Abstract
RNA viruses in aquatic environments remain poorly studied. Here, we analysed the RNA virome from approximately 10 l water from Yangshan Deep-Water Harbour near the Yangtze River estuary in China and identified more than 4,500 distinct RNA viruses, doubling the previously known set of viruses. Phylogenomic analysis identified several major lineages, roughly, at the taxonomic ranks of class, order and family. The 719-member-strong Yangshan virus assemblage is the sister clade to the expansive class Alsuviricetes and consists of viruses with simple genomes that typically encode only RNA-dependent RNA polymerase (RdRP), capping enzyme and capsid protein. Several clades within the Yangshan assemblage independently evolved domain permutation in the RdRP. Another previously unknown clade shares ancestry with Potyviridae, the largest known plant virus family. The ‘Aquatic picorna-like viruses/Marnaviridae’ clade was greatly expanded, with more than 800 added viruses. Several RdRP-linked protein domains not previously detected in any RNA viruses were identified, such as the small ubiquitin-like modifier (SUMO) domain, phospholipase A2 and PrsW-family protease domain. Multiple viruses utilize alternative genetic codes implying protist (especially ciliate) hosts. The results reveal a vast RNA virome that includes many previously unknown groups. However, phylogenetic analysis of the RdRPs supports the previously established five-branch structure of the RNA virus evolutionary tree, with no additional phyla.
Subject terms: Water microbiology, Computational biology and bioinformatics, Metagenomics, Microbial ecology, Phylogeny
Metagenomic analysis of a single RNA virome from the Yangshan Deep-Water Harbour in China enabled the recovery of more than 4,500 distinct RNA viruses, doubling the known set of RNA viruses to date, and provided insights into their biology.
Main
Metagenomics and metaviromics (that is, sequencing of DNA or RNA from virus particle fractions isolated from diverse environments or organisms) have led to rapid progress in virus discovery1–9. The International Committee on Taxonomy of Viruses has approved formal classification of viruses characterized solely by metagenomics10. The rapid advances in metaviromics have substantially expanded the known diversity of RNA viruses, yielding vast amounts of sequences for comprehensive studies on RNA virus evolution11–17.
Metagenomic investigation of various aquatic environments provides access to viromes of diverse prokaryotes and unicellular eukaryotes that could harbour ancient lineages of the RNA viruses2. Rich RNA viromes have been described in aquatic environments as diverse as Antarctic seas and wastewater11,14,15,18–20. Although metaviromic analyses do not typically identify the virus hosts, some of the marine RNA virome components have been phylogenetically anchored through similarity to viruses with known hosts. Perhaps the best characterized group of such viruses is the family Marnaviridae, which combines picorna-like viruses of diatoms and other stramenopiles21–26 with a growing number of species defined by metagenomics as probably infecting related aquatic unicellular eukaryotes27,28 (hereafter referred to as ‘protists’).
Another key development has been meta-transcriptome sequencing of invertebrate holobionts, doubling the size of the known RNA virome29–33. The high diversity of the invertebrate RNA virome suggests that RNA viruses of land plants and vertebrates evolved from viruses infecting invertebrates2. The known RNA viromes of plants, fungi, protists and bacteria have also expanded through meta-transcriptome sequencing, albeit not as massively as the invertebrate virome19,34–41.
A comprehensive phylogenetic analysis of RdRP, the only universally conserved protein of RNA viruses, produced a phylogenetic tree comprised of five major branches42. The deepest branch 1 includes the only known group of positive-sense (+)RNA viruses of prokaryotes, the leviviruses and their eukaryote-infecting descendants (narna- and ourmia-like viruses). The remaining four branches consist mostly of RNA viruses that infect eukaryotes. Branch 2 includes the assemblage of +RNA viruses denoted ‘picornavirus-like supergroup’, along with some of the smallest +RNA viruses in the Solemoviridae family and the largest +RNA viruses of the order Nidovirales. Branch 2 also contains two families of double-stranded RNA (dsRNA) viruses, Partitiviridae and Picobirnaviridae. Branch 3 consists solely of +RNA viruses, including the ‘Alphavirus supergroup’, a variety of viruses with small genomes resembling tombusviruses and nodaviruses, and the ‘Flavivirus supergroup’. Branch 4 consists of diverse dsRNA viruses, including the large families Reoviridae and Totiviridae, and the only known family of prokaryotic dsRNA viruses, Cystoviridae. Finally, branch 5 includes all known negative-sense (−)RNA viruses. A comprehensive virus ‘megataxonomy’ has been recently proposed and subsequently formally approved by the International Committee on Taxonomy of Viruses, in which the five major branches of the RdRPs correspond to five phyla in the kingdom Orthornavirae43,44. Despite these advances, a pressing question remains: would the current view of the RNA virome change substantially with deeper sampling, or are we getting close to an effectively complete coarse-grain picture of the global RNA virome? Is it likely that additional phyla of RNA viruses remain to be discovered?
Here we report an extensive analysis of an RNA virome in water samples from Yangshan Deep-Water Harbour near Shanghai, China, where the Yangtze River meets the East China Sea (Fig. 1). This analysis of the RNA virome from a single, albeit complex, aquatic habitat doubles the known diversity of RNA viruses, identifying several previously unrecognized groups of +RNA viruses (roughly, at the class, order or family taxonomic ranks). Despite the discovery of numerous virus groups, phylogenetic analysis of the RdRPs shows that a substantial majority of the identified viruses belong to already established phyla of RNA viruses44.
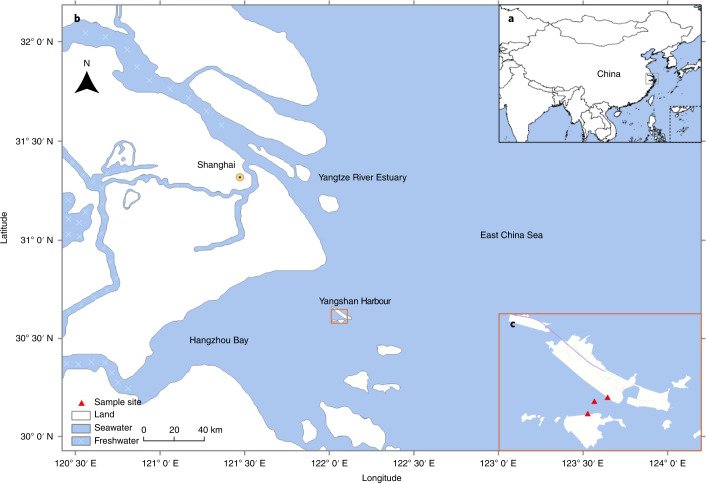
Fig. 1. The Yangshan Deep-Water Harbour.
a–c, Map of the Yangshan Deep-Water Harbour at three scales. a, Location within China. b, Magnified view of the region bounded by the grey box in a. c, Expanded view of the region marked by the orange box in b, showing the Yangshan Deep-Water Harbour. Red triangles in c mark seawater sampling sites. Map data: Google, 2014; Mapabc.com, 2014; CNES/Astrium, 2014 TerraMetics.
Results
Diversity of RNA viruses in the Yangshan harbour virome
RNA virome analysis performed using complementary DNA derived from approximately 10 l of samples from Yangshan Deep-Water Harbour yielded 4,593 nearly full-length RNA virus RdRPs that formed 2,192 clusters at 75% amino acid identity which represents virus diversity at a level between species and genus. Among the RdRP sequences from GenBank (October 2018), 2,021 comparable clusters were detected. Thus, the 10 l water sample analysed here more than doubles the known diversity of RNA viruses.
Phylogenetic analysis assigned 85% of the RdRPs from the Yangshan RNA virome to 9 clades and one complex assemblage, each comprising more than 100 RdRps from several clusters (Fig. 2 and Supplementary Dataset 1). Seven of these clades blended into those defined previously, whereas two previously unknown clades and the assemblage were dominated by viruses from the Yangshan virome (Fig. 2). All these clades represented +RNA viruses of the phyla Lenarviricota, Pisuviricota and Kitrinoviricota, whereas no members of Negarnaviricota were found. Only six dsRNA viruses (Duplornaviricota) were identified, but were not further analysed. No enveloped +RNA viruses of the families Flaviviridae and Togaviridae were detected. Common +RNA viruses of terrestrial vertebrates and plants (for example, members of Picornaviridae, Caliciviridae, Virgaviridae or Potyviridae) were also absent from the Yangshan virome.
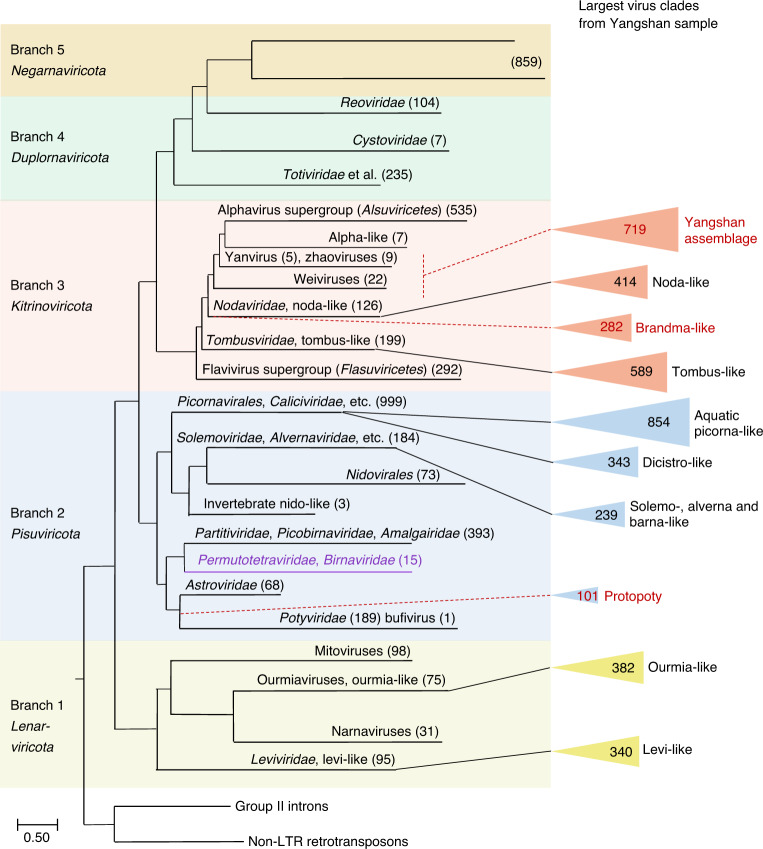
Fig. 2. Schematic phylogenetic tree of the RNA virus RdRPs.
The reverse transcriptases of group II introns and non-long-terminal-repeat (non-LTR) transposons were used as an outgroup to root the tree. The overall tree topology encompassing five major RdRP branches (highlighted by different background colours) has been described previously42. These branches correspond to RNA virus phyla, which are shown under the branch numbers. The positions of the largest clades of viruses identified in this study are indicated and represented by triangles, the areas of which are roughly proportional to the number of viruses in each clade (shown inside the triangle). The numbers in parentheses correspond to previously identified viruses included in the analysis in ref. 42. Provisional names of the previously undescribed virus clades are shown in red. Purple text denotes a virus lineage with permuted RdRPs (Permutotertraviridae and Birnaviradae) which was not included in the previous study42.
The largest RdRP group in the Yangshan virome (854 members; Supplementary Datasets 1 and 2) belongs to the ‘Aquatic picorna-like’ clade (order Picornavirales)30,42 in the phylum Pisuviricota (Fig. 2). This clade contains the Marnaviridae and other protist-infecting viruses as well as viruses identified in holobionts of molluscs, annelids and other marine invertebrates whose diets include protists. The largest of the 323 broad RdRP clusters in the Yangshan virome—OV.1 (where OV indicates Ocean Viruses, an operational term for RdRP clusters), with 653 members (Supplementary Datasets 1 and 2)—fell entirely into the Marnaviridae, vastly expanding the diversity of this family and highlighting the need for a taxonomic upgrade28. Given that isolated Marnaviridae infect diatoms and other aquatic Stramenopile protists21,22,26,39,45, most OV.1 members are likely to infect related unicellular eukaryotes. The genome organizations of the previously recognized marnaviruses and those from the Yangshan virome are nearly uniform: they encode either one or two polyproteins encompassing the same set of protein domains (Supplementary Dataset 2).
Pisuviricota accommodated another large clade with 343 RdRPs related to those of Dicistroviridae (order Picornavirales; Fig. 2), which infect marine and terrestrial arthropods30,46. Although these and previously recognized dicistroviruses share the same genome organization, they form sister groups in the RdRP phylogeny (for example, OV.9 and OV.13 in Supplementary Datasets 1 and 2), suggestive of distinct host ranges. The third largest clade within Pisuriviricota (239 RdRPs, including OV.12 and OV.27) joined the lineage that includes plant Solemoviridae, fungal Barnaviridae and protist Alvernaviridae.
The fourth clade (101 members) of the Yangshan virome RdRPs within Pisuviricota (including OV.16 and OV.23; Supplementary Datasets 1 and 2) is a sister group to Potyviridae, the largest family of plant viruses47,48. Because the marine virome appears to be ancestral to the terrestrial plant virome49, these aquatic relatives of the potyviruses probably resemble the common ancestor, and were accordingly dubbed Protopotyviruses (Fig. 2). Protopotyviruses share with potyviruses the conserved tandem of a chymotrypsin-like protease and the RdRP, but lack the SF2 helicase and the papain-like protease characteristic of potyviruses (Supplementary Dataset 2). Given the evolutionary affinity between the potyvirus SF2 helicase and the homologous helicase of flavi-like viruses50, this is likely to be a late acquisition in potyviruses. Most of the protopotyvirus genomes encode a single-jelly-roll capsid protein (SJR-CP), likely inherited from the common ancestor of all eukaryotic RNA viruses42. In contrast, filamentous potyviruses encode a distinct capsid protein51, which is homologous to nucleocapsid proteins of (−)RNA viruses52,53. These findings are consistent with the ancestral status of protopotyviruses with respect to potyviruses.
More than 1,700 Yangshan virome RdRPs belong to Kitrinoviricota; this was unexpected, given that so far, Kitrinoviricota consisted largely of viruses of terrestrial plants and animals30,42,54. Two virus groups from the Yangshan virome fell within the Tombus-like (589 members) and Noda-like (414 members) clades of Kitrinoviricota (Fig. 2). Nodaviridae is not monophyletic with respect to the Yangshan nodavirus-like group: the nematode-infecting Orsay-like viruses55 as well as Sclerophthora macrospora virus A56 and Plasmopara halstedii virus A57, both of which infect oomycetes, are nested within the diversity of the Yangshan RdRPs (OV.3 in Supplementary Dataset 2). Oomycetes, particularly those that parasitize diatoms58, are the plausible hosts for the noda-like viruses in the Yangshan virome, although free-living marine nematodes could not be ruled out as hosts59. Unlike the known members of Nodaviridae, most of the noda-like viruses identified in the Yangshan virome have monopartite genomes, which appears compatible with an ancestral state. None of the major Yangshan virome clades among Kitrinoviricota joined the ‘Alphavirus supergroup’ (class Alsuviricetes) comprising viruses that infect mostly plants, as well as animals and fungi.
A previously unknown, highly diverse assortment of RdRPs (719 members; hereafter, the Yangshan assemblage) consists of several clades positioned between the noda-like viruses and Alsuviricetes within Kitrinoviricota (Figs. 2 and 3). This assemblage includes three previously described small groups of viruses—namely, Weiviruses, Yanviruses and Zhaoviruses—and several unclassified viruses. The largest clade within the Yangshan assemblage (OV.2, hereafter the Yan-like clade) consists of 431 Yangshan RdRPs, all 5 previously described Yanviruses30 and several uncharacterized viruses, including the solitary RNA virus isolated from an acidic hot spring in Yellowstone National Park dominated by archaea60 (Fig. 3).
Fig. 3. The Yangshan virus assemblage.
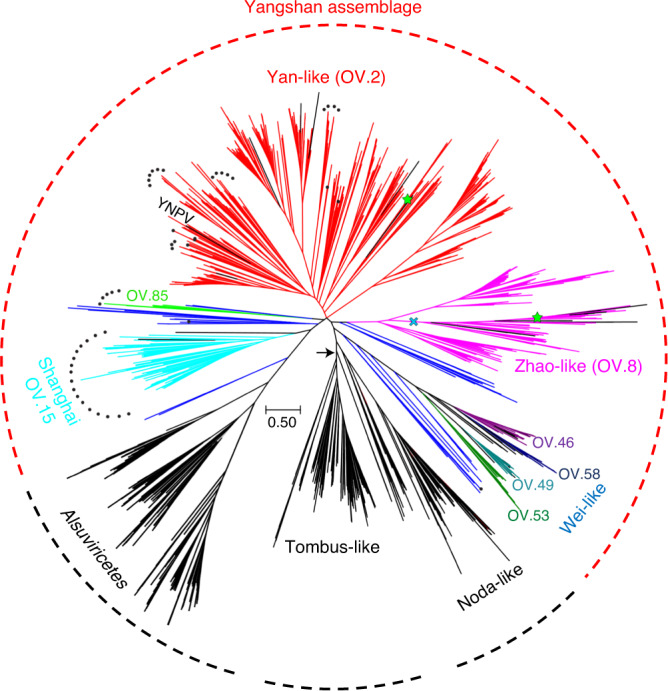
The rootless tree represents the subtree of branch 3 (Kitrinoviricota) in which the Yangshan virus assemblage belongs. The positions of RdRPs of previously reported viruses are shown in black. Dark blue represents assorted small clusters of the Yangshan assemblage. Other colours represent clusters (as indicated) of which Yan-like (red), Zhao-like (magenta) and Shanghai (electric blue) correspond to eponymous major clades, whereas the Wei-like clade encompasses four clusters. The permuted RdRPs are marked by black dots and RdRP groups with non-standard genetic codes are marked with green stars. The blue cross marks a virus group within the Zhao-like clade that includes viruses using protist genetic codes and encoding a capping enzyme similar to that of nodaviruses. The arrow points to a root position as in the tree in Fig. 2. YNPV, Yellowstone National Park virus60.
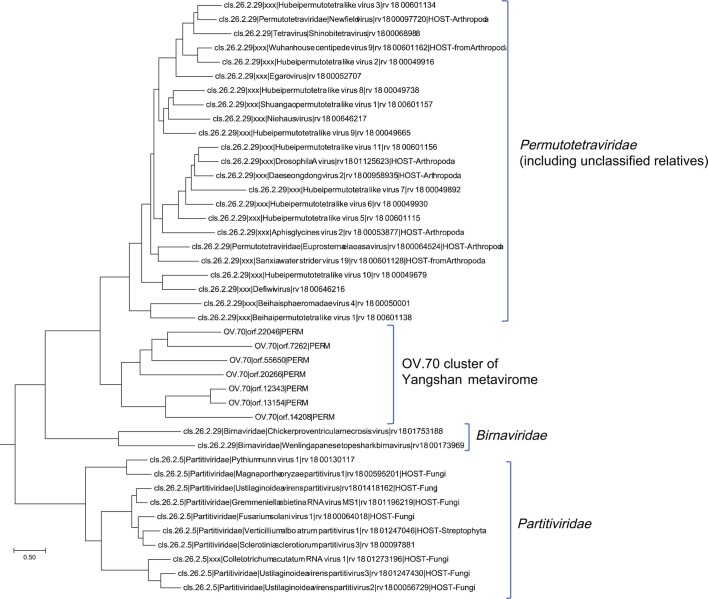
The Yan clade is a hotspot of RdRP domain permutation that apparently occurred on ten independent occasions within this clade alone. Previously, such permutations had been detected in the Permutotertraviridae and Birnaviridae61–63, but were excluded from our previous analysis due to interference with multiple RdRP alignments caused by the permutation42. Here we developed a procedure for swapping domains in the permuted RdRPs to restore the original domain order and included these reconstructed RdRP sequences in the phylogenetic analysis (Fig. 2). In the resulting trees, permutotetraviruses and birnaviruses formed a well-supported clade within Pisuviricota that was far removed from the Yangshan assemblage (Extended Data Fig. 1), again pointing to convergent evolution of this trait in diverse viruses.
Extended Data Fig. 1. Phylogenetic tree of the permuted RdRps of Permutotetraviridae, Birnaviridae and a related OV.70 cluster of seven RdRps identified in the Yangshan virome.
Note that this lineage of permuted RdRPs is confidently lodged as additional clade in Branch 2 (Pisuviricota) as a sister to Partitiviridae lineage.
Of the 387 long Yan-like contigs, 220 encode a distinct SJR-CP and 100 encode a capping enzyme. A HHpred comparison of the profile created from the sequence alignment of Yan-like virus capping enzymes against the profile database exclusively retrieved the capping enzymes of Alsuviricetes (PF01660.17; Vmethyltransf; P = 99.8; Extended Data Fig. 2), in support of the placement of the Yan-like clade near the base of Alsuviricetes (Fig. 3). The profile–profile comparisons also showed that the SJR-CP protein of Yan-like viruses has a two-domain organization, including the shell and projection domains, similar to the capsid proteins of certain nodaviruses and tombusviruses (Extended Data Fig. 3), solidifying the position of the Yan-like clade in the tree.
Extended Data Fig. 2. Results of the HHpred search seeded with the putative capping enzyme of Yan-like viruses.
H(h), α-helix; E(e), β-strand; C(c), coil. The single-letter designations of the amino acid residues are coloured according to their physical properties.
Extended Data Fig. 3. Results of the HHpred search seeded with the putative capsid protein of Yan-like viruses.
H(h), α-helix; E(e), β-strand; C(c), coil. The single-letter designations of the amino acid residues are colored according to their physical properties.
Another major clade within the Yangshan assemblage (OV.8, hereafter the Zhao-like clade) consists of 113 members (Fig. 3; Supplementary Datasets 1 and 2) and includes a previously orphan cluster of 9 Zhaoviruses identified in marine invertebrates30 along with ‘ciliovirus’ and ‘brinovirus’ from a San Francisco wastewater virome64. The Zhaoviruses, ‘ciliovirus’ and ‘brinovirus’, together with 36 Yangshan virome viruses, form a separate group within the Zhao-like clade. This group is distinguished by using ciliate and other protist genetic codes (see below) and by encoding a capping enzyme similar to the distinct capping enzyme of nodaviruses (Fig. 4 and Supplementary Dataset 2).
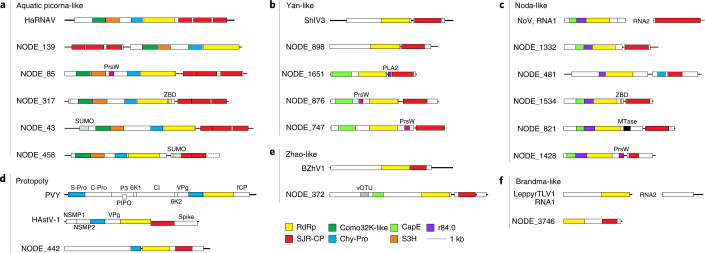
Fig. 4. Diversity of domain organizations in the major clades of marine RNA viruses in the Yangshan virome.
a, Aquatic picorna-like virus clade (as in Fig. 2). b, Yan-like virus clade (as in Fig. 3). c, Noda-like virus clade (as in Figs. 2 and 3). d, Protopotyvirus clade (as in Fig. 2). e, Zhao-like virus clade (as in Fig. 3). f, Brandma-like virus clade (as in Fig. 2). Each panel contains a genome map(s) of a phylogenetically close reference virus(es) at the top and viruses discovered in this study, identified as ‘NODE_NN’. NODE numbers correspond to contig gene IDs listed in Supplementary Datasets 3 and 5. Functional domains are colour coded and the colour key for the recurrent domains is shown at the bottom of the figure. HaRNAV, Heterosigma akashiwo RNA virus; PVY, potato virus Y; HAstV-1, human astrovirus 1; ShIV3, Shahe isopoda virus 3; BZhV1, Beihai zhaovirus-like virus 1; NoV, Nodamura virus; LeppyrTLV1, Leptomonas pyrrhocoris tombus-like virus 1; PrsW, PrsW-family protease; ZBD, zinc-binding domain; PLA2, phospholipase A2; vOTU, viral ovarian tumour protease; MTase, methyltransferase; Chy-Pro, chymotrypsin-like protease; S-Pro, serine protease; C-Pro, cysteine protease; VPg, viral genome-linked protein; CP, capsid protein; fCP, filamentous capsid protein; NSMP, non-structural mature protein; CapE, capping enzyme; S3H, superfamily 3 helicase; Como32K-like, comovirus 32K-like protease; r84.0, functionally uncharacterized domain conserved in RNA viruses; P3, protein 3; 6K1 and 6K2, 6 kDa proteins 1 and 2; PIPO, pretty interesting Potyviridae open reading frame protein; Spike, spike protein; CI, cylindrical inclusion protein.
The third major clade in the Yangshan assemblage, denoted ‘Shanghai’, harbours 74 Yangshan RdRPs and the unclassified ‘eunivirus’ (KF412900), which was identified in a wastewater virome (Fig. 3; OV.15). The signature of this clade is the domain permutation of the RdRP that apparently occurred at the base of this clade. Finally, the Wei-like clade with 57 Yangshan RdRPs (clusters OV.46, OV.49, OV.53, OV.58, OV.192, OV.233, OV.250 and OV.262) also includes 15 Weiviruses30. The phylogenomic diversity within the Yangshan assemblage seems to justify the establishment of a virus class, subdivided into multiple orders and families.
The last large clade within Kitrinoviricota (‘Brandma-like’ viruses) combines 282 RdRPs (cluster OV.4; Supplementary Datasets 1 and 2) with several previously reported orphan viruses from diverse sources (Fig. 2 and Supplementary Dataset 2). Most Brandma-like viruses have small, 4–5-kb genomes that encode only two recognizable domains, RdRp and SJR-CP (Supplementary Dataset 2). The Brandma-like viruses form a sister group to the Noda-like viruses (Fig. 2).
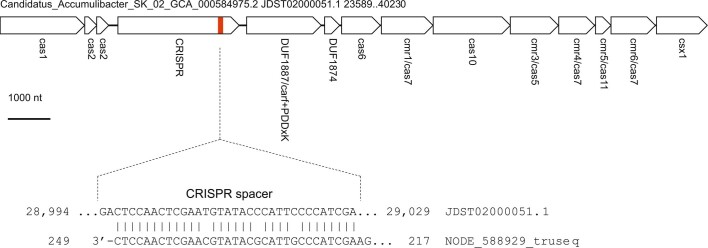
Finally, two large clusters of the Yangshan RdRP belong to Lenarviricota, grouping with the +RNA bacteriophages of the Leviviridae and levi-like viruses (340 members), or with ourmia-like viruses (382 members), the eukaryote-infecting descendants of +RNA bacteriophages2,30,37 (Fig. 2 and Supplementary Datasets 1 and 2). Such strong representation of the levi-like phages and ourmia-like viruses in an aquatic RNA virome is expected, and so is the absence of the other clades of Lenarviricota, namely, mito- and narnaviruses, common capsid-less +RNA agents of fungi65. Our search for RNA virus sequences homologous to bacterial clustered regularly interspaced short palindromic repeats (CRISPR) spacers yielded a single match between one of the Yangshan RNA virome contigs bearing a levi-like RdRP and the reverse transcriptase-associated type III-B CRISPR locus of the bacterium Candidatus Accumulibacter sp. SK-02 (Extended Data Fig. 4). To our knowledge, CRISPR spacers matching RNA virus genomes have not been reported previously. Although caution is warranted in the interpretation of this solitary RNA virus protospacer, this finding suggests that CRISPR–CRISPR-associated protein (Cas) systems can target RNA viruses66.
Extended Data Fig. 4.
A nucleotide sequence match between a Yangshan RNA virome contig bearing a levi-like RdRP (bottom line) and the type III-B CRISPR spacer locus of the bacterium Candidatus Accumulibacter sp. SK-02.
Overall, each of the three phyla of +RNA viruses42,44 is well represented in the complex Yangshan virome (Fig. 2). Among the largest (more than 100 members) clusters of the discovered RdRPs, four (Yan-like, Zhao-like, Brandma-like and Protopoty) form distinct clades within Pisuviricota and Kitrinoviricota (Figs. 2 and 3), each assimilating a handful of previously identified viruses of uncertain evolutionary provenance that now find their ‘phylogenetic homes’.
In addition to the RdRPs that could be assigned to previously identified clades at different depths of the phylogenetic tree, we attempted to detect putative highly divergent RdRPs using complementary approaches (see Methods) and identified 13 singleton RdRP sequences. The further expansion of the global RNA virome is expected to allow more confident assignment of these divergent RdRPs to additional clades, as was the case with the Yan-like, Zhao-like, Brandma-like and Protopoty clades.
Distinct domain architectures of virus proteins in the Yangshan virome
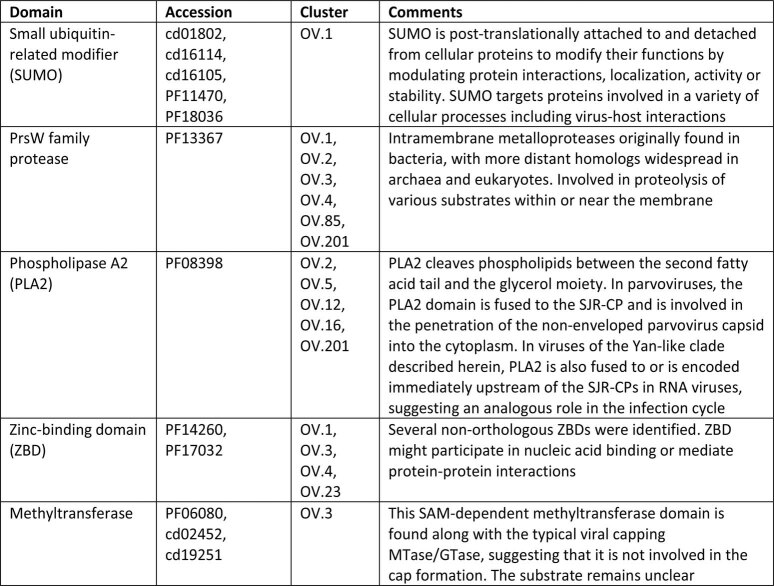
Analysis of the domain content of the longer Yangshan contigs indicates that the genome organizations are typically similar within clusters enriched in Yangshan viruses and closely resemble the genome organizations of the previously known viruses from the same clades. Nevertheless, we identified several domains that have not been previously observed in any RNA viruses (Fig. 4 and Supplementary Dataset 2), including small ubiquitin-like modifier (SUMO), PrsW-like protease and phospholipase A2 (Extended Data Fig. 5). In addition, many Yangshan virome clusters included viruses that appear to have relatively recently acquired other domains, in particular, Zn2+-binding and methyltransferase domains, as well as conserved domains of unknown function. Collectively, these observations reveal dynamic acquisition of multiple functional domains that might be involved in distinct virus–host interactions.
Extended Data Fig. 5. Protein domains identified in the Yangshan RNA virome that were not previously observed in known RNA viruses.
In the virome contigs, the nucleotide sequences encoding these domains were linked to those encoding RdRP thus demonstrating that they belonged to RNA virus genomes.
Alternative genetic codes in RNA viruses
The RdRPs in the Yangshan virome were identified in end-to-end six-frame translations of the contigs. Mapping the RdRP core domain profile to the best-matching frame established the RdRP core boundaries for each contig. In 98.7% of the contigs, the RdRP core translations obtained with the standard genetic code contained no stop codons. The remaining RdRP-coding regions, however, contained up to 26 stop codons (Supplementary Dataset 3), suggesting alternative genetic codes. These contigs were translated using all 26 known variants of the genetic code, and the code that yielded the longest protein including the RdRP core was selected for each contig. Viruses using alternative codes were identified in the Yan-like and Zhao-like clades within the Yangshan assemblage where the use of alternative codes is mostly confined to two distinct, smaller lineages (Fig. 3). Outside the Yangshan assemblage, alternative genetic codes were detected among the Ourmia-like viruses in Lenarviricota, Aquatic picorna-like and Dicistro-like viruses in Pisuviricota and Tombus-like and Noda-like viruses in Kitrinoviricota (Supplementary Dataset 3). The viruses with alternative codes probably infect protists and particularly, ciliates.
Discussion
Analysis of metaviromic samples from the single, mixed marine and freshwater habitat described here roughly doubles the known diversity of RNA viruses—as defined by an RdRP-sequence similarity threshold that falls between the species and genus ranks42. This discovery reveals the richness of complex aquatic environments and calls for in-depth study of similar biomes and viromes.
Most of the previously unknown viruses join the major lineages of RNA viruses, now established as phyla of the kingdom Orthornavirae42,44. Nevertheless, several major taxa are expected to emerge from this analysis, probably in the ranks of class (Yangshan assembly), order (for example, Picorna-like aquatic and Protopoty clades or Yan-like, Zhao-like, Wei-like and Shanghai viruses in the Yangshan assembly) and family (such as the Zhao-like subclade highlighted in Fig. 3).
We show that diversity is the defining factor for obtaining a reliable phylogeny of RNA viruses; once virus groups fill up with multiple, diverse RdRP sequences, most sequences that originally appeared as orphans coalesce into distinct clades and move up the tree. This trend is exemplified by several clades in the Yangshan assemblage (Fig. 3).
Our findings expand the understanding of the structural, functional and evolutionary plasticity of the +RNA viruses. We identified multiple virus lineages with RdRP domain permutation that is far more common than previously appreciated and is a recurrent variation in RdRP evolution rather than an ancestral configuration as has been suggested62. Previously unknown cases of domain recruitment by +RNA viruses were detected, suggesting unsuspected facets of virus–host interactions.
The Yangshan RNA virome analysis clarifies some critical stages in the evolution of +RNA viruses. Thus, the viruses of the Yangshan assemblage are probably evolutionary intermediates between simple, tombus-like viruses at the base of Kitrinoviricota and the more complex viruses of the expansive class Alsuviricetes. Similarly, Protopotyviruses seem to be the missing evolutionary link between simple, ancestral Pisuviricota and the more complex potyviruses. Likewise, recently discovered ‘plastroviruses’ appear to be evolutionary intermediates between astro-like and poty-like viruses67. Further identification of such missing links is expected to yield detailed scenarios for the origin of major groups of RNA viruses.
Inference of virus host range is a weak link in metaviromics. In the case of the Yangshan virome, clues come from the assignment of the largest cluster of Yangshan viruses to the family Marnaviridae, which is so far thought to include only protist viruses, and from the alternative genetic codes in several virus groups in the Yangshan assemblage, which also points to protist hosts. Additionally, in an attempt to characterize the Yangshan virome more comprehensively, we searched the DNA fraction of the Yangshan virome for signature proteins of different groups of DNA viruses. The overwhelming majority of the identified contigs belonged to various DNA bacteriophages and protist viruses, providing further support of the host assignments of RNA viruses (Extended Data Fig. 6). Thus, multiple lines of indirect evidence indicate that a substantial fraction—probably the majority—of the viruses in the Yangshan extracellular aquatic RNA virome infect unicellular eukaryotes. In particular, it is possible that the virus genome obtained from a Yellowstone National Park hot spring, for which an archaeal host has been proposed60, actually belongs to a protist virus. Apart from protists, some viruses in the Yangshan virome, such as dicistro-like viruses, are likely to infect marine arthropods, whereas for levi-like viruses, bacterial hosts can be confidently inferred.
Extended Data Fig. 6.
DNA virus sequences in the Yangshan virome.
The Yangshan virome could also shed light on RNA virus ecology. Quantitative analysis of contig occurrence revealed several extremely abundant viruses that are likely to reflect virus blooms on the most abundant hosts (Extended Data Fig. 10; Supplementary Dataset 4). The ecological composition of the Yangshan biome could also be relevant to the dominance of non-enveloped +RNA viruses in the extracellular RNA virome, to the exclusion of (−)RNA viruses. According to RdRP phylogenetic tree, Negarnaviricota are nested within Duplornaviricota, which are themselves lodged within the +RNA virus radiation (Fig. 2), implying more recent origin of (−)RNA viruses. Given that the greatest diversity of Negarnaviricota is found in invertebrates29, it has been suggested that this virus phylum evolved during the explosive Cambrian diversification of invertebrates2,49. This scenario is supported by the near absence of (−)RNA viruses in protists. A similar logic applies to the absence of the enveloped viruses of the Alsuviricetes and Flasuviricetes in the Yangshan virome: none of these viruses are known to infect protists. However, we cannot rule out that some unidentified technical bias in the procedures employed in this work also contributed to the dominance of +RNA viruses in the Yangshan virome.
Thus, a virome from a single, complex aquatic habitat doubles the known diversity of RNA viruses, points to unexpected features of virus biology and evolution, and is bound to substantially expand the taxonomy of RNA viruses. Nevertheless, the recently developed megataxonomic structure of the global RNA virome that includes five phyla of the kingdom Orthornavirae42,44 withstood the challenge from this data and might be approaching stability.
Methods
Sampling site, water sample collection and preparation
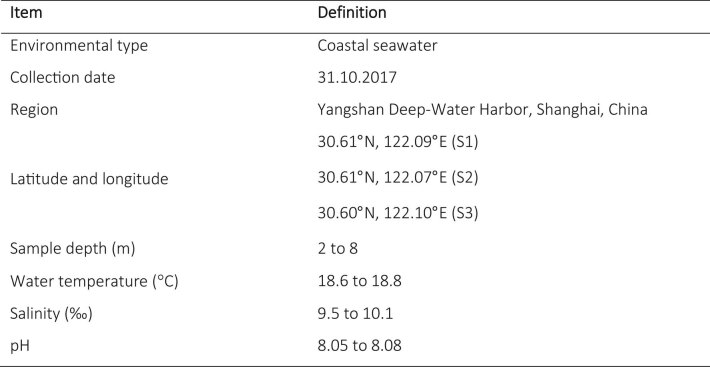
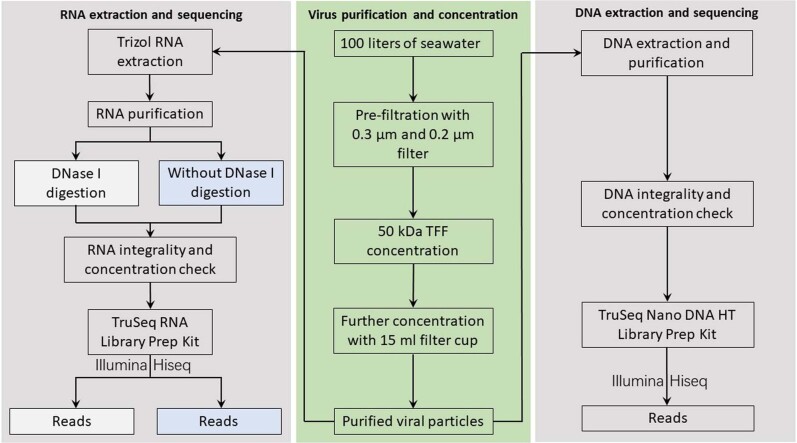
One-hundred litres of seawater were collected from three distinct sites in Yangshan Deep-Water Harbour, Shanghai, China on October 31 2017 (Extended Data Fig. 7). The samples were collected at the depths of 2–8 m from 3 sites in the Yangshan Deep-Water Harbour (>40 m depth) located between the Yangtze River estuary and Hangzhou Bay of East China Sea (Fig. 1 and Extended Data Fig. 7). The salinity of the harbour water (approximately 10‰, varying depending on currents) was intermediate between that of Yangtze River (0.2‰) and East China Sea (approximately 30‰), potentially contributing to the complexity of this aquatic habitat, which probably harbours freshwater-, estuary- and seawater-specific organisms, with the potential presence of some benthic organisms. The water samples were initially settled at 4 °C for 12 h, and viruses were isolated using tangential-flow-filtration procedures as previously described68 (Extended Data Fig. 8). The concentrated viral particles were stored at −80 °C before use. The absence of bacterial or cellular contamination in the filtrate was confirmed by transmission electron microscopy.
Extended Data Fig. 7.
General characteristics of the oceanic RNA virome of Yangshan Deep Water Harbor.
Extended Data Fig. 8.
Schematic of purification and sequencing of the oceanic RNA and DNA viromes. TFF, tangential flow filtration.
Virus nucleic acid extraction
One millilitre of concentrated virus (approximately 1010–1011 virus particles isolated from 10 l of seawater) was used for extraction of either DNA using Purification Resin and Mini Column (Promega)69, or RNA by using TRIzol LS Reagent (Invitrogen) and the Fast Total RNA Kit (Generay Biotech) (Extended Data Fig. 8). The integrity and concentration of nucleic acids were measured with NanoDrop 2000 (Thermo) and Qubit 3 analyser (Invitrogen). Virus RNA extracts (approximately 1.3 µg total) were subsequently divided into two parallel fractions. One was incubated with 1 μl DNase I (Thermo) at 37 °C for 10 min, and the other remained untreated.
High-throughput DNA and RNA sequencing
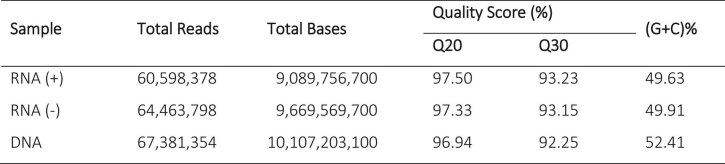
Two different RNA library-priming approaches (random-hexamer priming and template-switching reverse transcription) were used. Two 150 bp paired-end libraries (cDNA from total RNA) were generated using random-hexamer priming with the TruSeq RNA Library Prep Kit (Illumina) for the virus RNA extracts with or without DNase I digestion. Two single-end libraries were generated for the DNase I treated viral RNA extract using template-switching reverse transcription with the SMARTER stranded total RNA-seq kit (Clontech): one without fragmentation, and one with 4 min fragmentation at 94 °C, according to manufacturer’s instructions. The TruSeq Nano DNA HT Library Prep Kit (Illumina) was used to generate a 150-bp paired-end DNA library from the virus DNA extracts (Extended Data Fig. 8). High-throughput sequencing was performed on the Illumina MiSeq platform with v3 chemistry, and subsequently on the Illumina HiSeq 2500 platform. Both the library preparation and high-throughput sequencing were performed by Biozeron (Shanghai). Sequencing parameters are shown in Extended Data Fig. 9.
Extended Data Fig. 9.
Sequencing data for each metaviromic cDNA library.
Computational subtraction
Sequencing adapters were first removed, and nucleotides with quality scores lower than 20 were trimmed from the ends of reads using the cutadapt tool (https://cutadapt.readthedocs.io/en/stable/). To obtain a ‘clean’ RNA dataset, DNA-matching reads were computationally subtracted from the pool of RNA reads before virus genome assembly using a k-mer based approach. All unique 30-mers present in the DNA library were collected and RNA reads with an exact match to any 30-mer in the DNA library (on either read in the mate-pair for the paired-end datasets) were then excluded prior to contig assembly. Then, 20- and 25-mers were also tested to ensure that the subtraction was not sensitive to the k-mer length. As anticipated by a priori calculations, while subtraction using 20-mers resulted in gross overfiltering, 25- and 30-mers resulted in very similar numbers of removed reads. We also repeated the subtraction separately for the RNA libraries with or without DNase I treatment using 30-mers from the DNA dataset, and found no substantial difference in the numbers of removed reads (about 50% in each case), thereby underscoring the importance of in silico DNA subtraction.
Contig assembly
Contigs from the paired-end random-priming library were assembled using SPADES v.3.11.1 in metagenomics mode, while contigs from the single-end template-switching library were assembled using SPADES v.3.7 in metagenomics mode (v.3.11.1 only supports assembly of paired-end reads in metagenomics mode). After assembly, the two sets of contigs were unified into a single set of non-redundant contigs by excluding any contig from the template-switching dataset that shared more than 90% of its 15-mer sub-sequences with any contig in the random-priming dataset.
RdRP identification, clustering and phylogenetic analysis
RdRp sequences were identified using PSI-BLAST, which was run against the six-frame end-to-end translations of all contig sequences. Multiple alignments of virus RdRPs and reverse transcriptases from group II intron and non-long-terminal-repeat retrotransposons42 were used to generate query position-specific scoring matrices. Sequences that covered at least 75% of the query profile length were considered to contain full-length RdRP cores. This analysis identified almost 75,000 contigs (7.8% of all contigs; 150–11,000 nucleotides size range) encoding predicted proteins with significant amino acid sequence similarity to previously identified RdRP. Of these, 4,593 proteins were operationally considered ‘full-length’ RdRP. Initial clustering of the identified full-length RdRPs was performed using MMSEQ270 with sequence similarity threshold of 0.5. When the same position-specific scoring matrices were employed to search the protein sequences from GenBank, 5,481 full-length, non-redundant (<90% identity) RdRP sequences were identified that formed 2,021 clusters. After the addition of 4,593 full-length sequences from the Yangshan dataset, the combined set of 10,074 sequences produced 4,213 clusters under the same clustering procedure, increasing the number of clusters by a factor of 2.08.
Multiple alignments of sequences within clusters were generated using MUSCLE71. Cluster-derived profiles were compared to existing profiles using the HHsearch program72 to broadly assign the Yangshan sequences to the five major branches of RdRPs42. Iterations of clustering using HHsearch and profile–profile alignments using HHalign were performed to refine the positions of the Yangshan sequences within the RdRP tree. The clusters were delineated such as to include sufficiently diverse sequences and to be significantly enriched with sequences from the metaviromic sample. This procedure yielded 323 clusters (OV.1 to OV.323 in Supplementary Dataset 1) containing from 1 to 653 sequences. Phylogenetic trees for the cluster alignments were generated using FastTree73 with the WAG evolutionary model and gamma-distributed site rates. Nearly monophyletic groups of Yangshan RdRPs (containing at least 90% of Yangshan metagenome sequences) or mixed, but shallow groups of Yangshan RdRPs (corresponding to the tree depth of less than 1.0 substitution per site) were considered to be distinct Yangshan clusters.
For further phylogenetic analysis, the full-length RdRPs of the Yangshan set were aligned with their previously identified homologues and subjected to additional clustering based on the resulting preliminary phylogenetic trees. The resulting clusters were then fitted into the previously constructed RdRP tree47 using a procedure that involved several iterations of aligning Yangshan RdRPs with those from GenBank, constructing preliminary trees, and extracting Yangshan RdRPs that grouped together. The overwhelming majority of the Yangshan sequences (4,348 of 4,593, or 95%) and all large clusters (31 clusters encompassing 22 or more sequences each) were affiliated with previously identified RdRP lineages (Fig. 2; Supplementary Dataset 1).
The RdRp permutations make permuted sequences unalignable with those of the canonical configuration. To incorporate them into the phylogenetic analysis, the following de-permutation procedure was performed: first, permuted sequence were identified, clustered using MMSEQ2 with sequence similarity threshold of 0.5 and aligned with each other. Profile–profile alignments between these clusters and their closest canonical configuration relatives were performed using the HHALIGN program; the boundaries of the permuted catalytic loop were determined by examining the alignment and the corresponding alignment fragment was transposed to the canonical location (typically the location of the gap against the canonically located loop). Then the de-permuted sequences were returned to the pool, replacing the permuted originals. This procedure was used to generate Extended Data Fig. 1.
In addition to the RdRPs that could be assigned to previously identified clades at different depths of the phylogenetic tree, we attempted to detect putative highly divergent RdRPs. First, all long RNA contigs (>1,200 nucleotides; 10,813 contigs altogether) from the virome were translated stop-to-stop in 6 frames, and any which encoded open reading frames for more than 400 amino acids were selected and clustered by sequence similarity. The 37 profiles constructed from the resulting cluster alignments of 10 or more sequences were used as queries to search sequence databases with HHPred search. No RdRPs were found among these clusters. Second, open reading frames derived from 33 of the longest contigs in our dataset were analysed one at a time using HHPred; this procedure resulted in the identification of 13 singleton RdRP sequences (this analysis is too time consuming to perform on all potential RdRP-bearing sequences).
DNA viruses in the Yangshan virome
The nucleotide sequences of DNA viruses were identified by comparing position-specific scoring matrices for the respective capsid proteins to the 6-frame translated sequences of the DNA metagenomic contigs using PSI-BLAST. The set of scoring matrices consists of 200 profiles derived from multiple alignments of capsid and coat proteins of eukaryotic, bacterial and archaeal DNA viruses. Of these, 98 alignments were taken from National Center for Biotechnology Information Conserved Domains Database74 and 102 were developed in-house75–77. PSI-BLAST searches initiated by these profiles were competed against other, unrelated PFAM profiles in the Conserved Domains Database. Significant (e-value < 0.0001) hits were recorded; contigs containing these hits were tentatively assigned to the respective virus group. Sampled sequences were manually curated using HHPred to verify or correct assignments.
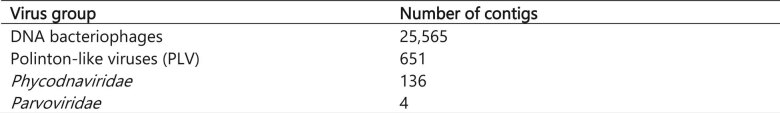
For many of the Polinton-like virus contigs, the best hits in the NR database are (erroneously) annotated as bacteria assembled from marine metagenomes (for example MAO23883.1/NZRF01000276.1, matching NODE_13251 contig). These ‘bacterial’ assemblies probably contain numerous fragments derived from the marine virome. All nucleo-cytoplasmic large DNA virus (NCDLV) contigs were found to be highly similar to Phycodnaviridae (for example YP_004062106.1/NC_014767.1 matching NODE_1923356 contig). Many of these also have close matches in ‘bacterial’ assemblies from marine metagenomes (MAB60321.1/NYUE01000104.1). All four parvovirus contigs showed only distant similarity (about 30% protein identity) to vertebrate parvoviruses (for example APQ44761.1/KY053092.1 matching NODE_10537 contig), suggesting that these are viruses of unidentified hosts rather than vertebrate virus contaminants.
Identification and annotation of protein domains
To identify protein domains, we performed sensitive profile–profile comparisons using HHsearch72. The identification procedure was run iteratively. First, profiles for each in silico-translated protein sequence were generated by performing one iteration against uniclust30_2018_08 database78 with HHblits79. The generated profiles were then compared against the previous generated RNA virus profile database42. Protein regions longer than 100 residues that did not display significant hits were extracted and clustered with CLANS80. Groups containing at least five members were identified using convex clustering algorithm implicated in CLANS, aligned with MUSCLE71, annotated when possible and added to the RNA virus profile database. In addition, extracted protein regions were searched against the European Bioinformatics Institute metagenomics database81, supplemented with the RNA virus protein sequences from the current study by performing one iteration of Jackhmmer82. Profiles with statistically significant hits (probability >95%) were annotated and added to the RNA virus profile database. Finally, domain identification procedure was repeated using the updated RNA virus profile database.
CRISPR-spacer search
CRISPR spacers (363,468 unique spacers) were matched against the set of oceanic virus contigs; 90% identity, 90% coverage criteria were used for matches, as previously described83.
Virus abundance
The abundances of viruses present in each virus cluster were calculated by mapping DNA-subtracted RNA sequencing reads back to RdRP-bearing contigs using bowtie284. A bowtie2 index was generated from the combined non-redundant contigs assembled from all RNA libraries, and bowtie2 was then used to map reads from each experiment back to these contigs. All RdRP-bearing contigs were more than 95% covered by mapped reads. The abundance of each contig was calculated as mapped reads per kilobase per million (RPKM) total reads in the library.
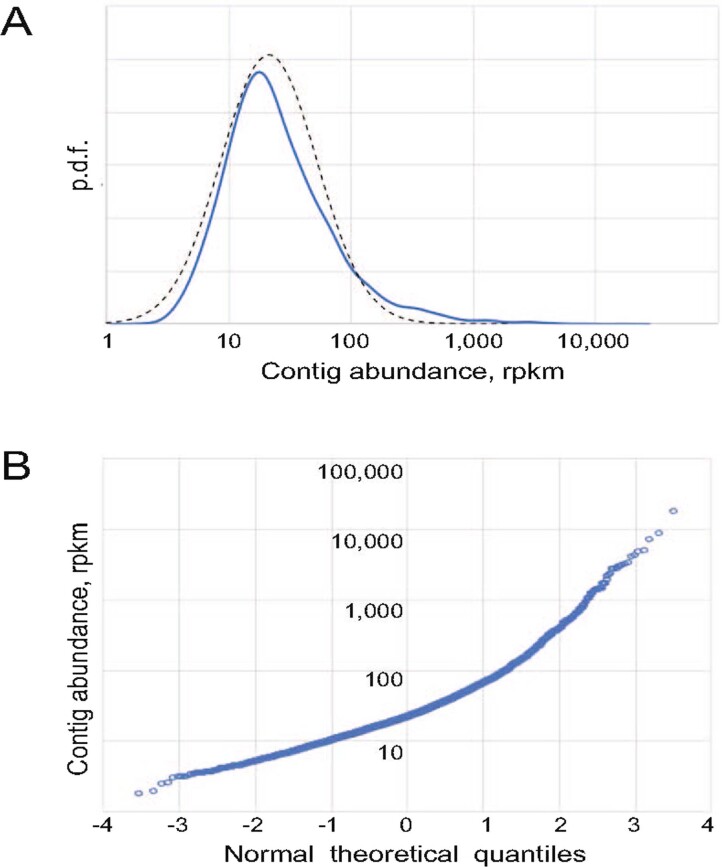
The distribution of contig abundances covers several orders of magnitude, is unimodal with a peak at ~18 RPKM and a median of 21.3 RPKM, and resembles a log-normal distribution (Extended Data Fig. 10a). However, the distribution is skewed such that the highly abundant assemblies are more abundant than expected from the log-normal distribution (Extended Data Fig. 10b).
Extended Data Fig. 10. Distribution of contig abundances.
(a) Probability density function (p.d.f.) for contig abundances (n = 4571 non-identical contigs. The dotted line plots the log-normal distribution with the same median and interquartile distance. (b) Quantile-quantile (Q-Q) plot of the distribution of contig abundances (n = 4571 non-identical contigs) versus the standard normal distribution. The figure shows that at the first approximation the distribution of contig abundances follows the log-normal distribution (typical in complex environments), but the deviations (a pronounced heavy tail of high values) hints on a dynamic environment producing superabundant viral blooms. Source data for panels (A) and (B) are presented in Source Data Figs. 1 and 2, respectively.
The top 20 contigs had at least 10× greater coverage than the median RPKM value (Supplementary Dataset 4). The most abundant virus, a member of OV.89 in the Tombus-like clade, was more than 800-fold over-represented compared to the median. The next three most abundant viruses were those from the Picorna-like aquatic/Marnaviridae and Zhao-like clades, all probably hosted by eukaryotic phytoplankton.
The contigs were then grouped by cluster or by clade to identify over-represented lineages (Supplementary Dataset 4). A pronounced correspondence between the diversity and abundance of the virus clusters was observed. The most abundant cluster was also the most diverse one (OV.1 of the Aquatic picorna-like/Marnaviridae clade), suggesting an overall prevalence of eukaryotic aquatic plankton. The Tombus-like clade was also well represented, largely, due to the most abundant virus mentioned above. The Yan-like and Zhao-like clades within the Yangshan assemblage contained several highly abundant viruses as well. Finally, several ourmia-like (OV.6) and levi-like viruses were prominent, particularly, the most abundant putative +RNA phage (OV.81).
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Operational clusters of the RdRP sequences identified in Yangshan RNA virome (OV.1 to OV.323; from largest to smallest) and used for the further phylogenetic analysis.
Clade-specific phylogenies for Yangshan RNA viruses. Each tree contains representatives of the indicated clade (for example, Ov1 and Ov2) as well as phylogenetically close reference viruses. Genome maps for the sequences and some reference viruses are shown on the right. The functional domains are colour-coded and the key is provided at the bottom of each panel. MP, movement protein; RBP, RNA-binding protein; GTase, guanylyltransferase; rXXX.0, uncharacterized conserved domains.
The RdRP core domains in the Yangshan RNA virome encoded in alternative genetic codes.
The relative abundance of the virus contigs in the Yangshan RNA virome.
The list of full-length RdRPs in the Yangshan RNA virome. Correspondence between the amino acid sequence IDs (‘orf.nnn’), contig IDs (‘NODE_xxx’), coordinates of the RdRP core in the nucleotide sequence and GenBank contig IDs are shown.
Acknowledgements
We thank N. Yutin for providing protein multiple-sequence analysis for DNA virus search. Y.I.W. and E.V.K. are supported through the Intramural Research Program of the US National Institutes of Health (National Library of Medicine). S.S. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-(2352-19)). A.F. was supported by National Institutes of Health awards R01GM37706 and R35GM130366. M.K. was supported by l’Agence Nationale de la Recherche grant ANR-17-CE15-0005-01 (ENVIRA). D.K. was funded by the European Social Fund under no. 09.3.3-LMT-K-712 ‘Development of Competences of Scientists, other Researchers and Students through Practical Research Activities’ measure. Y.W. was supported by the National Natural Science Foundation of China (nos. 41376135, 31570112 and 41876195).
Extended data
Source data
Source data for Extended Data Fig. 10.
Author contributions
Y.I.W. performed RdRP identification, clustering and phylogenetic analysis, DNA virus identification and CRISPR-spacer search; S.S. and M.B. performed nucleotide sequencing, contig assembly and analysed contig abundance; Y.W. and S.W. performed sample collection and preparation, isolation of nucleic acids and nucleotide sequencing; D.K. and M.K. identified and annotated protein domains and generated virus genome maps; A.F., V.V.D. and E.V.K supervised the study and wrote the manuscript, which was edited and approved by all authors.
Data availability
The sequence data analysed in this work are publicly available at the National Center for Biotechnology Information (NCBI) sequence databases under Bioproject PRJNA605028, accession JAAOEH000000000 (RNA virome) and Bioproject PRJNA610033, accession JAAOEI000000000 (DNA virome). Additional data (including alignments, trees and domain assignment) are available with no restrictions at ftp://ftp.ncbi.nih.gov/pub/wolf/_suppl/yangshan. Limited quantities of the remaining biological materials are available upon request. Source data are provided with this paper.
Code availability
Custom software code is available at ftp://ftp.ncbi.nih.gov/pub/wolf/_suppl/yangshan with no restrictions.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yuri I. Wolf, Sukrit Silas.
Contributor Information
Valerian V. Dolja, Email: doljav@oregonstate.edu
Eugene V. Koonin, Email: koonin@ncbi.nlm.nih.gov
Extended data
is available for this paper at 10.1038/s41564-020-0755-4.
Supplementary information
is available for this paper at 10.1038/s41564-020-0755-4.
References
- 1.Zhang YZ, Chen YM, Wang W, Qin XC, Holmes EC. Expanding the RNA virosphere by unbiased metagenomics. Annu. Rev. Virol. 2019;6:119–139. doi: 10.1146/annurev-virology-092818-015851. [DOI] [PubMed] [Google Scholar]
- 2.Dolja VV, Koonin EV. Metagenomics reshapes the concepts of RNA virus evolution by revealing extensive horizontal virus transfer. Virus Res. 2018;244:36–52. doi: 10.1016/j.virusres.2017.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lefeuvre P, et al. Evolution and ecology of plant viruses. Nat. Rev. Microbiol. 2019;17:632–644. doi: 10.1038/s41579-019-0232-3. [DOI] [PubMed] [Google Scholar]
- 4.Obbard DJ. Expansion of the metazoan virosphere: progress, pitfalls, and prospects. Curr. Opin. Virol. 2018;31:17–23. doi: 10.1016/j.coviro.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 5.Brum JR, Sullivan MB. Rising to the challenge: accelerated pace of discovery transforms marine virology. Nat. Rev. Microbiol. 2015;13:147–159. doi: 10.1038/nrmicro3404. [DOI] [PubMed] [Google Scholar]
- 6.Backstrom D, et al. Virus genomes from deep sea sediments expand the ocean megavirome and support independent origins of viral gigantism. mBio. 2019;10:e02497-18. doi: 10.1128/mBio.02497-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao L, Rosario K, Breitbart M, Duffy S. Eukaryotic circular rep-encoding single-stranded DNA (CRESS DNA) viruses: ubiquitous viruses with small genomes and a diverse host range. Adv. Virus Res. 2019;103:71–133. doi: 10.1016/bs.aivir.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Chow CE, Suttle CA. Biogeography of viruses in the sea. Annu. Rev. Virol. 2015;2:41–66. doi: 10.1146/annurev-virology-031413-085540. [DOI] [PubMed] [Google Scholar]
- 9.Gregory AC, et al. Marine DNA viral macro- and microdiversity from pole to pole. Cell. 2019;177:1109–1123. doi: 10.1016/j.cell.2019.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simmonds P, et al. Consensus statement: virus taxonomy in the age of metagenomics. Nat. Rev. Microbiol. 2017;15:161–168. doi: 10.1038/nrmicro.2016.177. [DOI] [PubMed] [Google Scholar]
- 11.Vlok M, Lang AS, Suttle CA. Marine RNA virus quasispecies are distributed throughout the oceans. mSphere. 2019;4:e00157-19. doi: 10.1128/mSphereDirect.00157-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greninger AL. A decade of RNA virus metagenomics is (not) enough. Virus Res. 2018;244:218–229. doi: 10.1016/j.virusres.2017.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janowski AB, et al. Statoviruses, a novel taxon of RNA viruses present in the gastrointestinal tracts of diverse mammals. Virology. 2017;504:36–44. doi: 10.1016/j.virol.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miranda JA, Culley AI, Schvarcz CR, Steward GF. RNA viruses as major contributors to Antarctic virioplankton. Environ. Microbiol. 2016;18:3714–3727. doi: 10.1111/1462-2920.13291. [DOI] [PubMed] [Google Scholar]
- 15.Ng TF, et al. High variety of known and new RNA and DNA viruses of diverse origins in untreated sewage. J. Virol. 2012;86:12161–12175. doi: 10.1128/JVI.00869-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waldron FM, Stone GN, Obbard DJ. Metagenomic sequencing suggests a diversity of RNA interference-like responses to viruses across multicellular eukaryotes. PLoS Genet. 2018;14:e1007533. doi: 10.1371/journal.pgen.1007533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi M, et al. The evolutionary history of vertebrate RNA viruses. Nature. 2018;556:197–202. doi: 10.1038/s41586-018-0012-7. [DOI] [PubMed] [Google Scholar]
- 18.Lopez-Bueno A, Rastrojo A, Peiro R, Arenas M, Alcami A. Ecological connectivity shapes quasispecies structure of RNA viruses in an Antarctic lake. Mol. Ecol. 2015;24:4812–4825. doi: 10.1111/mec.13321. [DOI] [PubMed] [Google Scholar]
- 19.Moniruzzaman M, et al. Virus–host relationships of marine single-celled eukaryotes resolved from metatranscriptomics. Nat. Commun. 2017;8:16054. doi: 10.1038/ncomms16054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosario K, Nilsson C, Lim YW, Ruan Y, Breitbart M. Metagenomic analysis of viruses in reclaimed water. Environ. Microbiol. 2009;11:2806–2820. doi: 10.1111/j.1462-2920.2009.01964.x. [DOI] [PubMed] [Google Scholar]
- 21.Lang AS, Culley AI, Suttle CA. Genome sequence and characterization of a virus (HaRNAV) related to picorna-like viruses that infects the marine toxic bloom-forming alga Heterosigma akashiwo. Virology. 2004;320:206–217. doi: 10.1016/j.virol.2003.10.015. [DOI] [PubMed] [Google Scholar]
- 22.Nagasaki K. Dinoflagellates, diatoms, and their viruses. J. Microbiol. 2008;46:235–243. doi: 10.1007/s12275-008-0098-y. [DOI] [PubMed] [Google Scholar]
- 23.Shirai Y, et al. Isolation and characterization of a single-stranded RNA virus infecting the marine planktonic diatom Chaetoceros tenuissimus Meunier. Appl. Environ. Microbiol. 2008;74:4022–4027. doi: 10.1128/AEM.00509-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomaru Y, Takao Y, Suzuki H, Nagumo T, Nagasaki K. Isolation and characterization of a single-stranded RNA virus infecting the bloom-forming diatom Chaetoceros socialis. Appl. Environ. Microbiol. 2009;75:2375–2381. doi: 10.1128/AEM.02580-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimura K, Tomaru Y. Discovery of two novel viruses expands the diversity of single-stranded DNA and single-stranded RNA viruses infecting a cosmopolitan marine diatom. Appl. Environ. Microbiol. 2015;81:1120–1131. doi: 10.1128/AEM.02380-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takao Y, Mise K, Nagasaki K, Okuno T, Honda D. Complete nucleotide sequence and genome organization of a single-stranded RNA virus infecting the marine fungoid protist Schizochytrium sp. J. Gen. Virol. 2006;87:723–733. doi: 10.1099/vir.0.81204-0. [DOI] [PubMed] [Google Scholar]
- 27.Gustavsen JA, Winget DM, Tian X, Suttle CA. High temporal and spatial diversity in marine RNA viruses implies that they have an important role in mortality and structuring plankton communities. Front. Microbiol. 2014;5:703. doi: 10.3389/fmicb.2014.00703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vlok M, Lang AS, Suttle CA. Application of a sequence-based taxonomic classification method to uncultured and unclassified marine single-stranded RNA viruses in the order Picornavirales. Virus Evol. 2019;5:vez056. doi: 10.1093/ve/vez056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li CX, et al. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. eLife. 2015;4:e05378. doi: 10.7554/eLife.05378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi M, et al. Redefining the invertebrate RNA virosphere. Nature. 2016;540:539–543. doi: 10.1038/nature20167. [DOI] [PubMed] [Google Scholar]
- 31.Shi M, et al. Divergent viruses discovered in arthropods and vertebrates revise the evolutionary history of the Flaviviridae and related viruses. J. Virol. 2016;90:659–669. doi: 10.1128/JVI.02036-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fauver JR, et al. West African Anopheles gambiae mosquitoes harbor a taxonomically diverse virome including new insect-specific flaviviruses, mononegaviruses, and totiviruses. Virology. 2016;498:288–299. doi: 10.1016/j.virol.2016.07.031. [DOI] [PubMed] [Google Scholar]
- 33.Webster CL, et al. The discovery, distribution, and evolution of viruses associated with Drosophila melanogaster. PLoS Biol. 2015;13:e1002210. doi: 10.1371/journal.pbio.1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grybchuk D, et al. Viral discovery and diversity in trypanosomatid protozoa with a focus on relatives of the human parasite Leishmania. Proc. Natl Acad. Sci. USA. 2018;115:E506–E515. doi: 10.1073/pnas.1717806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marzano SY, et al. Identification of diverse mycoviruses through metatranscriptomics characterization of the viromes of five major fungal plant pathogens. J. Virol. 2016;90:6846–6863. doi: 10.1128/JVI.00357-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kotta-Loizou I, Coutts RH. Studies on the virome of the entomopathogenic fungus Beauveria bassiana reveal novel dsRNA elements and mild hypervirulence. PLoS Pathog. 2017;13:e1006183. doi: 10.1371/journal.ppat.1006183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krishnamurthy SR, Janowski AB, Zhao G, Barouch D, Wang D. Hyperexpansion of RNA bacteriophage diversity. PLoS Biol. 2016;14:e1002409. doi: 10.1371/journal.pbio.1002409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roossinck MJ. Evolutionary and ecological links between plant and fungal viruses. N. Phytol. 2018;221:86–92. doi: 10.1111/nph.15364. [DOI] [PubMed] [Google Scholar]
- 39.Culley A. New insight into the RNA aquatic virosphere via viromics. Virus Res. 2018;244:84–89. doi: 10.1016/j.virusres.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 40.Coy SR, Gann ER, Pound HL, Short SM, Wilhelm SW. Viruses of eukaryotic algae: diversity, methods for detection, and future directions. Viruses. 2018;10:487. doi: 10.3390/v10090487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Callanan J, et al. Expansion of known ssRNA phage genomes: from tens to over a thousand. Sci. Adv. 2020;6:eaay5981. doi: 10.1126/sciadv.aay5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolf YI, et al. Origins and evolution of the global RNA virome. mBio. 2018;9:e02329-18. doi: 10.1128/mBio.02329-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuhn JH, et al. Classify viruses—the gain is worth the pain. Nature. 2019;566:318–320. doi: 10.1038/d41586-019-00599-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koonin EV, et al. Global organization and proposed megataxonomy of the virus world. Micobiol. Mol. Biol. Rev. 2020;84:e0061-19. doi: 10.1128/MMBR.00061-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kranzler CF, et al. Silicon limitation facilitates virus infection and mortality of marine diatoms. Nat. Microbiol. 2019;4:1790–1797. doi: 10.1038/s41564-019-0502-x. [DOI] [PubMed] [Google Scholar]
- 46.Valles SM, et al. ICTV virus taxonomy profile: Dicistroviridae. J. Gen. Virol. 2017;98:355–356. doi: 10.1099/jgv.0.000756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Revers F, Garcia JA. Molecular biology of Potyviruses. Adv. Virus Res. 2015;92:101–199. doi: 10.1016/bs.aivir.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 48.Gibbs AJ, Hajizadeh M, Ohshima K, Jones RAC. The Potyviruses: an evolutionary synthesis is emerging. Viruses. 2020;12:132. doi: 10.3390/v12020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dolja, V. V., Krupovic, M. & Koonin, E. V. Deep roots and splendid boughs of the global plant virome. Annu. Rev. Phytopathol.58, 10.1146/annurev-phyto-030320-041346 (2020). [DOI] [PubMed]
- 50.Koonin EV, Dolja VV, Krupovic M. Origins and evolution of viruses of eukaryotes: the ultimate modularity. Virology. 2015;479-480:2–25. doi: 10.1016/j.virol.2015.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dolja VV, Boyko VP, Agranovsky AA, Koonin EV. Phylogeny of capsid proteins of rod-shaped and filamentous RNA plant viruses: two families with distinct patterns of sequence and probably structure conservation. Virology. 1991;184:79–86. doi: 10.1016/0042-6822(91)90823-t. [DOI] [PubMed] [Google Scholar]
- 52.Agirrezabala X, et al. The near-atomic cryoEM structure of a flexible filamentous plant virus shows homology of its coat protein with nucleoproteins of animal viruses. eLife. 2015;4:e11795. doi: 10.7554/eLife.11795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zamora M, et al. Potyvirus virion structure shows conserved protein fold and RNA binding site in ssRNA viruses. Sci. Adv. 2017;3:eaao2182. doi: 10.1126/sciadv.aao2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dolja VV, Koonin EV. Common origins and host-dependent diversity of plant and animal viromes. Curr. Opin. Virol. 2011;1:322–331. doi: 10.1016/j.coviro.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Felix MA, et al. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 2011;9:e1000586. doi: 10.1371/journal.pbio.1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yokoi T, Yamashita S, Hibi T. The nucleotide sequence and genome organization of Sclerophthora macrospora virus A. Virology. 2003;311:394–399. doi: 10.1016/s0042-6822(03)00183-1. [DOI] [PubMed] [Google Scholar]
- 57.Heller-Dohmen M, Gopfert JC, Pfannstiel J, Spring O. The nucleotide sequence and genome organization of Plasmopara halstedii virus. Virol. J. 2011;8:123. doi: 10.1186/1743-422X-8-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scholz B, et al. Zoosporic parasites infecting marine diatoms—a black box that needs to be opened. Fungal Ecol. 2016;19:59–76. doi: 10.1016/j.funeco.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meldal BH, et al. An improved molecular phylogeny of the Nematoda with special emphasis on marine taxa. Mol. Phylogenet. Evol. 2007;42:622–636. doi: 10.1016/j.ympev.2006.08.025. [DOI] [PubMed] [Google Scholar]
- 60.Bolduc B, et al. Identification of novel positive-strand RNA viruses by metagenomic analysis of archaea-dominated Yellowstone hot springs. J. Virol. 2012;86:5562–5573. doi: 10.1128/JVI.07196-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ferrero DS, Buxaderas M, Rodriguez JF, Verdaguer N. The structure of the RNA-dependent RNA polymerase of a permutotetravirus suggests a link between primer-dependent and primer-independent polymerases. PLoS Pathog. 2015;11:e1005265. doi: 10.1371/journal.ppat.1005265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gorbalenya AE, et al. The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J. Mol. Biol. 2002;324:47–62. doi: 10.1016/S0022-2836(02)01033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sabanadzovic S, Ghanem-Sabanadzovic NA, Gorbalenya AE. Permutation of the active site of putative RNA-dependent RNA polymerase in a newly identified species of plant alpha-like virus. Virology. 2009;394:1–7. doi: 10.1016/j.virol.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 64.Greninger AL, DeRisi JL. Draft genome sequences of ciliovirus and brinovirus from San Francisco wastewater. Genome Announc. 2015;3:e00651-15. doi: 10.1128/genomeA.00651-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hillman BI, Cai G. The family Narnaviridae: simplest of RNA viruses. Adv. Virus Res. 2013;86:149–176. doi: 10.1016/B978-0-12-394315-6.00006-4. [DOI] [PubMed] [Google Scholar]
- 66.Schmidt F, Cherepkova MY, Platt RJ. Transcriptional recording by CRISPR spacer acquisition from RNA. Nature. 2018;562:380–385. doi: 10.1038/s41586-018-0569-1. [DOI] [PubMed] [Google Scholar]
- 67.Lauber C, Seifert M, Bartenschlager R, Seitz S. Discovery of highly divergent lineages of plant-associated astro-like viruses sheds light on the emergence of potyviruses. Virus Res. 2019;260:38–48. doi: 10.1016/j.virusres.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 68.Sun G, et al. Efficient purification and concentration of viruses from a large body of high turbidity seawater. MethodsX. 2014;1:197–206. doi: 10.1016/j.mex.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Henn MR, et al. Analysis of high-throughput sequencing and annotation strategies for phage genomes. PLoS ONE. 2010;5:e9083. doi: 10.1371/journal.pone.0009083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Steinegger M, Soding J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 2017;35:1026–1028. doi: 10.1038/nbt.3988. [DOI] [PubMed] [Google Scholar]
- 71.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Soding J. Protein homology detection by HMM–HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 73.Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marchler-Bauer A, et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yutin N, Wolf YI, Raoult D, Koonin EV. Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol. J. 2009;6:223. doi: 10.1186/1743-422X-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yutin N, Shevchenko S, Kapitonov V, Krupovic M, Koonin EV. A novel group of diverse Polinton-like viruses discovered by metagenome analysis. BMC Biol. 2015;13:95. doi: 10.1186/s12915-015-0207-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yutin N, et al. Discovery of an expansive bacteriophage family that includes the most abundant viruses from the human gut. Nat. Microbiol. 2018;3:38–46. doi: 10.1038/s41564-017-0053-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mirdita M, et al. Uniclust databases of clustered and deeply annotated protein sequences and alignments. Nucleic Acids Res. 2017;45:D170–D176. doi: 10.1093/nar/gkw1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Remmert M, Biegert A, Hauser A, Soding J. HHblits: lightning-fast iterative protein sequence searching by HMM–HMM alignment. Nat. Methods. 2011;9:173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- 80.Frickey T, Lupas A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004;20:3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- 81.Mitchell AL, et al. EBI metagenomics in 2017: enriching the analysis of microbial communities, from sequence reads to assemblies. Nucleic Acids Res. 2018;46:D726–D735. doi: 10.1093/nar/gkx967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eddy SR. Accelerated profile HMM searches. PLoS Comput. Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shmakov SA, et al. The CRISPR spacer space is dominated by sequences from species-specific mobilomes. mBio. 2017;8:e01397-17. doi: 10.1128/mBio.01397-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Operational clusters of the RdRP sequences identified in Yangshan RNA virome (OV.1 to OV.323; from largest to smallest) and used for the further phylogenetic analysis.
Clade-specific phylogenies for Yangshan RNA viruses. Each tree contains representatives of the indicated clade (for example, Ov1 and Ov2) as well as phylogenetically close reference viruses. Genome maps for the sequences and some reference viruses are shown on the right. The functional domains are colour-coded and the key is provided at the bottom of each panel. MP, movement protein; RBP, RNA-binding protein; GTase, guanylyltransferase; rXXX.0, uncharacterized conserved domains.
The RdRP core domains in the Yangshan RNA virome encoded in alternative genetic codes.
The relative abundance of the virus contigs in the Yangshan RNA virome.
The list of full-length RdRPs in the Yangshan RNA virome. Correspondence between the amino acid sequence IDs (‘orf.nnn’), contig IDs (‘NODE_xxx’), coordinates of the RdRP core in the nucleotide sequence and GenBank contig IDs are shown.
Data Availability Statement
The sequence data analysed in this work are publicly available at the National Center for Biotechnology Information (NCBI) sequence databases under Bioproject PRJNA605028, accession JAAOEH000000000 (RNA virome) and Bioproject PRJNA610033, accession JAAOEI000000000 (DNA virome). Additional data (including alignments, trees and domain assignment) are available with no restrictions at ftp://ftp.ncbi.nih.gov/pub/wolf/_suppl/yangshan. Limited quantities of the remaining biological materials are available upon request. Source data are provided with this paper.
Custom software code is available at ftp://ftp.ncbi.nih.gov/pub/wolf/_suppl/yangshan with no restrictions.