0.3 × 10−5 | 473 ± 41 | 38 ± 3 |
| W105D | 0.32 × 103 ± 0.07 × 103 | 3.5 × 10−5 ± 0.3 × 10−5 | 477 ± 48 | 110 ± 12 |
| W105A | 0.85 × 103 ± 0.07 × 103 | 7.7 × 10−5 ± 0.8 × 10−5 | 219 ± 23 | 90 ± 10 |
| W105Y | 3.8 × 103 ± 0.3 × 103 | 3.6 × 10−5 ± 0.2 × 10−5 | 463 ± 20 | 9.6 ± 0.8 |
| W105S | 0.92 × 103 ± 0.08 × 103 | 4.4 × 10−5 ± 0.6 × 10−5 | 386 ± 53 | 47 ± 6 |
Effects of W105 mutations on the enzymatic activity of KPC-2.
The impact of the W105 substitutions on the enzymatic activity of KPC-2 was evaluated using nitrocefin and meropenem as substrates (Table 2). The W105 substitution appeared to have a substrate-dependent effect on KPC-2 hydrolysis. The catalytic efficiency of KPC-2 against nitrocefin (characterized by kcat/Km) was significantly impaired for W105 mutants in which the tryptophan was replaced with a nonaromatic residue. For example, the W105D mutant was 14-fold less proficient in nitrocefin hydrolysis than the wild-type KPC-2. Overall, the W105 substitutions had slightly less effect on meropenem hydrolysis, with only a 7-fold difference in kcat/Km between the wild-type KPC-2 and the least efficient mutant, the W105S variant. It is also worth noting that when nitrocefin and meropenem were compared, W105 mutations increased the Km values for both antibiotics but had less influence on kcat. A notable exception is W105D in meropenem hydrolysis, where the kcat value was 4 times higher than that of the wild type (WT). As W105 forms extensive nonpolar interactions with β-lactam substrates (44), W105 mutations would most certainly affect substrate binding. For carbapenems like meropenem, their unique hydroxyethyl group hinders the attack of the catalytic water on the acyl-enzyme linkage, as demonstrated by the usually low deacylation rate of carbapenems compared with those of other β-lactams (44, 46–49). It is possible that some W105 mutations, especially W105D, may induce alternative conformations of the hydroxyethyl group and enable the catalytic water to access and break the acyl-enzyme bond.
TABLE 2.
Impact of KPC-2 W105 mutations on nitrocefin and meropenem hydrolysis
| Enzyme mutation | Nitrocefin |
Meropenem |
||||
|---|---|---|---|---|---|---|
| Km (μM) | kcat (s−1) | kcat/Km (s−1 μM−1) | Km (μM) | kcat (s−1) | kcat/Km (s−1 μM−1) | |
| WT | 43 ± 4 | 190 ± 6 | 4.40 ± 0.25 | 15 ± 2 | 3.8 ± 0.1 | 0.26 ± 0.02 |
| W105F | 71 ± 3 | 192 ± 13 | 2.70 ± 0.07 | 16 ± 2 | 1.8 ± 0.1 | 0.12 ± 0.01 |
| W105N | 160 ± 14 | 135 ± 17 | 0.84 ± 0.04 | 59 ± 5 | 3.1 ± 0.1 | 0.053 ± 0.003 |
| W105L | 357 ± 18 | 290 ± 4 | 0.81 ± 0.03 | 55 ± 4 | 2.5 ± 0.1 | 0.046 ± 0.002 |
| W105V | 236 ± 9 | 198 ± 8 | 0.84 ± 0.06 | 59 ± 7 | 4.2 ± 0.3 | 0.073 ± 0.007 |
| W105D | 558 ± 68 | 172 ± 10 | 0.31 ± 0.02 | 122 ± 4 | 15.7 ± 0.2 | 0.129 ± 0.003 |
| W105A | 299 ± 29 | 238 ± 3 | 0.80 ± 0.07 | 65 ± 10 | 2.8 ± 0.2 | 0.044 ± 0.004 |
| W105Y | 79 ± 5 | 247 ± 9 | 3.12 ± 0.11 | 19 ± 1 | 2.6 ± 0.04 | 0.14 ± 0.01 |
| W105S | 293 ± 11 | 169 ± 3 | 0.58 ± 0.02 | 64 ± 8 | 2.2 ± 0.2 | 0.035 ± 0.001 |
Effects of W105 mutations on vaborbactam potency.
To study the microbiological impact of W105 substitutions, plasmids carrying mutant KPC-2 genes were transformed into Pseudomonas aeruginosa (PAM1154 strain lacking the major efflux pump, MexAB-OprM) (50, 51). The evaluation of the effect of the W105 substitutions on resistance to various β-lactam antibiotics demonstrated multiple antibiotic-specific effects. The W105D mutant revealed a behavior unique among the other mutants; it lost resistance to all β-lactams except carbapenems (Table S2).
The effect of mutations on vaborbactam potency was investigated next by the evaluation of the concentration response of meropenem potentiation by vaborbactam against the strains producing various W105 mutations. Vaborbactam potency was defined as the 50% potentiation value (PV50) of antibiotic potentiation (22). The PV50 is the concentration of a β-lactamase inhibitor (BLI) that is required to reduce the antibiotic MIC to the middle of the MIC range, where the maximal MIC is the MIC for a KPC-producing strain with no inhibitor added and the minimal MIC is the MIC in the presence of an inhibitor at a concentration required to achieve maximum inhibition (or, simply, a MIC for a vector-alone strain). There appeared to be no significant variation in PV50s, which ranged from 0.65 μg/ml to 1.49 μg/ml (0.73 μg/ml for the wild type KPC-2) (Table 3; Fig. S1). However, close examination of the meropenem MIC in the absence of vaborbactam, vaborbactam PV50, and the fold reduction of meropenem MIC in the presence of vaborbactam at 1.25 μg/ml (concentration of vaborbactam inside the interval of PV50s) revealed interesting correlations with the mutants’ enzymatic activity and inhibition by vaborbactam. Overall, for mutants with catalytic efficiencies (kcat/Km) similar to that of the WT, such as the W105F, W105Y, and W105D mutants, the MIC of meropenem in the absence of vaborbactam was approximately 32 μg/ml, the same as for the WT. The other mutants, which had lower kcat/Km values, especially the W105L, W105A, and W105S mutants, had MICs up to 4 times lower. For the potentiation effect of vaborbactam, mutants with inactivation rates or Kd values similar to those of the WT, such as the W105F, W105Y, and W105N mutants, had ∼10% decreases in PV50s compared to the WT (no statistical significance) and exhibited WT-level fold reductions in meropenem MIC (64- to 256-fold) at a vaborbactam concentration of 1.25 μg/ml. Meanwhile, those with lower inactivation rates or higher Kd constants, such as the W105D, W105L, W105A, W105V, and W105S mutants, displayed 20 to 100% higher PV50s than the WT (statistically significant difference with P values of <0.05 for the W105D, W105L, and W105S mutants) and lower MIC reduction (16- to 32-fold). This is particularly evident for W105D, because the meropenem MIC for the strain expressing the W105D enzyme in the absence of vaborbactam was the same as that for the WT, yet the PV50 of vaborbactam was increased ∼2-fold, and the meropenem MIC in the presence of 1.25 μg/ml vaborbactam was 8 times higher than that for the WT (Table 3).
TABLE 3.
MIC of meropenem in combination with vaborbactam against P. aeruginosa expressing KPC-2 W105 mutants
| Enzyme mutation | Vaborbactam concn (μg/ml) |
PV50a (μg/ml) | Decrease in MICb | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.16 | 0.31 | 0.63 | 1.25 | 2.5 | 5 | 10 | |||
| WT | 32 | 32 | 16 | 2 | 0.25 | 0.063 | 0.063 | ≤0.031 | 0.73 ± 0.02 | 128 |
| W105F | 32 | 16 | 8 | 1 | 0.125 | 0.063 | ≤0.031 | ≤0.031 | 0.65 ± 0.11 | 256 |
| W105N | 16 | 8 | 4 | 1 | 0.25 | 0.063 | 0.063 | ≤0.031 | 0.65 ± 0.01 | 64 |
| W105L | 8 | 8 | 4 | 2 | 0.5 | 0.125 | 0.063 | ≤0.031 | 1.06 ± 0.11 | 16 |
| W105V | 8 | 4 | 4 | 1 | 0.25 | 0.125 | 0.063 | ≤0.031 | 0.87 ± 0.11 | 16 |
| W105D | 32 | 16 | 8 | 4 | 2 | 0.5 | 0.125 | ≤0.031 | 1.49 ± 0.09 | 16 |
| W105A | 8 | 8 | 4 | 2 | 0.25 | 0.125 | ≤0.031 | ≤0.031 | 0.87 ± 0.1 | 32 |
| W105Y | 32 | 16 | 8 | 2 | 0.25 | ≤0.031 | ≤0.031 | ≤0.031 | 0.68 ± 0.05 | 128 |
| W105S | 8 | 8 | 4 | 2 | 0.25 | 0.125 | ≤0.031 | ≤0.031 | 1.00 ± 0.02 | 32 |
Data are averages of three measurements. Differences from the WT are statistically significant (P < 0.05) for W105D, W105L, and W105S.
Fold decrease in meropenem MIC with vaborbactam at 1.25 μg/ml.
Influence of S130G mutation on vaborbactam inhibition.
S130 plays an important role in the catalytic mechanism of class A β-lactamases, including KPC-2 (12), and mutations at this position significantly affect interactions with substrates and inhibitors, including avibactam (52, 53). The effect of the S130G substitution on vaborbactam potentiation of various antibiotics was studied in microbiological experiments with P. aeruginosa PAM1154 expressing the KPC-2 WT and S130G mutant (Table S2). Consistent with earlier studies, KPC-2 S130G lost the ability to hydrolyze the majority of β-lactam antibiotics, as suggested by the drastic decreases in the MICs of most compounds (52–54). P. aeruginosa producing the KPC-2 S130G variant demonstrated resistance to the penicillins (i.e., carbenicillin and piperacillin) but not to other β-lactams. Subsequently, carbenicillin and piperacillin were chosen to study the concentration response to vaborbactam using a checkerboard method (Table 4; Fig. S2). As previously reported, cells producing KPC-2 S130G were highly resistant to avibactam potentiation of penicillins (54). The PV50 of avibactam for carbenicillin and piperacillin potentiation was increased >40- to 100-fold. Carbenicillin and piperacillin MICs against the KPC-2 WT-producing strain were reduced by 64- to 256-fold in the presence of avibactam at 4 μg/ml, and those against the strain that produced KPC-2 S130G were reduced only 2-fold. Notably, the S130G substitution did not significantly affect antibiotic potentiation by vaborbactam.
TABLE 4.
Concentration response of carbenicillin and piperacillin potentiation by vaborbactam and avibactam against KPC-2-producing strains of P. aeruginosa
| Plasmid | BLI | Antibiotic | Antibiotic MIC (μg/ml) in the presence of BLI at (μg/ml): |
PV50 (μg/ml) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 4 | 8 | 16 | 32 | ||||
| WT | Vaborbactam | Carbenicillin | 1,024 | 64 | 16 | 8 | 4 | 2 | 1 | 1.9 |
| S130G | Vaborbactam | Carbenicillin | 128 | 4 | 2 | 1 | ≤0.5 | ≤0.5 | ≤0.5 | <1 |
| WT | Avibactam | Carbenicillin | 1,024 | 64 | 64 | 32 | 16 | 8 | 2 | 4.6 |
| S130G | Avibactam | Carbenicillin | 128 | 128 | 128 | 64 | 64 | 64 | 32 | >150 |
| WT | Vaborbactam | Piperacillin | 128 | 1 | 0.5 | 0.25 | ≤0.13 | ≤0.13 | ≤0.13 | <1 |
| S130G | Vaborbactam | Piperacillin | 128 | 2 | 0.5 | 0.25 | 0.25 | ≤0.13 | ≤0.13 | <1 |
| WT | Avibactam | Piperacillin | 128 | 2 | 1 | 0.5 | 0.25 | ≤0.13 | ≤0.13 | <1 |
| S130G | Avibactam | Piperacillin | 128 | 128 | 64 | 64 | 64 | 32 | 16 | >100 |
Detailed kinetic studies confirmed the significant effect of the S130G substitution on the acylation efficiency of avibactam (52, 54, 55); it was decreased more than 1,000-fold (Table 5). Conversely, vaborbactam inactivated the KPC-2 S130G variant with an approximately 10-fold-higher efficiency than its inactivation of the KPC-2 WT (6.7 × 104 ± 0.3 × 104 versus 5.8 × 103 ± 0.3 × 103 M−1 s−1) (Tables 1 and 5). Another unexpected impact of S130G on vaborbactam kinetics was an almost 100-fold increase in the rate of recovery of enzymatic activity (4.5 ± 1.0 × 10−3 versus 4.3 ± 0.6 × 10−5 s−1) (Tables 1 and 5). Of note, due to an increase in the rate of onset of inhibition (k2/K), the Kd value remained in the nanomolar range, providing a possible explanation as to why the S130G mutation did not impact the antibiotic potentiation activity of vaborbactam.
TABLE 5.
Kinetic parameters of KPC-2 inhibition by avibactam and vaborbactama
| Enzyme mutation | Inhibitor | k2/K (M−1 s−1) | koff (s−1) | Residence time (min) | Kd (nM) |
|---|---|---|---|---|---|
| WT | Avibactam | 2.3 × 104 ± 0.2 × 104 | 3.3 × 10−4 ± 0.1 × 10−4 | 51 ± 2 | 14 ± 1 |
| S130G | Avibactam | 8.6 ± 1.3 | ND | ND | ND |
| WT | Vaborbactam | 5.8 × 103 ± 0.3 × 103 | 4.3 × 10−5 ± 0.6 × 10−5 | 394 ± 50 | 7.4 ± 0.9 |
| S130G | Vaborbactam | 6.7 × 104 ± 0.3 × 104 | 4.5 × 10−3 ± 1.0 × 10−3 | 3.8 ± 1.0 | 67 ± 13 |
ND, not determined.
Vaborbactam mode of action.
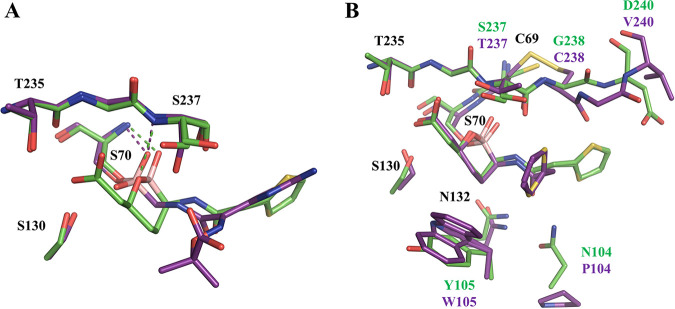
Previous boronic acid inhibitors of SBLs are transition-state analogs that mimic the intermediates found in either the acylation or deacylation steps of catalysis (56). The overall placement of the two vaborbactam boronate oxygen atoms resembles the acylation transition state rather than the deacylation transition state of class A β-lactamases, with the endocyclic oxygen positioned near the location of the β-lactam ring nitrogen of the substrate rather than the catalytic water. A distinct feature of the vaborbactam complexes is their interactions with the oxyanion hole, consisting of the NH groups of S70 and S/T237 in CTX-M-14 and KPC-2. Typically, substrates and inhibitors engage it with a single acceptor (oxygen atom or hydroxyl) (57–61). Vaborbactam, on the other hand, inserts the exocyclic oxygen into the oxyanion hole while establishing an additional HB with the backbone amide group of residue 237 through the endocyclic oxygen. The structural basis for this difference is illustrated by comparison with previous boronic acid acylation transition state analogs, such as compound 1 (Fig. 1), in complex with CTX-M-14 (19). In contrast to the linear structure of compound 1, the cyclic structure of vaborbactam restrains the orientation and hydrogen bonding capability of the endocyclic oxygen. Unlike the corresponding oxygen in compound 1, the endocyclic oxygen of vaborbactam is unable to form a HB with S130, which is compensated by a less favorable HB with S237 backbone N (3.1 Å, in comparison to 2.7 Å for the HB between the corresponding oxygen of compound 1 and S130) (Fig. 4A). In addition, whereas the carboxylate group of vaborbactam mimics the C-3/4 carboxylate group of β-lactams, the carboxylate group of compound 1 is part of the unique side chain taken from ceftazidime. As a result, it does not interact in the β-lactam carboxylate recognition site formed by S130, T235, and S237; instead, it is rotated out of the active site.
FIG 4.
Comparison of boronic acid complex structures. (A) Superimposition of CTX-M-14–vaborbactam (PDB code 6V7H; green) with CTX-M-14–boronic acid inhibitor compound 1 (PDB code 4UA9; purple). (B) Superimposition of CTX-M-14–vaborbactam (PDB code 6V7H; green) with KPC-2–vaborbactam (PDB code 6V7I; purple).
Previous studies have shown that K73 and S130 form a relay network to protonate the β-lactam ring nitrogen (38). The protonation states of catalytic residues in the complex with compound 1 represent a reaction stage during the collapse of the acylation transition state, where a charged K73 is poised to transfer a proton to S130, which in turn serves as an acid and HB donor to the ring nitrogen, mimicked by the boronic acid oxygen (19, 39). The lack of an HB between S130 and K73 in the vaborbactam complex makes it resemble the acylation transition state less and raises the question of the K73 protonation state. In one recently determined ultrahigh-resolution crystal structure, K73 was shown to be neutral (PDB code 4UAA) (19). It is therefore interesting to observe that K73 is still positively charged, resembling the complex with compound 1. Meanwhile, E166 is in a neutral state, as observed in all complex structures (19). Such information regarding residue protonation states will be useful for future modeling of these boronate compounds, which are currently being intensively studied and developed as broad-spectrum inhibitors active against all four classes of β-lactamases (62–64).
Structural basis and biological implication of vaborbactam binding kinetics.
The high binding affinity and long residence time of vaborbactam for class A β-lactamases is mostly derived from the covalent bond between the catalytic serine and the boron atom. However, it is unclear what extent of influence other factors, such as conformational entropy and noncovalent interactions, might have on the binding kinetics, including the inactivation and dissociation rates, as well as the stability of the covalent bond itself. The analysis of W105 KPC-2 mutants, together with the comparison of KPC-2 and CTX-M-15 from previous experiments, offers further insights into the interactions between vaborbactam and class A β-lactamases.
A recent study showed that the inactivation rate and koff rates of vaborbactam are approximately 3 and 50 times lower, respectively, for KPC-2 than CTX-M-15 (31). Another recent study reported similar trends of the binding kinetics for these two enzymes, albeit with some differences in the exact numbers, possibly due to different experimental conditions (65). The previously determined CTX-M-15 complex structure (PDB code 4XUZ) with vaborbactam is nearly identical to our CTX-M-14 model in the active site (30). Comparing the complex crystal structures of KPC-2 and CTX-M-14/15 illustrates several differences in vaborbactam binding between the two proteins, due to some unique structural features of the KPC-2 active site (Fig. 4B). In particular, unlike CTX-M enzymes, KPC-2 and related class A carbapenemases possess a disulfide bridge between the adjacent C69 and C238, which has been demonstrated to be important for activity (54, 66, 67). The disulfide bridge results in an outward shift of G239 and V240 on the β3 strand, which restrains the conformation of the thiophene moiety of vaborbactam. Consequently, vaborbactam adopts a more compact configuration in KPC-2. This configuration can be accommodated in CTX-M-15 active site, although a more extended side chain conformation is assumed due to more favorable interactions with the CTX-M-15 residues.
The unique vaborbactam conformation and the presence of W105 also lead to more extensive interactions between the inhibitor and residue 105 in KPC-2, in comparison to the equivalent Y105 in CTX-M-15. These nonpolar interactions suggest that W105 may potentially influence the binding kinetics of the inhibitor. The W105 mutants, particularly those with smaller side chains, exhibited lower inactivation rates, demonstrating that W105 contributes to the recruitment of vaborbactam to the enzyme active site. The ∼20-fold decrease of k2/K in the W105D mutant may also have resulted from unfavorable electrostatic interactions between the aspartate side chain and the vaborbactam boronate or carboxylate groups. Interestingly, the W105Y mutation decreased the inactivation rate only slightly, indicating that Y105 of CTX-M-15 may play a similar role in vaborbactam binding. The faster inactivation rate of CTX-M-15 by vaborbactam is likely due to the fewer constraints on vaborbactam conformation in the CTX-M-15 active site, allowing easier access to S70 than in KPC-2.
Despite the effect on the inactivation rates, the W105 mutations, including W105D, had a negligible influence on the koff values. This indicates that noncovalent interaction may not be a significant contributing factor in determining the koff rate. The ∼50-fold difference in koff values between KPC-2 and CTX-M-15 thus appears to have most likely originated from various stabilities of the covalent bond between S70 and the boron group, which is unique in its reversibility. We hypothesize that this is again likely due to the unique C69-C238 disulfide bond. The two cysteine residues are located next to S70 and T237, the residues making up the oxyanion hole that interacts with the two boronate oxygen atoms. The disulfide bond rigidifies the oxyanion hole and stabilizes the interactions between the boronate group and S70. In fact, the temperature factors of S70 and T237 backbone nitrogen atoms are 8.93 and 10.03 Å2, respectively, in the KPC-2 complex structure, which are significantly lower than the average values of the protein atoms (15.07 Å2). In comparison, the temperature factors of S70 and S237 backbone nitrogen atoms are 4.12 and 7.58 Å2, respectively, in CTX-M-15, the latter of which is slightly higher than the average value of 7.40 Å2. As temperature factors serve as an indicator of atomic movement, this variation at least supports our hypothesis that the oxyanion hole seems to be more stable in KPC-2 (68–71).
Previous studies have also suggested that residence time, or koff, correlates better with in vivo drug activity than Kd (32–37). These experiments were mostly focused on animal models. The study of the W105 mutations demonstrates that the inactivation rate, or the on rate, still matters in cell-based activity. There was no apparent correlation (r2 = 0.06) between the koff rate or residence time of vaborbactam and vaborbactam potency (PV50) of meropenem potentiation (Fig. S3A). At the same time, better correlation was observed between the kon rate and/or Kd versus vaborbactam potency (r2 = 0.5 to 0.7) (Fig. S3B and C). Specifically, the W015D mutation had little effect on the koff rate or residence time of vaborbactam and on the activity of meropenem hydrolysis. However, it led to an ∼20-fold decrease in the inactivation rate, an ∼2-fold increase in PV50, and an ∼8-fold increase in meropenem MIC at a few critical vaborbactam concentrations (e.g., 1.25 and 2.5 μg/ml) that significantly enhanced meropenem’s cell-based activity against bacteria expressing WT KPC-2. These results echo another recent study suggesting that the overall binding affinity is likely a better predictor of drug development success than residence time (72).
Our studies on the effect of the S130G mutation on vaborbactam and avibactam binding to KPC-2 have also offered further insight into key differences in protein interactions between these two compounds, despite both acting as covalent inhibitors. Like β-lactam antibiotics, avibactam forms an acyl-enzyme bond with S70, and the acylation process involves a series of proton transfers catalyzed by active-site residues such as K73 and S130. In contrast, vaborbactam relies on the unique reactivity of the boron group to form the covalent bond with S70. The S130G mutation would therefore significantly hinder the acylation reaction of avibactam, while having much less impact on vaborbactam. Interestingly, the KPC-2 complex with vaborbactam also reveals some minor steric clashes between S130Cβ and the carbon atom of the ligand’s six-membered ring, with a distance of 3.5 Å. This may at least partly explain why the S130G substitution actually increased the inactivation efficiency of vaborbactam, possibly by relieving steric hinderance in the active site. However, the mutation also eliminates a favorable HB between S130 and the inhibitor for both avibactam and vaborbactam and increases the flexibility of the active site. Such effects may reduce the stability of the covalent complex and lead to more rapid dissociation of the inhibitor than is seen with the WT enzyme.
Conclusion.
Bacterial antibiotic resistance to β-lactam antibiotics is a significant health threat worldwide. Of notable health concern is the class A β-lactamase KPC-2, due to its broad substrate profile, which includes extended-spectrum cephalosporins and carbapenems. Our biochemical and structural analyses have provided important insights into the binding kinetics and molecular interactions underlying the clinical utility of vaborbactam, a unique cyclic boron-based inhibitor, in countering bacterial resistance caused by KPC-2 and its mutants. They highlight the unique structural features of the KPC-2 active site, particularly as a result of the C69-C238 disulfide bond, which contribute to the difference in binding kinetics between KPC-2 and CTX-M enzymes. In addition, the CTX-M-14 structure illustrates for the first time the protonation states of key catalytic residues K73 and E166 in a cyclic boronate complex crystal structure. These results offer valuable information for the modeling and development of future boronate inhibitors.
MATERIALS AND METHODS
Generation of KPC-2 mutants.
Mutations in the KPC-2 gene cloned into either pUCP24 or pET28a were introduced using the Quik-Change Lightning site-directed mutagenesis kit (Agilent Technologies). The presence of desired mutations was confirmed by DNA sequencing of the entire KPC-2 gene.
Determination of MICs and vaborbactam potentiation experiments.
For microbiological studies, KPC-2 WT and its mutant genes were cloned in the pUCP24 shuttle vector. Resulting plasmids were transformed into P. aeruginosa PAM1154. MICs of various antibiotics were determined by standard broth microdilution method using cation-adjusted Mueller-Hinton broth (MHB) medium (73). Potentiation of antibiotic activity by vaborbactam in bacterial strains carrying WT and mutant KPC-2 genes was performed using a standard checkerboard assay (74). BLI potency was defined as the PV50, i.e., the minimal concentration of a BLI that is required to reduce antibiotic MIC to the middle of the MIC range where the highest MIC is the MIC for a KPC-producing strain with no inhibitor added and the lowest MIC is the MIC for the vector only strain, corresponding to the complete inhibition of KPC. To determine PV50s of antibiotic activity potentiation by BLIs, MICs obtained in checkerboard experiments were converted to log2 values and plotted against log10 of BLI concentrations. Prism software was used to calculate PV50s using a four-parameter dose response inhibition equation. The resulting numbers as well as BLI concentrations were imported in Prism software. PV50s were calculated using a dose-response – inhibition, variable slope (four parameters) equation (22). PV50s are presented as averages from three experiments. Statistical significance was calculated based on the t test. All microbiological studies were performed with 15 μg/ml of gentamicin in the media.
Purification of KPC-2 wild-type and mutant proteins for biochemical studies.
For protein expression, the full KPC-2 gene coding sequence with its Shine-Dalgarno (SD) box was cloned into the pET28a vector, which produced a construct with periplasmic KPC-2 secretion and a 6× His tag on its C terminus. The recombinant plasmids were transformed into BL21(DE3) E. coli pLysS strain. Two milliliters of overnight culture was inoculated in 1 liter of LB medium with 50 μg/ml of kanamycin and 20 μg/ml of chloramphenicol and grown at 37°C with shaking at 300 rpm until an optical density at 600 nm (OD600) of 0.7 to 0.8 was reached. IPTG (isopropyl-β-d-thiogalactopyranoside) was added to 0.2 mM, and cells continued to grow for an additional 3 h. Cells were harvested by centrifugation, and the cell pellet was resuspended in 40 ml of ice-cold 50 mM Tris-HCl (pH 8.0), 500 mM sucrose, 1 mM EDTA, and 1 tablet of complete protease inhibitor (Roche). The suspension was incubated on ice with six cycles of 15 s vortexing and a 5-min pause between them. The suspension was centrifuged for 30 min at 30,000 × g, supernatant was collected and sonicated for 30 s to reduce viscosity, and MgCl2 and imidazole were added to 5 mM. Lysate was loaded by gravity flow onto a 1-ml column with HisPur cobalt resin (Thermo Scientific) pre-equilibrated with 50 mM sodium phosphate (pH 7.4)–300 mM NaCl–5 mM imidazole buffer. Column was washed with 40 ml of the same buffer, and 6× His tagged protein was eluted with 50 mM sodium phosphate (pH 7.4)–300 mM NaCl–70 mM imidazole buffer. All wash and elution fractions were analyzed by 8 to 16% SDS-PAGE. Fractions containing target protein were pooled, concentrated, and dialyzed against 50 mM sodium phosphate (pH 7.0). Purity of all proteins was at least 95% as determined by SDS-PAGE. Protein preparations were aliquoted and stored at −20°C until further use.
Determination of Km and kcat values for nitrocefin cleavage by KPC-2 and mutant proteins.
Purified protein was mixed with various concentrations of nitrocefin in 50 mM sodium phosphate (pH 7.0)–0.1 mg/ml bovine serum albumin (reaction buffer), and substrate cleavage was monitored at 490 nm every 10 s for 10 min on a SpectraMax plate reader at 37°C. Initial rates of nitrocefin cleavage were calculated and used to obtain Km and kcat values with Prism software (GraphPad).
Determination of Km and kcat values for meropenem cleavage by KPC-2 and mutant proteins.
Purified protein was mixed with various concentrations of meropenem in reaction buffer, and substrate cleavage was monitored at 294 nm every 30 s for 30 min on SpectraMax plate reader at 37°C. Initial rates of meropenem cleavage were calculated and used to obtain Km and kcat values with Prism software (GraphPad).
Determination of k2/K inactivation constant for KPC-2 WT and mutant proteins inhibition by vaborbactam/avibactam.
Inactivation kinetic parameters were determined by the reporter substrate method for a slow, tight binding inhibitor kinetic scheme (75).
where E is free enzyme, I is inhibitor, EI is noncovalent enzyme-inhibitor complex, and EI* is covalent enzyme-inhibitor complex.
Protein was quickly mixed with 100 μM nitrocefin (NCF) and various concentrations of vaborbactam/avibactam in reaction buffer, and absorbance at 490 nm was measured immediately every 2 s for 180 s on SpectraMax plate reader (Molecular Devices) at 37°C. The resulting progression curves of OD490 versus time at various vaborbactam and avibactam concentrations were imported into Prism software (GraphPad), and pseudo-first-order rate constants (kobs) were calculated using the following equation: P = V0(1 − e−kobs × t)/kobs, where V0 is the uninhibited KPC-2 rate, P is absorption signal, e is the mathematical constant, and t is time.
kobs values calculated at various vaborbactam concentrations were fitted in the following equation: kobs = k−2 + k2/K × [I]/{1 + [NCF]/Km(NCF)}, where k2/K is the inactivation constant, [I] is the inhibitor concentration, [NCF] is the nitrocefin concentration, and Km(NCF) is the Michaelis constant of NCF for KPC-2.
Determination of koff rates of enzyme activity recovery after KPC-2 and mutant inhibition by vaborbactam.
Purified enzyme at 1 μM in reaction buffer was mixed with vaborbactam at an 8-fold higher concentration than its stoichiometry ratio (unpublished data). After 30 min incubation at 37°C, the reaction mixture was diluted 30,000-fold in reaction buffer, and 100 μl of diluted enzyme was mixed with 100 μl of 400 μM nitrocefin in reaction buffer. Absorbance at 490 nm was recorded every minute during 4 h at 37°C. Resulting reaction profiles were fitted into the following equation using Prism software (GraphPad) to obtain koff values: P = Vs × t + (Vo − Vs)(1 − e−koff × t)/koff, where Vs is uninhibited enzyme velocity, measured in the reaction with enzyme and no inhibitor, and Vo is completely inhibited enzyme velocity, measured in the reaction with no enzyme and NCF only.
Crystallization experiments.
CTX-M-14 for crystallization was expressed and purified as previously described (19). Vaborbactam was dissolved in dimethyl sulfoxide (DMSO) as a 200 mM stock solution and stored at −20°C. CTX-M-14 crystals were grown by mixing 15 mg/ml of protein solution with 1.0 M potassium phosphate (pH 8.3). The CTX-M-14–vaborbactam complex was obtained through crystal soaking for 20 h with 10 mM vaborbactam in 1.0 M potassium phosphate (pH 8.3). Crystals were cryoprotected in 1.0 M potassium phosphate (pH 8.3) and 30% (wt/vol) sucrose before flash cooling with liquid nitrogen. KPC-2 for crystallization was expressed and purified as previously described (44). Prior to crystallization setup, 5 mM vaborbactam was mixed with 11.4 mg/ml 6× His-tagged KPC-2 and incubated at room temperature for 5 min. KPC-2–vaborbactam crystals were grown by mixing protein solution with 2 M ammonium sulfate and 5% (vol/vol) ethanol. Crystals were cryoprotected in 2 M ammonium sulfate, 5% (vol/vol) ethanol, and 20% (vol/vol) glycerol and flash cooled with liquid nitrogen.
Data collection, structure determination, and refinement.
Diffraction data for the vaborbactam complexes with CTX-M-14 and KPC-2 were collected at the Advanced Photon Source (APS) beamline 19-ID. Diffraction data were indexed and integrated with iMosflm (76) and scaled with SCALA (77) from the CCP4 suite (78). Phasing was performed using molecular replacement with the program Phaser (79) with the CTX-M-14 structure (PDB code 4UA6) and KPC-2 structure (PDB code 5UL8). Structure refinement was performed using Phenix.refine (80) in the Phenix suite (81) and model building in WinCoot (82). The program eLBOW in the Phenix suite was used to obtain geometry restraint information for vaborbactam (83). The final model qualities were assessed using MolProbity (84). Figures were prepared using PyMOL 2.3.4 (Schrödinger).
Data availability.
The atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB) under accession codes 6V7H (CTX-M-14–vaborbactam) and 6V7I (KPC-2–vaborbactam).
Supplementary Material
ACKNOWLEDGMENTS
This project was funded in whole or in part with federal funds from the Department of Health and Human Services, Office of the Assistant Secretary for Preparedness and Response, Biomedical Advanced Research and Development Authority (BARDA), under contract no. HHSO100201400002 with Rempex Pharmaceuticals, a wholly owned subsidiary of The Medicines Company. It was also partly supported by NIH (AI103158, to Y.C.). Structural results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center (SBC) beamline 19-ID at the Advanced Photon Source. Argonne is operated by the University of Chicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research, under contract DE-AC02-06CH11357.
O.A.P. performed X-ray crystallographic experiments and structural refinements. R.T., M.T., and O.L. carried out the microbiological and biochemical experiments. O.A.P., R.T., M.T., O.L., and Y.C. designed the experiments. O.A.P. and Y.C. wrote the manuscript. The manuscript was written through contributions of all authors. All authors approved the final version of the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Mitic N, Miraula M, Selleck C, Hadler KS, Uribe E, Pedroso MM, Schenk G. 2014. Catalytic mechanisms of metallohydrolases containing two metal ions. Adv Protein Chem Struct Biol 97:49–81. doi: 10.1016/bs.apcsb.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Nicolau DP. 2008. Carbapenems: a potent class of antibiotics. Expert Opin Pharmacother 9:23–37. doi: 10.1517/14656566.9.1.23. [DOI] [PubMed] [Google Scholar]
- 3.Bonfiglio G, Russo G, Nicoletti G. 2002. Recent developments in carbapenems. Expert Opin Invest Drugs 11:529–544. doi: 10.1517/13543784.11.4.529. [DOI] [PubMed] [Google Scholar]
- 4.Therrien C, Levesque RC. 2000. Molecular basis of antibiotic resistance and beta-lactamase inhibition by mechanism-based inactivators: perspectives and future directions. FEMS Microbiol Rev 24:251–262. doi: 10.1111/j.1574-6976.2000.tb00541.x. [DOI] [PubMed] [Google Scholar]
- 5.Majiduddin FK, Materon IC, Palzkill TG. 2002. Molecular analysis of beta-lactamase structure and function. Int J Med Microbiol 292:127–137. doi: 10.1078/1438-4221-00198. [DOI] [PubMed] [Google Scholar]
- 6.van den Akker F, Bonomo RA. 2018. Exploring additional dimensions of complexity in inhibitor design for serine beta-lactamases: mechanistic and intra- and inter-molecular chemistry approaches. Front Microbiol 9:622. doi: 10.3389/fmicb.2018.00622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rudgers GW, Huang W, Palzkill T. 2001. Binding properties of a peptide derived from beta-lactamase inhibitory protein. Antimicrob Agents Chemother 45:3279–3286. doi: 10.1128/AAC.45.12.3279-3286.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vandavasi VG, Langan PS, Weiss KL, Parks JM, Cooper JB, Ginell SL, Coates L. 2017. Active-site protonation states in an acyl-enzyme intermediate of a class A beta-lactamase with a monobactam substrate. Antimicrob Agents Chemother 61:e01636-16. doi: 10.1128/AAC.01636-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacoby GA, Munoz-Price LS. 2005. The new beta-lactamases. N Engl J Med 352:380–391. doi: 10.1056/NEJMra041359. [DOI] [PubMed] [Google Scholar]
- 10.Bush K, Jacoby GA. 2010. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother 54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown NG, Shanker S, Prasad BV, Palzkill T. 2009. Structural and biochemical evidence that a TEM-1 beta-lactamase N170G active site mutant acts via substrate-assisted catalysis. J Biol Chem 284:33703–33712. doi: 10.1074/jbc.M109.053819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drawz SM, Bonomo RA. 2010. Three decades of beta-lactamase inhibitors. Clin Microbiol Rev 23:160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papp-Wallace KM, Endimiani A, Taracila MA, Bonomo RA. 2011. Carbapenems: past, present, and future. Antimicrob Agents Chemother 55:4943–4960. doi: 10.1128/AAC.00296-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Queenan AM, Bush K. 2007. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev 20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jorgensen JH, McElmeel ML, Fulcher LC, Zimmer BL. 2010. Detection of CTX-M-type extended-spectrum beta-lactamase (ESBLs) by testing with MicroScan overnight and ESBL confirmation panels. J Clin Microbiol 48:120–123. doi: 10.1128/JCM.01507-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rawat D, Nair D. 2010. Extended-spectrum beta-lactamases in Gram negative bacteria. J Glob Infect Dis 2:263–274. doi: 10.4103/0974-777X.68531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitchel B, Rasheed JK, Endimiani A, Hujer AM, Anderson KF, Bonomo RA, Patel JB. 2010. Genetic factors associated with elevated carbapenem resistance in KPC-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 54:4201–4207. doi: 10.1128/AAC.00008-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noguchi JK, Gill MA. 1988. Sulbactam: a beta-lactamase inhibitor. Clin Pharm 7:37–51. [PubMed] [Google Scholar]
- 19.Nichols DA, Hargis JC, Sanishvili R, Jaishankar P, Defrees K, Smith EW, Wang KK, Prati F, Renslo AR, Woodcock HL, Chen Y. 2015. Ligand-induced proton transfer and low-barrier hydrogen bond revealed by X-ray crystallography. J Am Chem Soc 137:8086–8095. doi: 10.1021/jacs.5b00749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zasowski EJ, Rybak JM, Rybak MJ. 2015. The beta-lactams strike back: deftazidime-avibactam. Pharmacotherapy 35:755–770. doi: 10.1002/phar.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Humphries RM, Hemarajata P. 2017. Resistance to ceftazidime-avibactam in Klebsiella pneumoniae due to porin mutations and the increased expression of KPC-3. Antimicrob Agents Chemother 61:e00537-17. doi: 10.1128/AAC.00537-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsivkovski R, Lomovskaya O. 2020. Potency of vaborbactam is less affected than avibactam in strains producing KPC-2 mutations that confer resistance to ceftazidime-avibactam. Antimicrob Agents Chemother 64:e01936-19. doi: 10.1128/AAC.01936-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crompton IE, Cuthbert BK, Lowe G, Waley SG. 1988. Beta-lactamase inhibitors. The inhibition of serine beta-lactamases by specific boronic acids. Biochem J 251:453–459. doi: 10.1042/bj2510453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weston GS, Blazquez J, Baquero F, Shoichet BK. 1998. Structure-based enhancement of boronic acid-based inhibitors of AmpC beta-lactamase. J Med Chem 41:4577–4586. doi: 10.1021/jm980343w. [DOI] [PubMed] [Google Scholar]
- 25.Tondi D, Calo S, Shoichet BK, Costi MP. 2010. Structural study of phenyl boronic acid derivatives as AmpC beta-lactamase inhibitors. Bioorg Med Chem Lett 20:3416–3419. doi: 10.1016/j.bmcl.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maveyraud L, Pratt RF, Samama JP. 1998. Crystal structure of an acylation transition-state analog of the TEM-1 beta-lactamase. Mechanistic implications for class A beta-lactamases. Biochemistry 37:2622–2628. doi: 10.1021/bi972501b. [DOI] [PubMed] [Google Scholar]
- 27.Ness S, Martin R, Kindler AM, Paetzel M, Gold M, Jensen SE, Jones JB, Strynadka NC. 2000. Structure-based design guides the improved efficacy of deacylation transition state analogue inhibitors of TEM-1 beta-Lactamase(,). Biochemistry 39:5312–5321. doi: 10.1021/bi992505b. [DOI] [PubMed] [Google Scholar]
- 28.Powers RA, Swanson HC, Taracila MA, Florek NW, Romagnoli C, Caselli E, Prati F, Bonomo RA, Wallar BJ. 2014. Biochemical and structural analysis of inhibitors targeting the ADC-7 cephalosporinase of Acinetobacter baumannii. Biochemistry 53:7670–7679. doi: 10.1021/bi500887n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen NQ, Krishnan NP, Rojas LJ, Prati F, Caselli E, Romagnoli C, Bonomo RA, van den Akker F. 2016. Crystal structures of KPC-2 and SHV-1 beta-lactamases in complex with the boronic acid transition state analog S02030. Antimicrob Agents Chemother 60:1760–1766. doi: 10.1128/AAC.02643-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hecker SJ, Reddy KR, Totrov M, Hirst GC, Lomovskaya O, Griffith DC, King P, Tsivkovski R, Sun D, Sabet M, Tarazi Z, Clifton MC, Atkins K, Raymond A, Potts KT, Abendroth J, Boyer SH, Loutit JS, Morgan EE, Durso S, Dudley MN. 2015. Discovery of a cyclic boronic acid beta-lactamase inhibitor (RPX7009) with utility vs class A serine carbapenemases. J Med Chem 58:3682–3692. doi: 10.1021/acs.jmedchem.5b00127. [DOI] [PubMed] [Google Scholar]
- 31.Lomovskaya O, Sun D, Rubio-Aparicio D, Nelson K, Tsivkovski R, Griffith DC, Dudley MN. 2017. Vaborbactam: spectrum of Beta-lactamase inhibition and impact of resistance mechanisms on activity in Enterobacteriaceae. Antimicrob Agents Chemother 61:e01443-17. doi: 10.1128/AAC.01443-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo D, Heitman LH, Ij AP. 2015. The role of target binding kinetics in drug discovery. ChemMedChem 10:1793–1796. doi: 10.1002/cmdc.201500310. [DOI] [PubMed] [Google Scholar]
- 33.Copeland RA, Pompliano DL, Meek TD. 2006. Drug-target residence time and its implications for lead optimization. Nat Rev Drug Discov 5:730–739. doi: 10.1038/nrd2082. [DOI] [PubMed] [Google Scholar]
- 34.Lu H, England K, Am Ende C, Truglio JJ, Luckner S, Reddy BG, Marlenee NL, Knudson SE, Knudson DL, Bowen RA, Kisker C, Slayden RA, Tonge PJ. 2009. Slow-onset inhibition of the FabI enoyl reductase from Francisella tularensis: residence time and in vivo activity. ACS Chem Biol 4:221–231. doi: 10.1021/cb800306y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tummino PJ, Copeland RA. 2008. Residence time of receptor-ligand complexes and its effect on biological function. Biochemistry 47:5481–5492. doi: 10.1021/bi8002023. [DOI] [PubMed] [Google Scholar]
- 36.Tonge PJ. 2018. Drug-target kinetics in drug discovery. ACS Chem Neurosci 9:29–39. doi: 10.1021/acschemneuro.7b00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walkup GK, You Z, Ross PL, Allen EK, Daryaee F, Hale MR, O'Donnell J, Ehmann DE, Schuck VJ, Buurman ET, Choy AL, Hajec L, Murphy-Benenato K, Marone V, Patey SA, Grosser LA, Johnstone M, Walker SG, Tonge PJ, Fisher SL. 2015. Translating slow-binding inhibition kinetics into cellular and in vivo effects. Nat Chem Biol 11:416–423. doi: 10.1038/nchembio.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewandowski EM, Lethbridge KG, Sanishvili R, Skiba J, Kowalski K, Chen Y. 2018. Mechanisms of proton relay and product release by class A beta-lactamase at ultrahigh resolution. FEBS J 285:87–100. doi: 10.1111/febs.14315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meroueh SO, Fisher JF, Schlegel HB, Mobashery S. 2005. Ab initio QM/MM study of class A beta-lactamase acylation: dual participation of Glu166 and Lys73 in a concerted base promotion of Ser70. J Am Chem Soc 127:15397–15407. doi: 10.1021/ja051592u. [DOI] [PubMed] [Google Scholar]
- 40.Lietz EJ, Truher H, Kahn D, Hokenson MJ, Fink AL. 2000. Lysine-73 is involved in the acylation and deacylation of beta-lactamase. Biochemistry 39:4971–4981. doi: 10.1021/bi992681k. [DOI] [PubMed] [Google Scholar]
- 41.Pan X, He Y, Lei J, Huang X, Zhao Y. 2017. Crystallographic snapshots of class A beta-lactamase catalysis reveal structural changes that facilitate beta-lactam hydrolysis. J Biol Chem 292:4022–4033. doi: 10.1074/jbc.M116.764340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Curley K, Pratt RF. 2000. The oxyanion hole in serine beta-lactamase catalysis: interactions of thiono substrates with the active site. Bioorg Chem 28:338–356. doi: 10.1006/bioo.2000.1184. [DOI] [PubMed] [Google Scholar]
- 43.Botos I, Wlodawer A. 2007. The expanding diversity of serine hydrolases. Curr Opin Struct Biol 17:683–690. doi: 10.1016/j.sbi.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pemberton OA, Zhang X, Chen Y. 2017. Molecular basis of substrate recognition and product release by the Klebsiella pneumoniae carbapenemase (KPC-2). J Med Chem 60:3525–3530. doi: 10.1021/acs.jmedchem.7b00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Papp-Wallace KM, Taracila M, Wallace CJ, Hujer KM, Bethel CR, Hornick JM, Bonomo RA. 2010. Elucidating the role of Trp105 in the KPC-2 beta-lactamase. Protein Sci 19:1714–1727. doi: 10.1002/pro.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Franceschini N, Segatore B, Perilli M, Vessillier S, Franchino L, Amicosante G. 2002. Meropenem stability to beta-lactamase hydrolysis and comparative in vitro activity against several beta-lactamase-producing Gram-negative strains. J Antimicrob Chemother 49:395–398. doi: 10.1093/jac/49.2.395. [DOI] [PubMed] [Google Scholar]
- 47.Tremblay LW, Fan F, Blanchard JS. 2010. Biochemical and structural characterization of Mycobacterium tuberculosis beta-lactamase with the carbapenems ertapenem and doripenem. Biochemistry 49:3766–3773. doi: 10.1021/bi100232q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fonseca F, Chudyk EI, van der Kamp MW, Correia A, Mulholland AJ, Spencer J. 2012. The basis for carbapenem hydrolysis by class A beta-lactamases: a combined investigation using crystallography and simulations. J Am Chem Soc 134:18275–18285. doi: 10.1021/ja304460j. [DOI] [PubMed] [Google Scholar]
- 49.Hazra S, Xu H, Blanchard JS. 2014. Tebipenem, a new carbapenem antibiotic, is a slow substrate that inhibits the beta-lactamase from Mycobacterium tuberculosis. Biochemistry 53:3671–3678. doi: 10.1021/bi500339j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Masuda N, Gotoh N, Ishii C, Sakagawa E, Ohya S, Nishino T. 1999. Interplay between chromosomal beta-lactamase and the MexAB-OprM efflux system in intrinsic resistance to beta-lactams in Pseudomonas aeruginosa. Antimicrob Agents Chemother 43:400–402. doi: 10.1128/AAC.43.2.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakae T, Nakajima A, Ono T, Saito K, Yoneyama H. 1999. Resistance to beta-lactam antibiotics in Pseudomonas aeruginosa due to interplay between the MexAB-OprM efflux pump and beta-lactamase. Antimicrob Agents Chemother 43:1301–1303. doi: 10.1128/AAC.43.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krishnan NP, Nguyen NQ, Papp-Wallace KM, Bonomo RA, van den Akker F. 2015. Inhibition of Klebsiella beta-lactamases (SHV-1 and KPC-2) by avibactam: a structural study. PLoS One 10:e0136813. doi: 10.1371/journal.pone.0136813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Helfand MS, Bethel CR, Hujer AM, Hujer KM, Anderson VE, Bonomo RA. 2003. Understanding resistance to beta-lactams and beta-lactamase inhibitors in the SHV beta-lactamase: lessons from the mutagenesis of SER-130. J Biol Chem 278:52724–52729. doi: 10.1074/jbc.M306059200. [DOI] [PubMed] [Google Scholar]
- 54.Papp-Wallace KM, Winkler ML, Taracila MA, Bonomo RA. 2015. Variants of beta-lactamase KPC-2 that are resistant to inhibition by avibactam. Antimicrob Agents Chemother 59:3710–3717. doi: 10.1128/AAC.04406-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winkler ML, Papp-Wallace KM, Taracila MA, Bonomo RA. 2015. Avibactam and inhibitor-resistant SHV beta-lactamases. Antimicrob Agents Chemother 59:3700–3709. doi: 10.1128/AAC.04405-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kalp M, Sheri A, Buynak JD, Bethel CR, Bonomo RA, Carey PR. 2007. Efficient inhibition of class A and class D beta-lactamases by Michaelis complexes. J Biol Chem 282:21588–21591. doi: 10.1074/jbc.C700080200. [DOI] [PubMed] [Google Scholar]
- 57.Beadle BM, Trehan I, Focia PJ, Shoichet BK. 2002. Structural milestones in the reaction pathway of an amide hydrolase: substrate, acyl, and product complexes of cephalothin with AmpC beta-lactamase. Structure 10:413–424. doi: 10.1016/s0969-2126(02)00725-6. [DOI] [PubMed] [Google Scholar]
- 58.Teotico DG, Babaoglu K, Rocklin GJ, Ferreira RS, Giannetti AM, Shoichet BK. 2009. Docking for fragment inhibitors of AmpC beta-lactamase. Proc Natl Acad Sci U S A 106:7455–7460. doi: 10.1073/pnas.0813029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vercheval L, Bauvois C, di Paolo A, Borel F, Ferrer JL, Sauvage E, Matagne A, Frere JM, Charlier P, Galleni M, Kerff F. 2010. Three factors that modulate the activity of class D beta-lactamases and interfere with the post-translational carboxylation of Lys70. Biochem J 432:495–504. doi: 10.1042/BJ20101122. [DOI] [PubMed] [Google Scholar]
- 60.Delmas J, Leyssene D, Dubois D, Birck C, Vazeille E, Robin F, Bonnet R. 2010. Structural insights into substrate recognition and product expulsion in CTX-M enzymes. J Mol Biol 400:108–120. doi: 10.1016/j.jmb.2010.04.062. [DOI] [PubMed] [Google Scholar]
- 61.Langan PS, Vandavasi VG, Weiss KL, Cooper JB, Ginell SL, Coates L. 2016. The structure of Toho1 beta-lactamase in complex with penicillin reveals the role of Tyr105 in substrate recognition. FEBS Open Bio 6:1170–1177. doi: 10.1002/2211-5463.12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krajnc A, Brem J, Hinchliffe P, Calvopina K, Panduwawala TD, Lang PA, Kamps JJAG, Tyrrell JM, Widlake E, Saward BG, Walsh TR, Spencer J, Schofield CJ. 2019. Bicyclic boronate VNRX-5133 inhibits metallo- and serine-β-lactamases. J Med Chem 62:8544–8556. doi: 10.1021/acs.jmedchem.9b00911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang YL, Liu S, Yu ZJ, Lei Y, Huang MY, Yan YH, Ma Q, Zheng Y, Deng H, Sun Y, Wu C, Yu Y, Chen Q, Wang Z, Wu Y, Li GB. 2019. Structure-based development of (1–(3′-mercaptopropanamido)methyl)boronic acid derived broad-spectrum, dual-action inhibitors of metallo- and serine-beta-lactamases. J Med Chem 62:7160–7184. doi: 10.1021/acs.jmedchem.9b00735. [DOI] [PubMed] [Google Scholar]
- 64.Liu B, Trout REL, Chu GH, McGarry D, Jackson RW, Hamrick JC, Daigle DM, Cusick SM, Pozzi C, De Luca F, Benvenuti M, Mangani S, Docquier JD, Weiss WJ, Pevear DC, Xerri L, Burns CJ. 2019. Discovery of taniborbactam (VNRX-5133): a broad-spectrum serine- and metallo-beta-lactamase inhibitor for carbapenem-resistant bacterial infections. J Med Chem 63:2789–2801. doi: 10.1021/acs.jmedchem.9b01518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsivkovski R, Lomovskaya O. 2019. Biochemical activity of vaborbactam. Antimicrob Agents Chemother 64:e01935-19. doi: 10.1128/AAC.01935-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ke W, Bethel CR, Thomson JM, Bonomo RA, van den Akker F. 2007. Crystal structure of KPC-2: insights into carbapenemase activity in class A beta-lactamases. Biochemistry 46:5732–5740. doi: 10.1021/bi700300u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smith CA, Nossoni Z, Toth M, Stewart NK, Frase H, Vakulenko SB. 2016. Role of the conserved disulfide bridge in class A carbapenemases. J Biol Chem 291:22196–22206. doi: 10.1074/jbc.M116.749648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Karplus PA, Schulz GE. 1985. Prediction of chain flexibility in proteins. Naturwissenschaften 72:212–213. doi: 10.1007/BF01195768. [DOI] [Google Scholar]
- 69.Ragone R, Facchiano F, Facchiano A, Facchiano AM, Colonna G. 1989. Flexibility plot of proteins. Protein Eng 2:497–504. doi: 10.1093/protein/2.7.497. [DOI] [PubMed] [Google Scholar]
- 70.Parthasarathy S, Murthy MR. 1999. On the correlation between the main-chain and side-chain atomic displacement parameters (B values) in high-resolution protein structures. Acta Crystallogr D Biol Crystallogr 55:173–180. doi: 10.1107/S0907444998006611. [DOI] [PubMed] [Google Scholar]
- 71.Carugo O. 2018. How large B-factors can be in protein crystal structures. BMC Bioinformatics 19:61. doi: 10.1186/s12859-018-2083-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Folmer RHA. 2018. Drug target residence time: a misleading concept. Drug Discov Today 23:12–16. doi: 10.1016/j.drudis.2017.07.016. [DOI] [PubMed] [Google Scholar]
- 73.Wiegand I, Hilpert K, Hancock RE. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 74.Anonymous. 2010. Synergism testing: broth microdilution checkerboard and broth macrodilution methods, p 5.12.1–5.12.2. In Garcia LS. (ed), Clinical microbiology procedures handbook, 3rd ed ASM Press, Washington, DC. [Google Scholar]
- 75.Waley SG. 1993. The kinetics of slow-binding and slow, tight-binding inhibition: the effects of substrate depletion. Biochem J 294:195–200. doi: 10.1042/bj2940195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. 2011. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr 67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Evans P. 2006. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 78.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. 2012. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moriarty NW, Grosse-Kunstleve RW, Adams PD. 2009. electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr D Biol Crystallogr 65:1074–1080. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB III, Snoeyink J, Richardson JS, Richardson DC. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB) under accession codes 6V7H (CTX-M-14–vaborbactam) and 6V7I (KPC-2–vaborbactam).