Enterococcus faecium has become a major opportunistic pathogen with the emergence of vancomycin-resistant enterococci (VRE). As part of the gut microbiota, they have to cope with numerous stresses, including effects of antibiotics and other xenobiotics, especially in patients hospitalized in intensive care units (ICUs) who receive many medications. The aim of this study was to investigate the impact of the most frequently prescribed xenobiotics for ICU patients on fitness, pathogenicity, and antimicrobial resistance of the vanB-positive E. faecium Aus0004 reference strain.
KEYWORDS: E. faecium, VRE, antibacterial, caspofungin, echinocandins
ABSTRACT
Enterococcus faecium has become a major opportunistic pathogen with the emergence of vancomycin-resistant enterococci (VRE). As part of the gut microbiota, they have to cope with numerous stresses, including effects of antibiotics and other xenobiotics, especially in patients hospitalized in intensive care units (ICUs) who receive many medications. The aim of this study was to investigate the impact of the most frequently prescribed xenobiotics for ICU patients on fitness, pathogenicity, and antimicrobial resistance of the vanB-positive E. faecium Aus0004 reference strain. Several phenotypic analyses were carried out, and we observed that caspofungin, an antifungal agent belonging to the family of echinocandins, had an important effect on E. faecium growth in vitro. We confirmed this effect by electron microscopy and peptidoglycan analysis and showed that, even at a subinhibitory concentration (1/4× MIC, 8 mg/liter), caspofungin had an impact on cell wall organization, especially with respect to the abundance of some muropeptide precursors. By transcriptome sequencing (RNA-seq), it was also shown that around 20% of the transcriptome was altered in the presence of caspofungin, with 321 and 259 significantly upregulated and downregulated genes, respectively. Since the fungal target of caspofungin (i.e., β-1,3-glucan synthase) was absent in bacteria, the mechanistic pathway of caspofungin activity was investigated. The repression of genes involved in the metabolism of pyruvate seemed to have a drastic impact on bacterial cell viability, while a decrease of glycerol metabolism could explain the conformational modifications of peptidoglycan. This is the first report of caspofungin antibacterial activity against E. faecium, highlighting the potential impact of nonantibiotic xenobiotics against bacterial pathogens.
INTRODUCTION
Infections are among the major threats for inpatients, especially for patients hospitalized in intensive care units (ICUs). They are responsible for an important increase of morbidity and mortality rates as well as a burst in medical costs since approximately 50% of ICU patients acquire an infection during their hospitalization, with 60% of deaths attributable to infections (1). While approximately 70% of noncardiac ICU patients receive antibiotics for preventive or curative indications during their stay, nonantibiotic molecules are also extensively used for patient care, including inotropic drugs, opioid and nonopioid analgesics, anxiolytics, anticoagulants, antacids, curares, or antifungal agents (2, 3). It is now well accepted that bacteria must cope with numerous environmental stresses to survive in their host, leading to significant physiological effects (4). Whereas numerous reports have been published in the literature concerning the impact of antibiotics (especially at subinhibitory concentrations) on bacterial cell physiology and pathogenicity (5), little is known about the direct effect of nonantibiotic molecules on bacterial pathogens. Nonetheless, it has been demonstrated that therapy with nonantibiotics may have significant effects upon the physiology and virulence of bacterial cells (6). For instance, catecholamine administration raised the growth rate and affected biofilm formation and enhanced survival traits in cases of antibiotic therapy in Pseudomonas aeruginosa in vivo and ex vivo models (7). Enhancement of virulence traits in the presence of catecholamines has been described in other bacterial pathogens such as Escherichia coli, Salmonella enterica serovar Typhimurium, Staphylococcus epidermidis, and Campylobacter jejuni (8–15). Recently, it was also reported that virulence of P. aeruginosa was enhanced in the presence of morphine, a major analgesic molecule largely prescribed in ICUs, using a murine intestinal colonization model (16). In ICUs, patients are commonly colonized or infected by a small contingent of multidrug-resistant (MDR) isolates gathered under the acronym “ESKAPE bugs” (for Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, P. aeruginosa, and Enterobacter spp.) (1, 17).
Initially considered to be part of the commensal gastrointestinal tract microbiota, E. faecium has become an important issue in the therapeutic field since MDR isolates have spread in hospital settings. Over the last decades, strains of vancomycin-resistant enterococci (VRE) have emerged within the E. faecium species, particularly with the worldwide spread of hospital-adapted isolates belonging to the clonal complex 17 (CC17) (18–20). Actually, these CC17 strains are part of a particular epidemic lineage of hospital-adapted strains described as clade A1. This particular A1 clade is genetically distant from the community-associated human lineage designated clade B (20). Since most reports concerning the influence of xenobiotics have described Gram-negative bacteria, the impact of nonantibiotic xenobiotics used in ICU patients on E. faecium isolates remained undocumented.
The aim of this study was to investigate the influence of highly prescribed nonantibiotic molecules in noncardiac ICUs, against a well-characterized clinical, CC17 hospital-adapted isolate of E. faecium (E. faecium Aus0004) (21). Since caspofungin (an antifungal agent considered to be a nonantibiotic molecule) was shown to have a significant impact on E. faecium, different approaches (in vitro tests, electron microscopy [EM], peptidoglycan analysis, and global transcriptomic analysis) were taken in order to decipher this unexpected effect.
RESULTS AND DISCUSSION
Impact of nonantibiotic molecules on E. faecium Aus0004 growth kinetics.
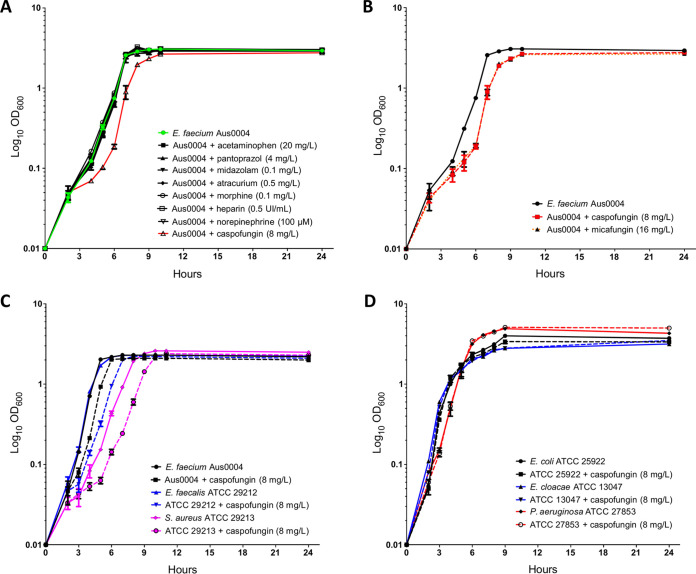
The effect of eight different nonantibiotic molecules (norepinephrine, morphine, acetaminophen, midazolam, unfractionated heparin, pantoprazole, atracurium, and caspofungin) extensively used in ICU patients was first phenotypically evaluated. The growth kinetics of E. faecium Aus0004 was monitored in tryptic soy broth (TSB) supplemented or not with xenobiotics at therapeutic free plasma concentrations (Fig. 1A). Of the eight molecules tested, only caspofungin had an important inhibitory effect on E. faecium Aus0004 since no growth was observed at the therapeutic concentration (data not shown). This induced us to determine the MIC of caspofungin for Aus0004, which was at 32 mg/liter (Table 1). Using a subinhibitory concentration (1/4× MIC, 8 mg/liter) of caspofungin, we found a significant impact on bacterial growth with a lag time extension of about 1 h (Fig. 1A). Note that there was no difference in exponential-growth rates under conditions that included the absence of caspofungin (−Cas) and presence of caspofungin (+Cas) (1.487 ± 0.034 h−1 and 1.444 ± 0.041 h−1, respectively; P = 0.2299, unpaired t test). In the light of these unexpected results, we decided to confirm if this effect was specific to caspofungin or observed with other molecules belonging to the echinocandin family (e.g., micafungin). The growth kinetics of E. faecium Aus0004 was then evaluated in TSB with or without micafungin at 16 mg/liter (corresponding to 1/4× MIC) (Table 1). A similar effect on bacterial growth (lag time extension) was observed with this second compound (Fig. 1B). These preliminary results demonstrated that echinocandins had a significant impact on E. faecium Aus0004 fitness in vitro.
FIG 1.
(A) Enterococcus faecium Aus0004 growth during 24 h in tryptic soy broth (TSB) supplemented or not with eight nonantibiotic molecules commonly used in ICU patients at the standard clinical concentrations (except for caspofungin used at 8 mg/liter, equivalent to 1/4× MIC). (B) E. faecium Aus0004 growth in TSB supplemented with caspofungin (8 mg/liter) and another echinocandin, micafungin (at a concentration of 1/4× MIC, 16 mg/liter). (C) Growth curves of E. faecium Aus0004, Enterococcus faecalis ATCC 29212, and Staphylococcus aureus ATCC 29213 in the presence or absence of caspofungin (8 mg/liter). (D) Growth in Luria-Bertani broth with or without addition of caspofungin (8 mg/liter) of Escherichia coli ATCC 25922, Enterobacter cloacae ATCC 13047, and Pseudomonas aeruginosa ATCC 27853.
TABLE 1.
MICs of several antibiotic and antifungal molecules for E. faecium Aus0004
| Antimicrobial | MIC (mg/liter) |
|---|---|
| Antibacterial | |
| Ampicillin | >256 |
| Erythromycin | >256 |
| Vancomycin | 8 |
| Teicoplanin | 1 |
| Daptomycin | 2 |
| Ciprofloxacin | 2 |
| Linezolid | 1 |
| Tigecycline | 0.06 |
| Antifungal | |
| Caspofungin | 32 |
| Micafungin | 64 |
| Voriconazole | >1,024 |
| Amphotericin B | >1,024 |
| Flucytosine | >1,024 |
In contrast to what it has previously been described in E. coli, S. Typhimurium and S. epidermidis (12, 22–24), we did not observe any effect of catecholamines on E. faecium growth kinetics. We hypothesized here the essential and critical impact of serum presence in the growth media, since, as previously described in Listeria monocytogenes, overgrowth observed with catecholamine was formerly linked with iron uptake, promoted by increased ferric reductase activity (25). Another important nonantibiotic molecule largely prescribed in ICU patients is morphine. In our study, no impact was observed on growth kinetics of E. faecium Aus0004 with this opioid analgesic, whereas it has been described that chronic exposition to morphine significantly increased proinflammatory interleukins serum and cecal levels that enhanced biofilm formation and adhesion in a P. aeruginosa murine infection model (16).
Interestingly, the impact of caspofungin on bacteria has been evaluated in only one study on S. aureus, where no impact was found on bacterial growth in vitro (26) whereas an important impact on biofilm formation kinetics was observed both in vitro and in vivo, when caspofungin was associated with moxifloxacin, a fluoroquinolone family antibiotic known to be effective against S. aureus. Note that in that study, caspofungin was used at a higher concentration (40 mg/liter) than that used in our study (8 mg/liter). As described in fungi, we can hypothesize that caspofungin, used at high-level concentrations, exhibits a paradoxical effect that is characterized by the resumption of growth of otherwise susceptible strains of S. aureus (27–29).
In vitro activity of caspofungin on Gram-positive and -negative bacterial pathogens.
The activity of caspofungin (at 8 mg/liter) was evaluated in vitro with representative Gram-positive and -negative bacterial species. The bacterial growth of Gram-positive bacteria (S. aureus and Enterococcus faecalis) was impacted (Fig. 1C), whereas that of Gram-negatives (P. aeruginosa, E. coli and Enterobacter cloacae) was not (Fig. 1D). Then, it appears that caspofungin has a significant impact only on in vitro bacterial growth of Gram-positive bacteria. This is likely due to the differences of bacterial cell wall composition between Gram-positive and -negative bacteria. Indeed, it is well known that Gram-negative bacteria have a thin peptidoglycan layer surrounded by an outer membrane enriched with lipopolysaccharide whereas Gram-positive bacteria lack the outer membrane and are surrounded by murein layers many times thicker and negatively charged. We could hypothesize here that the outer membrane of Gram-negative bacteria would be nonpermeable (30) to caspofungin, a high-molecular-weight negatively charged molecule.
Bactericidal activity of caspofungin against E. faecium Aus0004.
In order to compare their antibacterial activities, MICs of different antibacterial and antifungal agents against E. faecium Aus0004 were determined using the broth microdilution (BMD) reference method. As expected, caspofungin remained less active than the antibiotics tested (i.e., vancomycin, teicoplanin, linezolid, daptomycin, and tigecycline) (Table 1). Interestingly, caspofungin was ≥32-fold more active against E. faecium Aus0004 than other antifungals tested (i.e., amphotericin B, voriconazole, and 5-fluoro-cytosine) (Table 1).
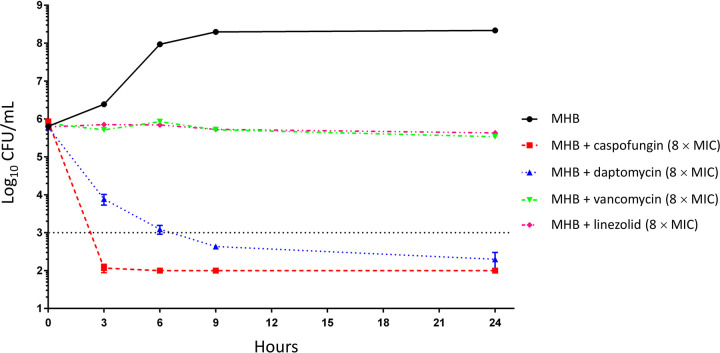
The next step was to determine if caspofungin had bacteriostatic or bactericidal activity against E. faecium Aus0004 by time-kill curve analysis (at 8× the MIC) using anti-Gram-positive antibiotics as comparators. Interestingly, we observed a rapid bactericidal effect (greater than −3 Log10 reduction) in the presence of caspofungin after only 3 h of incubation (Fig. 2). This effect was sustained during the 24-h period of the experiment without any regrowth (Fig. 2). Note that the bactericidal activity of caspofungin was even more rapid and pronounced than that of daptomycin, known as a model of rapid bactericidal antibiotic (31, 32). Both vancomycin and linezolid did not exhibit bactericidal activity (Fig. 2). These data are surprising since caspofungin and other β-1,3-glucan synthase inhibitors are known to be fungistatic molecules (33).
FIG 2.
Time-kill curves of different anti-Gram-positive antibiotics and caspofungin against E. faecium Aus0004 (8× MIC) in Mueller-Hinton broth (MHB).
Alterations of E. faecium Aus0004 cell wall under caspofungin exposure.
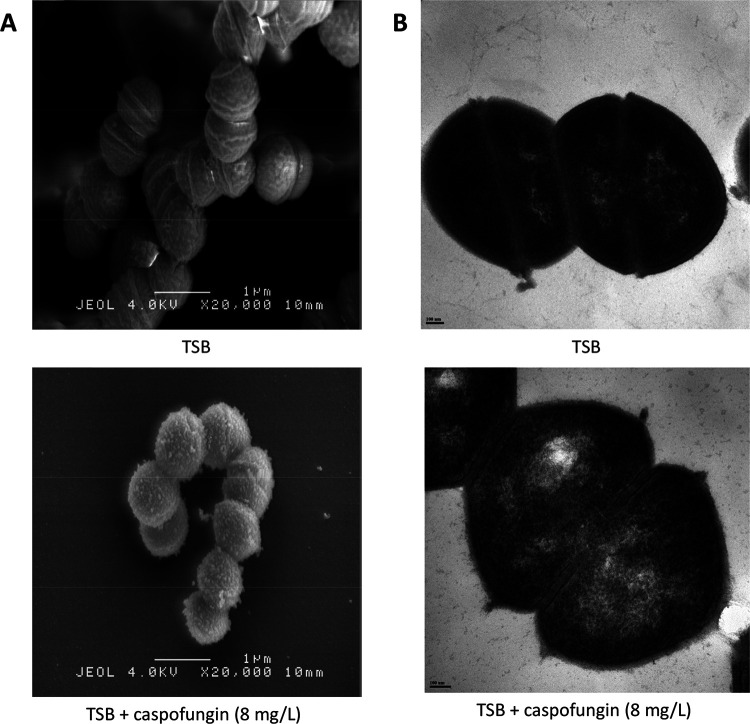
The impact of caspofungin (at 8 mg/liter) on cell wall components was first visualized using an ultrastructural morphology analysis by scanning electron microscopy and transmission electron microscopy (SEM and TEM, respectively). SEM experiments revealed that caspofungin was responsible for serious morphological abnormalities with roughened surface and extrusions all around the cell surface, suggesting a strong effect on bacterial envelope (Fig. 3A). TEM experiments did not reveal any change of the cell wall thickness in the presence of caspofungin (Fig. 3B).
FIG 3.
Cell wall analysis of E. faecium Aus0004 in the presence of caspofungin (8 mg/liter) by electron microscopy. (A) Scanning electron microscopy (SEM) images of E. faecium Aus0004 cells grown in tryptic soy broth (TSB) (top) or TSB plus caspofungin (bottom). Magnification, ×20,000. Morphological abnormalities on cell surface (roughened surface, extrusions) are easily visible. (B) Transmission electron microscopy (TEM) images of E. faecium Aus0004 cells grown in TSB (top) or TSB plus caspofungin (bottom).
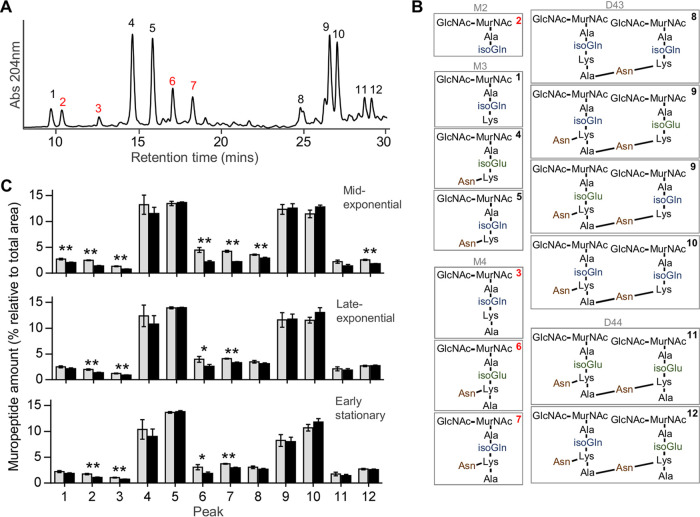
Analysis of the chemical composition of the peptidoglycan of E. faecium Aus0004 cultures growing in the presence of subinhibitory concentration of caspofungin (8 mg/liter) at different growth time points (mid-exponential, late-exponential, and early stationary phases) by ultraperformance liquid chromatography (UPLC) and tandem mass spectrometry (MS/MS) analysis showed a significant decrease of four muropeptides (Fig. 4). Interestingly, relative abundances of those muropeptides decreased by between 20% and 60% depending on the type of muropeptides and the incubation time of the cells (Fig. 4). This dramatic drop is consistent with an important impact of caspofungin on the peptidoglycan composition of E. faecium. Since peptidoglycan biosynthesis is essential during growth of the cell (34), the defects in bacterial growth (Fig. 1) and the bactericidal effect (Fig. 2) observed in the presence of caspofungin might be related to peptidoglycan modifications induced by the antifungal drug.
FIG 4.
Peptidoglycan modifications produced by caspofungin exposure. (A and B) Representative UPLC chromatogram of E. faecium Aus0004 peptidoglycan of a nontreated sample (A) and structure of the muropeptides determined for each indicated peak that was confirmed by MS analysis (B) (M, monomer; D, dimer; the numbers refer to the length of the stem peptide). Peaks representing significant changes after caspofungin treatment are labeled in red. (C) Quantification of the abundance of the muropeptides determined under two sets of growth conditions: tryptic soy broth (TSB) (gray bars, untreated samples) and TSB plus caspofungin 8 mg/liter (black bars, treated samples). The experiment was realized at 3 different times of growth (mid-exponential phase, late-exponential phase, and early stationary phase). Statistical analysis was performed using Student's t test. Asterisks represent significant P values from comparisons of treated samples to untreated samples (*, P < 0.05; **, P < 0.005). Each experiment was performed in triplicate.
Whole-transcriptome analysis of the response of E. faecium Aus0004 to caspofungin.
We used a global transcriptomic approach that incorporated transcriptome sequencing (RNA-seq) to decipher the impact of caspofungin at subinhibitory concentration (1/4× MIC, 8 mg/liter) on different metabolic pathways in E. faecium Aus0004. Total RNAs were isolated in biological duplicate from E. faecium Aus0004 grown to the late-exponential phase in both cases (same optical densities [OD]), following rRNA depletion. Between 7 and 16 million reads were obtained for each cDNA library, of which more than 97% mapped to the genome of E. faecium Aus0004 (see Table S1 in the supplemental material). Less than 2% of reads mapped to sequences of the three plasmids whereas less than 0.6% of reads corresponded to rRNA genes, confirming the high efficacy of rRNA depletion (Table S1). The reproducibility of the duplicate RNA-seq experiments was satisfactory (r2 > 0.97) under both sets of conditions (see Fig. S1 in the supplemental material).
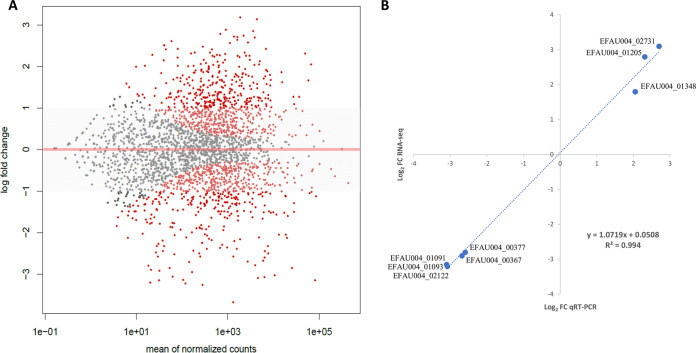
Differential gene expression (DGE) analysis was performed only for chromosomal genes (except for rRNA and tRNA genes) and not for plasmidic genes because of the low number of reads and the absence of significant changes in gene expression (data not shown). The fold change (FC) of expression of each annotated gene in the chromosome of E. faecium Aus0004 between cells grown in the presence (+Cas) or absence (−Cas) of caspofungin (8 mg/liter) is presented as a MA plot representation (Fig. 5A) (see also Table S2 in the supplemental material). To assess the reliability of RNA-seq in determining DGEs, we determined by reverse transcription-quantitative PCR (RT-qPCR) mRNA levels of three upregulated genes (hupA, sodA, and EFAU004_02731) and five downregulated genes (dexB, glpK, pdhD, pdhB, and EFAU004_02122) (Table 2). Those genes were chosen according to their levels of expression FCs and their putative functions. The ratios of the transcripts from −Cas and +Cas samples determined by RNA-seq and compared to those obtained by RT-qPCR showed excellent concordance, with a Pearson correlation value of 0.994 (Fig. 5B). Therefore, this confirmed that RNA-seq was a reliable method for global transcriptomic analysis in E. faecium under the conditions tested in this study.
FIG 5.
Transcriptional response of E. faecium Aus0004 growth with caspofungin. (A) Global analysis of transcript levels in E. faecium Aus0004 by RNA-seq represented by an MA plot (caspofungin 8 mg/liter versus control) generated by the DESeq2 R package. Log2 fold change values representing expression of each chromosomal gene are shown on the y axis versus the mean of normalized counts that is shown on the x axis (Log10 scale). Points corresponding to an adjusted P value of less than 0.1 are indicated in red. (B) Validation of RNA-seq results by quantitative reverse transcription-PCR (qRT-PCR) analysis of 10 genes. Mean log2 ratios of values determined in the qRT-PCR experiments are plotted against the mean log2 ratios of the RNA-seq experiments.
TABLE 2.
Selected genes used for RNA-seq validation by qRT-PCR experimentsa
| Gene no. | Gene name |
Product name | Gene start position |
Gene end position |
RNA-seq fold change |
Adjusted P value |
|---|---|---|---|---|---|---|
| EFAU004_00367 | dexB | Glucan 1,6-alpha-glucosidase | 364710 | 366333 | −9.6 | 1.20E−35 |
| EFAU004_00377 | glpK | Glycerol kinase | 379384 | 380881 | −6.0 | 5.60E−31 |
| EFAU004_01091 | pdhD | Dihydrolipoyl dehydrogenase | 1113699 | 1115106 | −6.4 | 4.70E−63 |
| EFAU004_01093 | pdhB | Transketolase | 1116781 | 1117759 | −8.6 | 4.32E−16 |
| EFAU004_01205 | hupA | DNA-binding protein HU | 1233614 | 1233890 | 4.9 | 3.76E−37 |
| EFAU004_01348 | sodA | Superoxide dismutase, Mn2+ | 1395234 | 1395843 | 4,1 | 2,33E−42 |
| EFAU004_02122 | l-Lactate oxidase | 2151833 | 2152937 | −8.4 | 8.70E−12 | |
| EFAU004_02731 | Zeta toxin | 2792658 | 2793333 | 6,5 | 1,75E−53 |
qRT-PCR, quantitative reverse transcription-PCR.
The analysis of transcriptomic data obtained by RNA-seq showed that 580 genes (20.3% of the chromosomal genes) had statistically significant alterations of their expression levels (FC greater than 2 or less than −2, adjusted P value < 0.1), with 321 upregulated genes and 259 downregulated genes (Table S2B and C). All the genes presenting modified amounts of mRNA between the two conditions were classified into functional categories using the COG and KEGG classifications (35) (Fig. S2 and S3). Among these 580 genes, more than 30% coded for proteins of unknown function or not found in other species, not allowing gene ontology analysis. In the presence of caspofungin, expression of genes coding for proteins involved in carbohydrate transport or metabolism and energy production or conversion was significantly repressed whereas expression of genes coding for proteins involved in transcription, replication, recombination and repair, and inorganic ion transport or metabolism was significantly upregulated (Fig. S2 and S3; see also Table S2B and C).
Impact of caspofungin on metabolism of E. faecium Aus0004.
Of the 20 genes whose expression was most highly repressed by caspofungin, 13 were found to be involved in carbohydrate transport or metabolism, in particular, genes coding for phosphotransferase systems (PTS), which are systems that mediate uptake and utilization of sugar as an energy source in bacteria (Table S2C). Since it has been demonstrated that some PTS genes act as regulatory factors promoting adaption to stressful metabolic conditions (36) and potentially enhance the possibility of bacterial survival, we hypothesized that the severe alteration of PTS transcript levels might play a role in the apparent lethality of caspofungin for E. faecium Aus0004. Interestingly, several genes involved in glycerol metabolism in E. faecium showed decreases in transcript levels. We observed downregulation of expression of genes composing the glpKOF operon (EFAU004_00377, EFAU004_00378, and EFAU004_00379) (fold changes, −6.0, −5.8, and −4.6, respectively) as well as genes composing the dhaKLM operon (EFAU004_00392, EFAU004_00393, and EFAU004_00394) (fold changes, −4.9, −4.2, and −5.1, respectively) (Table S2C). It is now widely assumed that glycerol is an essential precursor for the synthesis of lipids and, in many Gram-positive bacteria, including enterococci, for the biosynthesis of lipoteichoic acids (37). Moreover, it has also been reported that glycerol metabolism pathways are under the regulation of PTS and that the transcript levels are highly impacted (36). There was also a strong repression of genes involved in pyruvate metabolism since the so-called pdhABCD operon (EFAU004_01091, EFAU004_01092, EFAU004_01093, and EFAU004_01094) seemed to be impacted, with important downregulation results seen in our transcriptomic analysis (fold changes, −4.1, −8.6, −6.9, and −6.4, respectively) (Table S2C). These enzymes allow pyruvate transformation into acetyl coenzyme A (acetyl-CoA), which then proceeds by a two-step reduction process to generate ATP or directly enter into fatty acid biosynthesis (38). Since pyruvate dehydrogenation seemed to be impacted, we hypothesized that the presence of caspofungin results in a lack of ATP. This decrease of ATP formation may explain the changes in bacterial fitness that occurred in the presence of caspofungin. All these data pointed out that the presence of caspofungin induced an important form of stress that modified the carbohydrate metabolism and the cross-connected metabolic pathways essential for E. faecium Aus0004 growth. Moreover, it was evidenced that caspofungin likely induced an oxidative stress, since the sodA gene (EFAU004_01348) coding for the manganese-dependent superoxide dismutase, a well-known protein involved in oxidative stress regulation in Enterococcaceae (39), was significantly upregulated (fold change, +4.1) (Table S2B). Interestingly, sodA was previously described as representing an important pathway with a role in tolerance of cell wall-active antibiotics in enterococci and S. aureus (40).
Impact of caspofungin on antimicrobial resistance of E. faecium Aus0004.
Regarding findings on alterations of E. faecium Aus0004 cell wall and metabolism in the presence of echinocandins, we addressed the role of caspofungin in antimicrobial resistance as previously described (5) for subinhibitory concentrations of antibiotics. Then, we determined MICs of vancomycin, teicoplanin, daptomycin, and ciprofloxacin against E. faecium Aus0004 by the Etest strip method using Mueller-Hinton (MH) plates supplemented or not with 8 mg/liter caspofungin. Interestingly, we observed a 4-fold increase in MICs of vancomycin in the presence of caspofungin whereas no change was found for daptomycin, teicoplanin, and ciprofloxacin (Table 2). Note that this impact was not observed with other antifungal agents and seemed to be specific to echinocandins (data not shown). Since E. faecium Aus0004 is a vanB-positive strain, we assumed that this increase in the vancomycin MIC (and not in that of teicoplanin) was due to an upregulation of the vanB operon. However, RNA-seq data and RT-qPCR (FC in expression of vanB gene, −1.8, with an adjusted P value of >0.1) did not confirm this hypothesis (Table S2A). Also, we observed a 4-to-8-fold increase in the MICs of vancomycin among vanA- and vanB-positive isolates (except for one strain) (Table 3). These findings are consistent with the impact of echinocandins on cell wall components of E. faecium.
TABLE 3.
Antimicrobial susceptibility testing of E. faecium strains with different phenotypes of susceptibility or resistance to glycopeptides
| E. faecium strain | van operon | MIC (mg/liter)a |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Vancomycin |
Teicoplanin |
Daptomycin |
Ciprofloxacin |
||||||
| MH | CAS8 | MH | CAS8 | MH | CAS8 | MH | CAS8 | ||
| UCN103 | vanA | 32 | 256 | 4 | 4 | 4 | 8 | 8 | 8 |
| UCN104 | vanA | 32 | 256 | 2 | 2 | 2 | 4 | >32 | >32 |
| UCN105 | vanA | 4 | 32 | 2 | 2 | 4 | 8 | >32 | >32 |
| Aus0004 | vanB | 8 | 32 | 1 | 1 | 2 | 8 | 2 | 2 |
| UCN 106 | vanB | 16 | 64 | 1 | 1 | 4 | 8 | >32 | >32 |
| UCN 107 | vanB | 16 | 64 | 0.5 | 0.5 | 4 | 8 | >32 | >32 |
| UCN 108 | vanB | 32 | 64 | 0.5 | 0.5 | 4 | 4 | >32 | >32 |
| UCN 109 | vanB | 64 | 256 | 0.25 | 0.5 | 2 | 4 | >32 | >32 |
| UCN 110 | 0.5 | 1 | 0.25 | 1 | 2 | 4 | 2 | 2 | |
| UCN 111 | 0.5 | 1 | 0.12 | 0.25 | 2 | 4 | 4 | 4 | |
| UCN 112 | 1 | 1 | 0.12 | 0.25 | 2 | 4 | 2 | 4 | |
| UCN 113 | 0.5 | 1 | 0.25 | 1 | 2 | 4 | 8 | 8 | |
| UCN 114 | 0.5 | 1 | 0.25 | 0.5 | 1 | 4 | >32 | >32 | |
MH, Mueller-Hinton; CAS8, MH plus caspofungin (8 mg/liter).
Impact of caspofungin on biofilm formation.
This is well known that bacterial biofilms are a significant medical challenge because they are difficult to treat using standard therapeutic approaches, given that they are a major barrier to antibiotic effectiveness, especially in MDR E. faecium isolates (41). In order to characterize the effect of caspofungin on biofilm production, we evaluated levels of static biofilm formation of E. faecium Aus0004 (used as a biofilm nonproducer) and E. faecium HM1070 ΔasrR (used as a biofilm producer) (42) in the presence of a subinhibitory caspofungin concentration (8 mg/liter). As previously described in fungal models (43), caspofungin significantly reduced the ability of HM1070 ΔasrR to form biofilms whereas no difference was observed for E. faecium Aus0004 (Fig. 6). These data substantiate previous findings concerning the impact of caspofungin against S. aureus bacterial biofilms (26). Interestingly, the authors of the latter study explained that this biofilm formation shutdown in S. aureus was mediated through inhibition of an ica operon by caspofungin and in particular, IcaA, a protein that shares homology with the β-1,3-glucan synthase, the caspofungin fungal target. Here, the protein that shares the most homology with IcaA (i.e., EFAU004_00389) seemed to be not statistically significantly impacted by the presence of caspofungin as retrieved in our transcriptomic analysis (Table S2A).
FIG 6.
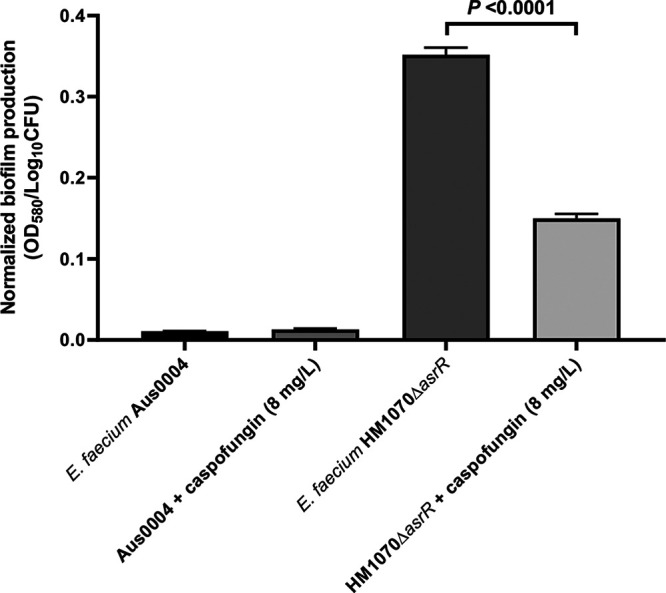
Normalized biofilm formation in the presence of caspofungin (8 mg/liter) in E. faecium Aus0004 strain (a biofilm nonproducer) and E. faecium HM1070 ΔasrR (a biofilm producer). Normalized biofilms were calculated by dividing the total biofilm value (OD580) by the bacterial growth value for each strain (expressed in Log10 values of CFU counts). Statistical comparison was performed using the unpaired t test.
Mechanism of action of caspofungin against E. faecium.
Since the fungal target of caspofungin in fungi is the β-1,3-glucan synthase, which does not exist in prokaryotes, we attempted to identify the bacterial target. To do this, we tried to select in vitro spontaneous caspofungin-resistant mutants by serial passages in agar containing a caspofungin concentration gradient. Unfortunately, after several sequential growth assays (i.e., 45 days of subcultures), we did not obtain any E. faecium colony harboring an increase in caspofungin MIC.
Conclusion.
We reported here that caspofungin seemed to have a strong bactericidal effect against E. faecium notwithstanding the lack of a protein similar to its fungal target, the β-1,3-glucan synthase. Interestingly, we showed that bacterial growth in the presence of a subinhibitory concentration of caspofungin altered the levels of transcripts to approximately 20% of the level seen with the E. faecium genome. Even though it is unlikely to be clinically relevant, greater knowledge of the observed antagonism with vancomycin would be helpful to understand the mechanism of action of caspofungin in E. faecium. The present article is the first report of caspofungin antibacterial activity against Gram-positive bacteria such as E. faecium, and further investigations of the effects of nonantibiotic xenobiotics against VRE should be conducted in the future.
MATERIALS AND METHODS
Bacterial isolates and growth conditions.
The bacterial strains used in this study are listed in Table S3 in the supplemental material. The main strain used was the vanB-positive reference strain E. faecium Aus0004, the complete genome sequence of which is available (GenBank accession number CP003351.1) (21). UCN strains were obtained from the French Reference Centre for Enterococci and used for detection of vanA/B genes as previously described (44). For biofilm formation experiments, E. faecium HM1070 ΔasrR was used as a positive control as previously described (42).
For growth experiments, E. faecium, S. aureus, and E. faecalis were cultured without shaking at 35°C in TSB whereas E. coli, E. cloacae and P. aeruginosa strains were cultured with shaking (200 rpm) at 35°C in Luria-Bertani (LB) broth. Bacteria were cultured in TSB with the addition of nonantibiotic molecules mostly prescribed in ICUs such as morphine (major analgesic), norepinephrine (vasoactive amine), pantoprazole (proton pump inhibitor), atracurium (neuromuscular blocking agent), paracetamol (minor analgesic), diazepam (benzodiazepine), unfractioned heparin (anticoagulant agent), and caspofungin (antifungal agent) at a concentration corresponding to the standard circulating blood level.
For phenotypic tests, a subinhibitory concentration of caspofungin corresponding to 1/4× MIC (8 mg/liter) was used for E. faecium Aus0004. Growth rates during the exponential phase were calculated for each condition (with or without caspofungin 8 mg/liter) and expressed as numbers of generations per hour (h−1).
We attempted to obtain mutants with decreased caspofungin susceptibility from E. faecium Aus0004 in vitro after serial passages on MH agar supplemented with increasing concentrations of caspofungin and by the gradient method on agar medium as previously described (45).
MIC determination.
MICs of five different antifungal agents (i.e., micafungin, 5-fluorocytosine, voriconazole, amphotericin B, and caspofungin) against E. faecium Aus0004 were determined by the broth microdilution (BMD) reference method in Mueller-Hinton (MH) broth. MICs of eight different antibiotic agents (i.e., ampicillin, erythromycin, vancomycin, teicoplanin, daptomycin, tigecycline, ciprofloxacin, and linezolid) against E. faecium Aus0004 were also determined by BMD according to Comité de l'antibiogramme de la Société Française de Microbiologie (CA-SFM)/EUCAST recommendations (www.sfm-microbiologie.org). Note that for daptomycin MIC determinations, calcium chloride (50 mg/liter) was added into MH broth. All determinations of MIC values were performed in three independent experiments.
MICs of four antibiotics (vancomycin, teicoplanin, daptomycin, and ciprofloxacin) were determined for E. faecium Aus0004 in the presence of 8 mg/liter of caspofungin in the MH medium using Etest strips following the manufacturer’s instructions (bioMérieux, Marcy-l’Etoile, France).
Time-kill curve experiments.
Time-kill curves were determined to determine antibacterial activity (using an antibiotic concentration equal to 8× the MIC) against E. faecium Aus0004, as previously described (46). Briefly, 16-h overnight cultures were inoculated 1:20 in 10 ml of fresh MH broth containing anti-Gram-positive antibiotics (vancomycin, linezolid, and daptomycin) or caspofungin and incubated at 35°C for 24 h. Bacterial survival was checked by CFU counts after 0, 3, 6, 9, and 24 h of incubation in three independent experiments by plating the cultures on BHI agar plates. For daptomycin assay, 50 mg/liter of calcium chloride was added to the MH broth.
Biofilm production assay.
The capacity of E. faecium Aus0004 and E. faecium HM1070 ΔasrR (a biofilm-positive strain) (42) to form biofilm in the presence of a subinhibitory concentration of caspofungin (8 mg/liter) was evaluated at 24 h. Briefly, bacteria that had been grown overnight were inoculated 1:100 in 10 ml of TSB with 0.25% glucose and apportioned into 96-microwell polystyrene plates (Nunc, Denmark). After 24 h of static incubation at 35°C, the plates were washed three times with phosphate-buffered saline (PBS) and stained with 1% crystal violet for 30 min. The wells were rinsed with distilled water and ethanol-acetone (80:20 [vol/vol]). After the wells had dried, optical density at 580 nm (OD580) was determined using a microplate reader (Multiskan Ascent; Thermo Electron Corporation). Biofilms were formed under static conditions, and each assay was performed in at least three independent experiments. Normalized biofilm values were calculated by dividing the total biofilm value (OD580) by the bacterial growth for each strain (expressed in Log10 values of CFU counts).
Cell wall analysis by electron microscopy.
For SEM experiments, E. faecium Aus0004 cells were cultured up to the late-exponential phase in TSB supplemented or not with a concentration of 8 mg/liter (1/4× MIC) of caspofungin and then pelleted by centrifugation, rinsed in PBS, and fixed with 2.5% glutaraldehyde–cacodylate buffer 0.1 M (pH 7.0) at 4°C for 15 h. The cells were rinsed in cacodylate buffer and then sedimented for 15 days on Thermanox coverslips (Thermo Fisher Scientific, Villebon-sur-Yvette, France) coated with poly-l-lysine and dehydrated in progressive baths of ethanol (70% to 100%). Bacterial cells were subjected to sputter coating with platinum and observed with a scanning electron microscope (Jeol 6400F; Jeol, Tokyo, Japan). For TEM experiments, the bacterial strain was cultured under the same conditions as were used for the SEM experiments, but after the PBS rinse, the cells were fixed with 2.5% glutaraldehyde–cacodylate buffer 0.1 M (pH 7.0) containing ruthenium red (0.4 mg/liter) for 15 h at 4°C. The cells were then rinsed and postfixed 1 h with 1% osmium tetroxide–cacodylate buffer 0.1 M (pH 7.0) in the presence of ruthenium red (0.4 mg/liter) at 4°C while protected from light. The cells were rinsed, pelleted in 1.5% agar with a low melting point (40°C), and then dehydrated in progressive ethanol baths (70% to 100%), embedded in resin (Embed 812; Electron Microscopy Sciences, Hatfield, PA, USA) and polymerized for 24 h at 60°C. Ultrathin sections were cut and stained with uranyl acetate and lead citrate. The cells were observed with a transmission electron microscope (Jeol 1011; Jeol, Tokyo, Japan), and images were taken with an Orius 200 charge-coupled-device (CCD) camera (Gatan France, Evry, France). Cell wall analysis by electron microscopy was performed in three independent experiments.
Determination of peptidoglycan composition and muropeptide analysis.
For peptidoglycan isolation, E. faecium Aus0004 cells were cultured in TSB with and without caspofungin (8 mg/liter) until mid-exponential phase, late-exponential phase, and early stationary phase and then pelleted, resuspended in PBS, and boiled while being stirred in 10% SDS for 1 h. Peptidoglycan isolation and digestion with muramidase were performed as previously described (47). Solubilized muropeptides were reduced by addition of 0.5 M sodium borate (pH 9.5) and sodium borohydride to reach a final concentration of 10 g/liter. Finally, samples were adjusted to pH 3.5 with phosphoric acid. UPLC analyses were performed on a Waters UPLC system equipped with an Acquity UPLC BEH C18 column (Water, USA) (130 Å, 1.7-μm pore size, 2.1 mm by 150 mm) and detected at A204. Muropeptides were separated mainly using a linear gradient from buffer A (phosphate buffer 50 mM [pH 4.35]) to buffer B (phosphate buffer 50 mM [pH 4.95], methanol 15% [vol/vol]) in a 40-min run. The identity of the muropeptides was assigned by MS/MS, and for quantification, the area of each peak in the chromatogram was analyzed. Peptidoglycan analysis was performed using three independent cultures for each strain.
RNA isolation and transcriptomic analysis.
E. faecium Aus0004 was cultured at 35°C until the late-exponential-growth phase (to the same optical densities) in TSB alone (−Cas media) or in TSB supplemented with a subinhibitory concentration of caspofungin (8 mg/liter) (+Cas media), corresponding to an incubation of 6 h 30 min or 7 h 30 min, respectively. Total RNA was extracted using a ZR fungal/bacterial RNA miniprep kit (Zymo Research, Irvine, CA, USA) in biological duplicate. Residual chromosomal DNA was removed by treating samples with the Turbo DNA-free kit (Life Technologies, Saint-Aubin, France). DNA-free RNA samples were quantified using a NanoDrop One spectrophotometer (Thermo Scientific, Villebon-sur-Yvette, France) and were then depleted of ribosomal RNAs (e.g., 23S, 16S, and 5S rRNAs) using a Ribo-Zero rRNA removal kit (Gram-positive bacteria) (Illumina-Epicentre, Madison, WI, USA) according to the manufacturer’s instructions. Finally, the samples were washed using an RNA Clean & Concentrator-5 kit (Zymo Research, Irvine, CA, USA). The rRNA depletion efficiency was evaluated by analyzing the samples using an Agilent 2100 bioanalyzer (Agilent Technologies, Les Ulis, France). cDNA libraries were prepared with a strand-specific NEXTflex rapid directional (dUTP-based) RNA-Seq kit (v2), and sequencing was performed using an Illumina HiSeq 2500 instrument (Illumina, San Diego, CA, USA) with the Run Rapid Single Read of 50 bp multiplexing protocol (ProfileXpert-LCMT, Lyon, France).
For bioinformatic analysis, reads were mapped against the genome sequence of E. faecium AUS0004 (GenBank accession numbers CP003351.1, CP003352.1, CP003353.1, and CP003354.1) using CLC Genomics Workbench software v10.0.1 (CLCbio; Qiagen, San Diego, CA, USA). Determinations of FC values and numbers of mapped reads per kilobase per million (RPKM) and statistical analyses were performed using the CLC Genomics Workbench and DESeq2 R package (48). Gene expression levels were identified using the log2 absolute fold change method (values higher than or lower than 2 were considered to represent induction or repression, respectively), and statistical significance was accepted in the cases in which the P value was <0.1. Mean expression and log2 FC values for each gene were plotted and visualized as an MA plot figure using the DESeq R package (48).
Validation of RNA-seq FC was done by RT-qPCR with specific primers for eight differentially expressed genes (Table S4). Total RNAs were extracted as described above, and residual DNA was removed by the use of a Turbo DNA-free kit. cDNAs were synthesized from total RNA (approximately 1 μg) using a QuantiFast reverse transcription kit (Qiagen, San Diego, CA, USA) according to the manufacturer’s instructions, and transcript levels were determined by the DeltaDelta threshold cycle (ΔΔCT) method using the atpA gene as a housekeeping control gene. Each experiment was performed in triplicate, including RNA-seq biological duplicate experiments.
Accession number(s).
Raw and processed data generated in this study have been submitted to the Gene Expression Omnibus (GEO) repository at the National Center for Biotechnology Information (NCBI) and are available under accession no. GSE100091.
Supplementary Material
ACKNOWLEDGMENTS
The technical assistance of Sébastien Galopin, Brigitte Belin, and Mamadou Godet was gratefully appreciated.
This work was supported by a grant from the Ministère de l’Enseignement Supérieur et de la Recherche to EA4655, Normandie Univ, UNICAEN, France.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Vincent JL, Rello J, Marshall J, Silva E, Anzueto A, Martin CD, Moreno R, Lipman J, Gomersall C, Sakr Y, Reinhart K. 2009. International study of the prevalence and outcomes of infection in intensive care units. JAMA 302:2323–2329. doi: 10.1001/jama.2009.1754. [DOI] [PubMed] [Google Scholar]
- 2.Biswal S, Mishra P, Malhotra S, Puri GD, Pandhi P. 2006. Drug utilization pattern in the intensive care unit of a tertiary care hospital. J Clin Pharmacol 46:945–951. doi: 10.1177/0091270006289845. [DOI] [PubMed] [Google Scholar]
- 3.Smythe MA, Melendy S, Jahns B, Dmuchowski C. 1993. An exploratory analysis of medication utilization in a medical intensive care unit. Crit Care Med 21:1319–1323. doi: 10.1097/00003246-199309000-00014. [DOI] [PubMed] [Google Scholar]
- 4.Wilde AD, Snyder DJ, Putnam NE, Valentino MD, Hammer ND, Lonergan ZR, Hinger SA, Aysanoa EE, Blanchard C, Dunman PM, Wasserman GA, Chen J, Shopsin B, Gilmore MS, Skaar EP, Cassat JE. 2015. Bacterial hypoxic responses revealed as critical determinants of the host-pathogen outcome by TnSeq analysis of Staphylococcus aureus invasive infection. PLoS Pathog 11:e1005341. doi: 10.1371/journal.ppat.1005341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sinel C, Cacaci M, Meignen P, Guérin F, Davies BW, Sanguinetti M, Giard J-C, Cattoir V. 2017. Subinhibitory concentrations of ciprofloxacin enhance antimicrobial resistance and pathogenicity of Enterococcus faecium. Antimicrob Agents Chemother 61:e0276-16. doi: 10.1128/AAC.02763-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maurice CF, Haiser HJ, Turnbaugh PJ. 2013. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 152:39–50. doi: 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freestone PP, Hirst RA, Sandrini SM, Sharaff F, Fry H, Hyman S, O'Callaghan C. 2012. Pseudomonas aeruginosa-catecholamine inotrope interactions: a contributory factor in the development of ventilator-associated pneumonia? Chest 142:1200–1210. doi: 10.1378/chest.11-2614. [DOI] [PubMed] [Google Scholar]
- 8.Chen C, Brown DR, Xie Y, Green BT, Lyte M. 2003. Catecholamines modulate Escherichia coli O157:H7 adherence to murine cecal mucosa. Shock 20:183–188. doi: 10.1097/01.shk.0000073867.66587.e0. [DOI] [PubMed] [Google Scholar]
- 9.Dowd SE. 2007. Escherichia coli O157:H7 gene expression in the presence of catecholamine norepinephrine. FEMS Microbiol Lett 273:214–223. doi: 10.1111/j.1574-6968.2007.00800.x. [DOI] [PubMed] [Google Scholar]
- 10.Bansal T, Englert D, Lee J, Hegde M, Wood TK, Jayaraman A. 2007. Differential effects of epinephrine, norepinephrine, and indole on Escherichia coli O157:H7 chemotaxis, colonization, and gene expression. Infect Immun 75:4597–4607. doi: 10.1128/IAI.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karavolos MH, Spencer H, Bulmer DM, Thompson A, Winzer K, Williams P, Hinton JCD, Khan CMA. 2008. Adrenaline modulates the global transcriptional profile of Salmonella revealing a role in the antimicrobial peptide and oxidative stress resistance responses. BMC Genomics 9:458. doi: 10.1186/1471-2164-9-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyte M, Freestone PP, Neal CP, Olson BA, Haigh RD, Bayston R, Williams PH. 2003. Stimulation of Staphylococcus epidermidis growth and biofilm formation by catecholamine inotropes. Lancet 361:130–135. doi: 10.1016/S0140-6736(03)12231-3. [DOI] [PubMed] [Google Scholar]
- 13.Cogan TA, Thomas AO, Rees LEN, Taylor AH, Jepson MA, Williams PH, Ketley J, Humphrey TJ. 2007. Norepinephrine increases the pathogenic potential of Campylobacter jejuni. Gut 56:1060–1065. doi: 10.1136/gut.2006.114926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu F, Wu C, Guo F, Cui G, Zeng X, Yang B, Lin J. 2015. Transcriptomic analysis of Campylobacter jejuni NCTC 11168 in response to epinephrine and norepinephrine. Front Microbiol 6:1060–1065. doi: 10.3389/fmicb.2015.00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green BT, Lyte M, Chen C, Xie Y, Casey MA, Kulkarni-Narla A, Vulchanova L, Brown DR. 2004. Adrenergic modulation of Escherichia coli O157:H7 adherence to the colonic mucosa. Am J Physiol Gastrointest Liver Physiol 287:G1238–G1246. doi: 10.1152/ajpgi.00471.2003. [DOI] [PubMed] [Google Scholar]
- 16.Babrowski T, Romanowski K, Fink D, Kim M, Gopalakrishnan V, Zaborina O, Alverdy J. 2013. The intestinal environment of surgical injury transforms Pseudomonas aeruginosa into a discrete hypervirulent morphotype capable of causing lethal peritonitis. Surgery 153:36–43. doi: 10.1016/j.surg.2012.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rice LB. 2010. Progress and challenges in implementing the research on ESKAPE pathogens. Infect Control Hosp Epidemiol 31(Suppl 1):S7–S10. doi: 10.1086/655995. [DOI] [PubMed] [Google Scholar]
- 18.Arias CA, Murray BE. 2012. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 10:266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cattoir V, Leclercq R. 2013. Twenty-five years of shared life with vancomycin-resistant enterococci: is it time to divorce? J Antimicrob Chemother 68:731–742. doi: 10.1093/jac/dks469. [DOI] [PubMed] [Google Scholar]
- 20.Lebreton F, van Schaik W, McGuire AM, Godfrey P, Griggs A, Mazumdar V, Corander J, Cheng L, Saif S, Young S, Zeng Q, Wortman J, Birren B, Willems RJL, Earl AM, Gilmore MS. 2013. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. mBio 4:e00534-13. doi: 10.1128/mBio.00534-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lam MMC, Seemann T, Bulach DM, Gladman SL, Chen H, Haring V, Moore RJ, Ballard S, Grayson ML, Johnson PDR, Howden BP, Stinear TP. 2012. Comparative analysis of the first complete Enterococcus faecium genome. J Bacteriol 194:2334–2341. doi: 10.1128/JB.00259-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyte M. 2004. Microbial endocrinology and infectious disease in the 21st century. Trends Microbiol 12:14–20. doi: 10.1016/j.tim.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 23.Freestone PP, Haigh RD, Lyte M. 2007. Blockade of catecholamine-induced growth by adrenergic and dopaminergic receptor antagonists in Escherichia coli O157:H7, Salmonella enterica and Yersinia enterocolitica. BMC Microbiol 7:8. doi: 10.1186/1471-2180-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freestone PPE, Haigh RD, Lyte M. 2007. Specificity of catecholamine-induced growth in Escherichia coli O157:H7, Salmonella enterica and Yersinia enterocolitica. FEMS Microbiol Lett 269:221–228. doi: 10.1111/j.1574-6968.2006.00619.x. [DOI] [PubMed] [Google Scholar]
- 25.Coulanges V, Andre P, Ziegler O, Buchheit L, Vidon DJ. 1997. Utilization of iron-catecholamine complexes involving ferric reductase activity in Listeria monocytogenes. Infect Immun 65:2778–2785. doi: 10.1128/IAI.65.7.2778-2785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siala W, Kucharíková S, Braem A, Vleugels J, Tulkens PM, Mingeot-Leclercq M-P, Van Dijck P, Van Bambeke F. 2016. The antifungal caspofungin increases fluoroquinolone activity against Staphylococcus aureus biofilms by inhibiting N-acetylglucosamine transferase. Nat Commun 7:13286. doi: 10.1038/ncomms13286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loiko V, Wagener J. 2017. The paradoxical effect of echinocandins in Aspergillus fumigatus relies on recovery of the β-1,3-glucan synthase Fks1. Antimicrob Agents Chemother 61:e01690-16. doi: 10.1128/AAC.01690-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walker LA, Gow NAR, Munro CA. 2010. Fungal echinocandin resistance. Fungal Genet Biol 47:117–126. doi: 10.1016/j.fgb.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall GS, Myles C, Pratt KJ, Washington JA. 1988. Cilofungin (LY121019), an antifungal agent with specific activity against Candida albicans and Candida tropicalis. Antimicrob Agents Chemother 32:1331–1335. doi: 10.1128/aac.32.9.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Machka K, Braveny I. 1987. Comparative in vitro activity of LY146032 (daptomycin) against Gram-positive cocci. Eur J Clin Microbiol 6:96–99. doi: 10.1007/BF02097210. [DOI] [PubMed] [Google Scholar]
- 32.Cattoir V, Giard J-C. 2014. Antibiotic resistance in Enterococcus faecium clinical isolates. Expert Rev Anti Infect Ther 12:239–248. doi: 10.1586/14787210.2014.870886. [DOI] [PubMed] [Google Scholar]
- 33.Georgopapadakou NH. 2001. Update on antifungals targeted to the cell wall: focus on beta-1,3-glucan synthase inhibitors. Expert Opin Invest Drugs 10:269–280. doi: 10.1517/13543784.10.2.269. [DOI] [PubMed] [Google Scholar]
- 34.Vollmer W, Blanot D, Pedro D, de Pedro MA. 2008. Peptidoglycan structure and architecture. FEMS Microbiol Rev 32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 35.Tatusov RL, Koonin EV, Lipman DJ. 1997. A genomic perspective on protein families. Science 278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 36.Deutscher J, Francke C, Postma PW. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev 70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hancock LE, Murray BE, Sillanpää J. 2014. Enterococcal cell wall components and structures In Gilmore MS, Clewell DB, Ike Y, Shankar N (ed), Enterococci: from commensals to leading causes of drug resistant infection. Massachusetts Eye and Ear Infirmary, Boston, MA: https://www.ncbi.nlm.nih.gov/books/NBK190431/. [Google Scholar]
- 38.Ramsey M, Hartke A, Huycke M. 2014. The physiology and metabolism of Enterococci In Gilmore MS, Clewell DB, Ike Y, Shankar N (ed), Enterococci: from commensals to leading causes of drug resistant infection. Massachusetts Eye and Ear Infirmary, Boston, MA: https://www.ncbi.nlm.nih.gov/books/NBK190432/. [PubMed] [Google Scholar]
- 39.Verneuil N, Mazé A, Sanguinetti M, Laplace J-M, Benachour A, Auffray Y, Giard J-C, Hartke A. 2006. Implication of (Mn)superoxide dismutase of Enterococcus faecalis in oxidative stress responses and survival inside macrophages. Microbiology 152:2579–2589. doi: 10.1099/mic.0.28922-0. [DOI] [PubMed] [Google Scholar]
- 40.Ladjouzi R, Bizzini A, Lebreton F, Sauvageot N, Rincé A, Benachour A, Hartke A. 2013. Analysis of the tolerance of pathogenic enterococci and Staphylococcus aureus to cell wall active antibiotics. J Antimicrob Chemother 68:2083–2091. doi: 10.1093/jac/dkt157. [DOI] [PubMed] [Google Scholar]
- 41.Willems RJ, Homan W, Top J, van Santen-Verheuvel M, Tribe D, Manzioros X, Gaillard C, Vandenbroucke-Grauls CM, Mascini EM, van Kregten E, van Embden JD, Bonten MJ. 2001. Variant esp gene as a marker of a distinct genetic lineage of vancomycin-resistant Enterococcus faecium spreading in hospitals. Lancet 357:853–855. doi: 10.1016/S0140-6736(00)04205-7. [DOI] [PubMed] [Google Scholar]
- 42.Lebreton F, van Schaik W, Sanguinetti M, Posteraro B, Torelli R, Le Bras F, Verneuil N, Zhang X, Giard J-C, Dhalluin A, Willems RJL, Leclercq R, Cattoir V. 2012. AsrR is an oxidative stress sensing regulator modulating Enterococcus faecium opportunistic traits, antimicrobial resistance, and pathogenicity. PLoS Pathog 8:e1002834. doi: 10.1371/journal.ppat.1002834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bachmann SP, VandeWalle K, Ramage G, Patterson TF, Wickes BL, Graybill JR, López-Ribot JL. 2002. In vitro activity of caspofungin against Candida albicans biofilms. Antimicrob Agents Chemother 46:3591–3596. doi: 10.1128/aac.46.11.3591-3596.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bourdon N, Lemire A, Fines-Guyon M, Auzou M, Périchon B, Courvalin P, Cattoir V, Leclercq R. 2011. Comparison of four methods, including semi-automated rep-PCR, for the typing of vancomycin-resistant Enterococcus faecium. J Microbiol Methods 84:74–80. doi: 10.1016/j.mimet.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 45.Szybalski W, Bryson V. 1952. Genetic studies on microbial cross resistance to toxic agents. I. Cross resistance of Escherichia coli to fifteen antibiotics. J Bacteriol 64:489–499. doi: 10.1128/JB.64.4.489-499.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moellering RC, Wennersten C, Weinberg AN. 1971. Studies on antibiotic synergism against enterococci. I. Bacteriologic studies. J Lab Clin Med 77:821–828. [PubMed] [Google Scholar]
- 47.Alvarez L, Hernandez SB, de Pedro MA, Cava F. 2016. Ultra-sensitive, high-resolution liquid chromatography methods for the high-throughput quantitative analysis of bacterial cell wall chemistry and structure. Methods Mol Biol 1440:11–27. doi: 10.1007/978-1-4939-3676-2_2. [DOI] [PubMed] [Google Scholar]
- 48.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.