Staphylococcus argenteus is a novel staphylococcal species associated with invasive disease. We report the first case of daptomycin/vancomycin-resistant S. argenteus, initially speciated as Staphylococcus aureus, that developed from repeated treatment with daptomycin for a complex vascular graft infection. Whole-genome sequencing of longitudinally collected isolates identified acquisition of MprF S337L, a mutation predicted to increase surface charge and repel cationic molecules.
KEYWORDS: Staphylococcus, Staphylococcus argenteus, antimicrobial resistance, antimicrobial stewardship, whole-genome sequencing
ABSTRACT
Staphylococcus argenteus is a novel staphylococcal species associated with invasive disease. We report the first case of daptomycin/vancomycin-resistant S. argenteus, initially speciated as Staphylococcus aureus, that developed from repeated treatment with daptomycin for a complex vascular graft infection. Whole-genome sequencing of longitudinally collected isolates identified acquisition of MprF S337L, a mutation predicted to increase surface charge and repel cationic molecules.
CASE PRESENTATION
A 72-year-old woman with a history of end-stage renal disease requiring intermittent hemodialysis; prior toxic epidermal necrolysis from vancomycin, piperacillin-tazobactam, and/or cefazolin; and multiple arteriovenous (AV) fistula occlusions underwent placement of a left arm prosthetic HeRO AV graft. The procedure was complicated by graft thrombosis; the graft was retained, and venous access was instead achieved via a left femoral tunneled dialysis catheter (TDC).
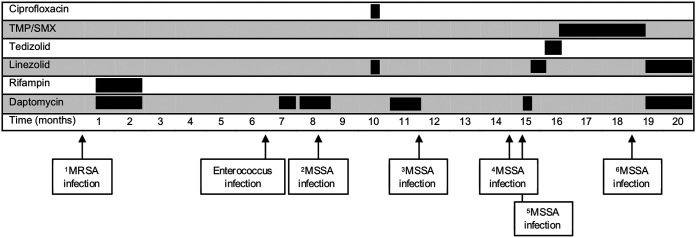
Six months later, the patient presented with left groin pain at her TDC site and fever found to be the result of methicillin-resistant Staphylococcus aureus (MRSA) bacteremia (Fig. 1). A transesophageal echocardiogram (TEE) showed no evidence of endocarditis. Her AV graft site exhibited mild erythema, swelling, and tenderness to palpation; but because surgical explantation was considered high risk, she underwent TDC exchange followed by 6 weeks of intravenous daptomycin (8 mg/kg Monday and Wednesday, 9 mg/kg Friday posthemodialysis [pHD]) in combination with rifampin. She was readmitted within another 6 months for an Enterococcus faecalis central line-associated bloodstream infection (CLABSI) (Fig. 1) and was treated with 2 weeks of daptomycin (8 mg/kg Monday and Wednesday, 12 mg/kg Friday pHD), following TDC exchange and blood culture sterilization.
FIG 1.
Overview of bloodstream infections and phenotypic antimicrobial susceptibility of Staphylococcus isolates. Time course of recurrent bacterial bloodstream infections experienced by the patient. S. argenteus isolate number is indicated by superscript.
A third episode of bacteremia occurred 1 month later. This time, methicillin-sensitive S. aureus (MSSA) was identified from blood cultures and speciated using matrix-assisted laser desorption/ionization–time of flight (MALDI-TOF) mass spectrometry (Fig. 1). A transthoracic echocardiogram (TTE) showed no evidence of endocarditis; and after removal of the TDC, blood cultures were sterilized, and she was discharged with a 4-week course of daptomycin (8 mg/kg Monday and Wednesday, 12 mg/kg Friday pHD). Unfortunately, just 3 months later, the patient experienced recurrent high-grade MSSA bacteremia (Fig. 1). A TTE was again negative for endocarditis, her TDC was removed, and she completed a fourth course of daptomycin (11 mg/kg pHD), this time for 4 weeks. Regrettably, MSSA bacteremia recurred yet again within 3 months (Fig. 1). Another TTE returned negative for endocarditis, her TDC was exchanged, blood cultures were sterilized, and she was discharged on a fifth course of daptomycin (11 mg/kg pHD). She had completed only 7 days of treatment before presenting with sepsis, now resulting from a daptomycin-resistant, vancomycin-intermediate staphylococcal infection.
Due to the emergence of this concerning resistance phenotype in the setting of prolonged daptomycin therapy, whole-genome sequencing (WGS) was performed on the patient’s cultured isolates (see Text S1 in the supplemental material). Whereas WGS confirmed that the initial isolate was MRSA, de novo genome assembly and phylogenetic analysis surprisingly revealed that the subsequent etiologic pathogen in isolates 2 through 6 (shown in Fig. 1) was not MSSA but instead a closely related Staphylococcus species, i.e., Staphylococcus argenteus, with an average nucleotide identity of 88.6% compared with S. aureus reference strain NC_007795.1. An unbiased single nucleotide polymorphism (SNP) analysis comparing the S. aureus isolates identified a single C-to-T transition resulting in an S337L mutation in MprF, previously described to confer daptomycin resistance in S. aureus isolates (1).
CHALLENGE QUESTION
A patient with multiply recurrent methicillin-resistant Staphylococcus aureus (MRSA) bacteremia who has received several courses of daptomycin therapy is admitted for a third recurrence. The Staphylococcus aureus isolate now demonstrates resistance to daptomycin and vancomycin. What is a possible mechanism underlying this resistance?
-
A.
Horizontal transfer of a gene encoding a daptomycin binding protein.
-
B.
Hypermethylation of the 30S ribosomal subunit.
-
C.
Development of a mutation in a protein modulating cell surface charge.
-
D.
Mutation in a bacterial topoisomerase gene.
TREATMENT AND OUTCOME
An additional TEE ruled out endocarditis, multiple Doppler ultrasound studies of the upper and lower extremities were negative for thrombophlebitis, and differential time-to-positivity (DTTP) blood cultures were inconsistent with CLABSI, clearly implicating the retained graft. At the recommendation of the consulting infectious disease service, this was finally removed; and after an additional TDC exchange, the patient was treated with linezolid for 2 weeks followed by tedizolid to complete a 4-week course. She was then given trimethoprim-sulfamethoxazole (TMP/SMX) suppression at one double-strength tablet daily and received an adjunctive TDC linezolid lock.
Despite graft removal, she experienced an additional episode of sepsis 3 months later. DTTP blood cultures this time implicated a CLABSI, but phenotypic testing no longer demonstrated daptomycin resistance. Even though MALDI-TOF identified S. aureus isolates, WGS revealed the same strain of S. argenteus but without the MprF S337L mutation. She was treated with 6 weeks of daptomycin (10 mg/kg pHD) plus linezolid and has not had another infection in more than a year. An overview of the clinical course, antibiotic regimen, and bacterial isolates analyzed by WGS is shown in Fig. 1. Antimicrobial susceptibility testing results are provided in Table 1.
TABLE 1.
Phenotypic antimicrobial susceptibility testing results of Staphylococcus aureus and Staphylococcus argenteus from the patient, which demonstrated evolution of daptomycin and vancomycin resistance
| Antimicrobial | MICa (μg/ml) in (isolate no. [date collected]): |
|||||
|---|---|---|---|---|---|---|
| S. aureus |
S. argenteus |
|||||
| 1 (2/2017) | 2 (10/2017) | 3 (1/2018) | 4 (4/2018) | 5 (5/2018) | 6 (9/2018) | |
| Clindamycin | >2 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 |
| Erythromycin | >4 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 |
| Linezolid | 2 | 2 | 2 | 2 | 2 | 2 |
| Nafcillin | >2 | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 | 0.5 |
| Rifampin | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 |
| TMP/SMX | ≤2 | ≤2 | ≤2 | ≤2 | ≤2 | ≤2 |
| Daptomycin | ≤0.5 | ≤0.5 | 1 | ≤0.5 | 4 | 1 |
| Vancomycin | 1 | 1 | 1 | 2 | 4b | 2 |
Unless otherwise noted, MICs were determined using the Trek Sensititre automated broth microdilution system. The Clinical and Laboratory Standards Institute (CLSI) breakpoint that defines daptomycin susceptibility in S. aureus isolates is ≤1 μg/ml.
E test.
S. argenteus is a divergent Staphylococcus species related to S. aureus originally identified in Aboriginal communities in Northern Australia (2). Given phylogenetic divergence characterized by <95% average nucleotide identity versus S. aureus isolates, unique peptidoglycan, and a different phenotypic morphology, S. argenteus was classified as a novel staphylococcal species in 2015 (2, 3). Although the spectrum of human disease caused by S. argenteus isolates is still incompletely understood, it has been identified worldwide as a cause of clinical syndromes similar to those of S. aureus, including skin and soft tissue infections, bone and joint infections, sepsis, and enterotoxin-induced gastroenteritis (4). This is the first report of S. argenteus infection in the United States.
S. argenteus isolates are coagulase and catalase positive and demonstrate β-hemolysis on blood agar (2). However, unlike S. aureus isolates, they lack a carotenoid pigment operon, resulting in colonies that are unpigmented, a phenotype reflected in the name argenteus, which is a Latin adjective for silver (2). Because S. argenteus infection has been described only relatively recently, many MALDI-TOF spectral databases used for microbiological identification are unable to differentiate it from S. aureus (5).
S. argenteus has its own virulence factors that are genetically dissimilar to those of S. aureus, and this species appears adept at acquiring toxin and antibiotic resistance genes from its environment, increasing its pathogenicity (6). That said, neither vancomycin nor daptomycin resistance has previously been reported in S. argenteus isolates. Despite genetic and microbiological differences, the spectra of infections caused by these two staphylococcal species appear to overlap, and growing evidence suggests that both induce severe disease in humans. Therefore, in a recent position paper by the European Society of Clinical Microbiology and Infectious Diseases Study Group for Staphylococci and Staphylococcal Diseases, the authors proposed identifying S. argenteus as S. aureus complex until further data are gathered about meaningful clinical differences to avoid confusion with lower or nonpathogenic staphylococci (7).
Here, we found that selection pressure from repeated courses of daptomycin in the absence of source control of an infected graft led to the emergence of both vancomycin and daptomycin resistance (8). It is likely that the graft was infected from the beginning, although no DTTP blood cultures were available from the patient’s first through fourth episodes of bacteremia to confirm this. Staphylococcal aggregation, biofilm formation on prosthetic material, and an endovascular infection with high bacterial burden may have all contributed to the development of resistance, although these mechanisms have been described in detail only in Staphylococcus aureus isolates (9). The patient’s prior severe drug allergies required an unorthodox approach to antibiotic selection, necessitating successive administration of daptomycin, an antibiotic with high risk of both MIC creep and development of resistance (10). Daptomycin pHD dosing has been controversial, but the dosing used in this case was adequate based on pharmacokinetic data collected in patients receiving thrice-weekly hemodialysis, lowering the likelihood that this was a contributing factor to the development of resistance (11).
Resistance developed due to a mutation not yet identified in S. argenteus isolates but previously characterized in S. aureus isolates resistant to vancomycin and daptomycin (8). MprF is an integral transmembrane protein responsible for lysylation of phosphatidylglycerol in the bacterial cell membrane, and the S337L mutation is proposed to enhance lysylation activity, increasing cell surface positive charge and repulsing cationic molecules, including endogenous antimicrobial peptides, vancomycin, and daptomycin (12). Notably, in the absence of continued daptomycin treatment, we observed that the S337L mutation was not maintained, suggesting a fitness cost to the bacterium, even though the patient likely remained colonized with S. argenteus isolates and ultimately experienced an additional episode of S. argenteus bacteremia.
In summary, this case highlights the pathogenicity of S. argenteus, an emerging virulent staphylococcal species with clinical features similar to those of S. aureus. In addition, it emphasizes that recurrent antibiotic exposure can select for resistant organisms, underscores the value of source control, and highlights the importance of antimicrobial stewardship.
Data availability.
Raw sequencing reads are available at NCBI BioProject accession number PRJNA562563.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Heart, Lung, and Blood Institute (grant K23HL138461-01A1 to C.L.) and the Chan Zuckerberg Biohub, San Francisco, CA.
We have no conflicts of interest to declare.
This Journal section presents a real, challenging case involving a multidrug-resistant organism. The case authors present the rationale for their therapeutic strategy and discuss the impact of mechanisms of resistance on clinical outcome. Two expert clinicians then provide a commentary on the case.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Ernst CM, Slavetinsky CJ, Kuhn S, Hauser JN, Nega M, Mishra NN, Gekeler C, Bayer AS, Peschel A. 2018. Gain-of-function mutations in the phospholipid flippase MprF confer specific daptomycin resistance. mBio 9:e01659-18. doi: 10.1128/mBio.01659-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tong SYC, Schaumburg F, Ellington MJ, Corander J, Pichon B, Leendertz F, Bentley SD, Parkhill J, Holt DC, Peters G, Giffard PM. 2015. Novel staphylococcal species that form part of a Staphylococcus aureus-related complex: the non-pigmented Staphylococcus argenteus sp. nov. and the non-human primate-associated Staphylococcus schweitzeri sp. nov. Int J Syst Evol Microbiol 65:15–22. doi: 10.1099/ijs.0.062752-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holt DC, Holden MTG, Tong SYC, Castillo-Ramirez S, Clarke L, Quail MA, Currie BJ, Parkhill J, Bentley SD, Feil EJ, Giffard PM. 2011. A very early-branching Staphylococcus aureus lineage lacking the carotenoid pigment staphyloxanthin. Genome Biol Evol 3:881–895. doi: 10.1093/gbe/evr078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thaipadungpanit J, Amornchai P, Nickerson EK, Wongsuvan G, Wuthiekanun V, Limmathurotsakul D, Peacock SJ. 2015. Clinical and molecular epidemiology of Staphylococcus argenteus infections in Thailand. J Clin Microbiol 53:1005–1008. doi: 10.1128/JCM.03049-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tunsjø HS, Kalyanasundaram S, Charnock C, Leegaard TM, Moen A. 2018. Challenges in the identification of methicillin-resistant Staphylococcus argenteus by routine diagnostics. APMIS 126:533–537. doi: 10.1111/apm.12843. [DOI] [PubMed] [Google Scholar]
- 6.Aung MS, San T, San N, Oo WM, Ko PM, Thet KT, Urushibara N, Kawaguchiya M, Sumi A, Kobayashi N. 2019. Molecular characterization of Staphylococcus argenteus in Myanmar: identification of novel genotypes/clusters in staphylocoagulase, protein A, alpha-haemolysin and other virulence factors. J Med Microbiol 68:95–104. doi: 10.1099/jmm.0.000869. [DOI] [PubMed] [Google Scholar]
- 7.Becker K, Schaumburg F, Kearns A, Larsen AR, Lindsay JA, Skov RL, Westh H. 2019. Implications of identifying the recently defined members of the Staphylococcus aureus complex S. argenteus and S. schweitzeri: a position paper of members of the ESCMID Study Group for Staphylococci and Staphylococcal Diseases (ESGS). Clin Microbiol Infect 25:1064–1070. doi: 10.1016/j.cmi.2019.02.028. [DOI] [PubMed] [Google Scholar]
- 8.Steed ME, Hall AD, Salimnia H, Kaatz GW, Kaye KS, Rybak MJ. 2013. Evaluation of daptomycin non-susceptible Staphylococcus aureus for stability, population profiles, mprF mutations, and daptomycin activity. Infect Dis Ther 2:187–200. doi: 10.1007/s40121-013-0021-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crosby HA, Kwiecinski J, Horswill AR. 2016. Staphylococcus aureus aggregation and coagulation mechanisms, and their function in host-pathogen interactions. Adv Appl Microbiol 96:1–41. doi: 10.1016/bs.aambs.2016.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boucher HW, Sakoulas G. 2007. Perspectives on daptomycin resistance, with emphasis on resistance in Staphylococcus aureus. Clin Infect Dis 45:601–608. doi: 10.1086/520655. [DOI] [PubMed] [Google Scholar]
- 11.Haselden M, Leach M, Bohm N. 2013. Daptomycin dosing strategies in patients receiving thrice-weekly intermittent hemodialysis. Ann Pharmacother 47:1342–1347. doi: 10.1177/1060028013503110. [DOI] [PubMed] [Google Scholar]
- 12.Jones T, Yeaman MR, Sakoulas G, Yang S-J, Proctor RA, Sahl H-G, Schrenzel J, Xiong YQ, Bayer AS. 2008. Failures in clinical treatment of Staphylococcus aureus infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob Agents Chemother 52:269–278. doi: 10.1128/AAC.00719-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing reads are available at NCBI BioProject accession number PRJNA562563.