Clonal outbreaks of fluconazole-resistant (FLZR) Candida parapsilosis isolates have been reported in several countries. Despite its being the second leading cause of candidemia, the azole resistance mechanisms and the clonal expansion of FLZR C. parapsilosis blood isolates have not been reported in Turkey. In this study, we consecutively collected C. parapsilosis blood isolates (n = 225) from the fifth largest hospital in Turkey (2007 to 2019), assessed their azole susceptibility pattern using CLSI M27-A3/S4, and sequenced ERG11 for all and MRR1, TAC1, and UPC2 for a selected number of C. parapsilosis isolates.
KEYWORDS: Candida, Candida parapsilosis, drug resistance mechanisms, fluconazole
ABSTRACT
Clonal outbreaks of fluconazole-resistant (FLZR) Candida parapsilosis isolates have been reported in several countries. Despite its being the second leading cause of candidemia, the azole resistance mechanisms and the clonal expansion of FLZR C. parapsilosis blood isolates have not been reported in Turkey. In this study, we consecutively collected C. parapsilosis blood isolates (n = 225) from the fifth largest hospital in Turkey (2007 to 2019), assessed their azole susceptibility pattern using CLSI M27-A3/S4, and sequenced ERG11 for all and MRR1, TAC1, and UPC2 for a selected number of C. parapsilosis isolates. The typing resolution of two widely used techniques, amplified fragment length polymorphism typing (AFLP) and microsatellite typing (MST), and the biofilm production of FLZR isolates with and without Y132F were compared. Approximately 27% of isolates were FLZR (60/225), among which 90% (54/60) harbored known mutations in Erg11, including Y132F (24/60) and Y132F+K143R (19/60). Several mutations specific to FLZR isolates were found in MRR1, TAC1, and UPC2. AFLP grouped isolates into two clusters, while MST revealed several clusters. The majority of Y132F/Y132F+K143R isolates grouped in clonal clusters, which significantly expanded throughout 2007 to 2019 in neonatal wards. Candida parapsilosis isolates carrying Y132F were associated with significantly higher mortality and less biofilm production than other FLZR isolates. Collectively, we documented the first outbreak of FLZR C. parapsilosis blood isolates in Turkey. The MRR1, TAC1, and UPC2 mutations exclusively found in FLZR isolates establishes a basis for future studies, which will potentially broaden our knowledge of FLZR mechanisms in C. parapsilosis. MST should be a preferred method for clonal analysis of C. parapsilosis isolates in outbreak scenarios.
INTRODUCTION
Bloodstream infections due to Candida species, i.e., candidemia, are associated with high morbidity and mortality, resulting in significant health care costs of $1.4 billion in the United States annually (1). Candida parapsilosis inhabits the gastrointestinal tract of 35% of healthy individuals (2) and ranks as the first to third cause of candidemia depending on geography, patients’ underlying condition, and age (3). The ability of C. parapsilosis to produce tenacious biofilms accounts for its persistence in clinical settings and poses the risk of future clonal outbreaks (3). The sibling species of C. parapsilosis, Candida orthopsilosis and Candida metapsilosis, have also been implicated in candidemia (4). Candida parapsilosis blood isolates were once thought to be universally fluconazole (FLZ) susceptible (FLZS), but a recent candidemia study conducted in South Africa indicates that over half of C. parapsilosis isolates are FLZ resistant (FLZR), and 44% of the latter are cross-resistant to voriconazole (VRZ) (5). Other studies performed in India (6), South Korea (7), Kuwait, Brazil (8), and the United States (9, 10) and a recent global study (11) confirmed the emergence of FLZR C. parapsilosis. Given that FLZ is the main antifungal drug used in developing countries (5, 12), the emergence of FLZR isolates undermines the efficacy of FLZ in the treatment of candidemia. Prolonged previous exposure to FLZ in clinical settings is believed to be a factor related to FLZ resistance (13), which is underlain by specific mutations in the ergosterol biosynthesis gene, ERG11, yielding amino acid substitutions such as Y132F and K143R, which alter the 3D conformation of Erg11 and reduce its affinity for FLZ (14). Moreover, gain-of-function (GOF) mutations in MRR1, TAC1, and UPC2 genes that cause overexpression of efflux pumps (Cdr1 and Mdr1) and Erg11 are also known to contribute to azole resistance in Candida species (14).
Candida parapsilosis infections may be spread by health care workers (3), and isolates from outbreaks can be more virulent than sporadic isolates (15). As C. parapsilosis is among the most genetically homogenous Candida species (3), the use of highly resolutive typing techniques is important to differentiate outbreak from nonoutbreak isolates. Amplified fragment length polymorphism typing (AFLP) (16) and microsatellite typing (MST) (6) have been used to explore the genotypic diversity of clinical C. parapsilosis isolates, but there is no study comparing the performance of the two techniques.
In this single-center study, we investigated the nature of an unusually high prevalence of C. parapsilosis blood isolates collected over 13 years (2007 to 2019) in Ege University Hospital, Izmir, Turkey, in order to track the evolution of C. parapsilosis azole resistance over time. We assessed the genetic relatedness of C. parapsilosis isolates using AFLP and MST and explored a recent hypothesis that FLZR C. parapsilosis isolates carrying the Y132F mutation may have a higher propensity to persist in clinical settings (15).
RESULTS
Patients’ clinical profiles.
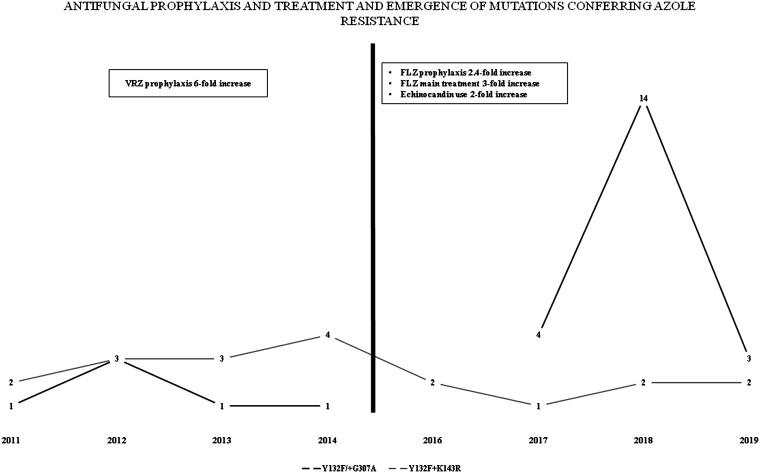
In total, 225 C. parapsilosis and 2 C. orthopsilosis isolates were recovered from 223 patients; 54.4% (n = 123) of the patients were men and 40.3% (n = 91) were women (no data for 12 cases). Children (<18 years old) (n = 95; 42%) and adults (≥18 years old) (n = 107; 47.3%) almost equally developed candidemia due to C. parapsilosis (no data for 24 cases). Prophylactic treatment with antifungals was not performed for 133 patients (58.8%); when it was performed, the most frequent choice was FLZ (n = 40; 17.7%) followed by VRZ (n = 9; 4%) (no data for 45 patients; 19.5%). Since fluconazole treatment, both prophylactic and targeted, was heavily used from 2015 onward, our analysis focused on microbiological changes observed before and after 2014. Accordingly, the use of FLZ for prophylaxis increased 2.4-fold (n = 25 [26.3%] versus 15 [11.7%]; 95% confidence interval [CI], 1.328 to 5.451; odds ratio [OR] = 2.690; P = 0.006) and that of VRZ decreased 6-fold (n = 1 [1.1%] versus 8 [6.2%]; 95% CI, 0.02 to 1.298; OR = 0.160; P = 0.086) in the second phase (2015 to 2019) compared to the first phase (2007 to 2014). Considering the targeted treatment, the majority of the patients (n = 137; 60.6%) received a single antifungal drug, whereas 8% of patients were treated by a combination of two drugs (n = 18; 8%), and 12.4% (n = 25) did not receive any antifungals (no data for 43 cases). FLZ was the most widely used single drug (n = 51; 22.6%), followed by caspofungin (n = 26; 11.5%), amphotericin B AMB (n = 25; 11.1%), anidulafungin (n = 16; 7.1%), micafungin (n = 11; 4.9%), and VRZ (n = 8; 3.5%). The use of FLZ was tripled (n = 35 [36.8%] versus 16 [12.5%]) and that of echinocandins doubled (n = 24 [25.2%] versus 16 [12.2%]) in the second phase compared to the first phase. The overall crude mortality rate was 38.1% (n = 86; no data for 40 patients). As we did not have mortality data for 32.1% of patients (n = 42) in the first phase, we could not compare the rates between the two phases. In terms of treatment, patients who received AMB formulations had the highest mortality (n = 15; 60%), and those treated with echinocandins (n = 27; 50.9%) and FLZ (n = 21; 42%) had similar mortality. The annual rate of C. parapsilosis isolation showed a sinusoidal trend, with peaks in 2012 and 2018 (see Fig. S1 in the supplemental material).
Antifungal susceptibility.
In total, 26.4% of the isolates were FLZR (n = 60); among them, 31.6% (n = 19) were cross-resistant to VRZ (Table S3). The rate of FLZR isolates doubled in the second phase (n = 34; 35.4%) compared to the first phase (n = 26; 19.6%) (Table 1). The number of VRZR isolates was comparable between the first and second phases (n = 11 [8.3%] versus 8 [9.4%]), whereas that of isolates with intermediate susceptibility to VRZ tripled in the second phase (n = 33 [34.7%] versus 10 [7.5%]). Overall, the frequency of FLZR isolates increased and was the highest in 2018, constituting almost half of the total number of isolates collected in that year (Table 1 and Fig. S2).
TABLE 1.
Isolates (n = 91) used for sequencing of genes implicated in azole resistancea
| Isolate group and cluster (n) | Ward (n) | No. of isolates | MIC (mg/liter) |
Mutation (n) in: |
Yr (n) | ||||
|---|---|---|---|---|---|---|---|---|---|
| FLZ | VRZ | Erg11p | Mrr1p | Tac1p | Upc2p | ||||
| Fluconazole-resistant isolates containing Y132F mutation | |||||||||
| A (18) | Pediatric surgery (5) | 19 | 32 or >32 | 0.06–4 | Y132F+K143R | G472V (1) | A21V (1) | L38I (3) | 2011 (2) |
| C (1) | Pediatric (4) | L926* (1) | Q965K+M966V (1) | L38I+A793S (3) | 2012 (3) | ||||
| Cardiac surgery (3) | P45H (1) | 2013 (3) | |||||||
| Pediatric oncology (2) | A793S (2) | 2014 (4) | |||||||
| Anesthesiology (2) | 2016 (2) | ||||||||
| Pediatric ICU (1) | 2017 (1) | ||||||||
| Infectious diseases (1) | 2018 (2) | ||||||||
| Neurology (1) | 2019 (2) | ||||||||
| A (2) | Pediatric (2) | 3 | 16–>32 | 0.5–2 | Y132F+G307A | L38I+Q371H (1) | 2016 (1) | ||
| C (1) | Pediatric surgery (1) | 2018 (2) | |||||||
| A (4) | Anesthesiology (7) | 24 | 8–32 | 0.06–2 | Y132F | G427V (1) | N7Y+L578M (1) | E7* (1) | 2011 (1) |
| B (11) | Pediatric (3) | Q1027R (2) | A21V (1) | L38I (1) | 2012 (3) | ||||
| C (1) | Thoracic surgery (3) | P150H (1) | L38I+A793S (2) | 2013 (1) | |||||
| D (4) | Chest diseases (3) | D603V+P803L (1) | Q372H (1) | 2014 (1) | |||||
| E (3) | General surgery (2) | L578M (1) | P45H (1) | 2017 (3) | |||||
| F (1) | Neurosurgery (2) | P45H+A793S (1) | 2018 (12) | ||||||
| Pediatric oncology (1) | A793S (5) | 2019 (3) | |||||||
| Pediatric Surgery (1) | |||||||||
| Emergency (1) | |||||||||
| Neurology (1) | |||||||||
| Isolates with other mutations causing fluconazole resistance (K143R and G458S) | |||||||||
| D (1) | Pediatric (2) | 2 | 16– | 0.5 | K143R | A21V (1) | L38I (1) | 2011 (1) | |
| E (1) | >32 | A793S (1) | 2013 (1) | ||||||
| D (3) | Pediatric surgery (2) | 4 | 16–>32 | 0.5–1 | G458S | Q371H (1) | 2016 (1) | ||
| E (1) | Pediatric ICU (1) | 2017 (1) | |||||||
| Pediatric ICU (1) | 2018 (2) | ||||||||
| C (1) | Pediatric oncology (1) | 1 | 16 | 0.25 | Q250K+R398I+G458S | P295L+Q1074* (1) | L390I (1) | 2014 (1) | |
| F (1) | Pediatric surgery (1) | 1 | 16 | 0.5 | G458S+T519A | A793S (1) | 2019 (1) | ||
| Other fluconazole-resistant isolates without Erg11 mutations causing fluconazole resistance | |||||||||
| A (1) | Pediatric surgery (3) | 6 | 8–>32 | 0.03-0.5 | R398I (3) | L419F (1) | G490R+S760R+A761G (1) | G342S (1) | 2013 (4) |
| C (3) | Cardiac surgery (1) | 2014 (1) | |||||||
| E (2) | Anesthesiology (1) | 2017 (1) | |||||||
| General surgery + organ transplant (1) | |||||||||
| Fluconazole-susceptible (dose dependent) isolates | |||||||||
| B (1) | Chest diseases (2) | 3 | 4 | 0.03–0.25 | R398I (2) | L578M (1) | A793S (1) | 2013 (1) | |
| C (2) | Pediatric (1) | 2014 (2) | |||||||
| Fluconazole-susceptible isolates | |||||||||
| Anesthesiology (5) | 28 | 0.06–2 | 0.0156–0.06 | R398I (13) | K606E (1) | N7I (2) | L38I (1) | 2009 (1) | |
| Chest diseases (5) | N7Y+A352V (1) | L38I+A793S (2) | 2012 (7) | ||||||
| Internal medicine (3) | F186I (2) | Q348P (1) | 2013 (10) | ||||||
| Pediatric surgery (3) | E312D (1) | P201S (1) | 2014 (3) | ||||||
| Pediatric (3) | L390I (1) | S577* (1) | 2018 (2) | ||||||
| Gastroenterology (2) | G490R (1) | A793S (5) | 2019 (5) | ||||||
| Pediatric oncology (2) | L574F (1) | ||||||||
| General surgery (2) | L578M (1) | ||||||||
| Cardiac surgery (2) | L578M+N602Y (1) | ||||||||
| Infectious diseases (1) | |||||||||
*, stop codon.
Sequencing of genes implicated in azole resistance.
Most FLZR isolates (90%; n = 54) carried Erg11 mutations known to cause FLZ resistance in C. parapsilosis (6) or C. orthopsilosis (17); among them, isolates carrying Y132F alone or in combination with other mutations constituted 90% (n = 54; 90%) (Table 1). Among FLZ resistance-related mutations, Y132F was the most prevalent (n = 24; 44.4%) followed by Y132F+K143R (n = 19; 35.1%), G458S (n = 6; 11.1%) (G458S alone, n = 4; G458S+T519A, n = 1; Q250K+R398I+G458S, n = 1), Y132F+G307A (n = 3; 5.5%), and K143R (n = 2; 3.7%) (Table 1; also Table S3). Heterozygosity was noted in five and six isolates carrying Y132F and Y132F+K143R, respectively, and the rest of the isolates were homozygotic for the mutations observed. Isolates with Y132F alone tripled in the second phase (n = 18 [75%] versus 6 [25%]), whereas those with Y132F+K143R were more prevalent in the first phase (n = 12 [63.1%] versus 7 [36.8%]) (Fig. 1). Isolates carrying Y132F+G307A (n = 3), G458S (n = 4), and G458S+T519A (n = 1) were detected only in the second phase, whereas those harboring K143R (n = 2) and G458S+R398I+Q250K (n = 1) were detected only in the first phase (Table 1; Table S3). Interestingly, 52% and 50% of FLZR isolates with or without ERG11 mutations, respectively, were recovered from pediatric wards. Thus, all isolates carrying K143R (n = 2), Y132F+G307A (n =3), and G458S (n = 6), 63.1% of Y132F+K143R isolates (n = 12), and 20.8% of isolates carrying only Y132F (n = 5) were detected in pediatric wards. Most VRZR isolates (n = 15; 78.94%) carried Y132F+K143R mutations, followed by Y132F (n = 3; 15.7%) and G458S (n = 1; 5.2%) (Table 1; also Table S3). Of note, most patients infected with isolates carrying Y132F (n = 17) died despite treatment with various antifungals; this rate was significantly higher (95% CI, OR = 6.8; P = 0.005) than that for patients infected with Y132F+K143R isolates (26.3%; n = 5). Among FLZR isolates carrying Erg11 mutations, 48.1% (n = 26), 16.6% (n = 9), and 11.1% (n = 6) also harbored nonsynonymous mutations in UPC2, TAC1, and MRR1, respectively (Table 1; also Table S3). All Mrr1 mutations were found in FLZR isolates, except for one FLZS isolate carrying K606E in Mrr1. Furthermore, P45H, Q371H, and Q372H in Upc2 and A21V, Q965K+M966V, P150H, D603V+P803L, and S760R+A761G in Tac1 were found exclusively in FLZR isolates (Table 1; also Table S3). Among FLZ- and VRZ-cross-resistant isolates, one (5.2%) harbored a unique Q965K+M966V (n = 1; 5.2%) mutation in Tac1, but none had mutations in Mrr1 or Upc2 specific to this phenotype.
FIG 1.
Frequency of mutations in ERG11 responsible for resistance and azole use for prophylaxis or treatment in 2007 to 2019.
Genotyping C. parapsilosis isolates.
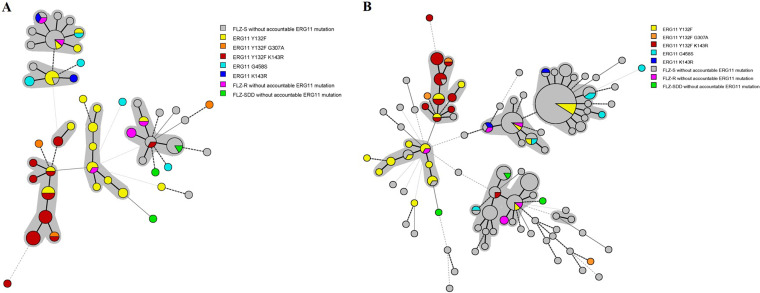
Compared to MST, AFLP revealed a higher degree of genetic similarity among the 225 isolates, and all isolates were clustered in only two major genotypes, whereas several clusters were revealed by MST (Fig. S3 to S6). Therefore, all interpretations regarding genetic relatedness of the isolates were inferred from MST data. The first isolates carrying Y132F (108FS) and Y132F+K143R (106FS and 107FS) were detected in 2011 in different patients and showed a high degree of similarity (Fig. S3); among them, 106FS and 107FS were 100% identical (Table 1 and Fig. S3). Surprisingly, 63% of the FLZR isolates carrying either Y132F or Y132F+K143R (n = 34) were clustered into two main clades located close to one another (Table 1, Fig. 2, and Fig. S3). Approximately 95% of the isolates carrying Y132F+K143R (n =18) and 16.6% of those harboring Y132F (n = 4) belonged to the same cluster and were recovered in 2011 to 2019. About 58% of the isolates carrying Y132F (n = 14) belonged to two distinct clusters (n = 11 [45.8%] and n = 3 [12.5%]) (Table 1, Fig. 2, and Fig. S3). As for their treatments, 48.8% of the patients infected with strains harboring Y132F alone and/or Y132F+K143R in Erg11 (n = 21) did not receive any azoles during their hospitalization periods. Among the FLZR isolates lacking ERG11 mutations responsible for resistance, 50% (n = 3; 34R, 37R, and 38R) were clustered together and were recovered in 2013 from different wards, whereas the remaining isolates were scattered among different genotypic clusters. Azole-cross-resistant isolates were grouped in clusters A (84.2%; n = 16), C (10.5%; n = 2), and E (5.25%; n = 1) (Table 1 and Fig. S3). Clonality was also detected among FLZS isolates; the largest cluster contained 63 isolates, followed by other clusters containing 20, 19, and 13 isolates (Fig. S5).
FIG 2.
Minimum spanning tree obtained by MST of 91 isolates with azole resistance (A) and all study isolates (B). Isolates carrying Y132F and Y132F+K143R formed distinct clusters. SDD, susceptible (dose dependent).
Biofilm formation.
Although the isolates significantly differed in biofilm-forming capacity, there was linear correlation between the results obtained with crystal violet and resazurin staining (r2 = 0.66, P < 0.001). Isolates carrying the Y132F mutation in Erg11p produced significantly less biofilm than the other FLZR isolates (P = 0.02) (Fig. S7).
DISCUSSION
A recent global study indicated that C. parapsilosis has the highest rate of FLZ resistance among Candida species, which has become a matter of growing concern, including in Europe (11). FLZ resistance rates significantly differ among countries and individual health care centers (11), emphasizing the need for active surveillance to prevent further expansion of FLZR C. parapsilosis in clinical settings. Interestingly, FLZR C. parapsilosis isolates are prevalent in three countries with the highest rate of FLZR C. auris (18), i.e., South Africa (5), India (6), and South Korea (7), further limiting the application of FLZ as first-line therapy. In this study, we report a clonal outbreak of FLZR C. parapsilosis in 2007 to 2019 in Ege University Hospital (Izmir, Turkey), which was especially characteristic of pediatric wards.
Overall, 26.5% and 8.3% of isolates were FLZR and VRZR, respectively; all of the latter were also cross-resistant to FLZ. Most FLZR isolates (90%) carried previously reported Erg11p mutations, and 5% of them carried a new one (G307A). Among the reported mutations, Y132 F and K143R have been detected in FLZR C. parapsilosis (6) and G458SS in FLZR C. orthopsilosis, a sibling species to C. parapsilosis (17). Although previous studies indicate that the Mrr1p mutations G583R and K873N (19) and L986P (20) are associated with FLZ and/or VRZ resistance, none of our isolates harbored them. Similar to Candida albicans (21), residues located near the C terminus of Mrr1 (926 and 1027) and Tac1 (760, 761, 803, 956, and 966) might contribute to azole resistance (21). Azole resistance mechanisms in C. parapsilosis, unlike those in C. albicans, are not well characterized, and we hope that the repertoire of mutations found in our study inspire heterologous expression studies in the future to broaden our knowledge on this growing problem.
The observed overall mortality rate in our study (38.1%) is similar to those reported in Brazil (22, 23), the United States (24), Portugal (25), and Italy (26) (30 to 46%) and 2.7 times higher than that reported in Taiwan (14%) (27). Importantly, we observed a link between mutations in genes implicated in azole resistance and mortality. Surprisingly, the mortality rate due to isolates with Y132F was 3 times higher than that caused by isolates with Y132F+K143R (OR = 6.8; P = 0.005). Experiments involving Galleria mellonella larvae infected with wild-type (WT) and C. parapsilosis isolates with Y132F showed a higher virulence when fluconazole was used for treatment (28). However, the impact of various ERG11 mutations on virulence of mutated and WT isolates has not been tested when Galleria infected with respective isolates is not treated with azoles. Although it has been shown that GOF mutation in Upc2 decreases the virulence of C. albicans (29), considering the increasing number of reported isolates with Y132F and considering that association of Tac1, Mrr1, and Upc2 with virulence is relatively unknown in C. parapsilosis, our findings may deserve detailed investigation in vivo.
C. parapsilosis can acquire azole resistance either by selective pressure due to azole use or by horizontal acquisition of azole-resistant C. parapsilosis isolates in antifungal-naive patients (7). Therefore, we analyzed patients’ treatment regimens and clonality of the isolates. Interestingly, the increasing FLZ use from 2015 onward paralleled an increasing frequency of isolation of C. parapsilosis isolates carrying Y132F. This finding is consistent with a study in Brazil showing that patients with clonal FLZR C. parapsilosis isolates carrying Y132F in Erg11p were previously exposed to FLZ (30). Moreover, significant positive correlation between nonsusceptibility of Candida species to azoles and FLZ use has been documented previously (31). Therefore, we speculate that the selective pressure exerted by azole use has partly resulted in emergence of FLZR C. parapsilosis in our hospital. To test the second idea, we assessed the genetic relatedness of FLZR isolates in relation to their treatment with azoles. Assessment of FLZR isolates by lineage using AFLP revealed two major clades with a high degree of similarity at the genome level. MST showed higher resolution and separated isolates into seven clusters, demonstrating that FLZR and FLZS isolates grouped in distinct clusters and accumulated over time. These observations suggest that AFLP does not have sufficient resolution to separate C. parapsilosis isolates and that MST should be the preferred method for clonal analysis of strains responsible for infection outbreaks. The overall high Simpson index value of the MST assay used here was demonstrated previously (32), and the fact that MST is being increasingly used for the genotyping assessment of clinically important fungi (33, 34), including C. parapsilosis (6), further suggests the reliability of this technique when dealing with outbreak scenarios. Moreover, the conspicuous grouping of FLZR and FLZS isolates into separate clusters and the finding that almost 50% of patients infected with FLZR isolates with Y132F/Y132F+K143R never received azoles may suggest an ongoing clonal outbreak of C. parapsilosis in our hospital that requires strict infection control and active environmental screening to identify and eradicate the source of infection.
It has been speculated that FLZR C. parapsilosis isolates with Y132F tend to be more clonal compared to other FLZR isolates and are more persistent in clinical settings (7). MST analysis found that both FLZR and FLZS isolates formed clonal clusters containing isolates recovered from 2007 to 2019. Moreover, biofilm formation, an index of persistence in hospital settings, was lower for isolates harboring Y132F than those without it, which is consistent with a study in Brazil showing that FLZR C. parapsilosis isolates with Y132F produced less biofilm than FLZS C. parapsilosis, C. orthopsilosis, and C. metapsilosis (30). Collectively, these observations argue against the notion that isolates with Y132F are more persistent than the other FLZR or FLZS C. parapsilosis isolates. Although it is tempting to attribute this phenomenon to a fitness cost posed by Y132F, isolates with the double mutation Y132F+K143R had a higher biofilm production. Therefore, dedicated studies are required to verify this finding in vivo and to identify the subcellular mechanisms involved.
Candida parapsilosis has been reported to respond to high concentrations of echinocandins when tested in vitro, which is attributed to a natural polymorphism in the FKS1 gene (35). However, in agreement with clinical studies conducted in the United States (36) and Spain (37), we did not observe a significant difference in the outcome for patients treated with FLZ or echinocandins. Of note, the comparative efficacy of azoles and echinocandins was not the focus of this study; such a study should take into account the severity of the disease and the underlying conditions. The alarming increase in the number of fatalities due to azole-resistant C. parapsilosis carrying the Y132F mutation, which could be further aggravated by clonal expansion in our hospital, together with the overall low resistance of C. parapsilosis to echinocandins (3, 11) reinforces the suitability of echinocandins for treatment of C. parapsilosis bloodstream infections. However, one should consider the possibility that the high in vitro MICs of anidulafungin for C. parapsilosis may hamper its efficacy in the clinical setting.
In conclusion, the observed clonal outbreak of fatal infections due to FLZR C. parapsilosis in our hospital, especially in pediatric wards, is worrisome and may be the consequence of inappropriate application of antifungal drugs and the lack of strict infection control measures, including hand hygiene. Therefore, active environmental surveillance followed by establishing strict infection control strategies and rigid sanitation standards are necessary to confine the spread of this pathogen. Moreover, implementation of appropriate antifungal therapy limiting the emergence of FLZR isolates is of paramount importance.
The retrospective nature of our study was one of its main limitations; as a result, we could not obtain clinical data for some patients. Also, the contribution of GOF mutations in TAC1, MRR1, and UPC2 to azole resistance, which has mainly been studied in C. albicans, is not fully recapitulated in C. parapsilosis (10); therefore, future studies in this direction are warranted.
MATERIALS AND METHODS
Study design, definitions, and identification.
The study included all patients with candidemia due to C. parapsilosis admitted to Ege University Hospital, Izmir, Turkey, from 2007 to 2019. Being among the five largest hospitals in Turkey with 1,816 beds, Ege University Hospital admits 67,000 inpatients and 1,200,000 outpatients annually. Although species identity was considered, neutropenic patients with fever and sepsis were treated with caspofungin or amphotericin B (AMB). Positive blood cultures (100 μl) were streaked on Sabouraud dextrose agar and incubated at 37°C for 24 to 48 h, and the single colonies obtained were stored at –80°C. All isolates were primarily identified using the API 20C AUX system (bioMérieux, Marcy l’Etoile, France) and further characterized using the MALDI Biotyper system (Bruker Daltonik, Bremen, Germany) with a full extraction method (38). This study was approved by the ethics committee of Ege University Hospital (approval number 20-2T/30).
Antifungal susceptibility testing.
Susceptibility to FLZ (Sigma-Aldrich, St. Louis, MO, USA) and VRZ (Sigma) was tested by the broth microdilution method according to CLSI document M27-A3 (39). Plates were incubated at 35°C for 24 h and MICs were determined by visual examination. MICs of ≥8 mg/liter and ≥1 mg/liter were considered to indicate resistance to FLZ and VRZ, respectively (40), while isolates for which FLZ and VRZ MICs were 4 mg/liter and 0.25 to 0.5 mg/liter were considered susceptible (dose dependent) and intermediate, respectively (40). Isolates with FLZ and VRZ MICs of ≤2 mg/liter and ≤0.12 mg/liter, respectively, were considered susceptible (40). Candida parapsilosis ATCC 22019 and Candida krusei ATCC 6258 were used for quality control.
DNA extraction, sequencing, and genotyping.
DNA extraction followed a cetyltrimethylammonium bromide (CTAB) protocol (38). Primers for ERG11, TAC1, UPC2, and MRR1 genes are shown in Table S1. Sequences were assembled using SeqMan Pro software (DNASTAR, Madison, WI, USA) and after curation were aligned to WT (41) ERG11 (GQ302972), MRR1 (HE605205), TAC1 (HE605204), and UPC2 (HE605206).
The genotypic diversity and genetic relatedness of the isolates were assessed by AFLP (42) and MST (32) as previously described.
Biofilm formation and quantification.
In vitro biofilm formation was assessed for 11 strains harboring Y132F and 9 strains lacking this mutation but resistant to FLZ (Table S2) (7, 43, 44). Biofilms were formed in 96-well microtiter plates for 24 h and stained with crystal violet or resazurin (CellTiter-Blue; Promega, Madison, WI, USA) as previously described (45). Absorbance (crystal violet) and fluorescence (resazurin) were measured using an Envision microtiter plate reader (Perkin Elmer, Waltham, MA, USA).
Statistical analysis.
Clinical and microbiological data were evaluated using SPSS v24 (SPSS Inc., Chicago, IL, USA). Biofilm formation was compared using an independent-samples t test. The association between two nominal variables of mutations and survival was assessed using Phi and Cramer’s V.
Data availability.
The sequences determined in this study for ERG11 (MK924157 to MK924381), MRR1 (MT429530 to MT429618), TAC1 (MK940393 to MK940481), and UPC2 (MT429619 to MT429707) were deposited in GenBank.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Major National R&D Projects of the National Health Department (2018ZX10101003), National Natural Science Foundation of China (31770161), Shanghai Science and Technology Committee (17DZ2272900 and 14495800500), Shanghai Municipal Commission of Health and Family Planning (2017ZZ01024-001), Shanghai Sailing Program (19YF1448000), and the Chinese Academy of Engineering (2019-XY-33).
We declare that this study was conducted in the absence of any financial relationship that could be considered a potential conflict of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Benedict K, Jackson BR, Chiller T, Beer KD. 2019. Estimation of direct healthcare costs of fungal diseases in the United States. Clin Infect Dis 68:1791–1797. doi: 10.1093/cid/ciy776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hallen-Adams HE, Suhr MJ. 2017. Fungi in the healthy human gastrointestinal tract. Virulence 8:352–358. doi: 10.1080/21505594.2016.1247140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tóth R, Nosek J, Mora-Montes HM, Gabaldon T, Bliss JM, Nosanchuk JD, Turner SA, Butler G, Vágvölgyi C, Gácser A. 2019. Candida parapsilosis: from genes to the bedside. Clin Microbiol Rev 32:e00111-18. doi: 10.1128/CMR.00111-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arastehfar A, Khodavaisy S, Daneshnia F, Najafzadeh MJ, Mahmoudi S, Charsizadeh A, Salehi MR, Zarrinfar H, Raeisabadi A, Dolatabadi S, Shahrabadi ZZ, Zomorodian K, Pan W, Hagen F, Boekhout T. 2019. Molecular identification, genotypic diversity, antifungal susceptibility, and clinical outcomes of infections caused by clinically underrated yeasts, Candida orthopsilosis, and Candida metapsilosis: an Iranian multicenter study (2014–2019). Front Cell Infect Microbiol 9:264. doi: 10.3389/fcimb.2019.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Govender NP, Patel J, Magobo RE, Naicker S, Wadula J, Whitelaw A, Coovadia Y, Kularatne R, Govind C, Lockhart SR, Zietsman IL, TRAC-South Africa Group. 2016. Emergence of azole-resistant Candida parapsilosis causing bloodstream infection: results from laboratory-based sentinel surveillance in South Africa. J Antimicrob Chemother 71:1994–2004. doi: 10.1093/jac/dkw091. [DOI] [PubMed] [Google Scholar]
- 6.Singh A, Singh PK, de Groot T, Kumar A, Mathur P, Tarai B, Sachdeva N, Upadhyaya G, Sarma S, Meis JF, Chowdhary A. 2019. Emergence of clonal fluconazole-resistant Candida parapsilosis clinical isolates in a multicentre laboratory-based surveillance study in India. J Antimicrob Chemother 74:1260–1268. doi: 10.1093/jac/dkz029. [DOI] [PubMed] [Google Scholar]
- 7.Choi YJ, Kim YJ, Yong D, Byun JH, Kim TS, Chang YS, Choi MJ, Byeon SA, Won EJ, Kim SH, Shin MG, Shin JH. 2018. Fluconazole-resistant Candida parapsilosis bloodstream isolates with Y132F mutation in ERG11 gene, South Korea. Emerg Infect Dis 24:1768–1770. doi: 10.3201/eid2409.180625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asadzadeh M, Ahmad S, Al-Sweih N, Khan Z. 2017. Epidemiology and molecular basis of resistance to fluconazole among clinical Candida parapsilosis isolates in Kuwait. Microb Drug Resist 23:966–972. doi: 10.1089/mdr.2016.0336. [DOI] [PubMed] [Google Scholar]
- 9.Grossman NT, Pham CD, Cleveland AA, Lockhart SR. 2015. Molecular mechanisms of fluconazole resistance in Candida parapsilosis isolates from a U.S. surveillance system. Antimicrob Agents Chemother 59:1030–1037. doi: 10.1128/AAC.04613-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berkow EL, Manigaba K, Parker JE, Barker KS, Kelly SL, Rogers PD. 2015. Multidrug transporters and alterations in sterol biosynthesis contribute to azole antifungal resistance in Candida parapsilosis. Antimicrob Agents Chemother 59:5942–5950. doi: 10.1128/AAC.01358-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castanheira M, Deshpande LM, Messer SA, Rhomberg PR, Pfaller MA. 2020. Analysis of global antifungal surveillance results reveals predominance of Erg11 Y132F alteration among azole-resistant Candida parapsilosis and Candida tropicalis and country-specific isolate dissemination. Int J Antimicrob Agents 55:105799. doi: 10.1016/j.ijantimicag.2019.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Chakrabarti A, Sood P, Rudramurthy SM, Chen S, Kaur H, Capoor M, Chhina D, Rao R, Eshwara VK, Xess I, Kindo AJ, Umabala P, Savio J, Patel A, Ray U, Mohan S, Iyer R, Chander J, Arora A, Sardana R, Roy I, Appalaraju B, Sharma A, Shetty A, Khanna N, Marak R, Biswas S, Das S, Harish BN, Joshi S, Mendiratta D. 2015. Incidence, characteristics and outcome of ICU-acquired candidemia in India. Intensive Care Med 41:285–295. doi: 10.1007/s00134-014-3603-2. [DOI] [PubMed] [Google Scholar]
- 13.Bennett JE, Izumikawa K, Marr KA. 2004. Mechanism of increased fluconazole resistance in Candida glabrata during prophylaxis. Antimicrob Agents Chemother 48:1773–1777. doi: 10.1128/aac.48.5.1773-1777.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cowen LE, Sanglard D, Howard SJ, Rogers PD, Perlin DS. 2014. Mechanisms of antifungal drug resistance. Cold Spring Harb Perspect Med 5:a019752. doi: 10.1101/cshperspect.a019752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhn DM, Mukherjee PK, Clark TA, Pujol C, Chandra J, Hajjeh RA, Warnock DW, Soll DR, Ghannoum MA. 2004. Candida parapsilosis characterization in an outbreak setting. Emerg Infect Dis 10:1074–1081. doi: 10.3201/eid1006.030873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tavanti A, Hensgens LAM, Mogavero S, Majoros L, Senesi S, Campa M. 2010. Genotypic and phenotypic properties of Candida parapsilosis sensu stricto strains isolated from different geographic regions and body sites. BMC Microbiol 10:203. doi: 10.1186/1471-2180-10-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morio F, Lombardi L, Binder U, Loge C, Robert E, Graessle D, Bodin M, Lass-Flörl C, Butler G, Le Pape P. 2019. Precise genome editing using a CRISPR-Cas9 method highlights the role of CoERG11 amino acid substitutions in azole resistance in Candida orthopsilosis. J Antimicrob Chemother 74:2230–2238. doi: 10.1093/jac/dkz204. [DOI] [PubMed] [Google Scholar]
- 18.Lone SA, Ahmad A. 2019. Candida auris—the growing menace to global health. Mycoses 62:620–637. doi: 10.1111/myc.12904. [DOI] [PubMed] [Google Scholar]
- 19.Branco J, Silva AP, Silva RM, Silva-Dias A, Pina-Vaz C, Butler G, Rodrigues AG, Miranda IM. 2015. Fluconazole and voriconazole resistance in Candida parapsilosis is conferred by gain-of-function mutations in MRR1 transcription factor gene. Antimicrob Agents Chemother 59:6629–6633. doi: 10.1128/AAC.00842-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Xiao M, Watts MR, Wang H, Fan X, Kong F, Xu YC. 2015. Development of fluconazole resistance in a series of Candida parapsilosis isolates from a persistent candidemia patient with prolonged antifungal therapy. BMC Infect Dis 15:340. doi: 10.1186/s12879-015-1086-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishimoto AT, Sharma C, Rogers PD. 2020. Molecular and genetic basis of azole antifungal resistance in the opportunistic pathogenic fungus Candida albicans. J Antimicrob Chemother 75:257–270. doi: 10.1093/jac/dkz400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brito LR, Guimarães T, Nucci M, Rosas RC, Paula Almeida L, Da Matta DA, Colombo AL. 2006. Clinical and microbiological aspects of candidemia due to Candida parapsilosis in Brazilian tertiary care hospitals. Med Mycol 44:261–266. doi: 10.1080/13693780500421476. [DOI] [PubMed] [Google Scholar]
- 23.Colombo AL, Nucci M, Park BJ, Nouér SA, Arthington-Skaggs B, da Matta DA, Warnock D, Morgan J, Brazilian Network Candidemia Study. 2006. Epidemiology of candidemia in Brazil: a nationwide sentinel surveillance of candidemia in eleven medical centers. J Clin Microbiol 44:2816–2823. doi: 10.1128/JCM.00773-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gudlaugsson O, Gillespie S, Lee K, van de Berg J, Hu J, Messer S, Herwaldt L, Pfaller M, Diekema D. 2003. Attributable mortality of nosocomial candidemia, revisited. Clin Infect Dis 37:1172–1177. doi: 10.1086/378745. [DOI] [PubMed] [Google Scholar]
- 25.Costa-de-Oliveira S, Pina-Vaz C, Mendonça D, Gonçalves Rodrigues A. 2008. A first Portuguese epidemiological survey of fungaemia in a university hospital. Eur J Clin Microbiol Infect Dis 27:365–374. doi: 10.1007/s10096-007-0448-4. [DOI] [PubMed] [Google Scholar]
- 26.Tumbarello M, Posteraro B, Trecarichi EM, Fiori B, Rossi M, Porta R, de Gaetano Donati K, La Sorda M, Spanu T, Fadda G, Cauda R, Sanguinetti M. 2007. Biofilm production by Candida species and inadequate antifungal therapy as predictors of mortality for patients with candidemia. J Clin Microbiol 45:1843–1850. doi: 10.1128/JCM.00131-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu PF, Liu WL, Hsieh MH, Hii IM, Lee YL, Lin YT, Ho MW, Liu CE, Chen YH, Wang FD. 2017. Epidemiology and antifungal susceptibility of candidemia isolates of non-albicans Candida species from cancer patients. Emerg Microbes Infect 6:e87. doi: 10.1038/emi.2017.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Souza ACR, Fuchs BB, Pinhati HMS, Siqueira RA, Hagen F, Meis JF, Mylonakis E, Colombo AL. 2015. Candida parapsilosis resistance to fluconazole: molecular mechanisms and in vivo impact in infected Galleria mellonella larvae. Antimicrob Agents Chemother 59:6581–6587. doi: 10.1128/AAC.01177-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lohberger A, Coste AT, Sanglard D. 2014. Distinct roles of Candida albicans drug resistance transcription factors TAC1, MRR1, and UPC2 in virulence. Eukaryot Cell 13:127–142. doi: 10.1128/EC.00245-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomaz DY, de Almeida JN Jr, Lima GME, Nunes MO, Camargo CH, Grenfell RC, Benard G, Del Negro G. 2018. An azole-resistant Candida parapsilosis outbreak: clonal persistence in the intensive care unit of a Brazilian teaching hospital. Front Microbiol 9:2997. doi: 10.3389/fmicb.2018.02997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Won EJ, Shin JH, Choi MJ, Lee WG, Park YJ, Uh Y, Kim SY, Lee MK, Kim SH, Shin MG, Suh SP, Ryang DW. 2015. Antifungal susceptibilities of bloodstream isolates of Candida species from nine hospitals in Korea: application of new antifungal breakpoints and relationship to antifungal usage. PLoS One 10:e0118770. doi: 10.1371/journal.pone.0118770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trobajo-Sanmartín C, Ezpeleta G, Pais C, Eraso E, Quindós G. 2018. Design and validation of a multiplex PCR protocol for microsatellite typing of Candida parapsilosis sensu stricto isolates. BMC Genomics 19:718. doi: 10.1186/s12864-018-5065-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cuomo CA, Alanio A. 2020. Tracking a global threat: a new genotyping method for Candida auris. mBio 11:e00259-20. doi: 10.1128/mBio.00259-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sewell TR, Zhu J, Rhodes J, Hagen F, Meis JF, Fisher MC, Jombart T. 2019. Nonrandom distribution of azole resistance across the global population of Aspergillus fumigatus. mBio 10:e00392-19. doi: 10.1128/mBio.00392-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Effron G, Katiyar SK, Park S, Edlind TD, Perlin DS. 2008. A naturally occurring proline-to-alanine amino acid change in Fks1p in Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis accounts for reduced echinocandin susceptibility. Antimicrob Agents Chemother 52:2305–2312. doi: 10.1128/AAC.00262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiotos K, Vendetti N, Zaoutis TE, Baddley J, Ostrosky-Zeichner L, Pappas P, Fisher BT. 2016. Comparative effectiveness of echinocandins versus fluconazole therapy for the treatment of adult candidaemia due to Candida parapsilosis: a retrospective observational cohort study of the Mycoses Study Group (MSG-12). J Antimicrob Chemother 71:3536–3539. doi: 10.1093/jac/dkw305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fernández-Ruiz M, Aguado JM, Almirante B, Lora-Pablos D, Padilla B, Puig-Asensio M, Montejo M, García-Rodríguez J, Pemán J, Ruiz Pérez de Pipaón M, Cuenca-Estrella M, CANDIPOP Project, GEIH-GEMICOMED (SEIMC), REIPI. 2014. Initial use of echinocandins does not negatively influence outcome in Candida parapsilosis bloodstream infection: a propensity score analysis. Clin Infect Dis 58:1413–1421. doi: 10.1093/cid/ciu158. [DOI] [PubMed] [Google Scholar]
- 38.Arastehfar A, Fang W, Pan W, Liao W, Yan L, Boekhout T. 2018. Identification of nine cryptic species of Candida albicans, C. glabrata, and C. parapsilosis complexes using one-step multiplex PCR. BMC Infect Dis 18:480. doi: 10.1186/s12879-018-3381-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard M27-A3, 3rd ed CLSI, Wayne, PA. [Google Scholar]
- 40.Pfaller MA, Diekema DJ. 2012. Progress in antifungal susceptibility testing of Candida spp. by use of Clinical and Laboratory Standards Institute broth microdilution. J Clin Microbiol 50:2846–2856. doi: 10.1128/JCM.00937-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar S, Stecher G, Tamura K. 2016. Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arastehfar A, Daneshnia F, Zomorodian K, Najafzadeh MJ, Khodavaisy S, Zarrinfar H, Hagen F, Shahrabadi ZZ, Lackner M, Mirhendi H, Salehi M, Roudbary M, Pan W, Kostrzewa M, Boekhout T. 2019. Low level of antifungal resistance in Iranian isolates of Candida glabrata recovered from blood samples in a multicenter study from 2015 to 2018 and potential prognostic values of genotyping and sequencing of PDR1. Antimicrob Agents Chemother 63:e02503-18. doi: 10.1128/AAC.02503-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Short B, Brown J, Delaney C, Sherry L, Williams C, Ramage G, Kean R. 2019. Candida auris exhibits resilient biofilm characteristics in vitro: implications for environmental persistence. J Hosp Infect 103:92–96. doi: 10.1016/j.jhin.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 44.Welsh RM, Bentz ML, Shams A, Houston H, Lyons A, Rose LJ, Litvintseva AP. 2017. Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic health care surface. J Clin Microbiol 55:2996–3005. doi: 10.1128/JCM.00921-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peeters E, Nelis HJ, Coenye T. 2008. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J Microbiol Methods 72:157–165. doi: 10.1016/j.mimet.2007.11.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequences determined in this study for ERG11 (MK924157 to MK924381), MRR1 (MT429530 to MT429618), TAC1 (MK940393 to MK940481), and UPC2 (MT429619 to MT429707) were deposited in GenBank.