Triazole antifungals are the primary therapeutic option against invasive aspergillosis. However, resistance to azoles has increased dramatically over the last decade. Azole resistance is known to primarily occur due to point mutations in the azole target protein Cyp51A, one of two paralogous 14-α sterol demethylases found in Aspergillus fumigatus. Despite the importance of Cyp51A, little is known about the function of its paralog, Cyp51B, and the behavior of these proteins within the cell or their functional interrelationship.
KEYWORDS: Cyp51A, Cyp51B, endoplasmic reticulum, azole resistance, voriconazole, cell wall stress, Aspergillus fumigatus, cell membrane stress
ABSTRACT
Triazole antifungals are the primary therapeutic option against invasive aspergillosis. However, resistance to azoles has increased dramatically over the last decade. Azole resistance is known to primarily occur due to point mutations in the azole target protein Cyp51A, one of two paralogous 14-α sterol demethylases found in Aspergillus fumigatus. Despite the importance of Cyp51A, little is known about the function of its paralog, Cyp51B, and the behavior of these proteins within the cell or their functional interrelationship. In this study, we addressed two important aspects of the Cyp51 proteins: (i) we characterized their localization patterns under normal growth versus stress conditions, and (ii) we determined how the proteins compensate for each other’s absence and respond to azole treatment. Both the Cyp51A and Cyp51B proteins were found to localize in distinct endoplasmic reticulum (ER) domains, including the perinuclear ER and the peripheral ER. Occasionally, the Cyp51 proteins concentrated in the peripheral ER network of tubules along the hyphal septa and at the hyphal tips. Exposure to voriconazole, caspofungin, and Congo red led to significant increases in fluorescence intensity in these alternative localization sites, indicative of Cyp51 protein translocation in response to cell wall stress. Furthermore, deletion of either Cyp51 paralog increased susceptibility to voriconazole, though a greater effect was observed following deletion of cyp51A, indicating a compensatory response to stress conditions.
INTRODUCTION
It is estimated that more than 2 million people suffer from life-threatening invasive fungal infections each year (1). Mortality due to invasive aspergillosis, caused largely by Aspergillus fumigatus, can reach 90% in some patient populations, with over 200,000 such infections estimated to occur each year (1, 2). Triazole antifungals are the guideline-recommended primary treatment for invasive aspergillosis (3), but resistance to these therapeutics is increasing (4–6). Resistance to triazoles by Aspergillus spp. has been linked to point mutations in Cyp51A, tandem repeats in the promoter of cyp51A, and overexpression of cyp51A, as well as other non-cyp51-mediated mechanisms (7, 8).
Cyp51A and its paralog, Cyp51B, are orthologous to Saccharomyces cerevisiae ERG11, the 14-α sterol demethylase responsible for catalyzing a key demethylation step in the synthesis of ergosterol, the primary sterol in the fungal cell membrane (9, 10). Direct inhibition of the Cyp51 proteins by azole antifungals leads to the depletion of ergosterol in the cell membrane, the accumulation of toxic sterol intermediates within the cell, and the progressive formation of carbohydrate patches in the cell wall (11, 12). Previous work in A. fumigatus has demonstrated that cyp51B is expressed in a constitutive manner, while cyp51A is inducible under a variety of conditions (13, 14). While cyp51A and cyp51B are individually dispensable for survival, genetic removal of both paralogs is lethal (13). Both Cyp51A and Cyp51B appear to localize to the perinuclear region in Aspergillus spp., most likely associating with the endoplasmic reticulum (ER) (15).
Several common nonsynonymous mutations in cyp51A alter the susceptibility of Aspergillus spp. to azoles, including mutations at Cyp51A G54, L98, Y121/T289, G138, M220, and G448 (16–21). Each common substitution affects susceptibility to a subset of azole drugs, depending upon the mutated residue (7, 22, 23). In contrast to the substantial focus on identifying cyp51A mutations conferring azole susceptibility, relatively little information is available on the contributions of either Cyp51 protein to cellular stress responses, including functional aspects of their response to triazole antifungals. This exclusive focus has been rationalized on the basis of the fact that Cyp51A mutations are responsible for most instances of azole resistance in A. fumigatus (24). However, azoles are known to bind and inhibit both Cyp51A and Cyp51B, with conformational elements of the azole-binding site thought to influence the interaction affinity and the inhibitory effect on Cyp51 enzymatic function (14, 15, 25). Modeling and structural analysis of the interactions between the A. fumigatus Cyp51 proteins and various azole drugs have provided further insights into the relationship between the conformation of the Cyp51A and Cyp51B binding sites and the binding affinity of specific azoles (14). Such analyses have identified specific residues responsible for key interactions between azoles and the Cyp51 proteins, including a hydrogen bond between Cyp51B Y122 and voriconazole which is absent during the interaction with fluconazole due to its smaller molecular size (14). Specific structural features of the lanosterol binding sites of Cyp51A and Cyp51B thus appear to be crucial factors contributing to azole targeting and efficiency.
In a recent study, we have shown dynamic changes in the localization pattern of the cell wall β-1,3-glucan synthase in response to caspofungin, the antifungal agent known to target Fks1 (26). Fks1 dynamically mislocalized from the hyphal tips to the vacuoles during hyphal growth inhibition by caspofungin and relocated to the hyphal tips during caspofungin-mediated paradoxical growth, revealing the mechanistic link between cell wall β-1,3-glucan synthesis and hyphal tip growth. Given the importance of the Cyp51 proteins for membrane ergosterol synthesis and maintenance of membrane structure and integrity, it is important to understand the response of these proteins to the antifungals targeting the cell membrane. Furthermore, the connection between azole exposure and the development of cell wall carbohydrate patches raises questions regarding the role of these proteins in responding to cell wall stressors (12). Although the Cyp51A and Cyp51B proteins are thought to localize to the ER, it is not clear how azole exposure or other cell surface stressors influence their localization patterns (15, 27). Furthermore, while expression of the cyp51 genes has been analyzed based on their transcription, little is known about the abundances of the proteins that they encode under standard conditions and conditions of azole stress (10). These gaps in knowledge compound with a lack of understanding about how the functions of the two Cyp51 proteins might overlap or help compensate for one another’s absence. In this study, we delved further into the function of these crucial Cyp51 proteins during targeted cell stress by observing their localization following exposure to antifungals and cell surface stressors. In addition, the protein levels of the Cyp51 paralogs, the antifungal susceptibility of the respective genetic deletion strains, and the impact of individual paralog deletions on voriconazole susceptibility were investigated to characterize the functional parameters of Cyp51A and Cyp51B in A. fumigatus. We report a strong association between the A. fumigatus Cyp51 proteins and the ER, hyphal septa, and apical tip that is influenced by exposure to voriconazole, caspofungin, and Congo red. Furthermore, A. fumigatus Cyp51A appears to compensate for the absence of Cyp51B through increased protein expression. These findings highlight the involvement of the Cyp51 proteins during responses to specific cell surface stresses and will be useful to our understanding of drug targeting strategies and in testing novel antifungals for their mechanisms of action.
RESULTS
Cyp51A and Cyp51B localize to the perinuclear and peripheral endoplasmic reticulum network in the apical and subapical hyphal compartments.
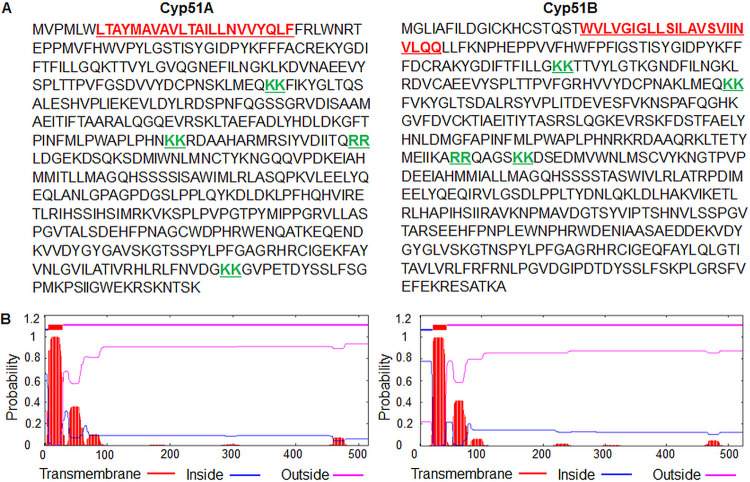
To verify the subcellular localization of the A. fumigatus Cyp51 proteins to the ER, we first analyzed their amino acid sequences for features associated with localization to specific cellular compartments. Most prominently, Cyp51A and Cyp51B each possess four dilysine/diarginine motifs (Fig. 1A). These motifs are known to promote the retention or redirection of a protein to the ER through retrograde vesicular trafficking (28, 29). Each of the Cyp51 proteins was also found to possess a single putative transmembrane domain at the N terminus, as determined through the use of TMHMM prediction software (Fig. 1B). This indicates the likelihood that the proteins are embedded within the membrane of one or more organelles, consistent with previous observations of Cyp51 proteins in Candida spp. (27). Based upon the observed ER localization signals in the Cyp51 proteins and previous work in Candida and Aspergillus species, we concluded that the Cyp51 proteins potentially localize predominantly at the ER (15, 30).
FIG 1.
(A) Sequence analysis of the 14-α sterol demethylase Cyp51A and Cyp51B paralogs in Aspergillus fumigatus reveals potential motifs for endoplasmic reticulum (ER) localization. Cyp51A and Cyp51B each contain four diarginine/dilysine motifs (underlined in green), indicating a potential link to the ER. (B) TMHMM server 2.0 prediction of one transmembrane region each (underlined in red in panel A) in Cyp51A and Cyp51B proteins.
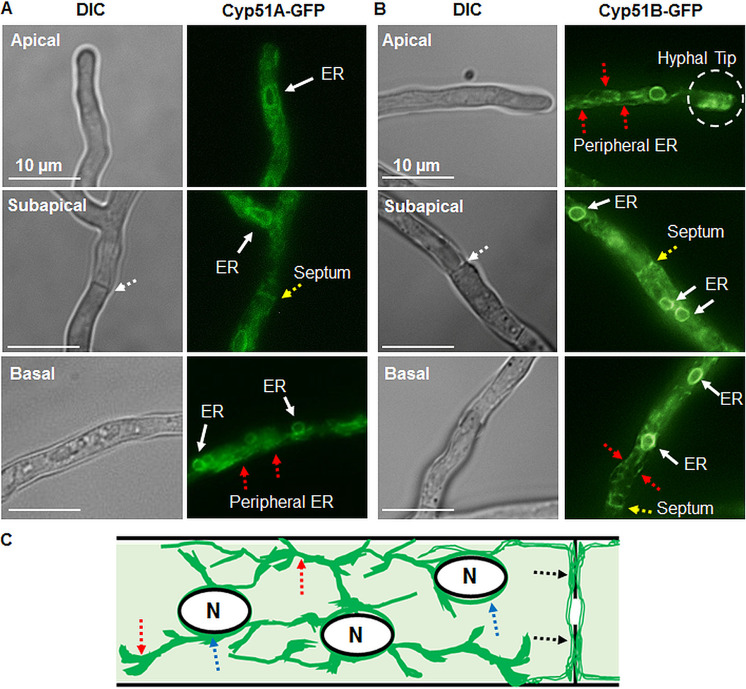
In order to confirm the in silico predictions, we generated two A. fumigatus strains expressing either the C-terminally green fluorescent protein (GFP)-labeled Cyp51A (Cyp51A-GFP) or Cyp51B (Cyp51B-GFP) fusion protein from their respective native loci. Fluorescence microscopy indicated a bright halo of fluorescence focused around the nuclei, while excluding the interior of the nuclei themselves, in the apical, subapical, and basal regions of the hyphae (Fig. 2A to C). This pattern is consistent with previous observations indicating the localization of other proteins to the perinuclear ER in Aspergillus spp. (15). In addition to this clear perinuclear localization, some fluorescence was observed in an elongated network of tubules throughout the hyphae in a manner consistent with localization to the peripheral ER (31). Costaining of the respective strains with ER-Tracker Red dye confirmed the localization of the Cyp51 proteins to the ER compartments (see Fig. S1 in the supplemental material). Cyp51A and Cyp51B also occasionally localized in the peripheral ER along the hyphal septa and at the apical tips of the hyphae (Fig. 2A and B). Both the septa and apical tips are active sites involved in the growth and organization of A. fumigatus hyphae.
FIG 2.
(A, B) Aspergillus fumigatus Cyp51A and Cyp51B proteins localize to the endoplasmic reticulum (ER) network, including the perinuclear and peripheral ER tubules in the apical, subapical, and basal regions of the hyphae. Red dotted arrows indicate the peripheral ER. The hyphal tip gradient of the ER observed at the hyphal tip is indicated by a dashed circle. Yellow arrows indicate peripheral ER tubules along the septum, and white arrows indicate the perinuclear ER. DIC, differential interference contrast. (C) Pictorial representation of a subapical region showing the Cyp51 protein ER localization patterns in a hyphal compartment. Dotted arrows in blue indicate the perinuclear ER, red dotted arrows indicate the peripheral ER, and black dotted arrows indicate peripheral ER tubular localization along the hyphal septum. N, nucleus.
The A. fumigatus Cyp51 proteins may play compensatory roles under normal growth conditions and in response to azoles.
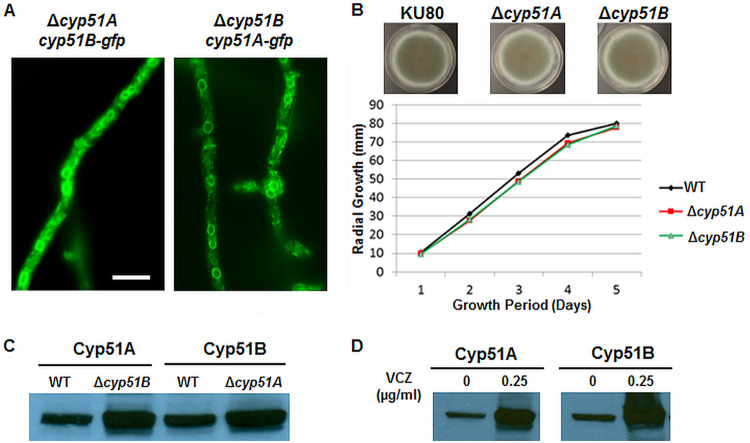
In addition to confirming the differential localization patterns of the Cyp51A and Cyp51B proteins, we analyzed the relationship and functional overlap between the two proteins. To accomplish this, the Cyp51 proteins were individually deleted to generate Δcyp51A and Δcyp51B strains. Furthermore, the Δcyp51 strains were transformed with C-terminally GFP-labeled versions of the remaining Cyp51 paralog (Δcyp51B::Cyp51A-GFP and Δcyp51A::Cyp51B-GFP), creating paired strains that could be used to analyze the localization of each Cyp51 paralog in the absence of the other. Paralog deletion had no noticeable impact on the localization of either Cyp51A or Cyp51B, each of which retained the typical ER localization pattern observed in the Cyp51A-GFP and Cyp51B-GFP strains (Fig. 3A).
FIG 3.
(A) GFP-tagged Cyp51A or Cyp51B localized in the ER, despite removal of their paralog. (B) Radial growth quantification of the Δcyp51A and Δcyp51B strains in comparison to that of the akuBKU80 (KU80) parent strain. (C) Western blot analysis of the Cyp51A and Cyp51B protein levels in the wild-type (WT), Δcyp51A, and Δcyp51B strains using anti-GFP antibody. (D) Cyp51A and Cyp51B protein expression increased in the presence of voriconazole (VCZ).
The potential compensatory functions of the respective Cyp51 proteins were examined in the context of hyphal growth, protein expression, and antifungal susceptibility. Deletion of Cyp51A or Cyp51B separately had no significant impact on radial growth, confirming that the Cyp51 proteins are individually dispensable for growth, likely due to functional compensation by the resident paralog (Fig. 3B). This possibility was analyzed by first examining Cyp51A and Cyp51B protein levels in the wild type and in the absence of Cyp51B (in the Δcyp51B strain) and Cyp51A (in the Δcyp51A strain) under normal growth conditions. As shown in Fig. 3C, the expression of Cyp51A was dramatically higher in the strain lacking the counterpart Cyp51B protein. Similarly, the expression of Cyp51B was increased in the strain lacking the Cyp51A protein. The observed elevated expression of the Cyp51A and Cyp51B proteins in response to the loss of each counterpart protein indicates a potential compensatory mechanism allowing for normal growth, despite selective impairment of an otherwise crucial gene. Next, we determined whether the levels of the Cyp51 proteins were altered during exposure to the Cyp51-targeting antifungal voriconazole. The Cyp51A-GFP- and Cyp51B-GFP-expressing strains were cultured in liquid medium for 22 h, followed by exposure to voriconazole at a sub-MIC of 0.125 μg/ml for 2 h. Western analysis revealed a higher baseline expression of Cyp51B than of Cyp51A, consistent with previous reports on the constitutive expression of Cyp51B, but the presence of voriconazole did not alter this expression pattern (data not shown) (14). We next treated the respective Cyp51A-GFP- and Cyp51B-GFP-expressing strains that had been cultured for 20 h with 0.25 μg/ml of voriconazole for 4 h. We noted that treatment for 4 h with 0.25 μg/ml voriconazole, which is the determined MIC of the antifungal, increased the expression of both the Cyp51 proteins in vivo (Fig. 3D).
The Cyp51A and Cyp51B deletion strains exhibit differential susceptibility to voriconazole.
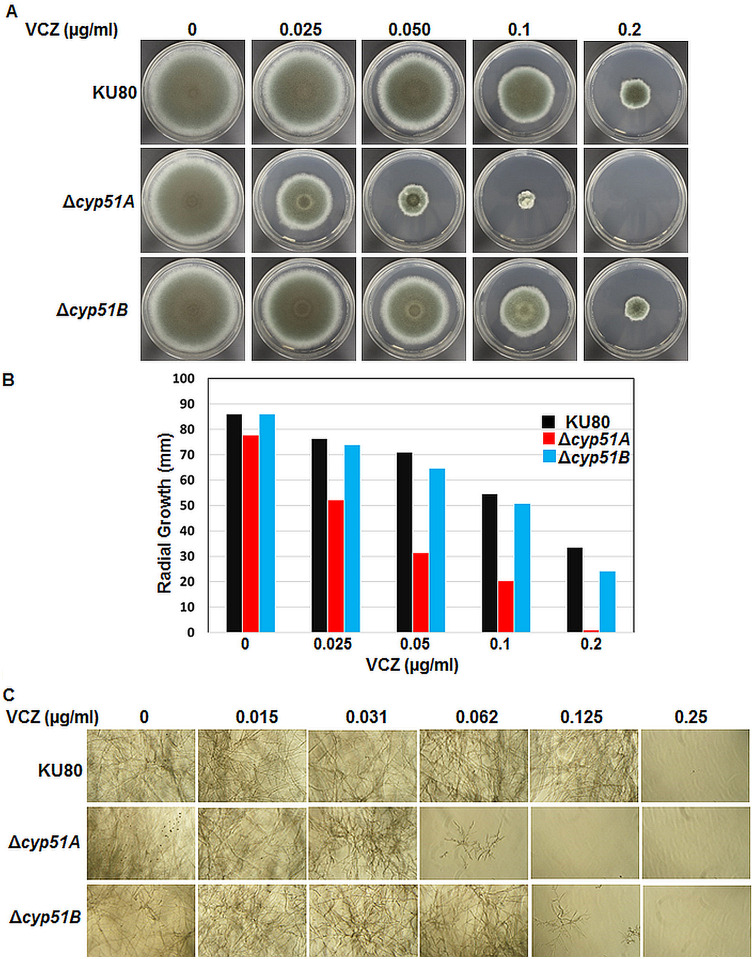
Previous studies have reported the deletion of cyp51A genes in two clinical isolates (CM-237 and CM-1252) of A. fumigatus (32) and the ΔakuBKU80 strain (33, 34). The CM-1252 strain and the ΔakuBKU80 strain with the cyp51A deletion showed sensitivity to voriconazole. Because we noted that both the Cyp51A and Cyp51B protein levels were increased by the presence of voriconazole at 0.25 μg/ml, the MIC for the wild-type strain, we next tested the Δcyp51A and Δcyp51B strains for susceptibility to voriconazole over a 5-day period of growth on glucose minimal medium (GMM) agar and also in RPMI 1640 liquid culture for 48 h. While the wild-type and the Δcyp51B strains showed susceptibility to voriconazole at 0.2 μg/ml, the Δcyp51A strain was more sensitive to voriconazole at 0.05 μg/ml and its growth was completely inhibited on GMM agar medium with 0.2 μg/ml voriconazole (Fig. 4A). In comparison to similarly treated wild-type and Δcyp51B strains, the Δcyp51A strain showed an approximately 50% and 40% growth reduction with 0.05 and 0.1 μg/ml voriconazole, respectively (Fig. 4B). Similarly, in the RPMI 1640 liquid cultures, after 48 h the Δcyp51A strain was more sensitive to 0.062 μg/ml voriconazole and the Δcyp51B strain was more sensitive to 0.125 μg/ml voriconazole than the wild-type strain, which exhibited growth inhibition only in the presence of concentrations of 0.25 μg/ml voriconazole and above (Fig. 4C). Taken together, the MICs of voriconazole for the respective strains, as determined by CLSI method, were 0.25 μg/ml for the wild type, 0.062 μg/ml for the Δcyp51A strain, and 0.125 μg/ml for the Δcyp51B strain. Although the Cyp51 paralogs appear to compensate for each other’s loss during growth under normal and azole stress conditions, the deletion of cyp51A results in a greater susceptibility to voriconazole than the deletion of cyp51B, indicating that voriconazole is more active on Cyp51B. This also indicates a probable role for Cyp51A in mitigating the consequences of azole exposure without being as directly impacted by the drug as Cyp51B. We cannot, however, exclude the possibility that the enzymatic activity of Cyp51A is a more major contributor to the synthesis of ergosterol than Cyp51B, which may have an impact on the observed differences in the azole susceptibility of the Δcyp51A and Δcyp51B strains.
FIG 4.
(A) The growth of the wild-type akuBKU80 (KU80), Δcyp51A, and Δcyp51B strains was assessed after 5 days on GMM agar with or without voriconazole. (B) Radial growth quantification at the end of the 5-day growth period in the presence or absence of increasing concentrations of voriconazole (0.025 to 0.2 μg/ml). Average values from experiments done in triplicate are depicted. (C) The akuBKU80 (KU80), Δcyp51A, and Δcyp51B strains (104 spores/ml) were cultured in RPMI 1640 liquid medium for 48 h in the presence or absence of increasing concentrations of voriconazole (0.015 to 0.25 μg/ml) and observed with an inverted microscope. All the assays were done in triplicate.
The Cyp51A and Cyp51B proteins display differential localization following exposure to different cell surface stressors.
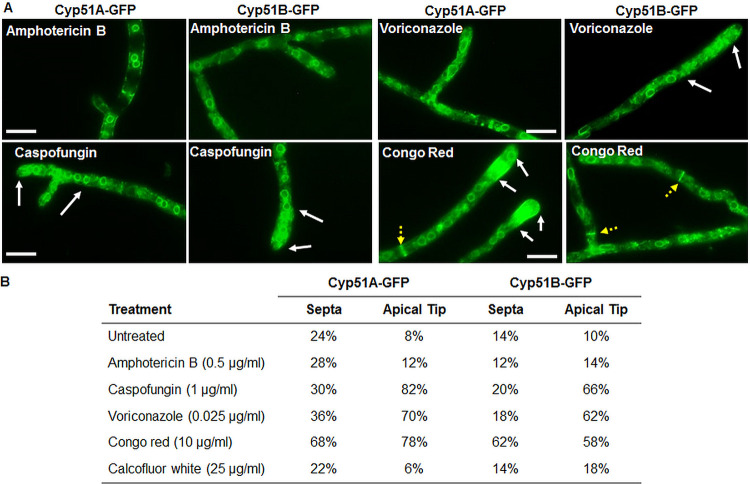
Given the differential azole susceptibility observed in the Δcyp51A and Δcyp51B strains, we next verified the impact of cell surface stress following exposure to the triazoles and other antifungals on the localization pattern of the Cyp51 proteins. The Cyp51A-GFP- and Cyp51B-GFP-expressing strains were exposed to either amphotericin B, caspofungin, voriconazole, Congo red, or calcofluor white for 2 h prior to imaging via fluorescence microscopy (Fig. 5A). Exposure to voriconazole, caspofungin, and Congo red resulted in concentrated GFP fluorescence at the hyphal tips, indicating a gradient-like distribution of the Cyp51 proteins in the ER network. Previous studies have reported such polarized hyphal gradients in the ER network distribution of the ER chaperone BipA and secretory proteins (35, 36). Interestingly, in comparison to the other antifungal treatments, exposure to Congo red resulted in a bright, concentrated GFP signal at several of the hyphal septa. Voriconazole exposure also revealed prominent, punctate GFP signals within the cytoplasm similar to stress granules observed in previous studies of Aspergillus spp. (37). Exposure neither to amphotericin B nor to calcofluor white had any notable effect on the baseline localization pattern of the Cyp51 proteins.
FIG 5.
(A) Localization of the Aspergillus fumigatus Cy51A and Cyp51B proteins was assessed following 2 h of exposure to subinhibitory concentrations of the antifungal agents amphotericin B, caspofungin, voriconazole, and Congo red. White arrows indicate fluorescent apical regions of the hyphae, and yellow dotted arrows indicate fluorescent septa observed with a high frequency in Congo red-treated cultures. (B) Quantification of fluorescent apical tips and septa following treatment with the listed antifungals. Treatment with Congo red resulted in a notable increase in the fluorescence of the septa and the apical regions of the hyphae, while treatment with voriconazole and caspofungin resulted in an increased prevalence of fluorescent apical segments.
Next, to assess the exclusive impact of each of these antifungal agents on the Cyp51A and Cyp51B localization patterns at the peripheral ER, we quantified the fluorescent septa and apical tips across the different stress conditions. Fifty apical tips and septa were observed in each strain following standard growth and exposure to the listed antifungals, with the final percentage of fluorescent structures reported for each condition (Fig. 5B). These counts confirmed our initial observations in support of the dynamic localization of the Cyp51 proteins, with significant increases in hyphal tip localization following exposure to voriconazole, caspofungin, and Congo red and significantly increased septal localization during Congo red exposure. The consistent translocation of Cyp51A and Cyp51B to the apical tip highlights a potential common mechanism by which Aspergillus attempts to compensate for weakening of the vulnerable site of elongation following interference from surface-targeting compounds. This is particularly relevant when considering this translocation in response to Congo red, which specifically targets and weakens the apical tips of the hyphae, leading to eventual tip lysis in filamentous fungi (38). Translocation to the septa following Congo red exposure and granular structures following voriconazole exposure may also reflect a response in which the cell attempts to reinforce critical architectural features or modulate ergosterol production to counteract the negative impacts of chemical stressors.
DISCUSSION
Inhibition of the ergosterol biosynthetic pathway impedes membrane production and hyphal extension through the impairment of membrane integrity (39). Despite the importance of the Cyp51 proteins in ergosterol biosynthesis and azole susceptibility, very little is known regarding the localization and expression of these proteins in response to cell surface stresses or selective Cyp51 impairment. Our findings in this study support the idea that both the A. fumigatus Cyp51A and Cyp51B proteins are involved in the response to stress conditions and that the overlapping functions of these proteins allow them to compensate for each other’s absence at the level of protein expression.
In addition to confirming a baseline perinuclear and peripheral ER localization pattern for the Cyp51 proteins, we also unexpectedly observed the occasional localization to the tubular ER near the septa or hyphal tips. These alternative Cyp51 ER localization compartments were observed only sporadically under standard conditions, potentially representing an important translocation step during the normal activity of Cyp51A and Cyp51B. Notably, growing apical tips require a constant supply of new cell membrane components, including ergosterol (40). The occasional localization of the Cyp51 proteins at the apical tip could represent a normal part of ergosterol biosynthesis proximal to the membrane, occasionally leading to a visibly elevated Cyp51-GFP signal during periods of intense growth. Localization to the hyphal septa could represent a process of membrane maintenance occurring throughout the hyphae, managing the ergosterol content as the hyphae age and elongate. Hyphal septa have been shown to play an essential role in the survival of A. fumigatus following exposure to caspofungin and to increase the cell survival time period following exposure to azoles (12, 41). Though much of this effect is due to the septum’s ability to contain membrane ruptures to single compartments, it is possible that azole inhibition also mitigates Cyp51-mediated maintenance activity around the septa. Thus, the localization of Cyp51 proteins to the septa could be a substantial step in A. fumigatus cell membrane maintenance.
We were able to demonstrate different patterns of Cyp51A and Cyp51B localization following treatment with various cell surface stressors. We found that exposure to subinhibitory concentrations of voriconazole, caspofungin, and Congo red resulted in the increased translocation of Cyp51 to compartments outside of the perinuclear ER. More specifically, exposure to all three compounds resulted in increased translocation to the apical tip of the hyphae, while exposure to Congo red also resulted in increased Cyp51 translocation to the septa. Both Congo red and caspofungin target and interfere with the fungal cell wall, while voriconazole targets the formation of membrane ergosterol (38, 39). The accumulation of Cyp51A and Cyp51B at the apical tip following exposure to these stressors thus may represent a general response by A. fumigatus to stresses placed upon the cell surface during apical growth, even in situations in which ergosterol biosynthesis is not the direct target of the stress. A similar, though more specific, process may be responsible for the substantial accumulation of Cyp51A and Cyp51B at the septa in response to Congo red exposure. The general induction of cell membrane/cell wall biosynthetic machinery following exposure to a cell wall-targeting compound would likely include the mobilization of proteins involved in ergosterol biosynthesis, potentially including a relocation of the Cyp51 proteins to the septa. Given that our observations are based specifically on cultures following 2 h of exposure to cell surface stressors, it is not clear whether this response occurs as part of a larger sequence of stress responses. Furthermore, any connections between the observed translocation and other known stress response pathways in Aspergillus have not been tested. Future work could delve deeper into these connections, revealing whether the dynamic translocation of Cyp51A and Cyp51B occurs as part of a general stress response or as a more specific reaction to individual compounds.
cyp51A and cyp51B are individually dispensable for survival, though the simultaneous removal of both paralogs results in a loss of viability in A. fumigatus. Previous studies have noted that cyp51A is expressed in an inducible manner (14). However, we noted the constitutive expression of the Cyp51A and Cyp51B proteins under control conditions, but their expression seemed to be induced in the presence of voriconazole. Historically, these observations have been explained as a compensation by each protein for the loss of the paralog (42). However, while the overall impact of Cyp51 deletions on viability has been explored previously, much less effort has been made to thoroughly define the consequences of individual Δcyp51A and Δcyp51B mutations on the expression and localization of the surviving paralog. We found that deletion of cyp51A or cyp51B had no noticeable impact on the localization of the remaining paralog, indicating that compensation by the proteins encoded by these genes does not occur through any major changes in localization. Importantly, we did observe that the protein-level expression of Cyp51A increases in the absence of its paralog, Cyp51B, and vice versa. Despite the apparent redundancy of the Cyp51 proteins during radial growth, both the Δcyp51A and Δcyp51B strains were more susceptible to voriconazole than the wild-type strain, indicating that the Cyp51 proteins are not fully redundant in the context of azole susceptibility. The increased susceptibility of the Δcyp51A strain compared to that of the Δcyp51B strain demonstrates that Cyp51A can act as a substantial mitigating factor for the damage caused by azole drugs. On the other hand, the less dramatic increase in susceptibility following deletion of Cyp51B could indicate that it responds to azole exposure in a less dynamic manner, though it remains important as a target of the drug. Our observations support the idea that Cyp51B may be the primary target of azole drugs, such as voriconazole (due to the observed azole hypersensitivity of the Δcyp51A strain), while Cyp51A is an important secondary target of azole drugs through its role as a compensatory protein for an impaired or suppressed Cyp51B. However, we also cannot exclude the possibility of Cyp51A protein being more important for the synthesis of ergosterol, leading to conferring an increase in azole tolerance. In this context, it would be interesting to further investigate how mutations in Cyp51B influence azole resistance in A. fumigatus.
In summary, our results indicate three significant findings: (i) the A. fumigatus Cyp51A and Cyp51B proteins localize to the tubular ER network along the septa and in the peripheral ER network at the apical tip of the hyphae, in addition to their typical localization at the perinuclear ER. The exact reason for this observed effect is currently unclear. (ii) The A. fumigatus Cyp51A and Cyp51B proteins show a gradient distribution in the peripheral ER at the apical tip and along the septum following exposure to cell surface stressors, such as voriconazole, caspofungin, and Congo red. These findings not only indicate a potential role for the Cyp51 proteins during general responses to cell surface stresses but also highlight an aspect of their functionality which could be targeted to impair the pathogen. (iii) Cyp51A protein expression increases in the absence of Cyp51B and vice versa. This finding sheds more light on the interplay between these two proteins, indicating the ability of one to compensate for the other’s absence at the protein level. However, the interplay between the two Cyp51 proteins is not equal in the context of azole resistance, in which the loss of Cyp51A has a greater impact on azole susceptibility than the loss of Cyp51B. While we highlight new and important aspects of the A. fumigatus Cyp51 proteins, future in-depth studies are required to better understand drug action and azole resistance mechanisms.
MATERIALS AND METHODS
Construction of cyp51 mutations in Aspergillus fumigatus.
C-terminal GFP labeling of Cyp51A and Cyp51B was accomplished by insertion of the coding region for each cyp51 gene upstream of egfp within a pUCGH vector. These vectors were then transformed into the akuBKU80 strain of A. fumigatus (43) and screened using hygromycin B as a selective agent, as described previously (44). Deletion of Cyp51A and Cyp51B (producing the Δcyp51A and Δcyp51B strains, respectively) was accomplished by replacing the cyp51A and cyp51B open reading frames with the A. parasiticus pyrG selectable marker in the akuBKU80 pyrG− genetic background (43). Target transformants were then identified by screening for prototrophic transformants, with subsequent confirmation via PCR amplification of the insert/locus junction and Southern blotting for the deletion strains being performed as described previously (44). Paired cyp51A::Cyp51B-GFP and cyp51B::Cyp51A-GFP strains were generated by transforming the Δcyp51A and Δcyp51B strains with the above-described GFP-labeled pUCGH vectors for Cyp51B and Cyp51A, respectively. Target transformants were again identified by screening with hygromycin B as a selective agent, followed by verification of the presence of the GFP tag via amplification of the labeled sequence using a forward primer targeting a genomic DNA sequence upstream of the labeled gene and a reverse primer targeting the egfp coding region. The strains created for this study are listed in Table 1.
TABLE 1.
Strains used in this study
| Strain | Parent strain | Genotype | Reference or source |
|---|---|---|---|
| akuBKU80 | CEA17 | Wild type | 43 |
| akuBKU80 pyrG− | CEA17 pyrG+ | pyrG | 43 |
| Δcyp51A | akuBKU80 pyrG− | cyp51A::pyrG | This study |
| Δcyp51B | akuBKU80 pyrG− | cyp51B::pyrG | This study |
| cyp51A-gfp | akuBKU80 | cyp51Apromo-cyp51A-egfp-hph | This study |
| cyp51B-gfp | akuBKU80 | cyp51Apromo-cyp51A-egfp-hph | This study |
| Δcyp51A::Cyp51B-GFP | akuBKU80 pyrG− | cyp51A::pyrG cyp51Bpromo-cyp51B-egfp-hph | This study |
| Δcyp51B::Cyp51A-GFP | akuBKU80 pyrG− | cyp51B::pyrG cyp51Apromo-cyp51A-egfp-hph | This study |
Radial growth and antifungal susceptibility testing.
Radial growth assays were performed for the Δcyp51A, Δcyp51B, Δcyp51B::Cyp51A-GFP, and Δcyp51A::Cyp51B-GFP strains by point inoculating conidia (104) on solid glucose minimal medium (GMM) agar (1% glucose, pH 6.5) and incubating for 5 days at 37°C. The antifungal susceptibility of the Δcyp51A and Δcyp51B strains was analyzed using standard microdilution techniques, testing in the presence of caspofungin, voriconazole, or amphotericin B (45). The A. fumigatus strain akuBKU80 (the wild type [WT]) and the Δcyp51A and Δcyp51B strains were cultured in the absence or presence of voriconazole (0.025 to 0.2 μg/ml) on GMM agar or RPMI 1640 liquid medium (0.015 to 0.25 μg/ml) in triplicate.
Fluorescence microscopy.
For fluorescence microscopic imaging of the Cyp51A-GFP and Cyp51B-GFP strains, conidia (104) were cultured in glass-bottomed petri dishes (35 mm) containing 3 ml of liquid GMM and incubated at 37°C for 16 h. To determine the impact of subinhibitory antifungal treatment and other cell stressors on the localization of the GFP-tagged Cyp51 proteins, relevant compounds were added individually to the plates after 14 h of incubation and the cultures were allowed to grow for an additional 2 h. Subinhibitory concentrations of amphotericin B (0.5 μg/ml), caspofungin (1 μg/ml), voriconazole (0.025 μg/ml), Congo red (10 μg/ml), and calcofluor white (25 μg/ml) were used for postexposure examination of Cyp51 protein localization. To quantify fluorescent septa and apical tips, 50 of each were counted for bright or background-level fluorescence in the Cyp51A-GFP, Cyp51B-GFP, Δcyp51B::Cyp51A-GFP, and Δcyp51A::Cyp51B-GFP strains following standard growth and exposure to the tested stressors. Fluorescence counts in the septa and apical tips were reported as a final percentage. ER staining was performed on cultures grown for 24 h using ER-Tracker Red dye (Thermo Fisher) according to the manufacturer’s instructions. Briefly, the cultures were incubated with the dye at a 1 μM concentration for 20 to 30 min at 30°C and observed using an Axio Observer 3 microscope (Carl Zeiss) equipped with ZEN lite imaging software.
Protein extraction and Western blot analysis.
Preparation of cell extracts and detection by Western blotting were performed as described earlier (44). Briefly, strains expressing the Cyp51A-GFP and Cyp51B-GFP fusion constructs were grown in GMM liquid medium as shaking cultures for a period of 24 h at 37°C. Cell extracts were then collected from these cultures and used to prepare total protein for each strain. Following quantification and standardization via the Bradford assay, 100 μg of total protein was resolved via electrophoresis through a 4 to 20% SDS-polyacrylamide gel using a Mini-Protean electrophoresis cell (Bio-Rad). Protein was then electroblotted onto a polyvinylidene difluoride membrane (Bio-Rad) and probed with anti-GFP primary antibody (1 μg/ml; GenScript) as a primary antibody. Peroxidase-labeled rabbit anti-IgG (1:5,000; Rockland) was used as a secondary antibody. Detection was carried out by use of the SuperSignal West Pico chemiluminescent substrate (Thermo Scientific).
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by NIH/NIAID R21 award AI127551 to P.R.J. and W.J.S.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci Transl Med 4:165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 2.Lin S-J, Schranz J, Teutsch SM. 2001. Aspergillosis case-fatality rate: systematic review of the literature. Clin Infect Dis 32:358–366. doi: 10.1086/318483. [DOI] [PubMed] [Google Scholar]
- 3.Patterson TF, Thompson GR III, Denning DW, Fishman JA, Hadley S, Herbrecht R, Kontoyiannis DP, Marr KA, Morrison VA, Nguyen MH, Segal BH, Steinbach WJ, Stevens DA, Walsh TJ, Wingard JR, Young J-A, Bennett JE. 2016. Practice guidelines for the diagnosis, and management of aspergillosis: 2016 update by the Infectious Diseases Society of America. Clin Infect Dis 63:e1–e60. doi: 10.1093/cid/ciw326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rivero-Menendez O, Alastruey-Izquierdo A, Mellado E, Cuenca-Estrella M. 2016. Triazole resistance in Aspergillus spp.: a worldwide problem? J Fungi 2:21. doi: 10.3390/jof2030021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baddley JW, Marr KA, Andes DR, Walsh TJ, Kauffman CA, Kontoyiannis DP, Ito JI, Balajee SA, Pappas PG, Moser SA. 2009. Patterns of susceptibility of Aspergillus isolates recovered from patients enrolled in the transplant-associated infection surveillance network. J Clin Microbiol 47:3271–3275. doi: 10.1128/JCM.00854-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiederhold NP, Patterson TF. 2015. Emergence of azole resistance in Aspergillus. Semin Respir Crit Care Med 36:673–680. doi: 10.1055/s-0035-1562894. [DOI] [PubMed] [Google Scholar]
- 7.Chowdhary A, Sharma C, Meis JF. 2017. Azole-resistant aspergillosis: epidemiology, molecular mechanisms, and treatment. J Infect Dis 216:S436–S444. doi: 10.1093/infdis/jix210. [DOI] [PubMed] [Google Scholar]
- 8.Perfect JR. 2017. The antifungal pipeline: a reality check. Nat Rev Drug Discov 16:603–616. doi: 10.1038/nrd.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lepesheva GI, Waterman MR. 2007. Sterol 14α-demethylase cytochrome P450 (CYP51), a P450 in all biological kingdoms. Biochim Biophys Acta 1770:467–477. doi: 10.1016/j.bbagen.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Li L, Lv Q, Yan L, Wang Y, Jiang Y. 2019. The fungal CYP51s: their functions, structures, related drug resistance, and inhibitors. Front Microbiol 10:691. doi: 10.3389/fmicb.2019.00691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dhingra S, Cramer RA. 2017. Regulation of sterol biosynthesis in the human fungal pathogen Aspergillus fumigatus: opportunities for therapeutic development. Front Microbiol 8:92. doi: 10.3389/fmicb.2017.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geißel B, Loiko V, Klugherz I, Zhu Z, Wagener N, Kurzai O, van den Hondel C, Wagener J. 2018. Azole-induced cell wall carbohydrate patches kill Aspergillus fumigatus. Nat Commun 9:3098. doi: 10.1038/s41467-018-05497-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu W, Sillaots S, Lemieux S, Davison J, Kauffman S, Breton A, Linteau A, Xin C, Bowman J, Becker J, Jiang B, Roemer T. 2007. Essential gene identification and drug target prioritization in Aspergillus fumigatus. PLoS Pathog 3:e24. doi: 10.1371/journal.ppat.0030024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hargrove TY, Wawrzak Z, Lamb DC, Guengerich FP, Lepesheva GI. 2015. Structure-functional characterization of cytochrome P450 sterol 14α-demethylase (CYP51B) from Aspergillus fumigatus and molecular basis for the development of antifungal drugs. J Biol Chem 290:23916–23934. doi: 10.1074/jbc.M115.677310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song J, Zhai P, Zhang Y, Zhang C, Sang H, Han G, Keller NP, Lu L. 2016. The Aspergillus fumigatus damage resistance protein family coordinately regulates ergosterol biosynthesis and azole susceptibility. mBio 7:e01919-15. doi: 10.1128/mBio.01919-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howard SJ, Arendrup MC. 2011. Acquired antifungal drug resistance in Aspergillus fumigatus: epidemiology and detection. Med Mycol 49:S90–S95. doi: 10.3109/13693786.2010.508469. [DOI] [PubMed] [Google Scholar]
- 17.Susan JH, Dasa C, Michael JA, Ahmed A, Matthew CF, Alessandro CP, Michel L, Maiken CA, David SP, David WD. 2009. Frequency and evolution of azole resistance in Aspergillus fumigatus associated with treatment failure. Emerg Infect Dis 15:1068–1076. doi: 10.3201/eid1507.090043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Linden JWM, Camps SMT, Kampinga GA, Arends JPA, Debets-Ossenkopp YJ, Haas PJA, Rijnders BJA, Kuijper EJ, van Tiel FH, Varga J, Karawajczyk A, Zoll J, Melchers WJG, Verweij PE. 2013. Aspergillosis due to voriconazole highly resistant Aspergillus fumigatus and recovery of genetically related resistant isolates from domiciles. Clin Infect Dis 57:513–520. doi: 10.1093/cid/cit320. [DOI] [PubMed] [Google Scholar]
- 19.Vermeulen E, Maertens J, Schoemans H, Lagrou K. 2012. Azole-resistant Aspergillus fumigatus due to TR46/Y121F/T289A mutation emerging in Belgium, July 2012. Euro Surveill 17(48):pii=20326. [PubMed] [Google Scholar]
- 20.Chowdhary A, Sharma C, Kathuria S, Hagen F, Meis JF. 2014. Azole-resistant Aspergillus fumigatus with the environmental TR46/Y121F/T289A mutation in India. J Antimicrob Chemother 69:555–557. doi: 10.1093/jac/dkt397. [DOI] [PubMed] [Google Scholar]
- 21.Parker JE, Warrilow AGS, Price CL, Mullins JGL, Kelly DE, Kelly SL. 2014. Resistance to antifungals that target CYP51. J Chem Biol 7:143–161. doi: 10.1007/s12154-014-0121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mellado E, Garcia-Effron G, Alcazar-Fuoli L, Cuenca-Estrella M, Rodriguez-Tudela JL. 2004. Substitutions at methionine 220 in the 14α-sterol demethylase (Cyp51A) of Aspergillus fumigatus are responsible for resistance in vitro to azole antifungal drugs. Antimicrob Agents Chemother 48:2747–2750. doi: 10.1128/AAC.48.7.2747-2750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Umeyama T, Hayashi Y, Shimosaka H, Inukai T, Yamagoe S, Takatsuka S, Hoshino Y, Nagi M, Nakamura S, Kamei K, Ogawa K, Miyazaki Y. 2018. CRISPR/Cas9 genome editing to demonstrate the contribution of Cyp51A Gly138Ser to azole resistance in Aspergillus fumigatus. Antimicrob Agents Chemother 62:e00894-18. doi: 10.1128/AAC.00894-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Rubio R, Cuenca-Estrella M, Mellado E. 2017. Triazole resistance in Aspergillus species: an emerging problem. Drugs 77:599–613. doi: 10.1007/s40265-017-0714-4. [DOI] [PubMed] [Google Scholar]
- 25.Liu M, Zheng N, Li D, Zheng H, Zhang L, Ge H, Liu W. 2016. cyp51A-based mechanism of azole resistance in Aspergillus fumigatus: illustration by a new 3D structural model of Aspergillus fumigatus CYP51A protein. Med Mycol 54:400–408. doi: 10.1093/mmy/myv102. [DOI] [PubMed] [Google Scholar]
- 26.Moreno-Velásquez SD, Seidel C, Juvvadi PR, Steinbach WJ, Read ND. 2017. Caspofungin-mediated growth inhibition and paradoxical growth in Aspergillus fumigatus involve fungicidal hyphal tip lysis coupled with regenerative intrahyphal growth and dynamic changes in β-1,3-glucan synthase localization. Antimicrob Agents Chemother 61:e00710-17. doi: 10.1128/AAC.00710-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noël T. 2012. The cellular and molecular defense mechanisms of the Candida yeasts against azole antifungal drugs. J Mycol Med 22:173–178. doi: 10.1016/j.mycmed.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 28.Cosson P, Letourneur F. 1994. Coatomer interaction with di-lysine endoplasmic reticulum retention motifs. Science 263:1629–1631. doi: 10.1126/science.8128252. [DOI] [PubMed] [Google Scholar]
- 29.Gaynor EC, Graham TR, Emr SD. 1998. COPI in ER/Golgi and intra-Golgi transport: do yeast COPI mutants point the way? Biochim Biophys Acta 1404:33–51. doi: 10.1016/S0167-4889(98)00045-7. [DOI] [PubMed] [Google Scholar]
- 30.Lamb DC, Kelly DE, Venkateswarlu K, Manning NJ, Bligh HFJ, Schunck W-H, Kelly SL. 1999. Generation of a complete, soluble, and catalytically active sterol 14α-demethylase−reductase complex. Biochemistry 38:8733–8738. doi: 10.1021/bi9825089. [DOI] [PubMed] [Google Scholar]
- 31.Markina-Iñarrairaegui A, Pantazopoulou A, Espeso EA, Peñalva MA. 2013. The Aspergillus nidulans peripheral ER: disorganization by ER stress and persistence during mitosis. PLoS One 8:e67154. doi: 10.1371/journal.pone.0067154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia-Effron G, Mellado E, Gomez-Lopez A, Alcazar-Fuoli L, Cuenca-Estrella M, Rodriguez-Tudela JL. 2005. Differences in interactions between azole drugs related to modifications in the 14-α sterol demethylase gene (cyp51A) of Aspergillus fumigatus. Antimicrob Agents Chemother 49:2119–2121. doi: 10.1128/AAC.49.5.2119-2121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-Rubio R, Alcazar-Fuoli L, Monteiro MC, Monzon S, Cuesta I, Pelaez T, Mellado E. 2018. Insight into the significance of Aspergillus fumigatus cyp51A polymorphisms. Antimicrob Agents Chemother 62:e00241-18. doi: 10.1128/AAC.00241-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mellado E, Garcia-Effron G, Buitrago MJ, Alcazar-Fuoli L, Cuenca-Estrella M, Rodriguez-Tudela JL. 2005. Targeted gene disruption of the 14-α sterol demethylase (cyp51A) in Aspergillus fumigatus and its role in azole drug susceptibility. Antimicrob Agents Chemother 49:2536–2538. doi: 10.1128/AAC.49.6.2536-2538.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maruyama J-I, Kikuchi S, Kitamoto K. 2006. Differential distribution of the endoplasmic reticulum network as visualized by the BipA–EGFP fusion protein in hyphal compartments across the septum of the filamentous fungus, Aspergillus oryzae. Fungal Genet Biol 43:642–654. doi: 10.1016/j.fgb.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 36.Kimura S, Maruyama J-I, Watanabe T, Ito Y, Arioka M, Kitamoto K. 2010. In vivo imaging of endoplasmic reticulum and distribution of mutant α-amylase in Aspergillus oryzae. Fungal Genet Biol 47:1044–1054. doi: 10.1016/j.fgb.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 37.Huang H-T, Maruyama J-I, Kitamoto K. 2013. Aspergillus oryzae AoSO is a novel component of stress granules upon heat stress in filamentous fungi. PLoS One 8:e72209. doi: 10.1371/journal.pone.0072209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roncero C, Durán A. 1985. Effect of calcofluor white and Congo red on fungal cell wall morphogenesis: in vivo activation of chitin polymerization. J Bacteriol 163:1180–1185. doi: 10.1128/JB.163.3.1180-1185.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghannoum MA, Rice LB. 1999. Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin Microbiol Rev 12:501–517. doi: 10.1128/CMR.12.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steinberg G. 2007. Hyphal growth: a tale of motors, lipids, and the Spitzenkörper. Eukaryot Cell 6:351–360. doi: 10.1128/EC.00381-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dichtl K, Samantaray S, Aimanianda V, Zhu Z, Prévost M-C, Latgé J-P, Ebel F, Wagener J. 2015. Aspergillus fumigatus devoid of cell wall β-1,3-glucan is viable, massively sheds galactomannan and is killed by septum formation inhibitors. Mol Microbiol 95:458–471. doi: 10.1111/mmi.12877. [DOI] [PubMed] [Google Scholar]
- 42.Mellado E, Diaz-Guerra TM, Cuenca-Estrella M, Rodriguez-Tudela JL. 2001. Identification of two different 14-α sterol demethylase-related genes cyp51A and cyp51B in Aspergillus fumigatus and other Aspergillus species. J Clin Microbiol 39:2431–2438. doi: 10.1128/JCM.39.7.2431-2438.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.da Silva Ferreira ME, Kress M, Savoldi M, Goldman MHS, Härtl A, Heinekamp T, Brakhage AA, Goldman GH. 2006. The akuBKU80 mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot Cell 5:207–211. doi: 10.1128/EC.5.1.207-211.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Juvvadi PR, Fortwendel JR, Rogg LE, Burns KA, Randell SH, Steinbach WJ. 2011. Localization and activity of the calcineurin catalytic and regulatory subunit complex at the septum is essential for hyphal elongation and proper septation in Aspergillus fumigatus. Mol Microbiol 82:1235–1259. doi: 10.1111/j.1365-2958.2011.07886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clinical and Laboratory Standards Institute. 2017. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi, 3rd ed CLSI document M38-A2 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.