Single nucleotide polymorphisms in the OATP1B1 transporter have been suggested to partially explain the large interindividual variation in rifampicin exposure. HEK293 cells overexpressing wild-type (WT) or OATP1B1 variants *1b, *4, *5, and *15 were used to determine the in vitro rifampicin intrinsic clearance. For OATP1B1*5 and *15, a 36% and 42% reduction in intrinsic clearance, respectively, compared to WT was found. We consider that these differences in intrinsic clearance most likely have minor clinical implications.
KEYWORDS: OATP1B1, SNP, drug transport, rifampicin
ABSTRACT
Single nucleotide polymorphisms in the OATP1B1 transporter have been suggested to partially explain the large interindividual variation in rifampicin exposure. HEK293 cells overexpressing wild-type (WT) or OATP1B1 variants *1b, *4, *5, and *15 were used to determine the in vitro rifampicin intrinsic clearance. For OATP1B1*5 and *15, a 36% and 42% reduction in intrinsic clearance, respectively, compared to WT was found. We consider that these differences in intrinsic clearance most likely have minor clinical implications.
TEXT
Tuberculosis (TB) is the leading cause of death from an infectious agent. In 2018, an estimated 10 million people developed TB and 1.45 million patients died (1). Rifampicin was first used clinically in 1966 and now is the keystone of TB treatment. Together with pyrazinamide, it enabled short-course TB chemotherapy (2). However, there is a large interindividual variability in the plasma pharmacokinetics (PK) of rifampicin (3), and low plasma concentrations of rifamycins (e.g., rifampicin and rifabutin) have been associated with treatment failure, relapse, and resistance (4). Both nongenetic factors (e.g., age, comorbidities, concomitant therapy) as well as genetic factors (e.g., sequence variants in genes encoding drug-metabolizing enzymes and transporters) contribute to this interindividual variability and influence the effect of TB drugs (5, 6).
The organic anion transporter polypeptide 1B1 (OATP1B1), located at the sinusoidal (basolateral) membrane of hepatocytes, mediates the uptake of a broad range of compounds, including rifampicin (7, 8). A large number of >45 nonsynonymous variants have been found in the solute carrier organic anion transporter gene (SLCO1B1) encoding OATP1B1 (9). Next to the wild-type haplotype, there are four common haplotypes resulting from three single nucleotide polymorphisms (SNP), which are summarized in Table 1. Their in vitro activity has been investigated for several (endogenous) substrates, often resulting in decreased activity, mainly for OATP1B1*5 and *15 (8, 10–14). The effect of SNPs on the transporter activity can be substrate specific (8), and therefore it is important to examine rifampicin-specific transport by OATP1B1 variants. Nonetheless, in vitro transport of rifampicin by different forms of OATP1B1 has only been investigated in one previous study, at only a single concentration, and therefore transport kinetics remain unknown (12). In clinical studies, decreased plasma exposure (∼40%) has been observed in patients carrying OATP1B1*4, probably indicating increased transporter activity (15, 16). In contrast, OATP1B1*1b was not associated with altered rifampicin exposure, nor was OATP1B1*5 (15–17). Rifampicin plasma exposure data related to OATP1B1*15 were not found in literature, but the *15 haplotype did show a significant association between rifampicin and susceptibility to drug-induced liver injury (DILI) in a Chinese population. In the same study, the in vitro uptake of the bile acid taurocholic acid (TCA) in OATP1B1 was measured, showing decreased uptake of TCA in *15-expressing cells compared to wild-type cells. This uptake could be further reduced by rifampicin acting as an inhibitor (18). Increased bile acids have previously been associated with drug-induced cholestasis and DILI (19, 20).
TABLE 1.
Literature overview of the tested haplotypes with corresponding nucleotide and amino acid changes, their allele frequencies in different ethnicities, residual in vitro activity for (endogenous) substrates (estrone-3-sulfate, estradiol-17β-d-glucuronide, pravastatin, atorvastatin, rosuvastatin, rifampicin), in vivo exposure to rifampicin, and transporter protein expression compared to OATP1B1 wild type in vivon
| Haplotype | Nucleotide change | Amino acid change | Allele frequency (%) |
Residual activity |
Protein expression (fold change) in vivo | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| European | East Asian | Central/south Asian | American | Sub-Saharan African | (Endogenous) substrates – in vitro | Rifampicin exposure – in vivo | ||||
| *1b | 388A>G | N130D | 30–47a,b | 59–86a,b | 42–52b | 55–71b | 72–84a,b | ↓↔↑ (∼35%–125%)c-h | ↔i | ↑ (∼1.5×)k,l |
| *4 | 463C>A | P155T | 13–23a,b | 0–3a,b | 5–10b | 0–6b | 2–10a,b | ↔c,g,h | ↓ (58–65%)i,j | ↑ (2.1×)l,m |
| *5 | 521T>C | V174A | 8–20a,b | 10–16a,b | 7–13b | 18–32b | 1–8a,b | ↓ (∼5%–80%)c-e,g,h | ↔i,j | ↔↓ (∼0.75×)k,l |
| *15 | 388A>G, 521T>C | N130D, V174A | 16b | 12b | 9b | 24b | 2b | ↓ (∼20%–55%)d,e | ? | ↔k,l |
Niemi et al. (8).
Pasanen et al. (14).
Tirona et al. (11).
Kameyama et al. (27).
Nozawa et al. (28).
Iwai et al. (29).
Tirona et al. (12).
Ho et al. (30).
Weiner et al. (15).
Kwara et al. (16).
Nies et al. (31).
Prasad et al. (22).
Genotype-based (without regard to haplotypes).
Arrows and percentages for the in vitro and in vivo activity represent the residual in vitro activity and in vivo plasma exposure of variants compared to wild-type OATP1B1 (set at 100%). For protein expression, the fold change relative to wild-type OATP1B1 is presented. Values in italic are the residual activity/fold change in protein expression compared to OAPT1B1 wild type, as far as they could be extracted from data presented in literature; for the activity, this is often measured at one concentration (i.e., no enzyme kinetics).
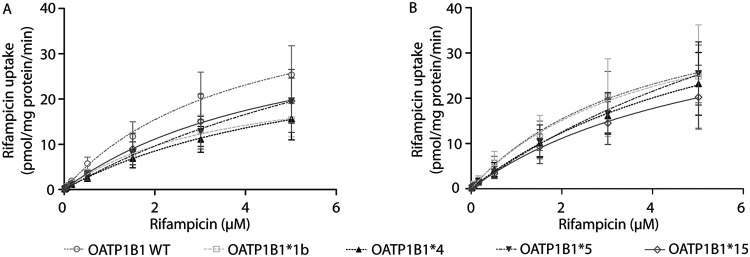
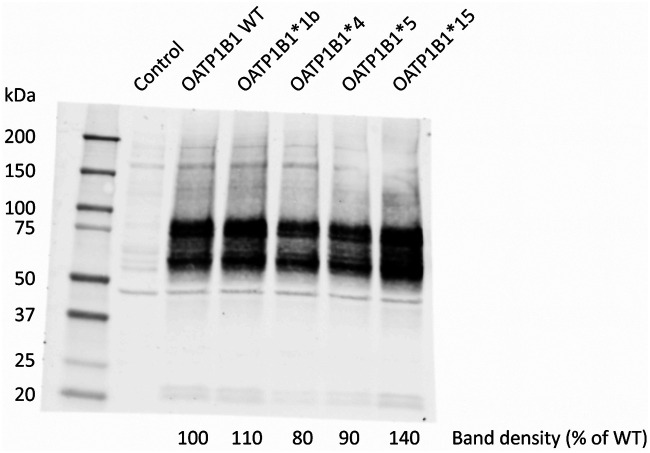
In this study, we aimed to identify the in vitro activity of OATP1B1 WT, *1b, *4, *5, and *15 for rifampicin transport. A detailed overview of the methods used can be found in the supplemental material file. In brief, human embryonic kidney 293 (HEK293) cells were transiently transduced with OATP1B1 WT, one of the SNP variants, or the vector control (i.e., background uptake). After confirmation of functional OATP1B1-mediated uptake with the model substrate [3H]-estradiol 17 β-d-glucuronide (E217βG; 18.9 nM) for OATP1B1 WT, *1b, *4, and *5 (see Fig. S1 in the supplemental material), cells were incubated with [3H]-rifampicin (46.6 Ci/mmol; Moravek Biochemicals, Brea, CA, USA). First, the time-dependent uptake of rifampicin in OATP1B1 WT was assessed, resulting in a linear (not saturated) uptake from 0 to 1 min (Fig. S2). Second, cells were incubated for 1 min with different concentrations of rifampicin (13.7 nM [3H]-rifampicin supplemented with 0.015, 0.05, 0.15, 0.5, 1.5, 3, and 5 μM rifampicin). Due to the high lipophilicity of rifampicin, there was high uptake via passive diffusion. Therefore, 5 μM was the highest concentration that could be tested in our system. As a result, it was not possible to reliably determine the maximum transport velocity (Vmax) and the substrate concentration at which half of this rate is obtained (Km). Hence, the intrinsic clearance (CLint, by definition, Vmax/Km, in our case determined with the slope of the linear part of the concentration-dependent uptake curve) was determined instead and used to compare the different OATP1B1 haplotypes. The mean intrinsic clearance was decreased to 64%, 51%, 49%, and 57% for *1b, *4, *5, and *15, compared to OATP1B1 WT, respectively (Table 2 and Fig. 1A). As a third step, we accounted for differences in transporter expression levels. Western blotting was performed using membrane fractions from the transduced HEK293 cells per virus batch using an equal amount of protein (25 μg). Proteins were separated on an SDS-PAGE gradient gel (4% to 20%), and OATP1B1 proteins were detected using a polyclonal anti-OATP2 antibody (1:1,000, which was a kind gift from B. Stieger, University Hospital Zurich, Zurich, Switzerland [21]) followed by an Alexa Fluor 680 goat anti-rabbit IgG secondary antibody (1:10,000; Life Technologies Invitrogen). The molecular mass of the glycosylated band was approximately 75 kDa; however, both the glycosylated and nonglycosylated band were used for quantification (Fig. 2). The mean expression levels of OATP1B1*1b, *4, and *5 were lower than OATP1B1 WT. Subsequently, intrinsic clearance values were normalized per experiment for the OATP1B1 protein expression, and significance was tested by four one-sample t tests comparing the experimental groups to the wild type (set at 100%). The mean normalized intrinsic clearance value of OATP1B1*5 and *15 were reduced to 64% (P = 0.04) and 58% (P = 0.01) of OATP1B1 WT, respectively, whereas the intrinsic clearance values of OATP1B1*1b and *4 were similar to OATP1B1 WT (Table 2, Fig. 1B).
TABLE 2.
Intrinsic clearance of rifampicin by different variants of OATP1B1f
| Haplotype |
CLint (mean± SD, μl/mg protein/min)a |
Relative transporter expression (mean ± SD, % of WT)b | Transporter expression corrected CLint (mean ± SD, % of WT)c |
|---|---|---|---|
| OATP1B1 WT | 11.3 ± 4.4 | 100 | 100 |
| OATP1B1*1b | 7.2 ± 2.7 | 77 ± 37 | 92 ± 30 |
| OATP1B1*4 | 5.8 ± 3.3 | 68 ± 13 | 73 ± 18 |
| OATP1B1*5 | 5.6 ± 2.7 | 78 ± 9 | 64 ± 14d |
| OATP1B1*15 | 6.5 ± 3.4 | 95 ± 41 | 58 ± 8e |
Slope of the concentration-dependent uptake curve based on the mean rifampicin uptake determined in three independent experiments.
Transporter expression was determined and normalized to wild-type expression per independent experiment (i.e., three times), after which, the mean relative transporter expression per variant was calculated.
Data per individual experiment were analyzed to determine the intrinsic clearance and normalized to the intrinsic clearance of the wild-type transporter per experiment. Subsequently, the CLint values were normalized for the expression of OATP1B1 protein per individual experiment, determined by Western blotTIN, an example of which is shown in Fig. 2. Significance was tested by four one-sample t tests comparing the experimental group to the wild type (100%).
P = 0.04.
P = 0.01.
SD, standard deviation.
FIG 1.
(A and B) Concentration-dependent uptake curve for OATP1B1-mediated uptake of rifampicin (A) and corrected for transporter expression by Western blotting (B). The transport velocity was determined by examining the uptake of rifampicin in OATP1B1 WT, *1b, *4, *5, and *15. The OATP1B1 mediated transport was obtained by subtracting the transport velocity in vector-transduced cells from those in OATP1B1 WT or variant-expressing cells. Each point and bar represents the mean ± standard error of the mean from three independent experiments. The solid/dotted lines represent the fitted lines.
FIG 2.
Quantification of OATP1B1 protein expression in HEK293 cells of one of the three independent experiments. Membrane fractions were obtained from HEK293 cells and separated by SDS-PAGE (4% to 20%). The applied amount of protein was 25 μg, and total protein staining (Ponceau) confirmed equal loading across the lanes. The OATP1B1 proteins were detected using a polyclonal anti-OATP2 antibody for OATP1B1 followed by an Alexa Fluor 680 goat anti-rabbit IgG antibody secondary antibody.
The lack of a difference in intrinsic clearance between OATP1B1*1b and WT is in agreement with literature, where this haplotype has not been associated with differences in rifampicin exposure in vivo (15, 16). In contrast, the amino acid change P155T (*4) has been associated with ∼40% lower rifampicin concentrations in vivo (15, 16). However, in this in vitro study, we did not observe an increased intrinsic clearance of rifampicin in OATP1B1*4-expressing cells compared to WT cells. Also, Tirona et al. did not find an increased activity of OATP1B1*4 in HeLa cells, although they studied only a single rifampicin concentration (12), nor was a difference observed in in vitro experiments of other (endogenous) substrates (8, 14). Prasad et al. reported that OATP1B1 expression in liver samples is about 2-fold higher in both the hetero- and homozygous c.463C>A (irrespective of haplotype) samples compared to CC samples (22). This increased transporter expression may be associated with decreased rifampicin plasma concentrations without a difference in observed transporter activity in our in vitro study.
We did find a reduction in intrinsic clearance for OATP1B1*5 (residual activity, 64%) and *15 (residual activity, 58%) compared to WT. This is in line with the reduction we observed in transport of the model substrate E217βG by OATP1B1*5 (residual activity, 44%; Fig. S1), as well as with previously reported residual transport of E217βG, estrone-3-sulfate, pravastatin, atorvastatin, and rosuvastatin by OATP1B1*5 and *15 (Table 1). Therefore, we hypothesize that the nucleotide change 521T>C is the SNP causing functional alteration of OATP1B1*15 in in vitro studies, including our study. However, in vivo, no association has been found between rifampicin plasma concentrations and OATP1B1*5 (15–17). For *15, no specific rifampicin exposure data have been reported in literature, even though an increased susceptibility to DILI has been described. We hypothesize that in vivo, other uptake transporters such as OATP1B3 may compensate for any reduced activity of OATP1B1*5 and *15 (23). Indeed, Vavricka et al. showed that rifampicin is transported by OATP1B3 and that the apparent Km value of rifampicin for OATP1B3 (2.3 μM) is slightly lower than for OATP1B1 (13 μM) (24). Furthermore, rifampicin is known for its (auto)inducing effect on metabolizing enzymes and drug transporters, including OATP1B1, probably inducing its net activity. Only limited data are available about whether SNPs impact the extent of OATP1B1 induction (25). Though we expect the impact of OATP1B1 SNPs on intrinsic transport activity and, consequently, rifampicin plasma exposure to be minor, it may still be a contributing factor to the high interindividual variability (up to 5-fold) in rifampicin plasma exposure observed in humans. This may also be relevant to the study and implementation of high-dose rifampicin, where the high interindividual variability can still result in relatively low individual exposures, causing treatment failures and preventing treatment shortening (26). Finally, we acknowledge that the applied overexpression system has its limitations, as it only considers differences in intrinsic activity between polymorphisms in OATP1B1, not including the impact of other transporters and metabolizing enzymes involved in rifampicin distribution and metabolism, and it cannot reflect possible SNP-associated protein expression differences in vivo.
In conclusion, we only found a reduction in the in vitro intrinsic clearance of rifampicin by OATP1B1*5 and *15. However, we do not expect these reductions to have significant clinical implications, as relevant compensatory mechanisms (e.g., OATP1B3 transport) could easily outweigh these effects in vivo.
Supplementary Material
ACKNOWLEDGMENTS
We thank H. Kidron (University of Helsinki, Helsinki, Finland) for kindly providing the OATP1B1*5 vector. We thank B. Stieger (University Hospital Zurich, Zurich, Switzerland) for providing the rabbit polyclonal anti-OATP2 antibody.
This project was supported by a Radboudumc RIHS Junior Researcher Round Grant 2017.
J.J.M.W.V.D.H. and J.B.K. are founders of PharmTox (Nijmegen, the Netherlands).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.WHO. 2019. Global tuberculosis report. World Health Organization, Geneva, Switzerland: License: CC BY-NC-SA 3.0 IGO. [Google Scholar]
- 2.Murray JF, Schraufnagel DE, Hopewell PC. 2015. Treatment of tuberculosis. A historical perspective. Ann Am Thorac Soc 12:1749–1759. doi: 10.1513/AnnalsATS.201509-632PS. [DOI] [PubMed] [Google Scholar]
- 3.Stott KE, Pertinez H, Sturkenboom MGG, Boeree MJ, Aarnoutse R, Ramachandran G, Requena-Mendez A, Peloquin C, Koegelenberg CFN, Alffenaar JWC, Ruslami R, Tostmann A, Swaminathan S, McIlleron H, Davies G. 2018. Pharmacokinetics of rifampicin in adult TB patients and healthy volunteers: a systematic review and meta-analysis. J Antimicrob Chemother 73:2305–2313. doi: 10.1093/jac/dky152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiner M, Benator D, Burman W, Peloquin CA, Khan A, Vernon A, Jones B, Silva-Trigo C, Zhao Z, Hodge T, Tuberculosis Trials Consortium. 2005. Association between acquired rifamycin resistance and the pharmacokinetics of rifabutin and isoniazid among patients with HIV and tuberculosis. Clin Infect Dis 40:1481–1491. doi: 10.1086/429321. [DOI] [PubMed] [Google Scholar]
- 5.McIlleron H, Abdel-Rahman S, Dave JA, Blockman M, Owen A. 2015. Special populations and pharmacogenetic issues in tuberculosis drug development and clinical research. J Infect Dis 211 Suppl 3:S115–25. doi: 10.1093/infdis/jiu600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans WE, McLeod HL. 2003. Pharmacogenomics: drug disposition, drug targets, and side effects. N Engl J Med 348:538–549. doi: 10.1056/NEJMra020526. [DOI] [PubMed] [Google Scholar]
- 7.Giacomini KM, Huang SM, Tweedie DJ, Benet LZ, Brouwer KL, Chu X, Dahlin A, Evers R, Fischer V, Hillgren KM, Hoffmaster KA, Ishikawa T, Keppler D, Kim RB, Lee CA, Niemi M, Polli JW, Sugiyama Y, Swaan PW, Ware JA, Wright SH, Yee SW, Zamek-Gliszczynski MJ, Zhang L, International Transporter Consortium. 2010. Membrane transporters in drug development. Nat Rev Drug Discov 9:215–236. doi: 10.1038/nrd3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niemi M, Pasanen MK, Neuvonen PJ. 2011. Organic anion transporting polypeptide 1B1: a genetically polymorphic transporter of major importance for hepatic drug uptake. Pharmacol Rev 63:157–181. doi: 10.1124/pr.110.002857. [DOI] [PubMed] [Google Scholar]
- 9.Lee HH, Ho RH. 2017. Interindividual and interethnic variability in drug disposition: polymorphisms in organic anion transporting polypeptide 1B1 (OATP1B1; SLCO1B1). Br J Clin Pharmacol 83:1176–1184. doi: 10.1111/bcp.13207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van de Steeg E, Greupink R, Schreurs M, Nooijen IH, Verhoeckx KC, Hanemaaijer R, Ripken D, Monshouwer M, Vlaming ML, DeGroot J, Verwei M, Russel FG, Huisman MT, Wortelboer HM. 2013. Drug-drug interactions between rosuvastatin and oral antidiabetic drugs occurring at the level of OATP1B1. Drug Metab Dispos 41:592–601. doi: 10.1124/dmd.112.049023. [DOI] [PubMed] [Google Scholar]
- 11.Tirona RG, Leake BF, Merino G, Kim RB. 2001. Polymorphisms in OATP-C: identification of multiple allelic variants associated with altered transport activity among European- and African-Americans. J Biol Chem 276:35669–35675. doi: 10.1074/jbc.M103792200. [DOI] [PubMed] [Google Scholar]
- 12.Tirona RG, Leake BF, Wolkoff AW, Kim RB. 2003. Human organic anion transporting polypeptide-C (SLC21A6) is a major determinant of rifampin-mediated pregnane X receptor activation. J Pharmacol Exp Ther 304:223–228. doi: 10.1124/jpet.102.043026. [DOI] [PubMed] [Google Scholar]
- 13.Yang F, Xiong X, Liu Y, Zhang H, Huang S, Xiong Y, Hu X, Xia C. 2018. CYP2C9 and OATP1B1 genetic polymorphisms affect the metabolism and transport of glimepiride and gliclazide. Sci Rep 8:10994. doi: 10.1038/s41598-018-29351-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pasanen MK, Neuvonen PJ, Niemi M. 2008. Global analysis of genetic variation in SLCO1B1. Pharmacogenomics 9:19–33. doi: 10.2217/14622416.9.1.19. [DOI] [PubMed] [Google Scholar]
- 15.Weiner M, Peloquin C, Burman W, Luo CC, Engle M, Prihoda TJ, Mac Kenzie WR, Bliven-Sizemore E, Johnson JL, Vernon A. 2010. Effects of tuberculosis, race, and human gene SLCO1B1 polymorphisms on rifampin concentrations. Antimicrob Agents Chemother 54:4192–4200. doi: 10.1128/AAC.00353-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwara A, Cao L, Yang H, Poethke P, Kurpewski J, Tashima KT, Mahjoub BD, Court MH, Peloquin CA. 2014. Factors associated with variability in rifampin plasma pharmacokinetics and the relationship between rifampin concentrations and induction of efavirenz clearance. Pharmacotherapy 34:265–271. doi: 10.1002/phar.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He YJ, Zhang W, Chen Y, Guo D, Tu JH, Xu LY, Tan ZR, Chen BL, Li Z, Zhou G, Yu BN, Kirchheiner J, Zhou HH. 2009. Rifampicin alters atorvastatin plasma concentration on the basis of SLCO1B1 521T>C polymorphism. Clin Chim Acta 405:49–52. doi: 10.1016/j.cca.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 18.Li LM, Chen L, Deng GH, Tan WT, Dan YJ, Wang RQ, Chen WS. 2012. SLCO1B1 *15 haplotype is associated with rifampin-induced liver injury. Mol Med Rep 6:75–82. doi: 10.3892/mmr.2012.900. [DOI] [PubMed] [Google Scholar]
- 19.Ma Z, Wang X, Yin P, Wu R, Zhou L, Xu G, Niu J. 2019. Serum metabolome and targeted bile acid profiling reveals potential novel biomarkers for drug-induced liver injury. Medicine (Baltimore, MD) 98:e16717. doi: 10.1097/MD.0000000000016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kock K, Ferslew BC, Netterberg I, Yang K, Urban TJ, Swaan PW, Stewart PW, Brouwer KL. 2014. Risk factors for development of cholestatic drug-induced liver injury: inhibition of hepatic basolateral bile acid transporters multidrug resistance-associated proteins 3 and 4. Drug Metab Dispos 42:665–674. doi: 10.1124/dmd.113.054304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reichel C, Gao B, Van Montfoort J, Cattori V, Rahner C, Hagenbuch B, Stieger B, Kamisako T, Meier PJ. 1999. Localization and function of the organic anion-transporting polypeptide Oatp2 in rat liver. Gastroenterology 117:688–695. doi: 10.1016/s0016-5085(99)70463-4. [DOI] [PubMed] [Google Scholar]
- 22.Prasad B, Evers R, Gupta A, Hop CE, Salphati L, Shukla S, Ambudkar SV, Unadkat JD. 2014. Interindividual variability in hepatic organic anion-transporting polypeptides and P-glycoprotein (ABCB1) protein expression: quantification by liquid chromatography tandem mass spectroscopy and influence of genotype, age, and sex. Drug Metab Dispos 42:78–88. doi: 10.1124/dmd.113.053819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niemi M, Kivisto KT, Diczfalusy U, Bodin K, Bertilsson L, Fromm MF, Eichelbaum M. 2006. Effect of SLCO1B1 polymorphism on induction of CYP3A4 by rifampicin. Pharmacogenet Genomics 16:565–568. doi: 10.1097/01.fpc.0000215070.52212.0e. [DOI] [PubMed] [Google Scholar]
- 24.Vavricka SR, Van Montfoort J, Ha HR, Meier PJ, Fattinger K. 2002. Interactions of rifamycin SV and rifampicin with organic anion uptake systems of human liver. Hepatology 36:164–172. doi: 10.1053/jhep.2002.34133. [DOI] [PubMed] [Google Scholar]
- 25.Niemi M, Backman JT, Fromm MF, Neuvonen PJ, Kivisto KT. 2003. Pharmacokinetic interactions with rifampicin: clinical relevance. Clin Pharmacokinet 42:819–850. doi: 10.2165/00003088-200342090-00003. [DOI] [PubMed] [Google Scholar]
- 26.Boeree MJ, Diacon AH, Dawson R, Narunsky K, Du Bois J, Venter A, Phillips PP, Gillespie SH, McHugh TD, Hoelscher M, Heinrich N, Rehal S, van Soolingen D, van Ingen J, Magis-Escurra C, Burger D, Plemper van Balen G, Aarnoutse RE, PanACEA Consortium. 2015. A dose-ranging trial to optimize the dose of rifampin in the treatment of tuberculosis. Am J Respir Crit Care Med 191:1058–1065. doi: 10.1164/rccm.201407-1264OC. [DOI] [PubMed] [Google Scholar]
- 27.Kameyama Y, Yamashita K, Kobayashi K, Hosokawa M, Chiba K. 2005. Functional characterization of SLCO1B1 (OATP-C) variants, SLCO1B1*5, SLCO1B1*15 and SLCO1B1*15+C1007G, by using transient expression systems of HeLa and HEK293 cells. Pharmacogenet Genomics 15:513–522. doi: 10.1097/01.fpc.0000170913.73780.5f. [DOI] [PubMed] [Google Scholar]
- 28.Nozawa T, Minami H, Sugiura S, Tsuji A, Tamai I. 2005. Role of organic anion transporter OATP1B1 (OATP-C) in hepatic uptake of irinotecan and its active metabolite, 7-ethyl-10-hydroxycamptothecin: in vitro evidence and effect of single nucleotide polymorphisms. Drug Metab Dispos 33:434–439. doi: 10.1124/dmd.104.001909. [DOI] [PubMed] [Google Scholar]
- 29.Iwai M, Suzuki H, Ieiri I, Otsubo K, Sugiyama Y. 2004. Functional analysis of single nucleotide polymorphisms of hepatic organic anion transporter OATP1B1 (OATP-C). Pharmacogenetics 14:749–757. doi: 10.1097/00008571-200411000-00006. [DOI] [PubMed] [Google Scholar]
- 30.Ho RH, Tirona RG, Leake BF, Glaeser H, Lee W, Lemke CJ, Wang Y, Kim RB. 2006. Drug and bile acid transporters in rosuvastatin hepatic uptake: function, expression, and pharmacogenetics. Gastroenterology 130:1793–1806. doi: 10.1053/j.gastro.2006.02.034. [DOI] [PubMed] [Google Scholar]
- 31.Nies AT, Niemi M, Burk O, Winter S, Zanger UM, Stieger B, Schwab M, Schaeffeler E. 2013. Genetics is a major determinant of expression of the human hepatic uptake transporter OATP1B1, but not of OATP1B3 and OATP2B1. Genome Med 5:1. doi: 10.1186/gm405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.