LETTER
The emergence of mobile colistin resistance (mcr) genes has raised a global concern (1–3), as colistin is considered a last-resort antimicrobial for treating fatal infections caused by multidrug-resistant or carbapenem-resistant Enterobacteriaceae. Ten mcr genes (mcr-1 to mcr-10) and a lot of variants have been identified since the finding of mcr-1 in 2016 in China (4–6). These mcr-positive bacteria have been found in humans, animals, meat, and the environment in more than 60 countries across six continents (5, 7). Very recently, mcr-10 was described in a nonconjugative IncFIA(HI1) plasmid recovered from a clinical Enterobacter roggenkampii strain 090065 in China (6). It shows 79.69% nucleotide identity to mcr-9 and confers a 4-fold increase in colistin MIC (from 1 to 4 mg/liter). Here, we report a conjugative plasmid harboring mcr-10 in E. roggenkampii of chicken origin in China.
Enterobacter spp. strain YK16 was isolated from anal swab of chicken in a large-scale chicken farm on 19 November 2019. It was identified by an automated system (BD Diagnostic Systems, Sparks, MD, USA). The antibiogram was determined by the disc diffusion method according to CLSI guidelines. YK16 was resistant to ampicillin, cefotaxime, cefoxitin, ceftazidime, aztreonam, and fosfomycin but susceptible to imipenem, colistin, and tigecycline.
The draft genome of YK16 was sequenced by the Illumina HiSeq platform and assembled by software SPAdes 3.12.0. The de novo assembly generated 67 contigs (total length of 5,052,815 bp) with an N50 of 268,013 bp. The draft genome of YK16 has a 98.28% average nucleotide identity (ANI) value with the type strain of E. roggenkampii DSM 16690 (GenBank accession number CP017184) determined by JSpeciesWS (http://jspecies.ribohost.com/jspeciesws/), indicating YK16 belongs to E. roggenkampii. YK16 harbors resistance genes blaMIR-3 (a chromosomal ampC gene intrinsic to Enterobacter species), fosA (mediating resistance to fosfomycin), and mcr-10 (79.77% identity to mcr-9) found by ResFinder 3.2 (https://cge.cbs.dtu.dk/services/ResFinder/). The mcr-10 gene in YK16 is the allele of mcr-10.1, showing 99.94% nucleotide identity to that from Enterobacter roggenkampii strain 090065 (GenBank accession number MN179494) with only one nonsense mutation (C1056T). Multilocus sequence typing (MLST) showed that YK16 belonged to Enterobacter cloacae complex ST1056 and strain 090065 belonged to ST515, indicating that these two mcr-10.1-harboring E. roggenkampii strains in China were not clonally related.
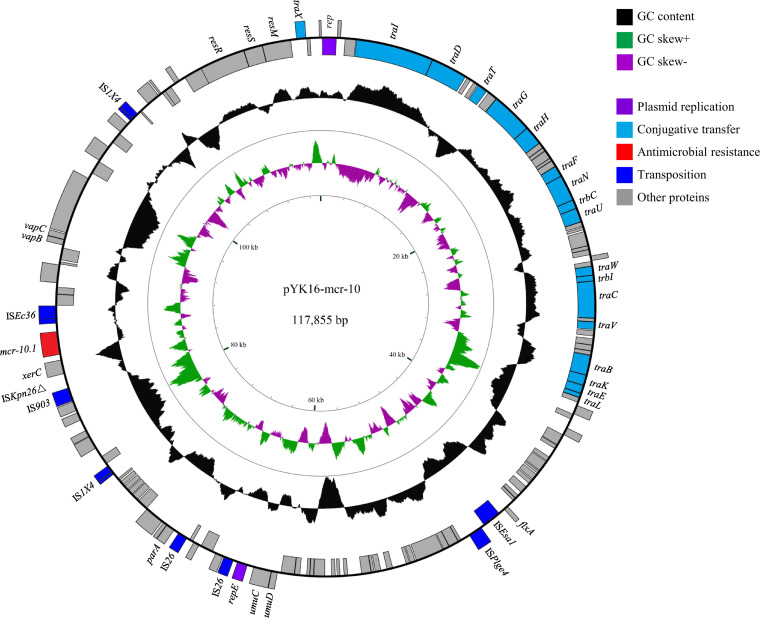
The complete sequence of the mcr-10.1-harboring plasmid, named pYK16-mcr-10, was assembled from two contigs (NODE_19 and NODE_26) by PCR linkage and sequencing. pYK16-mcr-10 is 117,855 bp in size with a GC content of 52.2%. PlasmidFinder 2.1 predicted that the replication initiation protein gene of pYK16-mcr-10 showed 82.82% nucleotide identity to that in the IncFII plasmid pECLA (GenBank accession number CP001919). pYK16-mcr-10 has the highest nucleotide identity (99.12%) with pIncFIB-1301491 (GenBank accession number CP031569) from E. hormaechei with 47% coverage. It consists of 133 predicted coding sequences (Fig. 1), including a tra module that might be associated with conjugative transfer. The genetic context of mcr-10.1 in pYK16-mcr-10 is IS903- ISKpn26Δ-xerC-mcr-10.1-ISEc36, which is similar to those described previously (6). A conjugation experiment was carried out by filter mating with the azide-resistant Escherichia coli J53 as the recipient. Transconjugants were selected on LB agar plates containing 2 mg/liter colistin and 200 mg/liter azide and confirmed by 16S rRNA gene sequencing for identification of E. coli and PCR for detection of mcr-10 using primers mcr-10-F (5′-GGACCGACCTATTACCAGCG-3′) and mcr-10-R (5′-GGCATTATGCTGCAGACACG-3′). pYK16-mcr-10 could be successfully transferred to E. coli J53 at a frequency of 2.8 × 10−6 transconjugants per donor (average of three independent determinations). The transconjugants decreased susceptibility to colistin (2 mg/liter) determined by microdilution.
FIG 1.
Circular genetic map of pYK16-mcr-10. Genes coding for plasmid replication, conjugative transfer, antimicrobial resistance, and transposition are indicated.
In conclusion, we reported mcr-10.1 in a conjugative plasmid recovered from E. roggenkampii of chicken origin in China, which might promote the horizontal transmission of mcr-10. The dissemination of mcr-10 in Enterobacteriaceae between humans and animals deserves more attention.
Data availability.
This whole-genome shotgun project of E. roggenkampii YK16 has been deposited at DDBJ/ENA/GenBank under accession number JABJWE000000000. The complete nucleotide sequence of plasmid pYK16-mcr-10 has been deposited into GenBank under accession number MT468575.
ACKNOWLEDGMENTS
This work was supported by the National Key R&D Program of China (grant number 2018YFD0500300), the China Agriculture Research System National System for Layer Production Technology (CARS-40-K14), and the Fundamental Research Funds for the Central Universities (SCU2019D013).
REFERENCES
- 1.Poirel L, Jayol A, Nordmann P. 2017. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev 30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun J, Zhang H, Liu YH, Feng Y. 2018. Towards understanding MCR-like colistin resistance. Trends Microbiol 26:794–808. doi: 10.1016/j.tim.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 3.El-Sayed Ahmed MAE-G, Zhong L-L, Shen C, Yang Y, Doi Y, Tian G-B. 2020. Colistin and its role in the era of antibiotic resistance: an extended review (2000–2019). Emerg Microbes Infect 9:868–885. doi: 10.1080/22221751.2020.1754133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 5.Ling Z, Yin W, Shen Z, Wang Y, Shen J, Walsh TR. 8 June 2020. Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9. J Antimicrob Chemother doi: 10.1093/jac/dkaa205. [DOI] [PubMed] [Google Scholar]
- 6.Wang C, Feng Y, Liu L, Wei L, Kang M, Zong Z. 2020. Identification of novel mobile colistin resistance gene mcr-10. Emerg Microbes Infect 9:508–516. doi: 10.1080/22221751.2020.1732231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Xu C, Zhang R, Chen Y, Shen Y, Hu F, Liu D, Lu J, Guo Y, Xia X, Jiang J, Wang X, Fu Y, Yang L, Wang J, Li J, Cai C, Yin D, Che J, Fan R, Wang Y, Qing Y, Li Y, Liao K, Chen H, Zou M, Liang L, Tang J, Shen Z, Wang S, Yang X, Wu C, Xu S, Walsh TR, Shen J. 4 June 2020. Changes in colistin resistance and mcr-1 abundance in Escherichia coli of animal and human origins following the ban of colistin-positive additives in China: an epidemiological comparative study. Lancet Infect Dis doi: 10.1016/S1473-3099(20)30149-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This whole-genome shotgun project of E. roggenkampii YK16 has been deposited at DDBJ/ENA/GenBank under accession number JABJWE000000000. The complete nucleotide sequence of plasmid pYK16-mcr-10 has been deposited into GenBank under accession number MT468575.