Serine β-lactamases are dominant causes of β-lactam resistance in Klebsiella pneumoniae isolates. Recently, this has driven clinical deployment of the β-lactam–β-lactamase inhibitor pairs ceftazidime/avibactam and meropenem/vaborbactam. We show that four steps, i.e., ompK36 and ramR mutation plus carriage of OXA-232 and KPC-3-D178Y variant β-lactamases, confer ceftazidime/avibactam and meropenem/vaborbactam resistance when both pairs are used together.
KEYWORDS: KPC, OXA-48, β-lactamase, porin
ABSTRACT
Serine β-lactamases are dominant causes of β-lactam resistance in Klebsiella pneumoniae isolates. Recently, this has driven clinical deployment of the β-lactam–β-lactamase inhibitor pairs ceftazidime/avibactam and meropenem/vaborbactam. We show that four steps, i.e., ompK36 and ramR mutation plus carriage of OXA-232 and KPC-3-D178Y variant β-lactamases, confer ceftazidime/avibactam and meropenem/vaborbactam resistance when both pairs are used together. These findings have implications for decision making about sequential and combinatorial use of these β-lactam–β-lactamase inhibitor pairs to treat K. pneumoniae infections.
INTRODUCTION
Klebsiella pneumoniae isolates can become resistant to third-generation cephalosporins (3GC) and/or carbapenems through acquisition of a wide range of serine β-lactamases (1). Most clinically significant are the CTX-M (2), KPC (3), and OXA-48-like (4) types. In response to the rise in KPC, which can confer 3GC and carbapenem resistance, serine β-lactamase inhibitors, including avibactam and vaborbactam, have recently been developed. Avibactam is used in combination with the 3GC ceftazidime (5). It inhibits CTX-M, KPC, and OXA-48-like β-lactamases (6, 7). Ceftazidime-avibactam (CAZ/AVI) nonsusceptibility can be caused by KPC changes, e.g., D179Y or V239G (8, 9), or by a P170S change in CTX-M (10). Vaborbactam is used in combination with the carbapenem meropenem (11). It has potent activity against KPC and, to a lesser extent, CTX-M (12). Meropenem-vaborbactam (MER/VAB) resistance in K. pneumoniae isolates is caused by loss of OmpK36 and OmpK35 porins in backgrounds carrying KPC or OXA-48-like enzymes (12, 13).
Given the appearance of K. pneumoniae clinical isolates resistant to MER/VAB or CAZ/AVI, it has been suggested that a combination of both given together would overcome isolates resistant to one or the other when used separately (14, 15). The aim of the work reported here was to identify steps that can generate resistance to MER/VAB and CAZ/AVI when used together.
First, a plasmid (pOXA-232) encoding the OXA-48-like carbapenemase OXA-232 was purified from K. pneumoniae clinical isolate KP11 and used to transform the ramR frameshift (Arg44FS) mutant clinical K. pneumoniae isolate KP21 using 8 μg · ml−1 piperacillin and 4 μg · ml−1 tazobactam as selection (16). This isolate was chosen because ramR mutation causes overproduction of the AcrAB-TolC efflux pump and reduces production of the OmpK35 porin, and this enhances the spectrum of resistance conferred by OXA-48 and other β-lactamases in K. pneumoniae isolates (17). However, despite its ramR mutation, the MIC of meropenem (CLSI microtiter assay [18]) in the presence of 8 μg · ml−1 vaborbactam (MedChemExpress) against KP21(pOXA-232) was 1 μg · ml−1 (Table 1), making it MER/VAB susceptible according to CLSI breakpoints (19). We therefore selected a spontaneous MER/VAB-resistant derivative. To do this, 100-μl aliquots of overnight cultures of KP21(pOXA-232) grown in nutrient broth were spread onto Mueller-Hinton agar containing 16 μg · ml−1 meropenem and 8 μg · ml−1 vaborbactam, which was then incubated for 24 h at 37°C.
TABLE 1.
MICs of meropenem with or without vaborbactam and of ceftazidime with or without avibactam against derivatives of K. pneumoniae clinical isolates
| K. pneumoniae derivative | MIC (μg · ml−1) ofa: |
|||
|---|---|---|---|---|
| Meropenem | MER/VAB CAZ/AVI | Ceftazidime | CAZ/AVI | |
| KP21[ramR] (pOXA-232)(pUBYT) | 1 | 1/8 | 2 | 0.5/4 |
| KP21[ramR] pOXA-232)(pCTX-M-14) | 1 | 1/8 | 32 | 1/4 |
| KP21[ramR] pOXA-232)(pCTX-M-14 P170S) | 1 | 1/8 | 256 | 4/4 |
| KP21[ramR] (pOXA-232)(pKPC-3) | 128 | 2/8 | >256 | 8/4 |
| KP21[ramR] (pOXA-232)(pKPC-3-D178Y) | 2 | 2/8 | >256 | 256/4 |
| KP21[ramR] (pOXA-232)(pKPC-3-V239G) | 64 | 4/8 | >256 | 128/4 |
| KP21[ramR] ompK35(pOXA-232)(pUBYT) | 4 | 4/8 | 2 | 0.5/4 |
| KP21[ramR] ompK35(pOXA-232)(pCTX-M-14) | 4 | 4/8 | 32 | 1/4 |
| KP21[ramR] ompK35(pOXA-232)(pCTX-M-14 P170S) | 2 | 2/8 | >256 | 4/4 |
| KP21[ramR] ompK35(pOXA-232)(pKPC-3) | 128 | 1/8 | >256 | 0.125/4 |
| KP21[ramR] ompK35(pOXA-232)(pKPC-3-D178Y) | 4 | 4/8 | >256 | 128/4 |
| KP21[ramR] ompK35(pOXA-232)(pKPC-3-V239G) | 64 | 4/8 | >256 | 32/4 |
| KP21[ramR] ompK36(pOXA-232)(pUBYT) | 256 | 256/8 | 2 | 0.5/4 |
| KP21[ramR] ompK36(pOXA-232)(pCTX-M-14) | 256 | 256/8 | 32 | 1/4 |
| KP21[ramR] ompK36(pOXA-232)(pCTX-M-14 P170S) | 256 | 256/8 | >256 | 8/4 |
| KP21[ramR] ompK36(pOXA-232)(pKPC-3) | >256 | 256/8 | >256 | 16/4 |
| KP21[ramR] ompK36(pOXA-232)(pKPC-3-D178Y) | 256 | 256/8 | >256 | >256/4 |
| KP21[ramR] ompK36(pOXA-232)(pKPC-3-V239G) | >256 | 256/8 | >256 | 128/4 |
| KP21[ramR] ompK36(pKPC-3) | >256 | 16/8 | >256 | 8/4 |
| KP21[ramR] ompK36(pKPC-3-D178Y) | 64 | 16/8 | >256 | 256/4 |
| KP21[ramR] ompK36(pKPC-3-V239G) | >256 | 32/8 | >256 | 256/4 |
| KP21 M[ramR ompK36] (pOXA-232)(pUBYT) | 256 | 256/8 | 2 | 1/4 |
| KP21 M[ramR ompK36] (pOXA-232)(pCTX-M-14) | 128 | 128/8 | 32 | 1/4 |
| KP21 M[ramR ompK36] (pOXA-232)(pCTX-M-14 P170S) | 256 | 128/8 | >256 | 8/4 |
| KP21 M[ramR ompK36] (pOXA-232)(pKPC-3) | >256 | 256/8 | >256 | 8/4 |
| KP21 M[ramR ompK36] (pOXA-232)(pKPC-3-D178Y) | 128 | 128/8 | >256 | 256/4 |
| KP21 M[ramR ompK36] (pOXA-232)(pKPC-3-V239G) | >256 | 256/8 | >256 | 64/4 |
| KP21 M[ramR ompK36] ompK35(pOXA-232)(pUBYT) | 256 | 256/8 | 2 | 1/4 |
| KP21 M[ramR ompK36] ompK35(pOXA-232)(pCTX-M-14) | 256 | 256/8 | 32 | 1/4 |
| KP21 M[ramR ompK36] ompK35(pOXA-232)(pCTX-M-14 P170S) | 256 | 256/8 | 256 | 8/4 |
| KP21 M[ramR ompK36] ompK35(pOXA-232)(pKPC-3) | >256 | 256/8 | >256 | 16/4 |
| KP21 M[ramR ompK36] ompK35(pOXA-232)(pKPC-3-D178Y) | 256 | 256/8 | >256 | >256/4 |
| KP21 M[ramR ompK36] ompK35(pOXA-232)(pKPC-3-V239G) | >256 | 256/8 | >256 | 128/4 |
| KP47 ompK36(pKPC-3) | >256 | 2/8 | >256 | 8/4 |
| KP47 ompK36(pKPC-3-D178Y) | 8 | 2/8 | >256 | 256/4 |
| KP47 ompK36(pOXA-232)(pKPC-3) | >256 | 16/8 | >256 | 16/4 |
| KP47 ompK36(pOXA-232)(pKPC-3-D178Y) | 16 | 16/8 | >256 | >256/4 |
Shading indicates nonsusceptibility (resistance or intermediate resistance) according to CLSI breakpoints (19).
Liquid chromatography-tandem mass spectrometry envelope proteomics, performed as described previously (17), confirmed that this putative MER/VAB-resistant mutant, KP21 M(pOXA-232), had undetectable OmpK36 porin levels. Whole-genome sequencing, performed as described previously (20), identified a single mutation resulting in a stop at codon 125 in ompK36 in KP21 M(pOXA-232). To confirm the effect of this ompK36 point mutation, we insertionally inactivated ompK36 in the parent strain KP21(pOXA-232) using the pKNOCK suicide plasmid (21). The ompK36 DNA fragment was amplified from K. pneumoniae strain Ecl8 genomic DNA using ompK36 KO FW (5′-CGTTCAGGCGAACAACACTG-3′) and RV (5′-AAGTTCAGGCCGTCAACCAG-3′) primers (22). The PCR product was ligated into the pKNOCK-Gm at the SmaI site. The recombinant plasmid was then transferred into KP21(pOXA-232) by conjugation. Mutants were selected using gentamicin (5 μg · ml−1) and confirmed by PCR using ompK36 full-length FW (5′-GAGGCATCCGGTTGAAATAG-3′) and RV (5′-ATTAATCGAGGCTCCTCTTAC-3′) primers. The MER/VAB MIC against the ompK36 point mutant KP21 M(pOXA-232) and against the insertionally inactivated mutant KP21 ompK36(pOXA-232) was 256/8 μg · ml−1, confirming resistance (Table 1). This finding reveals that OmpK35 downregulation seen in ramR mutants mimics the effect of OmpK35 loss previously shown to be essential alongside OmpK36 loss for MER/VAB resistance in K. pneumoniae isolates producing an OXA-48-like enzyme (12, 13).
Both KP21(pOXA-232) ompK36 mutants remained susceptible to ceftazidime as expected, since OXA-232 only weakly hydrolyses ceftazidime (23) (Table 1). Aiming to increase ceftazidime MICs, we introduced the low-copy-number recombinant plasmid pCTX-M-14. To make pCTX-M-14, blaCTX-M-14 was amplified along with its native promoter from human urinary Escherichia coli isolated from a primary care setting (24) using CTX-M-14 FW (5′-CCGGAATTCAATACTACCTTGCTTTCTGA-3′) and RV (5′-CCGGAATTCCGTAGCGGAACGTTCATCAG-3′) primers and ligated into pUBYT at the EcoRI site (25). Carriage of pUBYT recombinants was selected using kanamycin (30 μg · ml−1).
Even though CTX-M-14 only weakly hydrolyses ceftazidime (26), in the ramR mutant KP21, carriage of pCTX-M-14 conferred ceftazidime resistance, as seen in KP21(pOXA-232)(pCTX-M-14), KP21 M(pOXA-232)(pCTX-M-14), and KP21 ompK36(pOXA-232)(pCTX-M-14) (Table 1). However, these derivatives remained ceftazidime susceptible in the presence of 4 μg · ml−1 avibactam (MedChemExpress), which is a potent inhibitor of CTX-M (5) (Table 1). Replacement of blaCTX-M-14 with blaCTX-M-14-P170S, encoding a variant associated with reduced CAZ/AVI susceptibility (10), representing the naturally occurring variant CTX-M-19 (27), drove the CAZ/AVI MIC against KP21 ompK36(pOXA-232) and KP21 M(pOXA-232) up to 8/4 μg · ml−1, which is one doubling dilution below the breakpoint for resistance. To enable this, CTX-M-14 site-directed mutagenesis was performed directly on the recombinant plasmid pCTX-M-14 using the QuikChange Lightning site-directed mutagenesis kit (Agilent, USA) with the CTX-M-14-P170S-FW primer (5′-TCTGGATCGCACTGAATCTACGCTGAATACCGC-3′).
Next, we insertionally inactivated ompK35 in KP21 M[ompK36](pOXA-232)(pCTX-M-14-P170S) using the method described above but with the ompK35 KO FW (5′-TCCCAGACCACAAAAACCCG-3′) and RV (5′-CCAGACCGAAGAAGTCGGAG-3′) primers and checked with ompK35 full-length FW (5′-CACTTCGATGTATTTAACCAG-3′) and RV (5′-ATGATGAAGCGCAATATTCTG-3′) primers. However, this did not further increase the CAZ/AVI MIC, confirming that ramR mutation phenotypically mimics OmpK35 loss (Table 1). Accordingly, it was not possible for us to generate derivatives resistant to both MER/VAB and CAZ/AVI using CTX-M-14-P170S, even in a ramR ompK36 ompK35 triple mutant background coproducing OXA-232.
KPC-3 is a carbapenemase that can confer ceftazidime resistance (9). To create the recombinant plasmid pKPC-3, blaKPC-3 was amplified from pKpQIL isolated from K. pneumoniae strain KP30 by PCR using KPC-3 FW (5′-CCGGAATTCGTAAAGTGGGTCAGTTTTCAG-3′) and RV (5′-GGCTCTGAAAATCATCTATTGGAATTCCGG-3′) primers and ligated into pUBYT at the EcoRI site (16). We also generated KPC-3 variants associated with CAZ/AVI resistance (8). KPC-3 site-directed mutagenesis was performed using a two-step PCR-based strategy: blaKPC-3-D178Y was constructed using KPC-3-D178Y-FW (5′-AGGCGATGCGCGCTATACCTCATCGCC-3′) and -RV (5′-GGCGATGAGGTATAGCGCGCATCGCCT-3′) primers, and blaKPC-3-V239G was constructed using KPC-3-V239G-FW (5′-GGAACCTGCGGAGGGTATGGCACGGCA-3′) and -RV (5′-TGCCGTGCCATACCCTCCGCAGGTTCC-3′) primers. The common flanking primers were KPC-3 FW and RV, as above.
When introduced into KP21(pOXA-232) (i.e., with wild-type ompK36), both pKPC-3-D178Y and pKPC-3-V239G conferred CAZ/AVI resistance, but pKPC-3 did not. The D178Y variant did this at the expense of reducing meropenem MIC into the intermediate-resistant zone, as reported previously (8) (Table 1). The V239G variant did not suffer from such a large drop in meropenem MIC, but MER/VAB MICs were the same (and in the susceptible range) against KP21(pOXA-232) derivatives producing KPC-3 or its D178Y or V239G variants (Table 1). Accordingly, it was not possible to generate derivatives resistant to both MER/VAB and CAZ/AVI using KPC-3 variants in a background having wild-type ompK36.
Next, we introduced plasmids encoding the KPC-3 variants associated with CAZ/AVI resistance into the ompK36 mutant MER/VAB-resistant derivatives KP21 M(pOXA-232) and KP21 ompK36(pOXA-232). As expected, the result was resistance to CAZ/AVI and MER/VAB when used separately (Table 1). OXA-232 production is not essential for this phenotype because KP21 ompK36(pKPC-3-D178Y) and KP21 ompK36(pKPC-3-V239G), lacking OXA-232, were also resistant to CAZ/AVI and MER/VAB when used separately (Table 1). Furthermore, we found that CAZ/AVI plus MER/VAB resistance can be conferred by production of wild-type KPC-3 in a ramR ompK36 double mutant carrying OXA-232 (Table 1). Clinical K. pneumoniae isolates with ompK35 and ompK36 mutation and elevated KPC-3 production that are CAZ/AVI resistant have recently been identified (28), which supports our finding that KPC-3 mutation is not essential.
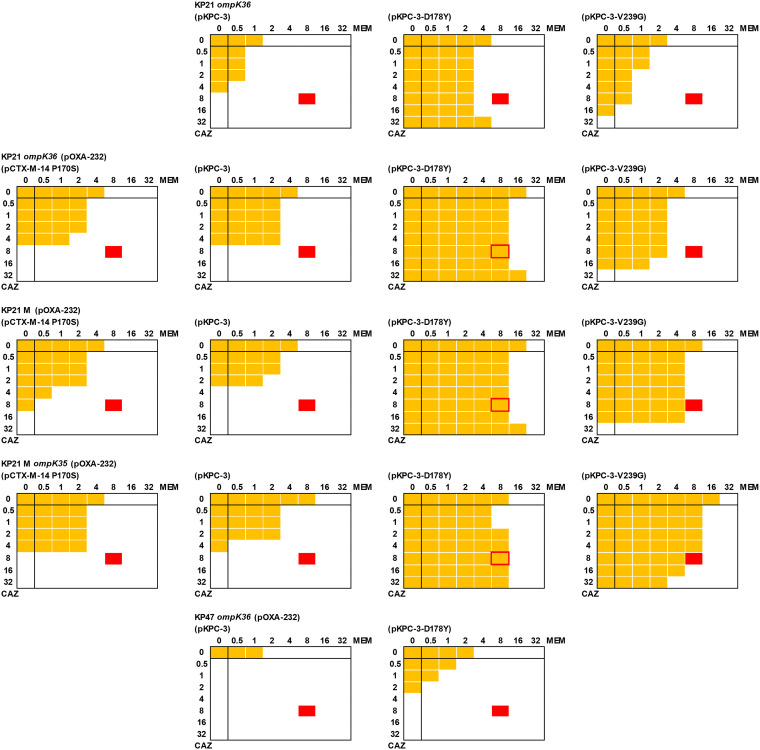
Finally, checkerboard assays were performed using an adapted microtiter MIC assay to test whether CAZ/AVI and MER/VAB would work synergistically against derivatives resistant to both pairs when used separately (Fig. 1). Briefly, a phosphate-buffered saline bacterial suspension was prepared to obtain a stock optical density at 600 nm (OD600) of 0.01. The final volume in each well of a 96-well cell culture plate (Corning Costar) was 200 μl and included 20 μl of the bacterial suspension. All wells contained cation-adjusted Muller-Hinton broth (CA-MHB) with avibactam (4 μg · ml−1) and vaborbactam (8 μg · ml−1) with serial dilutions of meropenem and ceftazidime. Bacterial growth (OD600) was determined after 20 h using a POLARstar Omega spectrophotometer (BMG Labtech). These assays confirmed that KP21 M(pOXA-232) and KP21 ompK36(pOXA-232) carrying pCTX-M14-P170S, pKPC-3, or pKPC-3-V239G were susceptible to meropenem in the presence of vaborbactam plus CAZ/AVI, suggesting that combined therapy would still work (Fig. 1). Further disruption of ompK35 did not alter this result. However, KP21 M(pOXA-232)(pKPC-3-D178Y) and KP21 ompK36(pOXA-232)(pKPC-3-D178Y) were resistant to meropenem (MIC, 16 μg · ml−1) and ceftazidime (MIC, ≥64 μg · ml−1) in the presence of vaborbactam plus avibactam.
FIG 1.
Checkerboard assays for ceftazidime and meropenem in the presence of avibactam and vaborbactam. Each image represents duplicate assays for an 8-by-8 array of wells in a 96-well plate. All wells contained CA-MHB, including avibactam (4 μg · ml−1) and vaborbactam (8 μg · ml−1). A serial dilution of meropenem (MEM; x axis) and ceftazidime (CAZ, y axis) was created from 32 μg · ml−1 in each plate as recorded. All wells were inoculated with a suspension of bacteria, made according to CLSI microtiter MIC guidelines (18), and the plate was incubated at 37°C for 20 h. Growth was recorded by measuring OD600, and growth above background (broth) is shown as a yellow block. Growth at 8 μg · ml−1 ceftazidime and 8 μg · ml−1 meropenem (indicated in red) in the presence of vaborbactam and avibactam defined resistance based on CLSI breakpoints (19). Bacterial suspensions used were as follows: top row, KP21[ramR] ompK36; second row, KP21[ramR] ompK36(pOXA-232); third row, KP21[ramR] M[ompK36](pOXA-232); fourth row, KP21[ramR] M[ompK36] ompK35(pOXA-232); and fifth row, KP47 ompK36(pOXA-232). In each case, bacteria also carried the following plasmids (where tested): first column, pCTX-M-14 P170S; second column, pKPC-3; third column, pKPC-3-D178Y; and fourth column, pKPC-3-V239G.
We have, therefore, generated K. pneumoniae derivatives resistant to CAZ/AVI and MER/VAB, when both pairs are used together. This was achieved in four steps relative to wild-type isolates: ramR frameshift mutation, acquisition of OXA-232, ompK36 frameshift mutation (or insertional inactivation), and acquisition of KPC-3-D178Y. When we constructed a derivative of clinical isolate KP47 (wild type for ramR [16]) carrying pOXA-232 and pKPC-3-D178Y and with ompK36 insertionally inactivated as described above, the derivative was resistant to CAZ/AVI and MER/VAB when used separately (Table 1) but not when used together (Fig. 1). When we constructed a derivative of KP21 ompK36 carrying pKPC-3-D178Y but not pOXA-232, it was also resistant to CAZ/AVI and MER/VAB when used separately (Table 1) but not when used together (Fig. 1). This confirms that ramR mutation and production of OXA-232 are both necessary for the dual-resistant phenotype. Because OXA-232 has a relatively low level of carbapenemase activity compared with the more commonly encountered OXA-48 (23), we are confident that this finding is representative of the wider family of OXA-48-like enzymes.
In conclusion, this work confirms the remarkable capacity of K. pneumoniae isolates to acquire resistance to the latest combination therapies available in the clinic by layering mechanisms. Of course, the order in which the four identified steps occur does not affect the result. So, it would be prudent to make every effort to identify, via molecular diagnostics, intermediate stages in this acquisition process. It would also be prudent to not rely on sequential use of CAZ/AVI and MER/VAB, which might select for the dual-resistant phenotype observed here.
ACKNOWLEDGMENTS
This work was funded by grant MR/S004769/1 to M.B.A. from the Antimicrobial Resistance Cross Council Initiative supported by the seven United Kingdom research councils and the National Institute for Health Research. Additional funding was provided by a bequest from the estate of the late Graham Ayliffe.
Genome sequencing was provided by MicrobesNG (http://www.microbesng.uk/), which is supported by the BBSRC (grant number BB/L024209/1).
We are grateful to Kate Heesom, School of Biochemistry, University of Bristol, for performing the proteomics analysis and to Jacqueline Findlay, School of Cellular & Molecular Medicine, University of Bristol, for purifying the plasmid pOXA-232.
We have no conflicts of interest to declare.
REFERENCES
- 1.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, Brisse S, Cao H, Wilksch J, Gorrie C, Schultz MB, Edwards DJ, Nguyen KV, Nguyen TV, Dao TT, Mensink M, Minh VL, Nhu NT, Schultsz C, Kuntaman K, Newton PN, Moore CE, Strugnell RA, Thomson NR. 2015. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A 112:E3574–E3581. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cantón R, González-Alba JM, Galán JC. 2012. CTX-M enzymes: origin and diffusion. Front Microbiol 3:110. doi: 10.3389/fmicb.2012.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-Venezia S, Carmeli Y, Brolund A, Giske CG. 2009. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother 53:3365–3370. doi: 10.1128/AAC.00126-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poirel L, Potron A, Nordmann P. 2012. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother 67:1597–1606. doi: 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]
- 5.Ehmann DE, Jahić H, Ross PL, Gu RF, Hu J, Kern G, Walkup GK, Fisher SL. 2012. Avibactam is a covalent, reversible, non-β-lactam β-lactamase inhibitor. Proc Natl Acad Sci U S A 109:11663–11668. doi: 10.1073/pnas.1205073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castón JJ, Lacort-Peralta I, Martín-Dávila P, Loeches B, Tabares S, Temkin L, Torre-Cisneros J, Paño-Pardo JR. 2017. Clinical efficacy of ceftazidime/avibactam versus other active agents for the treatment of bacteremia due to carbapenemase-producing Enterobacteriaceae in hematologic patients. Int J Infect Dis 59:118–123. doi: 10.1016/j.ijid.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 7.Wang DY, Abboud MI, Markoulides MS, Brem J, Schofield CJ. 2016. The road to avibactam: the first clinically useful non-β-lactam working somewhat like a β-lactam. Future Med Chem 8:1063–1084. doi: 10.4155/fmc-2016-0078. [DOI] [PubMed] [Google Scholar]
- 8.Shields RK, Chen L, Cheng S, Chavda KD, Press EG, Snyder A, Pandey R, Doi Y, Kreiswirth BN, Nguyen MH, Clancy CJ. 2017. Emergence of ceftazidime-avibactam resistance due to plasmid-borne blaKPC-3 mutations during treatment of carbapenem-resistant Klebsiella pneumoniae infections. Antimicrob Agents Chemother 61:e02097-16. doi: 10.1128/AAC.02097-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woodford N, Tierno PM Jr, Young K, Tysall L, Palepou MF, Ward E, Painter RE, Suber DF, Shungu D, Silver LL, Inglima K, Kornblum J, Livermore DM. 2004. Outbreak of Klebsiella pneumoniae producing a new carbapenem-hydrolyzing class A β-lactamase, KPC-3, in a New York medical center. Antimicrob Agents Chemother 48:4793–4799. doi: 10.1128/AAC.48.12.4793-4799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Both A, Büttner H, Huang J, Perbandt M, Belmar Campos C, Christner M, Maurer FP, Kluge S, König C, Aepfelbacher M, Wichmann D, Rohde H. 2017. Emergence of ceftazidime/avibactam non-susceptibility in an MDR Klebsiella pneumoniae isolate. J Antimicrob Chemother 72:2483–2488. doi: 10.1093/jac/dkx179. [DOI] [PubMed] [Google Scholar]
- 11.Hecker SJ, Reddy KR, Totrov M, Hirst GC, Lomovskaya O, Griffith DC, King P, Tsivkovski R, Sun D, Sabet M, Tarazi Z, Clifton MC, Atkins K, Raymond A, Potts KT, Abendroth J, Boyer SH, Loutit JS, Morgan EE, Durso S, Dudley MN. 2015. Discovery of a cyclic boronic acid β-lactamase inhibitor (RPX7009) with utility vs class A serine carbapenemases. J Med Chem 58:3682–3692. doi: 10.1021/acs.jmedchem.5b00127. [DOI] [PubMed] [Google Scholar]
- 12.Lomovskaya O, Sun D, Rubio-Aparicio D, Nelson K, Tsivkovski R, Griffith DC, Dudley MN. 2017. Vaborbactam: spectrum of β-lactamase inhibition and impact of resistance mechanisms on activity in Enterobacteriaceae. Antimicrob Agents Chemother 61:e01443-17. doi: 10.1128/AAC.01443-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun D, Rubio-Aparicio D, Nelson K, Dudley MN, Lomovskaya O. 2017. Meropenem-vaborbactam resistance selection, resistance prevention, and molecular mechanisms in mutants of KPC-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 61:e01694-17. doi: 10.1128/AAC.01694-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pogue JM, Bonomo RA, Kaye KS. 2019. Ceftazidime/avibactam, meropenem/vaborbactam, or both? Clinical and formulary considerations. Clin Infect Dis 68:519–524. doi: 10.1093/cid/ciy576. [DOI] [PubMed] [Google Scholar]
- 15.Athans V, Neuner EA, Hassouna H, Richter SS, Keller G, Castanheira M, Brizendine KD, Mathers AJ. 2018. Meropenem-vaborbactam as salvage therapy for ceftazidime-avibactam-resistant Klebsiella pneumoniae bacteremia and abscess in a liver transplant recipient. Antimicrob Agents Chemother 63:e01551-18. doi: 10.1128/AAC.01551-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wan Nur Ismah WAK, Takebayashi Y, Findlay J, Heesom KJ, Jiménez-Castellanos JC, Zhang J, Graham L, Bowker K, Williams OM, MacGowan AP, Avison MB. 2017. Prediction of fluoroquinolone susceptibility directly from whole-genome sequence data by using liquid chromatography-tandem mass spectrometry to identify mutant genotypes. Antimicrob Agents Chemother 62:e01814-17. doi: 10.1128/AAC.01814-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiménez-Castellanos JC, Wan Nur Ismah WAK, Takebayashi Y, Findlay J, Schneiders T, Heesom KJ, Avison MB. 2018. Envelope proteome changes driven by RamA overproduction in Klebsiella pneumoniae that enhance acquired β-lactam resistance. J Antimicrob Chemother 73:88–94. doi: 10.1093/jac/dkx345. [DOI] [PubMed] [Google Scholar]
- 18.Clinical and Laboratory Standards Institute. 2015. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard—10th ed. CLSI document M07-A10. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 19.Clinical and Laboratory Standards Institute. 2019. Performance standards for antimicrobial susceptibility testing; 29th informational supplement. CLSI document M100-S29. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 20.Dulyayangkul P, Wan Nur Ismah WAK, Douglas EJA, Avison MB. 2020. Mutation of kvrA causes OmpK35 and OmpK36 porin downregulation and reduced meropenem-vaborbactam susceptibility in KPC-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 64:e02208-19. doi: 10.1128/AAC.02208-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexeyev MF. 1999. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of Gram-negative bacteria. Biotechniques 26:824–828. doi: 10.2144/99265bm05. [DOI] [PubMed] [Google Scholar]
- 22.George AM, Hall RM, Stokes HW. 1995. Multidrug resistance in Klebsiella pneumoniae: a novel gene, ramA, confers a multidrug resistance phenotype in Escherichia coli. Microbiology 141:1909–1920. doi: 10.1099/13500872-141-8-1909. [DOI] [PubMed] [Google Scholar]
- 23.Potron A, Rondinaud E, Poirel L, Belmonte O, Boyer S, Camiade S, Nordmann P. 2013. Genetic and biochemical characterisation of OXA-232, a carbapenem-hydrolysing class D β-lactamase from Enterobacteriaceae. Int J Antimicrob Agents 41:325–329. doi: 10.1016/j.ijantimicag.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 24.Findlay J, Gould VC, North P, Bowker KE, Williams MO, MacGowan AP, Avison MB. 2020. Characterization of cefotaxime-resistant urinary Escherichia coli from primary care in south-west England 2017–18. J Antimicrob Chemother 75:65–71. doi: 10.1093/jac/dkz397. [DOI] [PubMed] [Google Scholar]
- 25.Takebayashi Y, Wan Nur Ismah WHK, Findlay J, Heesom KJ, Zhang J, Williams OM, MacGowan AP, Avison MB. 2017. Prediction of cephalosporin and carbapenem susceptibility in multi-drug resistant Gram-negative bacteria using liquid chromatography-tandem mass spectrometry. BioRxiv:138594. doi: 10.1101/138594. [DOI]
- 26.Ma L, Ishii Y, Chang FY, Yamaguchi K, Ho M, Siu LK. 2002. CTX-M-14, a plasmid-mediated CTX-M type extended-spectrum β-lactamase isolated from Escherichia coli. Antimicrob Agents Chemother 46:1985–1988. doi: 10.1128/AAC.46.6.1985-1988.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poirel L, Naas T, Le Thomas I, Karim A, Bingen E, Nordmann P. 2001. CTX-M-type extended-spectrum β-lactamase that hydrolyzes ceftazidime through a single amino acid substitution in the omega loop. Antimicrob Agents Chemother 45:3355–3361. doi: 10.1128/AAC.45.12.3355-3361.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coppi M, Di Pilato V, Monaco F, Giani T, Conaldi PG, Rossolini GM. 2020. Ceftazidime-avibactam resistance associated with increased blaKPC-3 gene copy number mediated by pKpQIL plasmid derivatives in ST258 Klebsiella pneumoniae. Antimicrob Agents Chemother 64:e01816-19. doi: 10.1128/AAC.01816-19. [DOI] [PMC free article] [PubMed] [Google Scholar]