Lactoferrin (LF) is an iron-binding glycoprotein with broad-spectrum antimicrobial activity. Previously, we discovered that LF synergistically enhanced the antifungal efficacy of amphotericin B (AMB) across a variety of yeast species and subsequently hypothesized that this synergy was enhanced by the presence of small peptides derived from the whole LF molecule. In this study, LF was digested with pepsin under a range of conditions. The resulting hydrolysates exhibited enhanced synergy with AMB compared to its synergy with undigested LF.
KEYWORDS: Candida, Cryptococcus, amphotericin, drug synergy, lactoferrin
ABSTRACT
Lactoferrin (LF) is an iron-binding glycoprotein with broad-spectrum antimicrobial activity. Previously, we discovered that LF synergistically enhanced the antifungal efficacy of amphotericin B (AMB) across a variety of yeast species and subsequently hypothesized that this synergy was enhanced by the presence of small peptides derived from the whole LF molecule. In this study, LF was digested with pepsin under a range of conditions. The resulting hydrolysates exhibited enhanced synergy with AMB compared to its synergy with undigested LF. Samples were analyzed using matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry, and 14 peptides were identified. The sequences of these peptides were predicted by matching their molecular weights to those of a virtual digest with pepsin. The relative intensities of predicted peptides in each hydrolysate were compared with the activity of the hydrolysate, and the structural and physicochemical properties of the peptides were assessed. From this, a 30-residue peptide was selected for synthesis and dubbed lactofungin (LFG). Pure LFG was highly synergistic with AMB, outperforming native LF in all fungal species tested. With potential for further structural and chemical improvements, LFG is an excellent lead for development as an antifungal adjuvant.
INTRODUCTION
There is an urgent need for novel antifungal therapies, due to the increasing incidence of invasive fungal infections worldwide and resistance of some fungal species to current therapies (1). Candida and Cryptococcus species are among the main causative agents of opportunistic mycoses and are associated with high mortality and morbidity rates (2). Amphotericin B (AMB), used for treatment of serious fungal infections, has high toxicity, and its administration requires hospitalization and monitoring of severe side effects that include chills, fever, headache, and nausea (3, 4). Combination therapy using synergents that potentiate the effect of AMB is a strategy that enables decreased drug dosages, thereby diminishing toxicity-related side effects, while maintaining efficacy equal to or greater than that of AMB alone.
5-Fluorocytosine (5FC) is used clinically to enhance the activity of AMB, with this combination generally leading to improved therapeutic outcome (5, 6). AMB plus 5FC (AMB+5FC) is the preferred induction therapy for HIV-associated cryptococcal meningitis, showing a higher cure rate and fewer relapses (7), and is used for azole-resistant Candida glabrata and for forms of invasive candidiasis that are difficult to treat, including infections of the central nervous system (CNS), endophthalmitis, and endocarditis (8). However, 5FC can be very expensive and is not available in many regions that need it the most, including most sub-Saharan African countries (9). Therefore, other antifungal agents capable of synergizing with AMB to reduce the necessary therapeutic dose are desirable.
Previously, we found the multifunctional milk protein lactoferrin (LF) to synergistically enhance the antifungal efficacy of AMB across a variety of yeast species. The activity of LF alone was primarily attributed to its ability to chelate iron, rendering it inaccessible to microbes. However, the synergistic activity was unaffected by iron chelation and is therefore likely due to smaller peptides derived from the large LF molecule (10). Structurally, LF is a globular glycoprotein that can range in size from 77 to 87 kDa and that contains two lobes, the N lobe and the C lobe, each containing two equal domains (11). Proteolytic digestion of LF produces a variety of diverse peptides from each of these domains. Many of these have been shown to have antimicrobial activity surpassing that of whole LF, in addition to immunomodulatory and other functions (12). Given the abundance of proteolytic enzymes in the gut, these peptides most likely contribute to the antimicrobial action of LF in the body (13).
Several studies have reported on the antimicrobial effects of LF hydrolysate produced by enzymatic digestion; however, these have primarily focused on its antibacterial properties (14–16). Antifungal activity of many purified or synthesized LF-derived peptides has also been demonstrated, but few of the peptides have been tested in combination with other antifungals, and their synergistic potential remains largely unexplored (17–19). In this study, we used both enzymatic and chemical proteolysis methods, under a range of different conditions, to generate LF digests and compared the activity of these with that of whole LF, both alone and in combination with AMB in Candida and Cryptococcus. MALDI-TOF mass spectrometry identified 14 predicted peptides among the digest samples, and based on a comparison of their putative properties, we synthesized and tested the most promising of these, herein termed lactofungin (LFG). The synergistic activity of LFG was compared to that of native, full-length LF, as well as those of the LF hydrolysates.
RESULTS
Simple digestion of LF can produce hydrolysates with improved synergism with AMB.
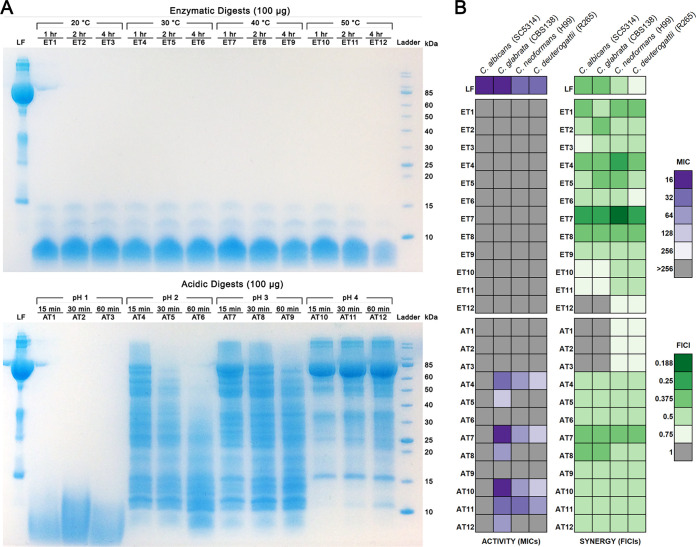
LF was digested using two methods: enzymatic hydrolysis, to simulate digestion that would occur in the human gut, and acidic hydrolysis. Enzymatic hydrolysis used pepsin and was carried out at a range of temperatures from 20 to 50°C to compare their degrees of efficacy. Acidic hydrolysis was carried out by adjusting pH to between 1 and 4 and heating samples to 100°C. In both cases, samples were taken at 3 different time points for each condition tested and the composition of the resulting digests was examined by SDS-PAGE (Fig. 1A). In the enzymatic digests, all remaining peptidic bands were less than 15 kDa in molecular mass, with the majority of the products being less than 10 kDa in molecular mass. The acidic digests were more variable. At pH 1, products in the hydrolysate were smaller than 15 kDa, while at pH 2 to 4, samples contained products ranging from larger than 85 kDa to smaller than 10 kDa. Not surprisingly, samples from each set of conditions became more digested and, thus, they possessed smaller products with increased digestion time.
FIG 1.
Enzymatic and acidic digests of LF and their antifungal activities alone and in combination with AMB. (A) Protein profiles of enzymatic (top) and acidic (bottom) digests of LF separated on SDS-PAGE gels. (B) Heatmap showing the MICs for each LF digest (left) and the FICIs for each digest combined with AMB based on abbreviated checkerboards (right).
To determine the activity of these digests compared to that of native LF, the antifungal activity of each sample was tested against two Candida and two Cryptococcus species: Candida albicans strain SC5314, Candida glabrata strain CBS138, Cryptococcus neoformans strain H99, and Cryptococcus deuterogattii strain R265. To compare their levels of synergistic activity, each digest was also tested in combination with AMB using an abbreviated diagonal-sampling checkerboard methodology. The resulting MICs and fractional inhibitory concentration indexes (FICIs) are shown in Fig. 1B, with full data for all combinations in Data Set S1 in the supplemental material. LF had an MIC of 16 for the Candida species and 32 for the Cryptococcus species; the interaction of LF with AMB was synergistic in both Candida species (FICI = 0.375) and C. neoformans (FICI = 0.5) but indifferent in C. deuterogattii (FICI = 0.75). The enzymatic digests had no activity on their own, with no MIC obtained in any species (>256 μg/ml). However, the majority of the enzymatic hydrolysates were either equally or significantly more synergistic with AMB than LF was, with FICIs ranging from 0.25 to 1 in Candida and 0.188 to 0.75 in Cryptococcus. For each condition, FICI values were best in digests taken at the earliest time point, indicating that the peptide(s) responsible for the synergistic activity are generated early and can become digested further over longer time periods. Some early-time-point acidic digests displayed activity on their own (MICs of 16 to 256 μg/ml); however, this never surpassed that of the full-length LF protein. Samples from digests at pH 2 and higher were synergistic with AMB in every strain, including C. deuterogattii, with FICIs ranging from 0.5 to 0.375, but never surpassed the activity of LF for Candida. Overall, the best digest was ET7, the sample digested enzymatically at 40°C for 1 h, which reduced the amount of AMB required by 2 to 8 times across all species compared to the results for native LF. The best acidic digest was AT7, the sample digested at pH 3.0 for 15 min, which reduced the amount of AMB required for Cryptococcus species by 2 to 4 times compared to the results for native LF.
MALDI-TOF mass spectrometry analysis predicts 14 peptides present in significant amounts in enzymatic digest samples.
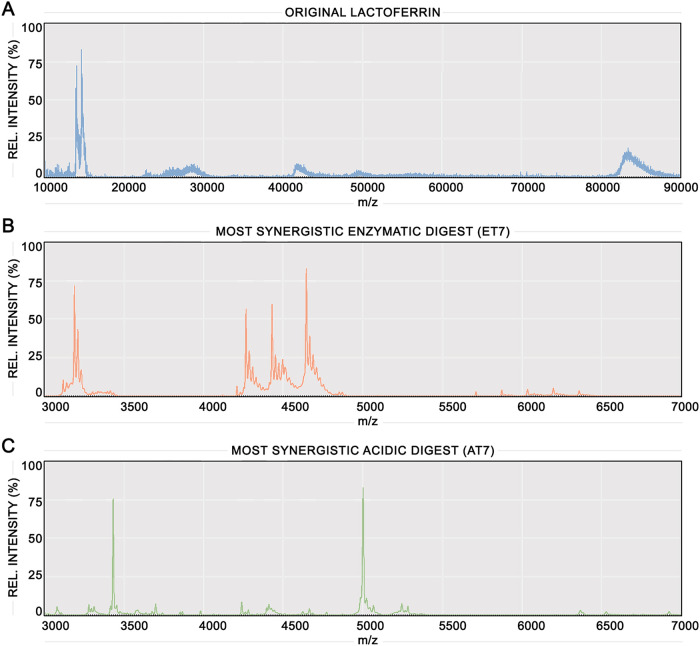
Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry was undertaken to characterize the composition of LF digests in depth. Figure 2A shows the mass spectrum of LF, with a major ion at ∼85 kDa corresponding to the whole LF protein. The ion at ∼42 kDa represents an isotope of whole LF, and minor peptides are present between 50 and 60 kDa, 20 and 30 kDa, and around ∼15 kDa. Figure 2B and C show the mass spectra of the most synergistic enzymatic digest (ET7; 40°C, 1 h) and the most synergistic acidic digest (AT7; pH 3.0, 15 min), with spectra for all digests in Data Set S2 in the supplemental material. A range of 3 to 7 kDa is shown for both spectra, as size exclusion filtration of the digests indicated that the active fractions were <10 kDa and no other peptides <10 kDa were present outside this range. ET7 and AT7 contain entirely different mass spectra, sharing no common peptides. ET7 has several peptides with high relative intensities that are concentrated between 3 and 3.5 and 4 and 5 kDa, and several more with lower relative intensities between 5.5 and 6.5 kDa. AT7 has two peaks with high relative intensities at ∼3.4 and ∼5 kDa and numerous peaks with lower relative intensities primarily between 3 and 5.5 kDa.
FIG 2.
Mass spectra of undigested LF and the most synergistic enzymatic and acidic digests. MALDI-TOF mass spectrum comparison of undigested LF control (A), the most synergistic enzymatic digest (ET7) (B), and the most synergistic acidic digest (AT7) (C).
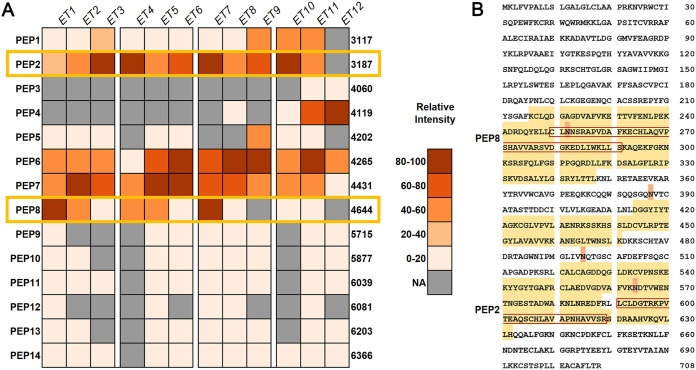
Deeper analysis was performed on the mass spectra for the enzymatic digest samples, as these exhibited the best synergism overall. Ions present at very low levels and ions only detected in a single sample were excluded, leaving 14 predicted peptides. These peptides were then identified by matching them by mass to peptides generated through a virtual pepsin digest of LF. A heatmap showing the presence and relative intensity of each peptide in each digest sample is shown in Fig. 3A. Based on the heatmap, two peptides, PEP2 and PEP8, were proposed to be responsible for the strongest synergistic antifungal activity, as their relative intensities aligned most closely with the pattern of synergy, generally being the best in the first time point and decreasing over time.
FIG 3.
Peptides predicted as most likely to be responsible for the synergistic interaction with AMB. (A) Heatmap showing the relative intensity of each major peptide identified in each enzymatic digest sample based on MALDI-TOF mass spectrometry data. Numbers to the right are the molecular weights of the predicted peptides. Yellow boxes mark the two peptides that were predicted to be most likely responsible for synergistic interactions based on their relative intensities and the synergy values of each digest. (B) Sequence coverage of predicted peptides shown against the entire lactoferrin sequence. Residues highlighted in yellow are present in at least one predicted peptide. Residues highlighted in orange are potential sites of glycosylation. Residues boxed in red are PEP2 and PEP8 as identified in panel A.
To narrow down the selection to a single best candidate, the structural and putative physicochemical properties of the predicted peptides were compared. Figure 3B shows the sequence of LF, with residues present in at least one peptide highlighted in yellow and known glycosylation sites highlighted in orange. The position within LF, amino acid sequence, size, weight, and putative physicochemical properties of all 14 peptides are shown in Table 1. Peptides ranged in size from 29 to 57 residues and 3,117 to 6,366 Da and comprised 300 of the 708 (42.4%) residues of the whole LF sequence. Antimicrobial peptides are often positively charged, and PEP2, the smaller of the two peptides, had a positive net charge of +1 while PEP8 had a neutral net charge of 0 (Table 1). Comparing their sequence locations within LF, outlined in red in Fig. 3B, there was a site of possible glycosylation within the sequence of PEP8 but not within PEP2. Their other putative properties were similar, with PEP2 and PEP8 being the only peptides with positive grand average of hydropathicity (GRAVY) values (0.14 and 0.07, respectively), indicating that they are more hydrophobic, while the other peptides are more hydrophilic, and they had Boman index values of 1.25 and 1.24, respectively, indicating intermediate protein binding potential. Based on this comparison, PEP2 was chosen as the best candidate for synthesis and further assessment and was named “lactofungin” (LFG).
TABLE 1.
Predicted peptides identified via mass spectrometry in enzymatic digests of LF and their putative physicochemical propertiesa
| Name | Position | Amino acid sequence | No. of amino acids | Mol wt | Net charge | GRAVYb | Boman indexc |
|---|---|---|---|---|---|---|---|
| PEP1 | 521–549 | CALCAGDDQGLDKCVPNSKEKYYGYTGAF | 29 | 3,117.47 | −1 | −0.48 | 1.32 |
| PEP2 | 590–619 | LLCLDGTRKPVTEAQSCHLAVAPNHAVVSR | 30 | 3,186.7 | +1 | 0.14 | 1.25 |
| PEP3 | 216–251 | KCLQDGAGDVAFVKETTVFENLPEKADRDQYELLCL | 36 | 4,059.58 | −4 | −0.34 | 1.78 |
| PEP4 | 263–299 | ECHLAQVPSHAVVARSVDGKEDLIWKLLSKAQEKFGK | 37 | 4,118.77 | +1 | −0.31 | 1.47 |
| PEP5 | 252–289 | NNSRAPVDAFKECHLAQVPSHAVVARSVDGKEDLIWKL | 38 | 4,201.77 | 0 | −0.28 | 1.79 |
| PEP6 | 661–699 | NDNTECLAKLGGRPTYEEYLGTEYVTAIANLKKCSTSPL | 39 | 4,264.79 | −1 | −0.5 | 1.61 |
| PEP7 | 550–588 | RCLAEDVGDVAFVKNDTVWENTNGESTADWAKNLNREDF | 39 | 4,430.74 | −5 | −0.81 | 2.79 |
| PEP8 | 249–290 | LCLNNSRAPVDAFKECHLAQVPSHAVVARSVDGKEDLIWKLL | 42 | 4,644.39 | 0 | 0.07 | 1.24 |
| PEP9 | 590–642 | LLCLDGTRKPVTEAQSCHLAVAPNHAVVSRSDRAAHVKQVLLHQQALFGKNGK | 53 | 5,714.61 | +4 | −0.18 | 1.52 |
| PEP10 | 238–289 | PEKADRDQYELLCLNNSRAPVDAFKECHLAQVPSHAVVARSVDGKEDLIWKL | 52 | 5,876.66 | −2 | −0.46 | 2.06 |
| PEP11 | 252–305 | NNSRAPVDAFKECHLAQVPSHAVVARSVDGKEDLIWKLLSKAQEKFGKNKSRSF | 54 | 6,038.88 | +4 | −0.61 | 2.23 |
| PEP12 | 414–470 | DGGYIYTAGKCGLVPVLAENRKSSKHSSLDCVLRPTEGYLAVAVVKKANEGLTWNSL | 57 | 6,080.97 | +2 | −0.1 | 1.08 |
| PEP13 | 653–708 | SETKNLLFNDNTECLAKLGGRPTYEEYLGTEYVTAIANLKKCSTSPLLEACAFLTR | 56 | 6,203.06 | −1 | −0.24 | 1.46 |
| PEP14 | 291–347 | SKAQEKFGKNKSRSFQLFGSPPGQRDLLFKDSALGFLRIPSKVDSALYLGSRYLTTL | 57 | 6,366.33 | +6 | −0.43 | 1.81 |
Molecular weight, net charge, and GRAVY were determined by using the ProtParam tool. Boman index was determined by using ADP3. Potentially synergistic peptides PEP2 and PEP8 are shown in bold.
Grand average of hydropathy (GRAVY) is a prediction of the average hydrophobicity of a peptide. Positive numbers indicate the peptide is more hydrophobic, while negative numbers indicate the peptide is more hydrophilic.
The Boman index is a prediction of the potential for a protein to bind to other proteins. Higher positive values indicate the ability to interact with a wide range of proteins.
Novel 30-residue peptide LFG is more synergistic with AMB than is whole LF.
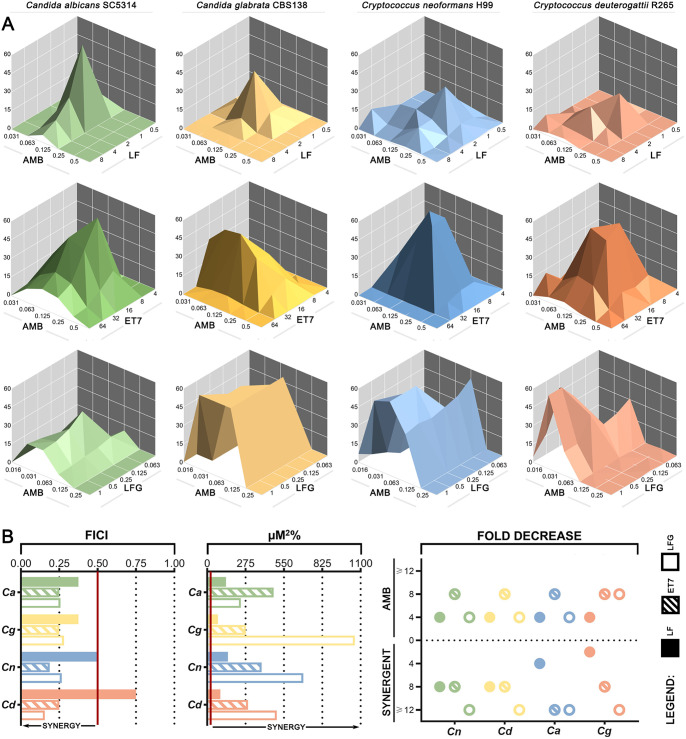
Initial testing of LFG found it to be inactive alone but with synergism similar to that of ET7. Full checkerboard assays were then conducted to compare the synergistic activities of undigested LF, the enzymatic digest ET7, and the synthesized peptide LFG with AMB. Figure 4A shows dose-response surfaces generated in MacSynery II in which significant synergy volumes are represented as peaks above the flat plane. LF+AMB plots have smaller and more irregular synergy peaks, while ET7+AMB synergy peaks are higher and cover a larger area, indicating stronger synergy. LFG+AMB plots show large raised areas that extend beyond the edges of the plot, indicating synergistic activity over a wide range of concentrations. Figure 4B summarizes the synergy values obtained using two models of synergy: FICI (Fig. 4B, left) which is based on Loewe additivity and assumes that both agents have the same mechanism of action, and μM2% (Fig. 4B, middle), calculated using MacSynergy II, which is based on Bliss independence and does not have this assumption. The reduction in the requirement for each agent expressed as fold decrease is also shown (Fig. 4B, right). Full synergy data are listed in Table 2.
FIG 4.
Comparison of LF, ET7, and LFG in their synergy with AMB. (A) Three-dimensional dose-response surfaces generated in MacSynergy II from full checkerboard data for the combination of LF+AMB (top), ET7+AMB (middle), or LFG+AMB (bottom). Significant synergy volumes, represented as peaks above the flat plane, are shown for C. albicans SC5314, C. glabrata CBS138, C. neoformans H99, and C. deuterogattii R265. (B) Summary of the interactions between LF and AMB, ET7 and AMB, and LFG and AMB in the same 4 species based on full checkerboard assays. Left, FICIs, based on the Loewe additivity model, with synergy to the left of the red cutoff line; middle, uM2%, based on the Bliss independence model and calculated by MacSynergy II, with synergy to the right of the red cutoff line; right, fold decrease in concentration of each agent required to inhibit growth when used in combination compared to when used alone.
TABLE 2.
Synergy data for undigested LF, the best enzymatic digest (ET7), and synthesized peptide LFG in combination with AMB
| Peptidic agent, species (molecular type) | Strain | Value fora: |
FICI | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Amphotericin B |
Peptidic agent |
|||||||||
| MICX | MICY | FICA | Fold Δ | MICX | MICY | FICB | Fold Δ | |||
| Undigested lactoferrin (LF) control | ||||||||||
| Candida albicans | SC5314 | 0.25 | 0.063 | 0.25 | 4 | 16 | 2 | 0.125 | 8 | 0.375 |
| Candida glabrata | CBS138 | 0.25 | 0.063 | 0.25 | 4 | 8 | 1 | 0.125 | 8 | 0.375 |
| Cryptococcus neoformans (VNI) | H99/WM148 | 0.25 | 0.063 | 0.25 | 4 | 16 | 4 | 0.25 | 4 | 0.5 |
| Cryptococcus deuterogattii (VGII) | R265 | 0.5 | 0.125 | 0.25 | 4 | 16 | 8 | 0.5 | 2 | 0.75 |
| Enzymatic treatment 7 (ET7) | ||||||||||
| Candida albicans | SC5314 | 0.25 | 0.031 | 0.125 | 8 | 256 | 32 | 0.125 | 8 | 0.25 |
| Candida glabrata | CBS138 | 0.25 | 0.031 | 0.125 | 8 | 256 | 32 | 0.125 | 8 | 0.25 |
| Cryptococcus neoformans (VNI) | H99/WM148 | 0.25 | 0.031 | 0.125 | 8 | 256 | 16 | 0.063 | 16 | 0.188 |
| Cryptococcus deuterogattii (VGII) | R265 | 0.5 | 0.063 | 0.125 | 8 | 256 | 32 | 0.125 | 8 | 0.25 |
| Lactofungin (LFG) | ||||||||||
| Candida albicans | SC5314 | 0.25 | 0.063 | 0.25 | 4 | >256 | 0.5 | 0.008 | 512 | 0.258 |
| Candida glabrata | CBS138 | 0.25 | 0.063 | 0.25 | 4 | >256 | 2 | 0.031 | 128 | 0.281 |
| Cryptococcus neoformans (VNI) | H99/WM148 | 0.25 | 0.063 | 0.25 | 4 | >256 | 1 | 0.016 | 256 | 0.266 |
| Cryptococcus deuterogattii (VGII) | R265 | 0.5 | 0.063 | 0.125 | 8 | >256 | 2 | 0.031 | 128 | 0.156 |
MICX is the MIC of the agent alone, MICY is the MIC of the agent in combination, each FIC is calculated as [MICX/MICY], and FICI is the sum of FICA and FICB. Where the MIC was >256 μg/ml, the value of 256 was used as the MIC for the purposes of FICI calculations.
In all instances, ET7 was again superior to the full-length LF protein, with FICIs from 0.188 to 0.25 compared to 0.375 to 0.5, μM2% synergy volumes from 278.09 to 475.51 compared to 77.46 to 148.10, and an 8-fold reduction in the requirement for AMB compared to a 4-fold reduction. Comparing LFG to the crude hydrolysate ET7, both were similarly potent according to the Loewe additivity model, with the LFG FICIs ranging from 0.156 to 0.28. While ET7 resulted in an 8-fold decrease in AMB for every strain, LFG matched this only for C. deuterogattii and resulted in a 4-fold decrease for both Candida species and C. neoformans. However, a much smaller amount of LFG (0.5 to 2 μg/ml) was needed to achieve this result compared to the required amount of ET7 (16 to 32 μg/ml). According to the Bliss independence model, LFG produced markedly larger synergy volumes in C. glabrata, C. neoformans, and C. deuterogattii, with μM2% ranging from 499.13 to 1,058.98 compared to 278.09 to 389.41, as it was effective over a larger range of concentrations. Although still more effective than LF against C. albicans, LFG had a smaller synergy volume (241.95) than ET7 (475.51), indicating that there may be one or more other peptides in ET7 that are more effective against C. albicans or that LFG interacts or synergizes with other digestion products in ET7 to exert its full anti-C. albicans activity.
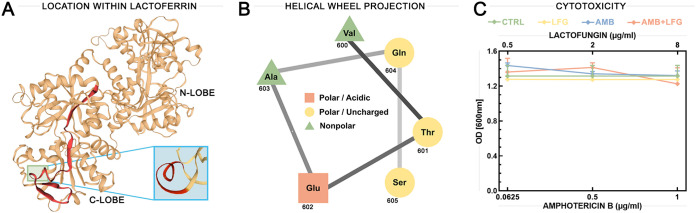
Figure 5A shows the location of LFG within the C lobe of the whole LF molecule and indicates the 310-helical region of LFG. A helical wheel projection (Fig. 5B) of the 310-helical section (VTEAQS) predicted that it is amphipathic, presenting distinct polar and nonpolar faces. The sequence of LFG was used for a BLAST query within the Collection of Anti-Microbial Peptides (CAMP) database for peptides with experimentally validated antimicrobial activity. Results are shown in Table 3, including alignment of LFG with the conserved areas of each peptide. A total of 14 matches from 9 organisms were returned, including birds, arthropods, mammals, and amphibians. These included peptides from the brevinin, escluentin, tachystatin, and defensin families. Two defensin peptides, gallinacin-2 from chickens and ostricacin-1 from ostriches, were recorded as inactive against C. albicans, while five peptides, brevinin-2-OA6, brevinin 2-OA8, and escluentin-1-OR3 from frogs (MICs of 4 to 11.2 μg/ml), microplusin from cattle ticks, and tachystatin-C (50% inhibitory concentration [IC50] = 0.9 μg/ml) from Japanese horseshoe crabs, were determined to be active against C. albicans.
FIG 5.
The predicted structure and cytotoxicity profile of LFG. (A) Overall structure of LF showing the position of LFG (red) within the C lobe. Green box indicates the location of the 310-helical region of LFG. Blue box shows this region magnified and from another angle, with the 310-helical residues highlighted in red. (B) Helical wheel projection of the 310-helical section of LFG, generated using the NetWheels online tool. (C) Cytotoxicity profiles of LFG alone and in combination with AMB against A549 cells, measured by MTT reduction assay. A549 cells were treated with 10% water as a vehicle control (CTRL) or with different concentrations of LFG (0.5, 2, and 8 μg/ml), AMB (0.0625, 0.5, and 1 μg/ml), or AMB+LFG combined for 24 h. Values shown are averages from six technical replicates for each treatment. OD, optical density. Points and error bars show the mean values ± 95% confidence intervals.
TABLE 3.
Peptides with experimentally validated antimicrobial activity that share structural similarity with LFGa
| Designation(s) | Alignmentb | Source | Taxonomy | Activity(ies) | Length | CAMP no. | Reference(s) |
|---|---|---|---|---|---|---|---|
| Lactofungin (LFG) | 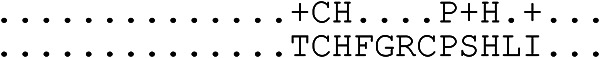 |
||||||
| Brevinin-2-OA6/brevinin-2-OA8 | 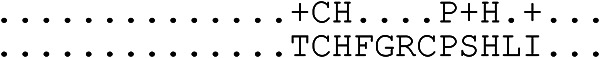 |
Odorrana andersonii (golden crossband frog) | Animalia, Amphibia | Antibacterial, antifungal | 33/33 | CAMPSQ3546/CAMPSQ3548 | 35 |
| Esculentin-1-OR3 | 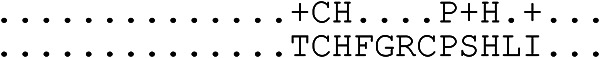 |
Odorrana rotodora | Animalia, Amphibia | Antibacterial, antifungal | 46 | CAMPSQ3570 | 35 |
| Tachystatin-C | 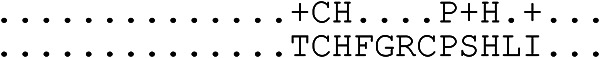 |
Tachypleus tridentatus (Japanese horseshoe crab) | Animalia | Antibacterial, antifungal | 41 | CAMPSQ931 | 36 |
| Beta-defensin Apl_AvBD-2 | 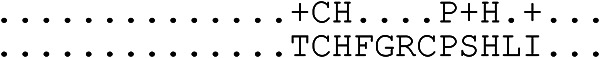 |
Anas platyrhynchos (domestic duck) | Animalia, Aves | Antimicrobial | 64 | CAMPSQ2742 | 60 |
| Beta-defensin 38 | 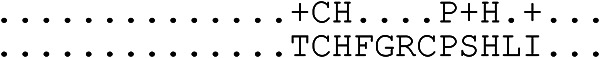 |
Mus musculus (mouse) | Animalia, Mammalia | Antibacterial | 42 | CAMPSQ592 | 61 |
| Chicken AvBD2/gallinacin 2 | 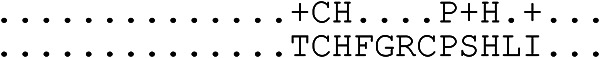 |
Gallus gallus (chicken) | Animalia, Aves | Antibacterial | 34/36 | CAMPSQ4340/CAMPSQ516 | 62, 63 |
| Microplusin | 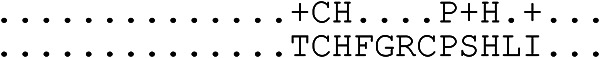 |
Rhipicephalus microplus (cattle tick) | Animalia, Arachnida | Antibacterial | 89/90 | CAMPSQ2454/CAMPSQ325 | 34 |
| Ostricacin-1 | 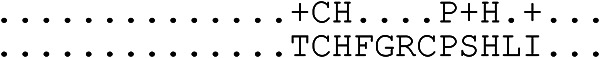 |
Struthio camelus (ostrich) | Animalia, Aves | Antibacterial | 36 | CAMPSQ973 | 64 |
| THP2 precursor/THP2/THP3 | 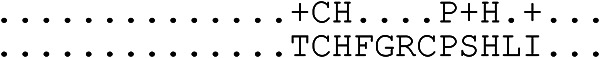 |
Meleagris gallopavo (common turkey) | Animalia, Aves | Antibacterial | 20/25/36 | CAMPSQ543/CAMPSQ61/CAMPSQ2754 | 65 |
Peptides identified from a BLAST query performed using the Collection of Anti-Microbial Peptides (CAMP) database.
Comparison of LFG (top sequence) and similar region of specified antimicrobial peptide.
Figure 5C shows the cytotoxicity profiles of LFG alone and in combination with AMB against A549 lung cancer cells. Treatment with up to 8 μg/ml of LFG alone resulted in no significant decrease in cell viability compared to the results for the vehicle control. Synergistic treatment with up to 1 μg/ml AMB and 8 μg/ml LFG resulted in no significant decrease in cell viability compared to the results for either LFG or AMB treatment alone or the vehicle control. These results indicate that LFG is not toxic to human cells at concentrations as high as 4 to 16 times the synergistic dose.
DISCUSSION
Peptides that contribute to the functionality of LF are good targets for AMP discovery.
As a natural product, LF has multiple biological functions, including antimicrobial, antitumor, antioxidant, and immunomodulatory activities (20, 21). When ingested, LF is digested in the stomach by digestive proteases, resulting in the production of peptides with their own suites of functions (22, 23). Therefore, it is likely that these peptides are essential for LF to exert its full range of antifungal action. Our previous research into the spectrum of activity of whole LF demonstrated that, while the primary mechanism of antifungal action was iron chelation, synergism with AMB was iron independent, with small peptides likely playing a major role (10). The current study therefore aimed to simulate in vivo digestion of lactoferrin, compare the antifungal activity of the resulting hydrolysate with that of the full-length LF protein, and identify potential antifungal peptides present in the hydrolysate that may be responsible for its synergistic interaction with AMB. In doing this, we predicted, synthesized, and tested a small novel LF-derived peptide that synergizes strongly with AMB in Cryptococcus and Candida, which we have called lactofungin (LFG).
The three other well-studied antimicrobial LF peptides, lactoferricin (LFcin), lactoferrampin (LFampin), and LF(1–11), were not detected in the enzymatic digests. LFampin (2,048 kDa) and LF(1–11) (1,334 kDa) are smaller than the smallest peptide detected from pepsin cleavage (3,117 kDa) (Table 1), and while LFcin is within the mass range (3,125 kDa) of peptides predicted, it has fewer residues (n = 25) than the smallest peptide detected (n = 29) (24). This indicates that LF may not have been digested for long enough for these smaller peptides to be generated, or they may have only been generated in very small amounts that were below the detection threshold. LFcin, LFampin, and LF(1–11) have all been shown to have antifungal activity surpassing that of LF (12), while none of the enzymatic digests had detectable antifungal activity on their own, which further confirms their absence.
LFG has structural features similar to those of other LF-derived peptides and sequence motifs in common with other AMPs.
AMPs can be broadly categorized into membrane-acting and non-membrane-acting peptides (25). LFcin, LFampin, and LF(1–11) all fit into the first category, being highly cationic peptides that bind to and disrupt the cell membrane (24). LFG is distinct from LFcin, LFampin, and LF(1–11) in that it is derived from the C lobe of LF, while the others all originate from the N lobe (12). Structurally, LFcin and LFampin both contain α-helical regions; however, LFcin only exists as an amphipathic α-helix within LF and becomes an amphipathic β-sheet hairpin in aqueous solution after pepsin digestion (26). Similarly, LFG within LF contains a 5-residue 310-helix that helical wheel projection shows has distinct polar and nonpolar faces, making LFG amphipathic (Fig. 5B). However, once separated after pepsin digestion, the even number of cysteine residues in LFG indicate that it may form disulfide bond-linked β-sheet structures or remain as a 310-helical structure (27). Most AMPs have either amphipathic helical or β-sheet secondary structures, with the former permitting more efficient interaction with biological membranes (28).
LFG has a positive GRAVY score of 0.14, making it hydrophobic (Table 1). Hydrophobicity is a crucial parameter affecting the capacity of AMPs to partition into the cell membrane (29). While containing hydrophobic residues, LFcin, LFampin, and LF(1–11) all have negative GRAVY scores, −0.576, −1.482, and −0.600, respectively, making them more hydrophilic. Although positive, the GRAVY score of LFG is relatively low, which may be beneficial, as very high levels of hydrophobicity are associated with mammalian cell toxicity and loss of antimicrobial specificity (30). Confirming this prediction, cytotoxicity assays showed that LFG has no significant toxicity to A549 cells at concentrations as high as 8 μg/ml. With a positive net charge of +1, LFG is not as strongly cationic as LFcin, LFampin, and LF(1–11), which have positive net charges of +8, +5, and +3, respectively. Net charge is an important parameter for antifungal activity, with cationic residues promoting an electrostatic attraction of the peptide to anionic phospholipids in the fungal membrane and yeast cell walls (31). Thus, the relatively low charge of LFG may contribute to its lack of antifungal activity when used alone. Most naturally occurring AMPs are not optimized for efficient activity and need to be improved through various strategies. In particular, it has been found that increasing the net charge and/or hydrophobicity can increase the ability of peptides to disrupt the microbial membrane (32, 33). Alteration of LFG through the insertion of positively charged residues and substitution or reduction of negatively charged residues may be a way to increase its efficacy and could result in antifungal activity when used alone.
In addition to similarities to LF-derived peptides, a homology search of AMPs with experimentally validated antimicrobial activity returned 14 peptides with sequence motifs in common with LFG, including frog skin-derived esculentins and brevinins, Japanese horseshoe crab-derived tachystatin, and avian and mammalian defensins (Table 3). Brevinins, esculentins, and defensins all function by binding the cell membrane and forming pores that cause leakage and collapse of the cell membrane (34, 35), while tachystatins function similarly through chitin binding (36). Five of these peptides (34, 35), brevinin-2-OA6, brevinin-2-OA8, esculentin-1-OR3, microplusin, and tachystatin-C, have validated antifungal activity against C. albicans reported at concentrations similar to those of the FIC of LFG determined in this study (35, 36), and these may therefore be useful to guide future research into the structure-activity relationship of LFG. Two peptides, gallinacin-2 and ostricacin-1, were recorded as inactive against C. albicans, while the remaining peptides have not been tested against fungal pathogens and therefore may potentially have antifungal in addition to antibacterial activity.
LFG is a stronger synergent than other LF-derived peptides and may have species-specific activity.
Although few studies have investigated synergy between other LF-derived peptides and antifungal drugs, comparing the results available indicates that LFG is much more potently synergistic. LFcin was reported as synergistic with several azoles against C. albicans planktonic cells at concentrations of 3.125 to 6.25 μg/ml for 80% growth inhibition but was ineffective when paired with AMB, nystatin, or 5FC (19), while another study reported LFcin as synergistic with AMB against C. albicans biofilms at concentrations of ≥64 μg/ml (18). LF(1–11) has also been seen to be synergistic with fluconazole against C. albicans at high concentrations of 100 to 200 μg/ml (17). In comparison, the synergistic range of LFG when paired with AMB was much lower, 0.5 to 2 μg/ml, for complete growth inhibition (Table 2). The current study focused on synergy with AMB, due to the previous finding that whole LF was consistently synergistic with AMB across multiple yeast species (10). However, values close to synergy (FICI = 0.75) for fluconazole, itraconazole, and voriconazole were previously achieved with whole LF in some species, indicating that LFG or other peptides present in LF hydrolysate may have the potential to synergize with azole antifungals. Further testing of LFG with other antifungals in a diverse range of strains is necessary to elucidate its full spectrum of synergistic activities and specificities.
The mechanisms of synergy between LF-derived peptides and antifungals are currently unknown (12). AMB functions by binding membrane ergosterol and disrupting cellular integrity, with studies suggesting that it also causes oxidative damage and can enter the cell and disrupt intracellular targets (37). If LFG does target the cell membrane, like many other AMPs, the mechanism of synergy with AMB could be due to the combined effect on different interaction sites in the membrane. Alternatively, if LFG acts on an intracellular target, damage to the cell by AMB that allowed increased uptake of LFG could facilitate the simultaneous inhibition of different fungal cell targets. Species-specific differences in the degrees of efficacy were seen in the dose-response surfaces for LFG+AMB treatment, with C. albicans being the least susceptible and C. glabrata the most. This result indicates that the mechanism of synergy may involve specific targets or metabolic pathways that differ between species. Previous transcriptomic analysis of LF+AMB synergy in Cryptococcus and Saccharomyces species showed markedly different responses to treatment between the two species, with metal- and stress-related transcripts decreasing in Saccharomyces while stress response processes increased in Cryptococcus (38, 39). Although belonging to the Candida genus, C. glabrata is more closely related to Saccharomyces than to C. albicans (40), and thus, its response to synergistic treatment may be similar to that of Saccharomyces and distinct from that of Cryptococcus, as well as from that of C. albicans.
The current status of AMPs as therapeutic molecules and the potential of LFG to be used as an adjuvant.
The only antifungal peptides that have received full FDA approval to date are the echinocandin family of β-glucan inhibitors, which are used for the treatment of candidemia and invasive aspergillosis (41). Although AMPs are thought to be less likely to induce resistance in the natural environment, resistance to echinocandin therapies is increasingly reported (42). Another limitation hampering clinical and commercial development of AMPs is the prohibitive production costs of peptides (43). Hence, the use of LFG as an adjuvant to AMB is an attractive strategy as it can both decrease the risk of developing resistance by acting on multiple targets and lower costs by reducing the required amount of each agent. Furthermore, LFG has the potential to increase the fungicidal effect of treatment and decrease toxicity. With an intermediate Boman index of 1.25, LFG may not interact with a very wide range of proteins within the cell, which may make it a more suitable therapeutic agent with fewer unintended side effects (44).
Several other AMPs are currently in preclinical and clinical trials as antifungal therapies, including two LF derivatives (45). LF(1–11) is being developed for the intravenous treatment of bacterial and fungal infections in stem cell transplant patients (46), and PLX01, an LF analogue, is being developed for the topical treatment of postsurgical adhesions (47). Most other antifungal peptides currently being developed are for topical treatment, including the dimeric peptide CZEN-002 for vaginal candidiasis (48), the histatin-5 analogue PAC113 for oral candidiasis (49), the cyclical cationic peptide novexatin for nail infections (45), the lipopeptide HB1275 for skin infections (50), and the polyarginine cationic peptide novamycin for various fungal infections (45). These peptides are smaller in mass and length than LFG, ranging from 971 to 3,061 Da with 7 to 25 residues, with net positive charges ranging from 2 to 14. Possessing diverse structural features, including α-helical regions, disulfide bridges, extended/random coils, and amphipathicity, they all primarily function through membrane disruption and/or immunomodulation. As a peptide derived from a multifunctional parent protein, LFG may too have immunomodulatory functions that may be revealed upon clinical use. The small size of peptides currently being developed indicates that LFG may need to be reduced to its most functional residues before being a viable candidate for therapeutic usage, which will be the subject of future work in our laboratories.
Conclusion.
We have shown that simple digestion of LF can produce an LF hydrolysate containing a mixture of peptides that synergize with AMB substantially more effectively than the full-length protein and have identified and tested LFG, a novel LF-derived peptide from this hydrolysate that exhibits further-improved synergy. Given its small size and favorable structural properties, LFG is a viable candidate for development as a future adjuvant to potentiate the effect of AMB, lowering the required dose, reducing toxicity, and improving efficacy. The application of rational design to modify physical and chemical properties of LFG to improve its activity, stability, and permeability, along with investigations to elucidate the spectrum of activity of LFG combined with other antifungal drugs and determine its mechanism of action, will help clarify its full potential.
MATERIALS AND METHODS
Antifungal agents.
Bovine lactoferrin (LF) used in this study was kindly supplied by two dairy companies, Bega Bionutrients (92.2% purity, 14.5% iron saturation) and Fonterra (>90% purity, 10.3% iron saturation); amphotericin B (AMB) was purchased from Sigma-Aldrich. Lactofungin (LFG) was synthesized by Pepmic Co. (Suzhou, China). Compounds were assayed at concentration ranges of 0.0039 to 4 μg/ml for AMB and LFG and 0.25 to 256 μg/ml for LF and its digests. Stock solutions were prepared in Milli-Q water, stored at −80°C, and used within 6 months.
Fungal strains and culture conditions.
Four yeast reference strains were used for antifungal testing: Cryptococcus neoformans strain H99, Cryptococcus deuterogattii strain R265, Candida albicans strain SC5314, and Candida glabrata strain CBS138. Strains were cultured from glycerol stocks maintained at −80°C, streaked for single colonies on Sabouraud dextrose agar (SDA) (10 g peptone, 40 g glucose, 15 g agar, 1 liter distilled water [dH2O]), and incubated at 30°C for 48 h. All assays were performed with technical duplicates, and at least three biological replicates were performed on separate days. Final inoculum concentration was confirmed by back-plating.
Generation of lactoferrin peptides by enzymatic and acidic digestion.
Two 5% (wt/vol) solutions of LF were prepared by dissolving 12 g of LF in 240 ml of ultrapure water and separating it into two 120-ml batches. For the enzymatic treatment, 3% (wt/wt) porcine pepsin was dissolved in the first LF solution and the pH was adjusted to 3 using 1 M HCl. The solution was then split into 12 10-ml aliquots in 15-ml falcon tubes, and individual tubes were incubated at 20°C, 30°C, 40°C, or 50°C in a water bath for 1, 2, or 4 h. To terminate the enzymatic reaction, samples were heated at 80°C in a water bath for 15 min. For the acidic treatment, the pH of the second LF solution was first adjusted to 4.0 using 1 M HCl and then to 3.0, 2.0, and 1.0, with three 10-ml aliquots being removed and placed in 15-ml falcon tubes at each stage. The samples were incubated at 100°C in a water bath for 15, 30, or 60 min. For both treatments, all samples were snap-frozen in liquid nitrogen to prevent further digestion. Samples were then thawed, and the pH adjusted to 7.0 using 1 M NaOH. Insoluble peptides were removed by centrifugation at 15,000 × g for 30 min, and the supernatants were stored at −80°C.
Digest characterization by SDS-PAGE.
The composition of digested LF samples was analyzed by SDS-PAGE on 10 to 20% Criterion TGX precast midi protein gels (Bio-Rad) with Tris-tricine running buffer (100 mM Tris, 100 mM tricine, 0.1% SDS). Samples were diluted 1:1 with Laemmli sample buffer (Bio-Rad) and β-mercaptoethanol, heated at 90°C for 5 min, and spun down prior to being loaded on gels. A 20-μl aliquot of each sample and 2 μl of broad-range unstained protein standard (New England Biolabs) were loaded, and gels were run at 200 V for 45 min. Gels were washed 3 times for 5 min in 200 ml of Milli-Q water to remove SDS, drained before being covered with 50 ml Bio-Safe Coomassie stain (Bio-Rad), and incubated at room temperature with gentle shaking for 1 h. Gels were then washed once again in 200 ml of Milli-Q water for 30 min and photographed using a digital camera.
Size exclusion filtration.
Digest samples were separated into two fractions using 10-kDa molecular weight cutoff (MWCO) spin filters (Corning). Samples were diluted to 10 mg/ml in Milli-Q water, and 6-ml amounts were added to spin filters and centrifuged at 10,000 × g for 20 min. The <10-kDa fraction was recovered from the concentrate pocket and adjusted to 6 ml, and then 6 ml Milli-Q water was added to the filter to recover the >10-kDa fraction.
Antifungal susceptibility testing by broth microdilution.
Antifungal susceptibility testing was performed by broth microdilution methodology in 96-well microtiter plates in accordance with CLSI guidelines for yeasts, with some modifications (51). Fungal inocula were prepared from colonies growing on agar plates to a final concentration of 0.5 × 103 to 2.5 × 103 CFU/ml. Cryptococcus strains were tested in YNB (Sigma-Aldrich) supplemented with 0.165 M MOPS (morpholinepropanesulfonic acid) and 0.5% d-glucose and incubated for 72 h, while Candida strains were tested in RPMI 1640 (Sigma-Aldrich) supplemented with 0.165 M MOPS and 2% d-glucose and incubated for 48 h. All strains were incubated without agitation at 35°C, and MICs were determined visually and defined as the lowest drug concentration at which growth was inhibited 100%. Each test plate included the reference strain Candida parapsilosis strain ATCC 22019.
Drug interaction testing by checkerboard assay.
Checkerboard assays were used to determine pairwise interactions between LF, its digests, or LFG and AMB. Serial 2-fold dilutions starting at 4 times the MIC of each test agent were prepared and plated in 96-well microtiter plates in the horizontal and vertical directions, respectively. Inoculum preparation, media, and incubation conditions followed those described above for antifungal susceptibility testing. Checkerboard results were assessed both visually, with 100% inhibition as the endpoint, and by absorbance at 600 nm (BioTek ELx800). Initial determination of interactions between the 24 LF digests and AMB used an abbreviated diagonal-sampling checkerboard method (52).
Models used for assessment of drug interactions.
Two models were used to assess drug interactions. The fractional inhibitory concentration index (FICI), which is based on the Loewe additivity mode, determines the fractional inhibitory concentration (FIC) of each drug in the pair as [MICX/MICY], where MICX is the MIC of the drug alone and MICY is the MIC of the drug in combination. FICI is then calculated as FICDRUG A + FICDRUG B. This model defines interactions as synergistic (≤0.5), indifferent (>0.5 to 4), or antagonistic (>4) (53). MacSynergy II is based on the Bliss independence model and uses the equation EAB = EA + EB(EAEB), where EAB is the additive effect of drugs A and B as predicted by their individual effects (EA and EB) (54). MacSynergy II generates a three-dimensional interaction surface by calculating the predicted indifferent effect and representing this as a flat plane, with peaks and troughs representing synergistic and antagonistic interactions, respectively. This model uses interaction volumes (μM2%) and defines positive volumes as synergistic and negative volumes as antagonistic. It additionally defines interactions within these categories as insignificant (≤25 μM2%), minor (>25 to 50 μM2%), moderate (>50 to 100 μM2%), or strong (>100 μM2%) as a rough estimate of significance based on the analysis of past drug interactions.
Lactoferrin characterization by mass spectrometry.
Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry was used to analyze the composition of LF and the digested LF samples. Two methods of sample preparation were used, which were optimized for larger protein and small peptide targets, respectively. For protein analysis preparation, a 1-μl aliquot of matrix solution A (sinapinic acid [SA] saturated in ethanol) was applied to the ground steel target in a thin layer. Samples were then mixed 1:1 with matrix solution B (SA saturated in 0.1% trifluoroacetic acid [TFA], 30% acetonitrile [TA30]), and 0.5-μl amounts were applied on top of the matrix solution A layer and allowed to dry. For peptide analysis preparation, samples were mixed 1:1 with matrix solution (α-cyano-4-hydroxycinnamic acid [HCCA] saturated in TA30), and 0.5-μl amounts were applied to the ground steel target and allowed to dry. Mass spectra were acquired on a Bruker Autoflex speed LRF mass spectrometer in linear mode using a laser power of 85% from the sum of 10,000 laser shots.
Peptide identification.
MALDI-TOF mass spectra were processed and analyzed using mMass (55). Spectra underwent baseline correction (precision, 75; relative offset, 25), smoothing (Savitzky-Golay, 5-m/z window, 5 cycles), peak picking (S/N threshold, 4; absolute intensity threshold, 0.0; relative intensity threshold, 0.5; picking height, 75), and deisotoping (maximum charge, 1; isotope mass tolerance, 0.15 m/z; isotope intensity tolerance, 50%). The UniProt sequence for bovine LF (lactotransferrin from Bos taurus; UniProt accession number P24627) was imported, a virtual protein digest was run using pepsin as the enzyme (mass charge, 1; maximum miscleavages, 5), and the peptides generated were matched to the peak list by mass (maximum tolerance, 5 Da).
Peptide characterization.
Putative physicochemical properties, including theoretical pI, aliphatic index, net charge, grand average of hydropathicity (GRAVY), and instability index, were determined by using the ProtParam tool (56). The hydrophobic ratio and Boman index were determined by using ADP3 (28). Structural models for each peptide were generated using the SWISS-MODEL homology-modeling server with bovine lactoferrin as a template (57). Helical wheel projections were generated for the helical structured region of LFG using the NetWheels online tool (58). A BLAST query was performed against LFG for similar antimicrobial peptides (AMPs) within the Collection of Anti-Microbial Peptides (CAMP) Database (59). The results were limited to peptides with experimentally validated antifungal activity.
Cell culture and cytotoxicity assays.
The cytotoxicity of LFG alone and in combination with AMB toward A549 lung cancer cells was determined by methyl thiazol tetrazolium (MTT) assay. A549 cells were grown in 25-cm2 cell culture flasks containing 5 ml advanced Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 2% serum, 1% glutamine, and 1% penicillin-streptomycin and maintained at 37°C with 5% CO2. Confluent flasks were washed with phosphate-buffered saline (PBS) and trypsinized, and cells were enumerated using a Countess automated cell counter with trypan blue stain to determine viability. After counting, 5,000 cells/well were transferred into a 96-well plate and incubated overnight to allow them to attach. Different concentrations of LFG (0.5, 2, and 8 μg/ml), AMB (0.0625, 0.5, and 1 μg/ml), or AMB+LFG, as well as a 10% water-only vehicle control, were then added. Six technical replicates for each condition were included on each plate. Plates were incubated at 37°C with 5% CO2 for 24 h, and then MTT (1 mg/ml) was added to each well and incubation was continued for a further 4 h. The supernatant was then removed, 100 μl of dimethyl sulfoxide (DMSO) was added to each well, and plates were incubated at room temperature with rotation for 5 min. The absorbance of formazan produced by the cells was then measured at 600 nm using a BioTek ELx800.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the facilities as well as scientific and technical assistance of the Mass Spectrometry Facility in the School of Chemistry at the University of Sydney. Special thanks to Nicholas Proschogo for assistance with mass spectrometry experiments and Aviva Levina for assistance with cell culture and cytotoxicity assays.
K.E.F. and D.A.C. conceived and designed the experiments. K.E.F. produced, collated, and analyzed the data and wrote the manuscript with assistance from D.A.C. and R.J.P.
This work was financially supported by the Australian National Health and Medical Research Council (grant number APP1021267). K.E.F. was financially supported by a research training program scholarship.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Krysan DJ. 2017. The unmet clinical need of novel antifungal drugs. Virulence 8:135–137. doi: 10.1080/21505594.2016.1276692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bongomin F, Gago S, Oladele RO, Denning DW. 2017. Global and multi-national prevalence of fungal diseases—estimate precision. J Fungi (Basel) 3:57. doi: 10.3390/jof3040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laniado-Laborín R, Cabrales-Vargas MN. 2009. Amphotericin B: side effects and toxicity. Rev Iberoam Micol 26:223–227. doi: 10.1016/j.riam.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Loyse A, Thangaraj H, Easterbrook P, Ford N, Roy M, Chiller T, Govender N, Harrison TS, Bicanic T. 2013. Cryptococcal meningitis: improving access to essential antifungal medicines in resource-poor countries. Lancet Infect Dis 13:629–637. doi: 10.1016/S1473-3099(13)70078-1. [DOI] [PubMed] [Google Scholar]
- 5.Campitelli M, Zeineddine N, Samaha G, Maslak S. 2017. Combination antifungal therapy: a review of current data. J Clin Med Res 9:451–456. doi: 10.14740/jocmr2992w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson MD, Perfect JR. 2010. Use of antifungal combination therapy: agents, order, and timing. Curr Fungal Infect Rep 4:87–95. doi: 10.1007/s12281-010-0018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perfect JR, Dismukes WE, Dromer F, Goldman DL, Graybill JR, Hamill RJ, Harrison TS, Larsen RA, Lortholary O, Nguyen MH, Pappas PG, Powderly WG, Singh N, Sobel JD, Sorrell TC. 2010. Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the Infectious Diseases Society of America. Clin Infect Dis 50:291–322. doi: 10.1086/649858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pappas PG, Kauffman CA, Andes D, Benjamin DK Jr, Calandra TF, Edwards JE Jr, Filler SG, Fisher JF, Kullberg BJ, Ostrosky-Zeichner L, Reboli AC, Rex JH, Walsh TJ, Sobel JD, Infectious Diseases Society of America. 2009. Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clin Infect Dis 48:503–535. doi: 10.1086/596757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rajasingham R, Rolfes MA, Birkenkamp KE, Meya DB, Boulware DR. 2012. Cryptococcal meningitis treatment strategies in resource-limited settings: a cost-effectiveness analysis. PLoS Med 9:e1001316. doi: 10.1371/journal.pmed.1001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandes KE, Weeks K, Carter DA. 2020. Lactoferrin is broadly active against yeasts and highly synergistic with amphotericin B. Antimicrob Agents Chemother 64:e02284-19. doi: 10.1128/AAC.02284-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson BF, Baker HM, Norris GE, Rice DW, Baker EN. 1989. Structure of human lactoferrin: crystallographic structure analysis and refinement at 2.8 A resolution. J Mol Biol 209:711–724. doi: 10.1016/0022-2836(89)90602-5. [DOI] [PubMed] [Google Scholar]
- 12.Fernandes KE, Carter DA. 2017. The antifungal activity of lactoferrin and its derived peptides: mechanisms of action and synergy with drugs against fungal pathogens. Front Microbiol 8:2. doi: 10.3389/fmicb.2017.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinha M, Kaushik S, Kaur P, Sharma S, Singh TP. 2013. Antimicrobial lactoferrin peptides: the hidden players in the protective function of a multifunctional protein. Int J Pept 2013:390230. doi: 10.1155/2013/390230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rastogi N, Nagpal N, Alam H, Pandey S, Gautam L, Sinha M, Shin K, Manzoor N, Virdi JS, Kaur P, Sharma S, Singh TP. 2014. Preparation and antimicrobial action of three tryptic digested functional molecules of bovine lactoferrin. PLoS One 9:e90011. doi: 10.1371/journal.pone.0090011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomita M, Bellamy W, Takase M, Yamauchi K, Wakabayashi H, Kawase K. 1991. Potent antibacterial peptides generated by pepsin digestion of bovine lactoferrin. J Dairy Sci 74:4137–4142. doi: 10.3168/jds.S0022-0302(91)78608-6. [DOI] [PubMed] [Google Scholar]
- 16.Yamauchi K, Tomita M, Giehl TJ, Ellison RT III. 1993. Antibacterial activity of lactoferrin and a pepsin-derived lactoferrin peptide fragment. Infect Immun 61:719–728. doi: 10.1128/IAI.61.2.719-728.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lupetti A, Paulusma-Annema A, Welling MM, Dogterom-Ballering H, Brouwer CP, Senesi S, Van Dissel JT, Nibbering PH. 2003. Synergistic activity of the N-terminal peptide of human lactoferrin and fluconazole against Candida species. Antimicrob Agents Chemother 47:262–267. doi: 10.1128/aac.47.1.262-267.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wakabayashi H, Abe S, Okutomi T, Tansho S, Kawase K, Yamaguchi H. 1996. Cooperative anti-Candida effects of lactoferrin or its peptides in combination with azole antifungal agents. Microbiol Immunol 40:821–825. doi: 10.1111/j.1348-0421.1996.tb01147.x. [DOI] [PubMed] [Google Scholar]
- 19.Wakabayashi H, Abe S, Teraguchi S, Hayasawa H, Yamaguchi H. 1998. Inhibition of hyphal growth of azole-resistant strains of Candida albicans by triazole antifungal agents in the presence of lactoferrin-related compounds. Antimicrob Agents Chemother 42:1587–1591. doi: 10.1128/AAC.42.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giansanti F, Panella G, Leboffe L, Antonini G. 2016. Lactoferrin from milk: nutraceutical and pharmacological properties. Pharmaceuticals 9:61. doi: 10.3390/ph9040061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez L, Calvo M, Brock JH. 1992. Biological role of lactoferrin. Arch Dis Child 67:657–661. doi: 10.1136/adc.67.5.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Britton JR, Koldovsky O. 1989. Gastric luminal digestion of lactoferrin and transferrin by preterm infants. Early Hum Dev 19:127–135. doi: 10.1016/0378-3782(89)90123-0. [DOI] [PubMed] [Google Scholar]
- 23.Troost FJ, Steijns J, Saris WH, Brummer RJ. 2001. Gastric digestion of bovine lactoferrin in vivo in adults. J Nutr 131:2101–2104. doi: 10.1093/jn/131.8.2101. [DOI] [PubMed] [Google Scholar]
- 24.Bruni N, Capucchio MT, Biasibetti E, Pessione E, Cirrincione S, Giraudo L, Corona A, Dosio F. 2016. Antimicrobial activity of lactoferrin-related peptides and applications in human and veterinary medicine. Molecules 21:752. doi: 10.3390/molecules21060752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pushpanathan M, Gunasekaran P, Rajendhran J. 2013. Antimicrobial peptides: versatile biological properties. Int J Pept 2013:675391. doi: 10.1155/2013/675391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunter HN, Demcoe AR, Jenssen H, Gutteberg TJ, Vogel HJ. 2005. Human lactoferricin is partially folded in aqueous solution and is better stabilized in a membrane mimetic solvent. Antimicrob Agents Chemother 49:3387–3395. doi: 10.1128/AAC.49.8.3387-3395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang G, Li X, Wang Z. 2016. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res 44:D1087–D1093. doi: 10.1093/nar/gkv1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahlapuu M, Hakansson J, Ringstad L, Bjorn C. 2016. Antimicrobial peptides: an emerging category of therapeutic agents. Front Cell Infect Microbiol 6:194. doi: 10.3389/fcimb.2016.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y, Guarnieri MT, Vasil AI, Vasil ML, Mant CT, Hodges RS. 2007. Role of peptide hydrophobicity in the mechanism of action of alpha-helical antimicrobial peptides. Antimicrob Agents Chemother 51:1398–1406. doi: 10.1128/AAC.00925-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jindal MH, Le CF, Mohn Yusof MY, Sekaran SD. 2014. Net charge, hydrophobicity and specific amino acids contribute to the activity of antimicrobial peptides. JUMMEC 17:1–7. doi: 10.22452/jummec.vol17no1.1. [DOI] [Google Scholar]
- 31.van der Weerden NL, Bleackley MR, Anderson MA. 2013. Properties and mechanisms of action of naturally occurring antifungal peptides. Cell Mol Life Sci 70:3545–3570. doi: 10.1007/s00018-013-1260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pasupuleti M, Walse B, Svensson B, Malmsten M, Schmidtchen A. 2008. Rational design of antimicrobial C3a analogues with enhanced effects against staphylocci using an integrated structure and function-based approach. Biochemistry 47:9057–9070. doi: 10.1021/bi800991e. [DOI] [PubMed] [Google Scholar]
- 33.Ringstad L, Andersson Nordahl E, Schmidtchen A, Malmsten M. 2007. Composition effect on peptide interaction with lipids and bacteria: variants of C3a peptide CNY21. Biophys J 92:87–98. doi: 10.1529/biophysj.106.088161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fogaca AC, Almeida IC, Eberlin MN, Tanaka AS, Bulet P, Daffre S. 2006. Ixodidin, a novel antimicrobial peptide from the hemocytes of the cattle tick Boophilus microplus with inhibitory activity against serine proteinases. Peptides 27:667–674. doi: 10.1016/j.peptides.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 35.Yang X, Lee WH, Zhang Y. 2012. Extremely abundant antimicrobial peptides existed in the skins of nine kinds of Chinese odorous frogs. J Proteome Res 11:306–319. doi: 10.1021/pr200782u. [DOI] [PubMed] [Google Scholar]
- 36.Osaki T, Omotezako M, Nagayama R, Hirata M, Iwanaga S, Kasahara J, Hattori J, Ito I, Sugiyama H, Kawabata S. 1999. Horseshoe crab hemocyte-derived antimicrobial polypeptides, tachystatins, with sequence similarity to spider neurotoxins. J Biol Chem 274:26172–26178. doi: 10.1074/jbc.274.37.26172. [DOI] [PubMed] [Google Scholar]
- 37.Ellis D. 2002. Amphotericin B: spectrum and resistance. J Antimicrob Chemother 49:7–10. doi: 10.1093/jac/49.suppl_1.7. [DOI] [PubMed] [Google Scholar]
- 38.Lai Y, Campbell L, Wilkins MR, Pang C, Chen S, Carter DA. 2016. Synergy and antagonism between iron chelators and antifungal drugs in Cryptococcus. Int J Antimicrob Agents 48:388–394. doi: 10.1016/j.ijantimicag.2016.06.012. [DOI] [PubMed] [Google Scholar]
- 39.Lai YW, Pang CNI, Campbell LT, Chen SCA, Wilkins MR, Carter DA. 2019. Different pathways mediate amphotericin-lactoferrin drug synergy in Cryptococcus and Saccharomyces. Front Microbiol 10:2195. doi: 10.3389/fmicb.2019.02195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gabaldon T, Carrete L. 2016. The birth of a deadly yeast: tracing the evolutionary emergence of virulence traits in Candida glabrata. FEMS Yeast Res 16:fov110. doi: 10.1093/femsyr/fov110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matejuk A, Leng Q, Begum MD, Woodle MC, Scaria P, Chou ST, Mixson AJ. 2010. Peptide-based antifungal therapies against emerging infections. Drugs Future 35:197. doi: 10.1358/dof.2010.035.03.1452077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perlin DS. 2015. Echinocandin resistance in Candida. Clin Infect Dis 61:S612–S617. doi: 10.1093/cid/civ791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfalzgraff A, Brandenburg K, Weindl G. 2018. Antimicrobial peptides and their therapeutic potential for bacterial skin infections and wounds. Front Pharmacol 9:281. doi: 10.3389/fphar.2018.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boman HG. 2003. Antibacterial peptides: basic facts and emerging concepts. J Intern Med 254:197–215. doi: 10.1046/j.1365-2796.2003.01228.x. [DOI] [PubMed] [Google Scholar]
- 45.Koo HB, Seo J. 2019. Antimicrobial peptides under clinical investigation. Pept Sci 111: doi: 10.1002/pep2.24122. [DOI] [Google Scholar]
- 46.van der Does AM, Hensbergen PJ, Bogaards SJ, Cansoy M, Deelder AM, van Leeuwen HC, Drijfhout JW, van Dissel JT, Nibbering PH. 2012. The human lactoferrin-derived peptide hLF1-11 exerts immunomodulatory effects by specific inhibition of myeloperoxidase activity. J Immunol 188:5012–5019. doi: 10.4049/jimmunol.1102777. [DOI] [PubMed] [Google Scholar]
- 47.Wiig M, Olmarker K, Hakansson J, Ekstrom L, Nilsson E, Mahlapuu M. 2011. A lactoferrin-derived peptide (PXL01) for the reduction of adhesion formation in flexor tendon surgery: an experimental study in rabbits. J Hand Surg Eur Vol 36:656–662. doi: 10.1177/1753193411410823. [DOI] [PubMed] [Google Scholar]
- 48.Fjell CD, Hiss JA, Hancock RE, Schneider G. 2011. Designing antimicrobial peptides: form follows function. Nat Rev Drug Discov 11:37–51. doi: 10.1038/nrd3591. [DOI] [PubMed] [Google Scholar]
- 49.Rothstein DM, Spacciapoli P, Tran LT, Xu T, Roberts FD, Dalla Serra M, Buxton DK, Oppenheim FG, Friden P. 2001. Anticandida activity is retained in P-113, a 12-amino-acid fragment of histatin 5. Antimicrob Agents Chemother 45:1367–1373. doi: 10.1128/AAC.45.5.1367-1373.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pirri G, Giuliani A, Nicoletto S, Pizzuto L, Rinaldi A. 2009. Lipopeptides as anti-infectives: a practical perspective. Open Life Sci 4:258–273. doi: 10.2478/s11535-009-0031-3. [DOI] [Google Scholar]
- 51.CLSI. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard third edition, vol 28. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 52.Cokol-Cakmak M, Bakan F, Cetiner S, Cokol M. 2018. Diagonal method to measure synergy among any number of drugs. J Vis Exp 2018:e57713. doi: 10.3791/57713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Odds FC. 2003. Synergy, antagonism, and what the chequerboard puts between them. J Antimicrob Chemother 52:dkg301. doi: 10.1093/jac/dkg301. [DOI] [PubMed] [Google Scholar]
- 54.Prichard MN, Shipman C Jr. 1990A three-dimensional model to analyze drug-drug interactions. Antiviral Res 14:181–205. doi: 10.1016/0166-3542(90)90001-N. [DOI] [PubMed] [Google Scholar]
- 55.Strohalm M, Hassman M, Kosata B, Kodicek M. 2008. mMass data miner: an open source alternative for mass spectrometric data analysis. Rapid Commun Mass Spectrom 22:905–908. doi: 10.1002/rcm.3444. [DOI] [PubMed] [Google Scholar]
- 56.Wilkins MR, Gasteiger E, Gooley AA, Herbert BR, Molloy MP, Binz PA, Ou K, Sanchez JC, Bairoch A, Williams KL, Hochstrasser DF. 1999. High-throughput mass spectrometric discovery of protein post-translational modifications. J Mol Biol 289:645–657. doi: 10.1006/jmbi.1999.2794. [DOI] [PubMed] [Google Scholar]
- 57.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T. 2018. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mól AR, Castro MS, Fontes W. 2018. NetWheels: a web application to create high quality peptide helical wheel and net projections. BioRxiv doi: 10.1101/416347. [DOI]
- 59.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soman SS, Arathy DS, Sreekumar E. 2009. Discovery of Anas platyrhynchos avian beta-defensin 2 (Apl_AvBD2) with antibacterial and chemotactic functions. Mol Immunol 46:2029–2038. doi: 10.1016/j.molimm.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 61.Zaballos A, Villares R, Albar JP, Martinez AC, Marquez G. 2004. Identification on mouse chromosome 8 of new beta-defensin genes with regionally specific expression in the male reproductive organ. J Biol Chem 279:12421–12426. doi: 10.1074/jbc.M307697200. [DOI] [PubMed] [Google Scholar]
- 62.Derache C, Labas V, Aucagne V, Meudal H, Landon C, Delmas AF, Magallon T, Lalmanach AC. 2009. Primary structure and antibacterial activity of chicken bone marrow-derived beta-defensins. Antimicrob Agents Chemother 53:4647–4655. doi: 10.1128/AAC.00301-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harwig SSL, Swiderek KM, Kokryakov VN, Tan L, Lee TD, Panyutich EA, Aleshina GM, Shamova OV, Lehrer RI. 1994. Gallinacins: cysteine-rich antimicrobial peptides of chicken leukocytes. FEBS Lett 342:281–285. doi: 10.1016/0014-5793(94)80517-2. [DOI] [PubMed] [Google Scholar]
- 64.Sugiarto H, Yu PL. 2006. Identification of three novel ostricacins: an update on the phylogenetic perspective of beta-defensins. Int J Antimicrob Agents 27:229–235. doi: 10.1016/j.ijantimicag.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 65.Evans EW, Beach GG, Wunderlich J, Harmon BG. 1994. Isolation of antimicrobial peptides from avian heterophils. J Leukoc Biol 56:661–665. doi: 10.1002/jlb.56.5.661. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.