The Swiss Centre for Antibiotic Resistance (ANRESIS) has recently noted an increase of extended-spectrum cephalosporin-resistant (ESC-R) Shigella sonnei isolates nationwide (3.8% in 2016 versus 37.5% in 2019). To understand this phenomenon, we analyzed 25 representative isolates (of which 14 were ESC-R) collected in Switzerland during 2016 to 2019. Whole-genome sequencing was achieved using both the Illumina and the Nanopore platforms. Both ESC-R and extended-spectrum cephalosporin-susceptible isolates belonged to sequence type 152 (ST152).
KEYWORDS: Shigella, ESBL, CTX-M, plasmid, WGS, MLST, cgST, core genome, plasmid-mediated resistance
ABSTRACT
The Swiss Centre for Antibiotic Resistance (ANRESIS) has recently noted an increase of extended-spectrum cephalosporin-resistant (ESC-R) Shigella sonnei isolates nationwide (3.8% in 2016 versus 37.5% in 2019). To understand this phenomenon, we analyzed 25 representative isolates (of which 14 were ESC-R) collected in Switzerland during 2016 to 2019. Whole-genome sequencing was achieved using both the Illumina and the Nanopore platforms. Both ESC-R and extended-spectrum cephalosporin-susceptible isolates belonged to sequence type 152 (ST152). The ESC-R isolates carried blaCTX-M-3 in IncI1-pST57 (n = 5), blaCTX-M-15 in IncFII (F2:A-:B-) (n = 5), blaCTX-M-15 in IncI1-pST16, and blaCTX-M-27, blaCTX-M-55, or blaCTX-M-134 in other IncFII plasmids (n = 1 each). Plasmids having the same bla and Inc group exhibited high degrees of genetic identity to each other but also to plasmids previously reported in other Enterobacterales. Core-genome analysis showed that there were 4 main clusters, each of which included strains that differed by <58 single nucleotide variants (SNVs) and that consisted of both blaCTX-M-positive and blaCTX-M-negative isolates. Moreover, most isolates belonging to the same cluster shared an identical core-genome sequence type (cgST). For instance, cluster 1 included 4 isolates of cgST113036, of which only 3 harbored the IncI1-pST57 blaCTX-M-3-positive plasmid. The 25 S. sonnei isolates were also subjected to phylogenetic comparison with deposited international strains. As a result, matching isolates (isolates that had the same cgST and that differed by <8 SNVs) have been reported in the United Kingdom, the United States, France, and the Netherlands. Overall, our results suggest that some common S. sonnei clusters can spread between continents and can be imported into other nations after international trips. Such clusters include, in part, isolates that do not possess blaESBL-harboring plasmids, indicating their tendency to acquire them from other Enterobacterales.
INTRODUCTION
Shigella flexneri is one of the most common causes of diarrhea in low- and middle-income countries and is associated with high morbidity and mortality rates. In contrast, Shigella sonnei is the leading species in high-income nations, with the majority of cases being described in returning travelers, men who have sex with men (MSM), and young children (1–3).
The emergence of antibiotic-resistant S. sonnei isolates is nowadays a matter of concern (4). The high rates of resistance to first-line options (e.g., ciprofloxacin and azithromycin) have made ceftriaxone the drug of choice for empirical treatment. However, there has also been a significant recent increase in extended-spectrum cephalosporin-resistant (ESC-R) isolates, especially in Asia (1, 5).
Usually, ESC-R S. sonnei isolates produce extended-spectrum β-lactamases (ESBL) of the CTX-M type, of which CTX-M-3, CTX-M-14, CTX-M-15, CTX-M-27, and CTX-M-55 are the most common (6). However, only a few studies have implemented whole-genome sequencing (WGS) to characterize the blaCTX-M-carrying plasmids in detail. So far, a blaCTX-M-3-harboring IncI1 plasmid from Italy (7), a blaCTX-M-14-IncB/O/K/Z plasmid from China (8), a blaCTX-M-15-harboring IncI1 plasmid from South Korea (9), a blaCTX-M-27-harboring IncFII plasmid from the United Kingdom (10), and a blaCTX-M-55-harboring IncI2 plasmid from China have been described in S. sonnei (11). For Switzerland, we note that the first two ESC-R S. sonnei strains (CTX-M-14 and CTX-M-15 producers) were isolated in 2009, but no WGS analyses were performed on the 8 strains detected during 2009 to 2014 (12).
Due to the ability of S. sonnei to acquire plasmids harboring multidrug resistance (MDR) genes (MDR plasmids) and the fact that it shows a higher prevalence in industrialized countries than S. flexneri (3), attention needs to be focused on the clonality of ESC-R S. sonnei isolates. Just recently (2018), in the European Union, 17 outbreaks due to S. sonnei have been documented (13).
Several authors have implemented multilocus sequence typing (MLST), which has revealed that sequence type 152 (ST152) is the most frequent lineage among extended-spectrum cephalosporin-susceptible (ESC-S) S. sonnei isolates (14–19). Other recent studies have also used core-genome analyses to investigate epidemiological events (2, 15, 20, 21), although only one survey in the United Kingdom analyzed exclusively ESBL-producing S. sonnei strains (10). Overall, these studies have shown that single nucleotide variant (SNV) analysis represents a high-resolution tool for determining clonality and tracking outbreaks at the community and global levels.
In this study, therefore, we used WGS to characterize the plasmids of ESC-R S. sonnei isolates detected in Switzerland. Moreover, to investigate the hidden epidemiological profile of contemporary circulating isolates, we implemented a core-genome analysis to determine the clonality of ESC-R and ESC-S S. sonnei strains.
RESULTS AND DISCUSSION
Rate of ESC-R S. sonnei and analyzed strains.
According to the Swiss Centre for Antibiotic Resistance (ANRESIS) database, 53, 39, 85, and 56 S. sonnei isolates were identified nationwide in 2016, 2017, 2018, and 2019, respectively, by participating laboratories. Of them, 2 (3.8%), 5 (12.8%), 12 (14.1%), and 21 (37.5%), respectively, were reported to be ESC-R.
Unfortunately, such results could not be compared to those of other countries. In fact, though the spread of ESC-R Shigella spp. is of concern, recent studies analyzing their trends are lacking (1, 4). Nevertheless, we note that in China the rate of ESC-R S. sonnei increased from 31.6% in 2012 to 64.3% in 2015 (22), whereas lower rates were recorded in other nations during point-prevalence surveys (e.g., in 2015, 12% in England/Wales and 0% in Nepal; in 2015 and 2016, 7.1% in New Zealand) (10, 23, 24). It is therefore difficult to interpret our data on the persistent increase in resistance to ESCs. In this context, we emphasize that the Swiss population is at greater risk of acquiring and importing MDR Shigella spp. from areas of endemicity due to its high propensity for international travel (12, 25). For this reason, in order to better understand this general epidemiological phenomenon, a molecular characterization of the strains is essential.
In the present study, we analyzed 14 ESC-R S. sonnei isolates and 11 ESC-S S. sonnei isolates collected in Switzerland during 2016 to 2019. Species identification (ID) and the antibiotic resistance phenotypes of all strains were confirmed by appropriate methods before further molecular analyses (see Materials and Methods and Table S1 in the supplemental material).
ARGs.
As shown in Table 1, both ESC-R and ESC-S S. sonnei isolates carried numerous antimicrobial resistance genes (ARGs) conferring resistance to different classes of antibiotics, including quinolones (e.g., qnrS1) and macrolides [e.g., erm(B) and mph(A)] (6). Those phenotypically resistant to ESCs mainly possessed blaCTX-M-3 (n = 5) or blaCTX-M-15 (n = 6) ESBL genes, but unique isolates harboring blaCTX-M-27, blaCTX-M-55, and blaCTX-M-134 were also detected.
TABLE 1.
Summary of the demographic and travel-related (if any) data along with the results of WGS analyses of the S. sonnei strains analyzed in the present studyh
| Strain | Yr of isolation | Age (yr)/sexa | Sample | Origin of infectiona | Groupb | Coresistanceb | STc | Antimicrobial resistance genes/plasmid replicons (pMLST, approximate size)c | No. (%) of called alleles among a total of 2,513d | No. (%) of allele matches in cgSTd | cgSTd |
|---|---|---|---|---|---|---|---|---|---|---|---|
| L4094 | 2018 | NA/NA | NA | NA | ESC-R | ST152 | blaCTX-M-3, aadA1, mdf(A), dfrA1/l1 (pST57, 86 kb), Col156, Col(BS512) | 2,503 (99.60) | 2,496 (99.32) | cgST113036 | |
| 1205-3131 | 2018 | 35/M | Stool | Unknown | ESC-R | ST152 | blaCTX-M-3, aadA1, mdf(A), dfrA1/l1 (pST57, 86 kb), Col156, Col(BS512) | 2,500 (99.48) | 2,492 (99.16) | cgST113036 | |
| 7111-69 | 2019 | 20/M | Stool | Turkey | ESC-R | ST152 | blaCTX-M-3, aadA1, mdf(A), dfrA1/l1 (pST57, 86 kb), Col156, Col(BS512) | 2,499 (99.44) | 2,491 (99.12) | cgST113036 | |
| LC-1477-18e | 2018 | 10/F | Stool | Albania | ESC-R | SXT | ST152 | blaCTX-M-3, aadA1, aph(3'')-Ib, aph(6)-Id, mdf(A), dfrA1, sul2, tet(A)/I1 (pST57, 85 kb), Col156, Col(BS512) | 2,505 (99.68) | 2,498 (99.40) | cgST118753 |
| 509-1022 | 2019 | 50/F | Stool | Unknown | ESC-R | SXT | ST152 | blaCTX-M-3, aadA1, aph(3″)-lb, aph(6)-ld, mdf(A), dfrA1, sul2, tet(A)/l1 (pST57, 88 kb), Col156, Col(BS512) | 2,503 (99.60) | 2,495 (99.28) | cgST115537 |
| 19-0822-3296 | 2019 | 5/F | Stool | Unknown | ESC-R | SXT | ST152 | blaCTX-M-3, aadA1, aph(3″)-lb, aph(6)-ld, mdf(A), dfrA1, sul2, tet(A)/l1 (pST57, 88 kb), Col156, Col(BS512) | 2,504 (99.64) | 2,495 (99.28) | cgST115537 |
| 6607-69 | 2017 | 60/F | Stool | Sri Lanka | ESC-R | SXT | ST1503f | blaCTX-M-15, aph(3″)-lb, aph(6)-ld, mdf(A), dfrA1, tet(A), sul2/I1 (pST16, 90 kb), FII, Col156, Col(BS512) | 2,504 (99.64) | 2,496 (99.32) | cgST64457 |
| 19-0821-3486 | 2019 | 45/M | Stool | Egypt | ESC-R | SXT | ST152 | blaCTX-M-15, aadA1, aph(3″)-lb, aph(6)-ld, mdf(A), dfrA1, qnrS1, sul2, tet(A)/FII (F2:A-:B-, 83 kb), Col156, Col(BS512) | 2,500 (99.48) | 2,493 (99.20) | cgST112958 |
| 0401930105 | 2019 | 50/M | Stool | Egypt | ESC-R | SXT | ST152 | blaCTX-M-15, aadA1, aph(3″)-lb, aph(6)-ld, mdf(A), dfrA1, qnrS1, tet(A), sul2/FII (F2:A-:B-, 83 kb), Col156, Col(BS512) | 2,502 (99.56) | 2,496 (99.32) | cgST112958 |
| 6904-27 | 2018 | 35/M | Stool | Local | ESC-R | SXT | ST152 | blaCTX-M-15, aadA1, aph(3″)-lb, aph(6)-ld, mdf(A), dfrA1, qnrS1, tet(A), sul2/FII (F2:A-:B-, 83 kb), Col156, Col(BS512) | 2,502 (99.56) | 2,497 (99.36) | cgST112958 |
| 19-1125-3493 | 2019 | 40/F | Stool | Unknown | ESC-R | SXT | ST152 | blaCTX-M-15, aadA1, aph(3″)-lb, aph(6)-ld, mdf(A), dfrA1, qnrS1, tet(A), sul2/FII (F2:A-:B-, 83 kb), Col156, Col(BS512) | 2,496 (99.32) | 2,489 (99.04) | cgST112958 |
| 19-0820-1561 | 2019 | 15/F | Stool | Nepal | ESC-R | SXT, CIP, AZT | ST152 | blaCTX-M-15, aadA5, mdf(A), mph(A), dfrA1, dfrA17, qnrS1, sul1/FII (F2: A-:B-, 83 kb), Col156, Col(BS512) | 2,493 (99.20) | 2,477 (98.57) | cgST117387 |
| 0401952027 | 2019 | 45/M | Stool | Egypt | ESC-R | SXT, AZT | ST152 | blaCTX-M-55, aadA1, aph(3″)-lb, aph(6)-ld, mdf(A), mph(A), dfrA1, sul2, tet(A)/I1, FII (F2:A-:B-, 74 kb), Col156, Col(BS512) | 2,498 (99.40) | 2,489 (99.04) | cgST20888 |
| 09163633 | 2019 | 50/M | Stool | Unknown | ESC-R | SXT | ST152 | blaCTX-M-27, aadA1, aph(3″)-lb, aph(6)-ld, mdf(A), dfrA1, sul2, tet(A)/FII (F2:A-:B-, 68 kb), B/O/K/Z, Col156, Col(BS512) | 2,497 (99.36) | 2,489 (99.04) | cgST67380 |
| 3123885 | 2019 | 30/M | Stool | Israel | ESC-R | SXT, CIP, AZT | ST152 | blaCTX-M-134, aph(6)-ld, mdf(A), mph(A), dfrA1, sul2, tet(A)/FII (F35:A-:B-, 69 kb), B/O/K/Z, Col156, Col(BS512) | 2,489 (99.04) | 2,480 (98.69) | cgST114011 |
| 7103-58g | 2018 | 10/M | Stool | Romania | ESC-S | SXT, CIP, AZT | ST152 | blaTEM-1B, aadA5, aph(3″)-lb, aph(6)-ld, mdf(A), mph(A), erm(B), dfrA1/A17, sul1/2, tet(A)/Col156, Col(BS512) | 2,489 (99.04) | 2,482 (98.77) | cgST107674 |
| 7103-28g | 2018 | 50/M | Stool | Romania | ESC-S | SXT, CIP, AZT | ST152 | blaTEM-1B, aadA5, aph(3″)-lb, aph(6)-ld, mdf(A), mph(A), erm(B), dfrA1/A17, sul1/2, tet(A)/Col156, Col(BS512) | 2,490 (99.08) | 2,481 (98.73) | cgST107674 |
| 6407-57 | 2017 | 40/M | Stool | Local | ESC-S | SXT, CIP, AZT | ST152 | blaTEM-1B, aadA5, aph(3″)-lb, aph(6)-ld, mdf(A), mph(A), erm(B), dfrA1/A17, sul1/2, tet(A)/I1, Col156, Col(BS512) | 2,492 (99.16) | 2,484 (98.85) | cgST108068 |
| 6110-62 | 2016 | 60/F | Stool | Brazil | ESC-S | SXT | ST152 | blaTEM-1B, aph(3″)-lb, aph(6)-ld, mdf(A), dfrA8, sul2/FII, Col156, Col(BS512) | 2,499 (99.44) | 2,492 (99.16) | cgST108083 |
| 6105-15 | 2016 | 35/M | Stool | Egypt | ESC-S | SXT | ST152 | aadA1, aph(3″)-lb, aph(6)-ld, mdf(A), dfrA1, sul2, tet(A)/FII, Col156, Col(BS512) | 2,497 (99.36) | 2,487 (98.97) | cgST37499 |
| 6101-40 | 2016 | 40/F | Stool | Western Africa | ESC-S | SXT | ST152 | aadA1, aph(3″)-lb, aph(6)-ld, mdf(A), dfrA1, sul2, tet(A)/FII, Col156, Col(BS512) | 2,492 (99.16) | 2,479 (98.65) | cgST98334 |
| 6412-75 | 2017 | 50/M | Blood, stool | India | ESC-S | CIP | ST152 | mdf(A), dfrA1/Col156, Col(BS512) | 2,495 (99.28) | 2,487 (98.97) | cgST101592 |
| 6502-32 | 2017 | 40/F | Stool | Dominican Republic | ESC-S | SXT | ST152 | aph(3″)-lb, aph(6)-ld, mdf(A), dfrA1, sul2, tet(A)/I1, Col156, Col(BS512) | 2,494 (99.24) | 2,484 (98.85) | cgST108763 |
| 7111-23 | 2019 | 25/F | Stool | Philippines | ESC-S | ST152 | sul2, dfrA14, aph(3″)-lb, aph(6)-ld, mdf(A)/I1, FII, Col156 | 2,495 (99.28) | 2,481 (98.73) | cgST108909 | |
| 7109-28 | 2019 | 30/M | Stool | Colombia | ESC-S | SXT | ST152 | aadA1, aph(3″)-lb, aph(6)-ld, sul2, mdf(A), dfrA1, qnrB19, qnrB5, qnrB81, tet(A)/I1, Col156 | 2,499 (99.44) | 2,486 (98.93) | cgST109254 |
| 7001-38 | 2018 | 50/F | Stool | Local | ESC-S | SXT | ST152 | aadA1, mdf(A), dfrA1/Col156, Col(BS512) | 2,503 (99.60) | 2,494 (99.24) | cgST113036 |
Age and sex are based on the information provided to the clinical laboratory analyzing routine samples. Age has been approximated at ±5 years. The origin of infection has been attributed to a foreign country if symptoms (i.e., diarrhea) occurred during or after the patients returned from a specific country.
Groups are based on the MICs obtained with the Sensititre GNX2F panel and interpreted according to the EUCAST 2019 criteria. Only key antibiotics to which nonsusceptibility is shown are reported (see Table S1 in the supplemental material for full MIC results).
Performed by implementing the tools of the Center for Genomic Epidemiology (CGE), specifically, MLST (v2.0; E. coli scheme 1), ResFinder (v3.2), PlasmidFinder (v2.0), and pMLST (v2.0), when available. The main bla genes and the associated plasmids carrying these genes are indicated in bold.
Core-genome results obtained with cgMLSTFinder (v1.1).
Strain LC-1477-18 was detected in Italy (7). It was added to the analysis as a control.
ST1503 is a single allele variant of ST152.
The patients from whom strains 7103-58 and 7103-28 were isolated are relatives.
M, male; F, female; ESC-R, extended-spectrum cephalosporin resistant; ESC-S, extended-spectrum cephalosporin susceptible; SXT, trimethoprim-sulfamethoxazole; CIP, ciprofloxacin; AZT, azithromycin; ST, sequence type; cgST, core-genome sequence type; NA, not available.
Although studies analyzing the prevalence of specific blaESBL genes in S. sonnei are lacking, blaCTX-M-15 and blaCTX-M-3 appear to be the most frequent worldwide (6). In particular, CTX-M-3 producers were described in Turkey, Switzerland, and Italy (7, 12, 26, 27), while those with CTX-M-15 have been found in various countries, including South Korea, where an outbreak was described (9). With regard to the other ESBLs, a CTX-M-27-producing S. sonnei clone was responsible for an outbreak in 2015 among MSM in England (10) and CTX-M-55 was reported in S. sonnei isolates from China and South Korea (11, 28), while CTX-M-134 was only recently described in Escherichia coli (29).
Since blaCTX-M genes are usually carried by plasmids that can be exchanged between different species of enterobacteria (e.g., from E. coli to S. sonnei in the human gut) (30), their characterization is crucial for understanding the expansion of ESC-R S. sonnei isolates.
IncI1 blaCTX-M-3-carrying plasmids.
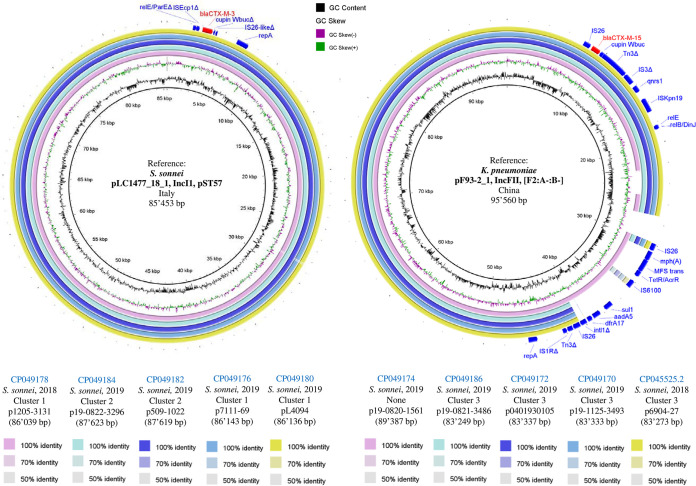
As shown in Fig. 1A, 5 S. sonnei isolates harbored 86- to 87-kb IncI1-pST57 blaCTX-M-3-carrying plasmids with a high degree of genetic identity to each other and to the Italian pLC1477_18_1 plasmid that we recently described (conjugation frequency, 1.2 × 10−4) (7).
FIG 1.
BLAST comparisons of the sequences of S. sonnei blaCTX-M-carrying plasmids against reference sequences. (A) Comparison of the sequences of five S. sonnei blaCTX-M-3-carrying IncI1 plasmids against the sequence of the S. sonnei IncI1 plasmid pLC1477_18-1 (GenBank accession number CP035009) as a reference sequence. (B) Comparison of the sequences of five S. sonnei blaCTX-M-15-carrying IncFII (F2:A-:B-) plasmids against the sequence of the K. pneumoniae IncFII (F2:A-:B-) plasmid pF93-2_1 (GenBank accession number CP026158) as a reference sequence. Rings were constructed using BRIG (BLAST Ring Image Generator; v0.95) software. Similarities with the reference plasmid are represented by the colored rings. Genome accession numbers are indicated in the key at the bottom. Red and blue arrows above the rings correspond to the gene features of interest. The delta symbol (Δ) next to the feature label corresponds to a partial/incomplete gene CDS. For each plasmid, we report the GenBank accession number, the species of isolation, the tree cluster from Fig. 2, the year, the plasmid name, and the plasmid size.
In all of these plasmids, blaCTX-M-3 was associated with a truncated ISEcp1 in the same element reported in the Italian plasmid. Considering the strong genetic similarity of the five IncI1-pST57 blaCTX-M-3-carrying plasmids collected in Switzerland, it is quite possible that other plasmids with similar genetic characteristics exist outside the country. For instance, an Enterobase S. sonnei strain from the United Kingdom recovered in 2019 (named strain 811053; BioSample accession number SAMN12881824) of the same core-genome sequence type (cgST) as our 509-1022 and 19-0822-3296 isolates (cgST115537) was found to harbor blaCTX-M-3 and contained at least one pST57 plasmid (Center for Genomic Epidemiology [CGE] analysis; data not shown). Similarly, an E. coli strain from a study conducted in the Netherlands 2013 to 2015 was reported to carry an IncI1-pST57 plasmid that possessed blaCTX-M-3 (31).
IncFII blaCTX-M-15-harboring plasmids.
As depicted in Fig. 1B, 5 other ESC-R S. sonnei isolates harbored IncFII (F2:A-:B-) blaCTX-M-15-carrying plasmids (83 to 89 kb) with a high degree of genetic identity to each other and to pF93-2_1 (GenBank accession number CP026158) from Klebsiella pneumoniae found in China in 2014. Globally, F2:A-:B- is the predominant F plasmid type carrying blaCTX-M genes among Enterobacteriaceae and is highly conjugative (32).
The 5 blaCTX-M-15-carrying plasmids identified in the present work shared an identical genetic environment around blaCTX-M-15, including both full (IS26 and ISKpn19) and partial (ΔTn3 and ΔIS3) transposable element-coding sequences (CDS), along with qnrS1 (Fig. 1B). The plasmid p19-0820-1561 also contained additional ARGs [mph(A), sul1, aadA5, and dfrA17] that were present only in pF93-2_1 in the form of the IS26-mph(A)-MFS transporter-tetR or acrR-IS6100-sul1-aadA5-dfrA17-intl1Δ-IS26-Tn3Δ-IS1RΔ unit. This element has been reported in multiple species, such as E. coli and K. pneumoniae (BLAST analysis; data not shown).
Other blaCTX-M-carrying plasmids.
The remaining 4 ESC-R S. sonnei isolates possessed unique blaCTX-M-positive plasmids (Table 1). In particular, a blaCTX-M-15 gene associated with ISEcp1 was carried in an 89-kb IncI1-pST16 plasmid (p6607-69), but without further ARGs. This plasmid showed a high degree of identity with others found in both S. sonnei and E. coli isolates (mostly from Asia), including some expressing CTX-M-55, which is a single amino acid variant of CTX-M-15 (Fig. S1). Likewise, one of our S. sonnei isolates carried blaCTX-M-55, but in this case, the gene was located in a 74-kb IncFII (F2:A-:B-) plasmid (p0401952027) and was flanked by two IS26 elements. This plasmid showed a high degree of genetic identity with others possessing blaCTX-M-55 or blaCTX-M-15 that came from E. coli or K. pneumoniae isolates detected in Europe or North America, but, interestingly, none of them cocarried the tetR or acrR-MFS trans-mph(A) unit between IS6100 and IS1RΔ (Fig. S2).
Another ESC-R S. sonnei isolate carried a 67-kb IncFII (F2:A-:B-) plasmid (p09163633) that harbored only blaCTX-M-27 and that showed a high degree of identity with the backbone of plasmids from E. coli and S. flexneri isolates. Nevertheless, only p09163633 possessed the IS26-IS903BΔ-blaCTX-M-27-ISEcp1Δ-IS26 unit (Fig. S3), which has been reported in multiple E. coli and K. pneumoniae isolates from Vietnam and China (BLAST analysis; data not shown) and which has also been described in a Japanese epidemic ST131 E. coli isolate (33). Moreover, it was also present in the 69-kb IncFII (F35:A-:B-) plasmid (p3123885) found in our last ESC-R S. sonnei isolate (Fig. S4), though this mobile genetic element (MGE) encoded the single amino acid variant CTX-M-134 instead of CTX-M-27 (29).
Coresistance to azithromycin.
Besides the specific ESBLs identified, 6 of the 25 S. sonnei isolates were macrolide resistant due to the presence of erm(B) and/or mph(A) ARGs (Table 1; Table S1). As mentioned above, 3 ESC-R isolates carried mph(A) in different IncFII plasmids coharboring blaCTX-M-15, blaCTX-M-55, or blaCTX-M-134. Of note, two of these plasmids carried the element IS26-mph(A)-MFS trans-tetR or acrR-IS6100 (Fig. 1B; Fig. S4) and the other one carried, with a slightly different arrangement, IS6100-tetR or acrR-MFS trans-mph(A)-IS1RΔ (Fig. S2). These two very similar elements have been found in many plasmids carried by E. coli, K. pneumoniae, and Salmonella enterica (BLAST analysis; data not shown) and have also been identified in the chromosome of a CTX-M-15-producing S. enterica serovar Haardt isolate recovered from Japanese food workers (34).
Overall, these findings are epidemiologically relevant, since coresistance to azithromycin and ESCs makes the treatment of shigellosis difficult (1, 5). Such MDR plasmids have rarely been reported in S. sonnei, though an IncFII (F2:A-:B-) plasmid possessing blaCTX-M-27, mph(A), and erm(B) was associated with the outbreak among MSM in England (10), while an IncB/O/K/Z plasmid coharboring blaCTX-M-14 and mph(A) was linked to a waterborne outbreak in China in 2015 (8). Having observed that at least three of our IncFII plasmids carried very similar macrolide resistance elements, we speculate that under a certain antibiotic selective pressure (e.g., azithromycin), mph(A) can be acquired via integration of transposable elements [e.g., IS26-mph(A)-MFS trans-tetR or acrR-IS6100] (35).
MLST and cgMLST.
Regardless of the presence of blaCTX-M genes, 24 S. sonnei isolates were of ST152, while 1 was of its single allele variant, ST1503 (Table 1). ST152 has been previously reported in ESC-S S. sonnei isolates in many countries (e.g., the United States [California], China, Germany, and Iran) (15–19). Recently, we also described the ST152 CTX-M-3-producing strain LC-1477-18, isolated in Italy from a girl who acquired the infection in Albania (7). Overall, since our S. sonnei isolates were acquired in different periods and/or in diverse geographic areas (Table 1), one could speculate that a unique clone (ST152) is spreading worldwide.
To better investigate the clonality of our S. sonnei isolates, we performed a core-genome MLST (cgMLST) analysis according to the E. coli scheme. The higher resolution of cgMLST resulted in multiple cgSTs: (i) three blaCTX-M-3-possessing isolates and one ESC-S isolate were of cgST113036, (ii) two blaCTX-M-3-positive isolates were of cgST115537, (iii) four blaCTX-M-15-harboring isolates were of cgST112958, and (iv) two ESC-S isolates carrying mph(A) and erm(B) were of cgST107674. The remaining isolates showed different cgSTs, but overall, high allele matches were maintained among the 2,513 isolates analyzed (i.e., >98.5% for ESBL producers and >98.6% for those ESC-S isolates) (Table 1). These results support the hypothesis that common ESC-R clones may spread in different countries and could be imported to other nations (e.g., Switzerland) after international trips. Based on the identification of clones including both ESBL producers and nonproducers, it can be also speculated that some ESC-S S. sonnei isolates may be well predisposed to acquire MDR plasmids from other Enterobacterales.
Core-genome analyses.
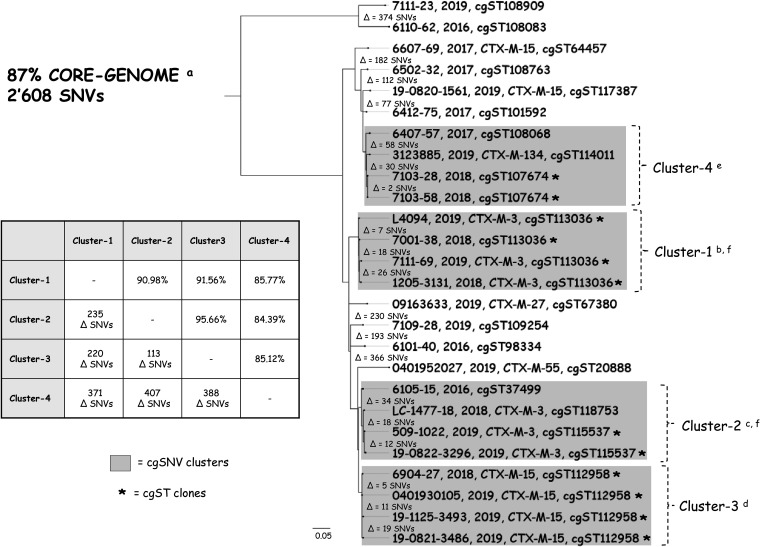
To determine the distance and clonality between our S. sonnei strains, a high-resolution SNV analysis was performed (87% core-genome coverage among all strains). As shown in Fig. 2, the 4 S. sonnei clones identified by cgMLST were also confirmed to be 4 independent SNV clusters, though several additional isolates were grouped within cluster 2 and cluster 4. To summarize: cluster 1 and cluster 2 included CTX-M-3-producing or ESC-S S. sonnei isolates, cluster 3 encompassed CTX-M-15 producers, and cluster 4 included the CTX-M-134 producer and the ESC-S S. sonnei isolates. Notably, strains belonging to the same cluster differed by only a limited number of SNVs (i.e., for cluster 1, the difference was 7 to 26 SNVs; for cluster 2, the difference was 12 to 34 SNVs; for cluster 3, the difference was 5 to 19 SNVs; and for cluster 4, the difference was 2 to 58 SNVs).
FIG 2.
Analysis of the core-genome phylogeny of 25 S. sonnei isolates together with that of Italian strain LC-1477-18. For each strain, we show the strain, the collection year, the main β-lactamase (if present), and the cgST. The assembled WGS of strains is presented in a core-genome SNV tree. The differences in the SNV value (e.g., Δ = 1 SNV) corresponds to the number of nonidentical SNVs of the core genome between two strains. The four main clusters (gray boxes) were defined when the nucleotide identity across two or more strains was ≥97.5% of shared SNVs (difference of ≤65 SNVs). The cluster matrix shows the maximum nucleotide identity (in percent) between all strains across two clusters (top right corner) and the number of SNVs not shared among all strains compared. The scale bar (0.05) represents the average number of nucleotide substitutions per site. Asterisks represent identical cgSTs, as determined by CGE’s cgMLSTFinder (v1.1) program. a, the core genome represents the maximum total coverage (87%) of the alignment among all 26 S. sonnei conserved sequences, which corresponded to 2,608 SNVs; b, cluster 1 strains shared 98.77% SNVs; c, cluster 2 strains shared 98.01% SNVs; d cluster 3, strains shared 99.08% SNVs; e, cluster 4 strains shared 97.54% SNVs; f, in cluster 1 and cluster 2, blaCTX-M-3 was consistently carried by the same IncI1-pST57 plasmid (Fig. 1A).
Together, these results corroborate the above-mentioned hypothesis on the dissemination of CTX-M-producing hyperepidemic S. sonnei clones. This is consistent with what has been observed by other authors for the fluoroquinolone-resistant international clones (e.g., global lineage III [GIII]) (36, 37). However, our data also indicate that MLST analysis alone has a limited resolution for studying the spread of such MDR pathogens. In fact, although almost all of our S. sonnei isolates were identified to be ST152, several clusters with different ARG and plasmid patterns could be differentiated using cgMLST and/or core-genome SNV analyses.
We also note that isolates included in cluster 1 and cluster 2 carried the same IncI1-pST57 blaCTX-M-3-harboring plasmid (Fig. 1). This finding was surprising, as the two bacterial groups were genetically different (i.e., a difference of 235 SNVs; Fig. 2). We do not have a clear explanation for the independent clustering, but it can be hypothesized that ESC-S S. sonnei isolates belonging to cluster 1 and cluster 2 acquired the pST57 plasmid from a common enterobacterial ancestor, including other Shigella spp.
Link with international isolates.
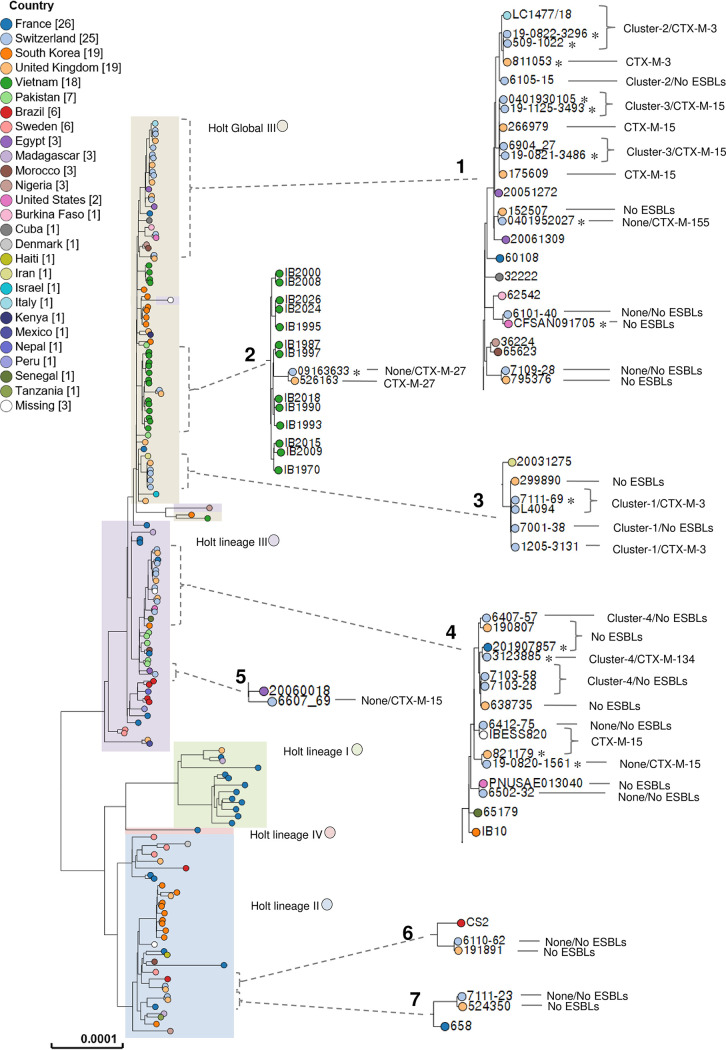
To explore the lineage origins of our Swiss S. sonnei isolates, we performed a search of the Enterobase database for strains of global lineage and matching cgST. A core-genome analysis was then performed using the results of the Enterobase phylogenetic analysis as a reference. Overall, we compared our collection (n = 25) to a subset of 114 strains of global lineage (38), 16 with a matching cgST, and the Italian LC-1477-18 isolate (7). The alignment of all strains (n = 156) resulted in 4,551 SNVs and 42% core-genome coverage among all isolates.
As depicted in Fig. 3 and Table S2, all S. sonnei isolates included in cluster 1 were related to an Iranian GIII strain detected in 2003 and were almost identical to another one found in the United Kingdom in 2016 (with the same cgST and a difference of 2 to 3 SNVs). We also note that one of our isolates (isolate 7111-69) had its clinical origin in Turkey (Table 1), the area where the first CTX-M-3-producing S. sonnei isolate was described (2001), and subsequently caused epidemic events (26, 27).
FIG 3.
Enterobase SNV tree dendrogram of the Swiss S. sonnei strains (n = 25), the global lineage of strains (n = 114), strains with matching cgST (n = 16), and the Italian LC-1477-18 strain as a reference. The combined single nucleotide polymorphism profiles of all 156 strains mapped to the reference sequence are represented in a RAxML tree, corresponding to a total of 9,850 SNVs. Country labels are represented by colored circles (missing country labels correspond to strains IBESS820 from the Netherlands, Ss046 from China, and 53G from South Korea). Holt lineages I, II, III, GIII, and IV are presented in colored boxes. Dashed black brackets with lines correspond to zoom-in sections of the tree where the present study’s strains are clustered. For the Swiss isolates, we show the cluster (if any)/ESBL (if any), while for the international strains with a matching cgST, we show only the ESBL (if any). Among these strains, those detected in 2019 are indicated with an asterisk. The scale bar represents the average number of nucleotide substitutions per site. See Table S2 in the supplemental material for more information regarding the Parsnp program SNV analysis results for zoom-in sections 1 to 7: section 1, compared to the Swiss isolates possessing the same cgST, strains 811053, 266979, 175609, 152507, CFSAN091705, and 795376 show 0 to 1, 0 to 4, 1 to 5, 2, 5, and 4 SNVs, respectively; section 2, compared to the Swiss isolate, strain 526163 has the same cgST and shows 2 SNVs; section 3, compared to the Swiss isolates, strain 299890 has the same cgST and shows 2 to 3 SNVs; section 4, compared to the Swiss isolates possessing the same cgST, strains 190807, 201907857, 638735, IBESS820, 821179, and PNUSAE013040 show 6, 1, 1, 0, 8, and 1 SNVs, respectively; section 5, 61 SNVs are found between the Swiss isolate and the one isolate found in Egypt; section 6, compared to the Swiss isolate, strain 191891 has the same cgST and shows 2 SNVs; section 7, compared to the Swiss isolate, strain 524350 has the same cgST and shows 4 SNVs.
The cluster 2 and cluster 3 isolates were part of a large group shared by two GIII strains of Egyptian origin reported in 2005 and 2006, LC-1477-18, and four isolates detected in the United Kingdom: one CTX-M-3 producer isolated in 2019, two CTX-M-15 producers detected in 2015 and 2016, and one ESC-S isolate found in 2015 (Fig. 3). These UK strains showed differences in SNVs of ≤5 compared to the sequences of cgST-matching isolates from Switzerland, indicating their commonality. Further evidence of their possible origin could be seen in isolates 6105-15, 0401930105, 19-0821-3486, and 0401952027, the origin of which was Egypt (Table 1), suggesting that these strains may have originated in that geographic area.
S. sonnei cluster 4 isolates were grouped with recently detected ESC-S strains in the United Kingdom, the United States, and France. Two additional CTX-M-15 producers, one from the United Kingdom (821179) and one from the Netherlands (IBESS820), were also highly related to the cluster 4 isolates (Fig. 3). The latter was identified in 2017 during a cross-sectional multicenter study (39) and was genetically identical to our ESC-S S. sonnei 6412-75 strain (they had the same cgST and a difference of 0 SNPs). In that study, the patient from whom strain IBESS820 was recovered was reported to have a history of travel to India, as did the Swiss patient with an infection caused by 6412-75 (Table 1). Despite these similarities, the Swiss S. sonnei isolate was ESBL negative; nevertheless, this finding highlights the great capacity of certain clones to acquire blaCTX-M-harboring plasmids.
In total, 12 of the 16 international S. sonnei strains that had the same cgST as our Swiss isolates were detected in the United Kingdom, while the remaining 4 were isolated in the United States, France, and the Netherlands. This indicates that common S. sonnei lineages have been circulating in Europe at least since 2015 and are now expanding in Switzerland. In fact, we note that all 10 Swiss ESC-R S. sonnei isolates detected in 2019 were linked to isolates detected in the same year in the United Kingdom and France (Table 1 and Fig. 3), and most of these were producers of CTX-M-3 or CTX-M-15, as in the case of those in Switzerland.
It can be speculated that the same common plasmids described in the present work (e.g., the IncI1-pST57 blaCTX-M-3-positive plasmid) are also carried by contemporary non-Swiss ESC-R S. sonnei isolates (as demonstrated for LC-1477-18). However, since the matching cgST isolates identified from the Enterobase database are in the form of whole-genome shotgun assemblies generated from short-read sequencing data, without the full characterization of blaCTX-M-carrying plasmids with long-read sequencing data, as in our study, this hypothesis cannot be fully corroborated.
Conclusions.
In this work, we present the first detailed molecular investigation of S. sonnei isolates detected in Switzerland. Hybrid WGS assemblies were implemented to accurately describe the blaCTX-M-harboring plasmids, while core-genome and phylogenetic analyses were used to study the clonality of the strains.
Based on our results, we conclude that most of the contemporary Swiss ESBL-producing S. sonnei isolates carry identical blaCTX-M-positive plasmids that often have their counterparts in other reported Enterobacterales worldwide. More importantly, due to transnational travel, common international clones of MDR S. sonnei are emerging in Switzerland, and this limits our therapeutic armamentarium. Overall, our findings underline the importance of continuously conducting epidemiological surveys using the WGS approach and linking the results with those from other countries (40).
MATERIALS AND METHODS
Epidemiological data.
Phenotypic data regarding the S. sonnei isolates detected in Switzerland during 2016 to 2019 were retrieved from the Swiss Centre for Antibiotic Resistance (ANRESIS) database (http://www.anresis.ch/), which collects information from 30 Swiss clinical laboratories. Strains were categorized as ESC-R when nonsusceptible (i.e., intermediate or resistant) to ceftazidime, ceftriaxone, and/or cefepime according to the criteria implemented for Enterobacterales by the routine clinical laboratories during the corresponding years. The research project was exempted from the requirement for ethical approval because no health-related personal data were used, while age, gender, and trip-related information (if available) were retrieved from the laboratory databases.
Strains, ID, and antimicrobial susceptibility tests.
All ESC-R and ESC-S S. sonnei isolates available at −80°C and collected during 2016 to 2019 at the Institute for Infectious Diseases, MCL Medizinische Laboratorien, and labormedizinisches zentrum Dr. Risch were analyzed.
The initial identification, obtained by implementing matrix-assisted laser desorption ionization–time of flight mass spectrometry (Bruker), was confirmed with the Type Strain Genome Server (TYGS) tools using the Genome BLAST Distance Phylogeny approach (https://tygs.dsmz.de/), based on genome data (see below). MICs were obtained by implementing Sensititre GNX2F microdilution panels (Thermo Fisher). For isolates possessing the erm(B) and/or mph(A) gene, MICs for azithromycin were obtained using the Etest (bioMérieux). Results were interpreted according to the EUCAST 2019 criteria (41).
Whole-genome sequencing (WGS).
WGS was performed using both the NovaSeq-6000 (Illumina) and the MinION (Oxford Nanopore) sequencing platforms as previously described (42–45). In brief, Illumina raw reads were quality filtered with the Trimmomatic (v0.36) trimmer, followed by whole-genome shotgun assembly with the SPAdes (v3.12.0) program. Adaptors from Nanopore raw reads were trimmed with the Porechop (v0.2.4) program and quality filtered with the Filtlong (v0.2.0) program. Long-read assemblies were done with the Canu (v1.7) program. The final hybrid assemblies were generated by aligning the paired-end Illumina reads to the Canu assemblies with the Bowtie2 (v2.3.4.1) program, followed by multiple rounds of polishing with the Pilon (v1.22) program.
Illumina SPAdes assemblies were used for whole-genome ID and analysis with the tools of the Center for Genomic Epidemiology (CGE; http://www.genomicepidemiology.org/): ResFinder, MLST with E. coli scheme 1, PlasmidFinder, and pMLST. Hybrid assemblies were used to characterize the blaESBL-carrying plasmids.
Annotations of both the Illumina and the hybrid assemblies were carried out by the NCBI Prokaryotic Genome Annotation Pipeline. All annotated features presented in Fig. 1 and Fig. S1 to S4 in the supplemental material were manually curated with the UniProt (https://www.uniprot.org/blast/) and ISfinder (https://isfinder.biotoul.fr/) tools and annotated accordingly.
Core-genome analyses.
All S. sonnei isolates underwent cgMLST with the CGE cgMLSTFinder (v1.1) program, using Illumina raw reads as the input and the species database set to E. coli Enterobase. These isolates also underwent core-genome SNV analysis as previously done (46). Briefly, the core-genome alignment was performed with the Parsnp (v1.2) program. All strains were treated as curated genomes (−C parameter), and the Italian ST152 CTX-M-3-producing S. sonnei strain (strain LC-1477-18; GenBank accession number JAATWD000000000) was used as a reference (7). The −C parameter was set to 200, and the other parameters were left at the default. Variants with no flags (PASS) were determined to be reliable and used for downstream SNV analysis with a custom R (v3.6.2) script. The Parsnp program-generated core-genome SNV phylogenetic tree was visualized with the FigTree (v1.4.4) program and set to midpoint rooted and nodes by decreasing order (Fig. 2).
An SNV tree dendrogram of the Swiss S. sonnei collection versus global lineage and matching cgST strains was created in the Enterobase Escherichia/Shigella database (https://enterobase.warwick.ac.uk/species/index/ecoli) (Fig. 3). The analyzed strains consisted of 156 total strains, of which 114 were of global lineages (38), 16 had matching cgSTs, 25 were from Switzerland, and 1 was the Italian LC-1477-18 strain, used as a reference. The following search queries were used to find the global lineage strains (date, 10 April 2020) in Enterobase: species equals “Shigella sonnei”; comment contains “Holt lineage”; and to find matching cgST strains (date, April 21, 2020), the experiment type was cgMLST V1 + HierCC V1 and ST were 108909, 108083, 64457, 108763, 117387, 101592, 108068, 114011, 107674, 113036, 67380, 109254, 98334, 20888, 37499, 118753, 115537, and 112958. The Illumina raw reads of our 25 S. sonnei isolates were uploaded to Enterobase for processing. The resulting genomes assembled by Enterobase were used to create an SNV project of 156 strains with default settings (minimum percentage of sites present, 95). The tree was visualized with the web-based browser.
An independent core-genome analysis (Table S2) with the Parsnp program was used to analyze the strain clusters identified in Fig. 3, which also included the 156 S. sonnei assemblies. As described above, the genome of the Italian LC-1477-18 strain was used as the reference genome, the Parsnp −C parameter was set to 300, and the rest of the parameters were set as the default.
Data availability.
The Illumina SPAdes assemblies were deposited in GenBank under BioProject accession number PRJNA578838. The hybrid assemblies (blaESBL-carrying plasmids and the corresponding chromosomes) were deposited in GenBank under BioProject accession number PRJNA578858.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NRP-72, National Research Program, Antimicrobial Resistance (Swiss National Science Foundation grant no. 177378 to A.E.), and by the Swiss Centre for Antibiotic Resistance (ANRESIS; to A.K.).
We thank Parham Sendi (Institute for Infectious Diseases, University of Bern) for the ethical advice. We also thank Maria V. Elzi, Carlo Casanova, and Thomas Büdel (Institute for Infectious Diseases, University of Bern) for technical support.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Kotloff KL, Riddle MS, Platts-Mills JA, Pavlinac P, Zaidi A. 2018. Shigellosis. Lancet 391:801–812. doi: 10.1016/S0140-6736(17)33296-8. [DOI] [PubMed] [Google Scholar]
- 2.Bardsley M, Jenkins C, Mitchell HD, Mikhail AFW, Baker KS, Foster K, Hughes G, Dallman TJ. 2020. Persistent transmission of shigellosis in England is associated with a recently emerged multidrug-resistant strain of Shigella sonnei. J Clin Microbiol 58:e01692-19. doi: 10.1128/JCM.01692-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson CN, Duy PT, Baker S. 2015. The rising dominance of Shigella sonnei: an intercontinental shift in the etiology of bacillary dysentery. PLoS Negl Trop Dis 9:e0003708. doi: 10.1371/journal.pntd.0003708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Puzari M, Sharma M, Chetia P. 2018. Emergence of antibiotic resistant Shigella species: a matter of concern. J Infect Public Health 11:451–454. doi: 10.1016/j.jiph.2017.09.025. [DOI] [PubMed] [Google Scholar]
- 5.Ingle DJ, Easton M, Valcanis M, Seemann T, Kwong JC, Stephens N, Carter GP, Goncalves da Silva A, Adamopoulos J, Baines SL, Holt KE, Chow EPF, Fairley CK, Chen MY, Kirk MD, Howden BP, Williamson DA. 2019. Co-circulation of multidrug-resistant Shigella among men who have sex with men in Australia. Clin Infect Dis 69:1535–1544. doi: 10.1093/cid/ciz005. [DOI] [PubMed] [Google Scholar]
- 6.Ranjbar R, Farahani A. 2019. Shigella: antibiotic-resistance mechanisms and new horizons for treatment. Infect Drug Resist 12:3137–3167. doi: 10.2147/IDR.S219755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luzzaro F, Clement M, Principe L, Viaggi V, Bernasconi OJ, Endimiani A. 2019. Characterisation of the first extended-spectrum β-lactamase (ESBL)-producing Shigella sonnei clinical isolate in Italy. J Glob Antimicrob Resist 17:58–59. doi: 10.1016/j.jgar.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Ma Q, Xu X, Luo M, Wang J, Yang C, Hu X, Liang B, Wu F, Yang X, Wang J, Liu H, Li W, Zhong Y, Li P, Xie J, Jia L, Wang L, Hao R, Du X, Qiu S, Song H, Sun Y. 2017. A waterborne outbreak of Shigella sonnei with resistance to azithromycin and third-generation cephalosporins in China in 2015. Antimicrob Agents Chemother 61:e00308-17. doi: 10.1128/AAC.00308-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JS, Kim J, Jeon SE, Kim SJ, Kim NO, Hong S, Kang YH, Han S, Chung GT. 2014. Complete nucleotide sequence of the IncI1 plasmid pSH4469 encoding CTX-M-15 extended-spectrum β-lactamase in a clinical isolate of Shigella sonnei from an outbreak in the Republic of Korea. Int J Antimicrob Agents 44:533–537. doi: 10.1016/j.ijantimicag.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Mook P, McCormick J, Bains M, Cowley LA, Chattaway MA, Jenkins C, Mikhail A, Hughes G, Elson R, Day M, Manuel R, Dave J, Field N, Godbole G, Dallman T, Crook P. 2016. ESBL-producing and macrolide-resistant Shigella sonnei infections among men who have sex with men, England, 2015. Emerg Infect Dis 22:1948–1952. doi: 10.3201/eid2211.160653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qu F, Ying Z, Zhang C, Chen Z, Chen S, Cui E, Bao C, Yang H, Wang J, Liu C, Mao Y, Zhou D. 2014. Plasmid-encoding extended-spectrum β-lactamase CTX-M-55 in a clinical Shigella sonnei strain, China. Future Microbiol 9:1143–1150. doi: 10.2217/fmb.14.53. [DOI] [PubMed] [Google Scholar]
- 12.Nüesch-Inderbinen M, Heini N, Zurfluh K, Althaus D, Hächler H, Stephan R. 2016. Shigella antimicrobial drug resistance mechanisms, 2004–2014. Emerg Infect Dis 22:1083–1085. doi: 10.3201/eid2206.152088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.European Food Safety Authority and European Centre for Disease Prevention and Control (EFSA and ECDC). 2019. The European Union One Health 2018 zoonoses report. https://www.ecdc.europa.eu/sites/default/files/documents/zoonoses-EU-one-health-2018-report.pdf. doi: 10.2903/j.efsa.2019.5926. [DOI] [PMC free article] [PubMed]
- 14.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozyreva VK, Jospin G, Greninger AL, Watt JP, Eisen JA, Chaturvedi V. 2016. Recent outbreaks of shigellosis in California caused by two distinct populations of Shigella sonnei with either increased virulence or fluoroquinolone resistance. mSphere 1:e00344-16. doi: 10.1128/mSphere.00344-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shahsavan S, Nobakht M, Rastegar-Lari A, Owlia P, Bakhshi B. 2016. Multi-locus sequence type analysis of Shigellas pp. isolates from Tehran, Iran. Iran J Microbiol 8:298–306. [PMC free article] [PubMed] [Google Scholar]
- 17.Shokoohizadeh L, Kaydani GA, Ekrami A. 2017. Molecular characterization of Shigella spp. isolates from a pediatric hospital in southwestern Iran. Gastroenterol Hepatol Bed Bench 10:319–322. [PMC free article] [PubMed] [Google Scholar]
- 18.Cao Y, Wei D, Kamara IL, Chen W. 2012. Multi-locus sequence typing (MLST) and repetitive extragenic palindromic polymerase chain reaction (REP-PCR), characterization of Shigella spp. over two decades in Tianjin China. Int J Mol Epidemiol Genet 3:321–332. [PMC free article] [PubMed] [Google Scholar]
- 19.Inouye M, Conway TC, Zobel J, Holt KE. 2012. Short read sequence typing (SRST): multi-locus sequence types from short reads. BMC Genomics 13:338. doi: 10.1186/1471-2164-13-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dallman TJ, Chattaway MA, Mook P, Godbole G, Crook PD, Jenkins C. 2016. Use of whole-genome sequencing for the public health surveillance of Shigella sonnei in England and Wales, 2015. J Med Microbiol 65:882–884. doi: 10.1099/jmm.0.000296. [DOI] [PubMed] [Google Scholar]
- 21.Abelman RL, M'Ikanatha NM, Figler HM, Dudley EG. 2019. Use of whole genome sequencing in surveillance for antimicrobial-resistant Shigella sonnei infections acquired from domestic and international sources. Microb Genom 5:e000270. doi: 10.1099/mgen.0.000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qian H, Liu G, Chen Y, Ma P, Kong X, Zhou L, Hong J, Bao C, Gu B. 2018. Increasing clinical resistance rate of Shigella sonnei to cefotaxime in Jiangsu Province, China, between 2012 and 2015. Ann Transl Med 6:207. doi: 10.21037/atm.2018.05.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shakya G, Acharya J, Adhikari S, Rijal N. 2016. Shigellosis in Nepal: 13 years review of nationwide surveillance. J Health Popul Nutr 35:36. doi: 10.1186/s41043-016-0073-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heffernan H, Woodhouse R, Hewison C, Sherwood J. 2018. Antimicrobial resistance among Shigella in New Zealand. N Z Med J 131:56–62. [PubMed] [Google Scholar]
- 25.Buhler S, Ruegg R, Steffen R, Hatz C, Jaeger VK. 2014. A profile of travelers—an analysis from a large Swiss travel clinic. J Travel Med 21:324–331. doi: 10.1111/jtm.12139. [DOI] [PubMed] [Google Scholar]
- 26.Acikgoz ZC, Eser OK, Kocagoz S. 2008. CTX-M-3 type β-lactamase producing Shigella sonnei isolates from pediatric bacillary dysentery cases. Jpn J Infect Dis 61:135–137. [PubMed] [Google Scholar]
- 27.Acikgoz ZC, Gulay Z, Bicmen M, Gocer S, Gamberzade S. 2003. CTX-M-3 extended-spectrum β-lactamase in a Shigella sonnei clinical isolate: first report from Turkey. Scand J Infect Dis 35:503–505. doi: 10.1080/00365540310013270. [DOI] [PubMed] [Google Scholar]
- 28.Kim JS, Kim S, Park J, Shin E, Yun YS, Lee DY, Kwak HS, Seong WK, Chung GT, Kim J. 2017. Plasmid-mediated transfer of CTX-M-55 extended-spectrum β-lactamase among different strains of Salmonella and Shigella spp. in the Republic of Korea. Diagn Microbiol Infect Dis 89:86–88. doi: 10.1016/j.diagmicrobio.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 29.Castanheira M, Costello S, Jones R. 2015. Molecular characterization of genes encoding CTX-M-134, TEM-207 and TEM-212 detected among clinical isolates from USA hospitals, poster P0982. Eur Congr Clin Microbiol Infect Dis (ECCMID), Copenhagen, Denmark. [Google Scholar]
- 30.Rashid H, Rahman M. 2015. Possible transfer of plasmid mediated third generation cephalosporin resistance between Escherichia coli and Shigella sonnei in the human gut. Infect Genet Evol 30:15–18. doi: 10.1016/j.meegid.2014.11.023. [DOI] [PubMed] [Google Scholar]
- 31.Liakopoulos A, van den Bunt G, Geurts Y, Bootsma MCJ, Toleman M, Ceccarelli D, van Pelt W, Mevius DJ. 2018. High prevalence of intra-familial co-colonization by extended-spectrum cephalosporin resistant Enterobacteriaceae in preschool children and their parents in Dutch households. Front Microbiol 9:293. doi: 10.3389/fmicb.2018.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rozwandowicz M, Brouwer MSM, Fischer J, Wagenaar JA, Gonzalez-Zorn B, Guerra B, Mevius DJ, Hordijk J. 2018. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J Antimicrob Chemother 73:1121–1137. doi: 10.1093/jac/dkx488. [DOI] [PubMed] [Google Scholar]
- 33.Matsumura Y, Johnson JR, Yamamoto M, Nagao M, Tanaka M, Takakura S, Ichiyama S, Kyoto-Shiga Clinical Microbiology Study Group. 2015. CTX-M-27- and CTX-M-14-producing, ciprofloxacin-resistant Escherichia coli of the H30 subclonal group within ST131 drive a Japanese regional ESBL epidemic. J Antimicrob Chemother 70:1639–1649. doi: 10.1093/jac/dkv017. [DOI] [PubMed] [Google Scholar]
- 34.Shigemura H, Sakatsume E, Sekizuka T, Yokoyama H, Hamada K, Etoh Y, Carle Y, Mizumoto S, Hirai S, Matsui M, Kimura H, Suzuki M, Onozuka D, Kuroda M, Inoshima Y, Murakami K. 2020. Food workers as a reservoir of extended-spectrum cephalosporin-resistant Salmonella in Japan. Appl Environ Microbiol 86:e00072-20. doi: 10.1128/AEM.00072-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Darton TC, Tuyen HT, The HC, Newton PN, Dance DAB, Phetsouvanh R, Davong V, Campbell JI, Hoang NVM, Thwaites GE, Parry CM, Thanh DP, Baker S. 2018. Azithromycin resistance in Shigella spp. in Southeast Asia. Antimicrob Agents Chemother 62:e01748-17. doi: 10.1128/AAC.01748-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chung The H, Rabaa MA, Pham Thanh D, De Lappe N, Cormican M, Valcanis M, Howden BP, Wangchuk S, Bodhidatta L, Mason CJ, Nguyen Thi Nguyen T, Vu Thuy D, Thompson CN, Phu Huong Lan N, Voong Vinh P, Ha Thanh T, Turner P, Sar P, Thwaites G, Thomson NR, Holt KE, Baker S. 2016. South Asia as a reservoir for the global spread of ciprofloxacin-resistant Shigella sonnei: a cross-sectional study. PLoS Med 13:e1002055. doi: 10.1371/journal.pmed.1002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chung The H, Boinett C, Pham Thanh D, Jenkins C, Weill FX, Howden BP, Valcanis M, De Lappe N, Cormican M, Wangchuk S, Bodhidatta L, Mason CJ, Nguyen TNT, Ha Thanh T, Voong VP, Duong VT, Nguyen PHL, Turner P, Wick R, Ceyssens PJ, Thwaites G, Holt KE, Thomson NR, Rabaa MA, Baker S. 2019. Dissecting the molecular evolution of fluoroquinolone-resistant Shigella sonnei. Nat Commun 10:4828. doi: 10.1038/s41467-019-12823-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holt KE, Baker S, Weill FX, Holmes EC, Kitchen A, Yu J, Sangal V, Brown DJ, Coia JE, Kim DW, Choi SY, Kim SH, da Silveira WD, Pickard DJ, Farrar JJ, Parkhill J, Dougan G, Thomson NR. 2012. Shigella sonnei genome sequencing and phylogenetic analysis indicate recent global dissemination from Europe. Nat Genet 44:1056–1059. doi: 10.1038/ng.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hendriks ACA, Reubsaet FAG, Kooistra-Smid A, Rossen JWA, Dutilh BE, Zomer AL, van den Beld MJC, IBESS Group. 2020. Genome-wide association studies of Shigella spp. and enteroinvasive Escherichia coli isolates demonstrate an absence of genetic markers for prediction of disease severity. BMC Genomics 21:138. doi: 10.1186/s12864-020-6555-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.European Centre for Disease Prevention and Control. 2016. Expert opinion on whole genome sequencing for public health surveillance. European Centre for Disease Prevention and Control, Stockholm, Sweden. doi: 10.2900/12442. [DOI] [Google Scholar]
- 41.European Committee on Antimicrobial Susceptibility Testing (EUCAST). 2019. Breakpoints tables for interpretation of MICs and zone diameters, version 9.0, valid from 1 January 2019. https://eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_9.0_Breakpoint_Tables.pdf.
- 42.Clement M, Ramette A, Bernasconi OJ, Principe L, Luzzaro F, Endimiani A. 2018. Whole-genome sequence of the first extended-spectrum β-lactamase-producing strain of Salmonella enterica subsp. enterica serovar Napoli. Microbiol Resour Announc 7:e00973-18. doi: 10.1128/MRA.00973-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernasconi OJ, Dona V, Pires J, Kuenzli E, Hatz C, Luzzaro F, Perreten V, Endimiani A. 2018. Deciphering the complete deletion of the mgrB locus in an unusual colistin-resistant Klebsiella pneumoniae isolate colonising the gut of a traveller returning from India. Int J Antimicrob Agents 51:529–531. doi: 10.1016/j.ijantimicag.2017.09.014. [DOI] [PubMed] [Google Scholar]
- 44.Clement M, Keller PM, Bernasconi OJ, Stirnimann G, Frey PM, Bloemberg GV, Sendi P, Endimiani A. 2019. First clinical case of in vivo acquisition of DHA-1 plasmid-mediated AmpC in a Salmonella enterica subsp. enterica isolate. Antimicrob Agents Chemother 63:e00992-19. doi: 10.1128/AAC.00992-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Endimiani A, Bernasconi OJ, Budel T, Campos-Madueno EI, Kuenzli E, Hatz C, Carattoli A. 2020. Whole-genome characterization of a Shewanella algae strain co-harboring blaCTX-M-15 and armA genes on a novel IncC plasmid. Antimicrob Agents Chemother 64:e00267-20. doi: 10.1128/AAC.00267-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Büdel T, Kuenzli E, Campos-Madueno EI, Mohammed AH, Hassan NK, Zinsstag J, Hatz C, Endimiani A. 20 June 2020. On the island of Zanzibar people in the community are frequently colonized with the same multidrug-resistant Enterobacterales found in poultry and retailed chicken meat. J Antimicrob Chemother doi: 10.1093/jac/dkaa198. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Illumina SPAdes assemblies were deposited in GenBank under BioProject accession number PRJNA578838. The hybrid assemblies (blaESBL-carrying plasmids and the corresponding chromosomes) were deposited in GenBank under BioProject accession number PRJNA578858.