Abstract
The vacuolar H+-ATPases (V-ATPases) are essential, ATP-dependent proton pumps present in a variety of eukaryotic cellular membranes. Intracellularly, V-ATPase-dependent acidification functions in such processes as membrane traffic, protein degradation, autophagy and the coupled transport of small molecules. V-ATPases at the plasma membrane of certain specialized cells function in such processes as bone resorption, sperm maturation and urinary acidification. V-ATPases also function in disease processes such as pathogen entry and cancer cell invasiveness, while defects in V-ATPase genes are associated with disorders such as osteopetrosis, renal tubular acidosis and neurodegenerative diseases. This review highlights recent advances in our understanding of V-ATPase structure, mechanism, function and regulation, with an emphasis on the signaling pathways controlling V-ATPase assembly in mammalian cells. The role of V-ATPases in cancer and other human pathologies, and the prospects for therapeutic intervention, are also discussed.
Keywords: vacuolar ATPase, acidification, proton transport, cancer metastasis, regulated assembly, nutrient sensing
1. V-ATPase structure and mechanism
The V-ATPase is a multi-subunit complex composed of two large domains: the soluble V1 domain, which hydrolyzes ATP, and the membrane-embedded V0 domain, which transports protons [1–10] (Fig. 1). V1 is composed of subunits A through H in a stoichiometry of A3B3CDE3FG3H, while V0 is composed of subunits a, c, c″, d, e, in a stoichiometry of ac9c″de (yeast also have a subunit c′ in place of one of the c subunits) [2,8–10]. Eukaryotic V-ATPases are structurally and mechanistically related to the F1FO ATP synthases (F-ATPases), but always perform primary active proton transport rather than using the proton motive force to drive ATP synthesis [1,2,11]. V-ATPases bear even greater similarity to the archaeal-type ATPases (A-ATPases), including the H+-ATPase from T. thermophilus, which functions in ATP synthesis, and the Na+-ATPase from E. hirae, which functions to transport sodium ions [2,12,13]. As with the F-ATPase and A-ATPase, the V-ATPase is a rotary machine, and therefore contains both stationary (stator) and rotational (rotor) subcomplexes [14–16]. The stator is made up of an A3B3 hexamer containing three catalytic ATP binding pockets, three peripheral EG stalks, and subunits C, H and a. The rotor is made up of a central stalk composed of subunits D and F connected to subunit d, and a proteolipid ring of subunits c, (cʹ) and cʺ. While the function of subunit e is unknown, it is essential for V-ATPase activity, and its positioning suggests it is a stator subunit [17–20].
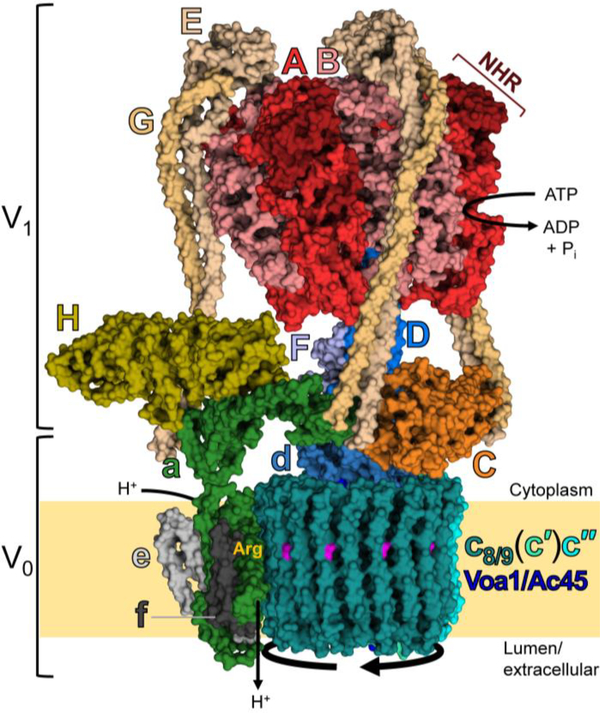
Fig. 1.
Structure and mechanism of the V-ATPase.
The V-ATPase is organized into two large domains: the peripheral V1 domain, which hydrolyzes ATP, and the integral V0 domain, which transports protons. V1 is composed of subunits A, B, C, D, E, F, G and H. The A3B3 hexamer contains nucleotide binding sites at each of the six AB interfaces, with three of the sites performing ATP hydrolysis. Subunit A also contains a 100 amino acid non-homologous region (NHR) which is not present in the ATP synthase β subunit. The V0 domain is composed of subunits a, c, c′ (in yeast), c″, d, e, f, and Voa1 (homologous to mammalian Ac45). The complex operates by a rotary mechanism, with ATP hydrolysis causing rotation of the central stalk and proteolipid ring. Protons enter an aqueous hemichannel in subunit a and protonate glutamate residues (magenta color) in each proteolipid subunit. After performing a full rotation, protons exit through a second hemichannel upon stabilization of the glutamate side chain by a critical arginine (yellow Arg) in subunit a. The complex is always oriented with V1 in the cytoplasm, so proton transport is driven into lumenal compartments or the extracellular space. This 6.6Å cryo-EM structure of the Stv1p-containing, yeast V-ATPase holoenzyme was determined by Vasanthakumar et al. (PDB ID: 6O7V) [46].
ATP hydrolysis causes conformational changes in the catalytic hexamer which drive rotation of the central stalk and the proteolipid ring [16,21–23]. Rotation of the proteolipid ring relative to subunit a is essential for proton transport [24]. This is because subunit a contains two aqueous hemichannels: a cytosol-facing hemichannel enables proton entry, while a lumen-facing hemichannel enables proton exit. As protons enter from the cytosol, they protonate a buried glutamate residue contained in each proteolipid subunit [25–28]. Rotation of the proteolipid ring causes the protonated glutamates to sequentially encounter a critical buried arginine residue in subunit a, enabling proton release and exit through the lumen-facing hemichannel [29–31].
The peripheral stator subunits, consisting of three EG stalks (designated EG1, EG2 and EG3) [32] and subunits C, H and a, stabilize the entire complex and prevent rotation of the stator during ATP hydrolysis. The EG C-termini bind the N-termini of the B subunits, while the EG N-termini bind to subunits C, H and the N-terminus of subunit a (aNT) [33–36]. Subunits C and H are each composed of two globular domains (Chead/Cfoot and HNT/HCT) connected by a flexible linker [37,38]. Chead interacts strongly with EG3 in a binary interface [39], while Cfoot interacts relatively weakly with EG2 and aNT in a ternary interface [40]. Another relatively weak ternary interaction is formed by subunit H, EG1, and aNT [41]. Upon disassembly (discussed below), subunit H undergoes a substantial conformational shift, bringing HCT in contact with subunit F of the central stalk, possibly to prevent rotation [42–44]. Moreover, a crystal structure of the yeast V1 has revealed that HCT may inhibit ATP hydrolysis by stabilizing one of the catalytic sites in the open state [45].
Additional putative V-ATPase subunits have recently been identified. A 3.9-Å cryo-EM structure of yeast V0 revealed the existence of an integral protein associated with subunit a and adjacent to subunit e, which has been designated subunit f [27]. Furthermore, a recently resolved 3.5-Å cryo-EM structure of yeast V0 revealed the presence of V0 assembly protein 1 (Voa1) within the center the proteolipid ring [28]. Voa1p functions in biosynthetic assembly of the V-ATPase in the endoplasmic reticulum, and was recently proposed to be the yeast homolog of Ac45, a protein which also functions in V-ATPase biosynthesis in higher eukaryotes [47–49]. Ac45 also associates with certain assembled V-ATPase complexes, and appears to be important for acidification of secretory vesicles in neuroendocrine cells, and for osteoclast function [50,51]. Proteins such as Ac45, which are not exclusively found within V-ATPase complexes, are classified as V-ATPase accessory subunits. Another accessory subunit present in higher eukaryotes (but not yeast) is the prorenin receptor (PRR), which associates with V0, and this interaction appears to be important for Wnt signaling [52].
2. V-ATPase function
2.1. Function of intracellular V-ATPases
Lumenal pH, which is chiefly controlled by V-ATPase-dependent proton transport, is a critical parameter for the function of various organelles. Thus, the acidic pH within sorting endosomes triggers the release of ligands internalized via receptor-mediated endocytosis, enabling the return of unoccupied receptors to the cell surface. Receptor recycling is important for controlling the rate of uptake of ligands such as low-density lipoprotein and transferrin [53]. Rates of recycling can also control the cell surface density of particular receptors, thus influencing the sensitivity of responses to hormones and growth factors such as insulin and epidermal growth factor (EGF). Dissociation from receptors causes signal termination and enables internalized ligands to reach their final cellular destinations, usually culminating in lysosomal degradation. Another type of receptor recycling occurs in late endosomes, where acidic pH triggers the release of newly synthesized proteases from the mannose 6-phosphate receptor (MPR). Ligand-receptor dissociation in this case enables both protease delivery to lysosomes and the return of MPRs to the trans-Golgi network [54]. Endosomal acidification also plays a role in the formation of certain carrier vesicles, which transport cargo within the endocytic and secretory pathways [55].
Several pathogens take advantage of the low pH within endosomes to gain entry into the cytoplasm. Exposure to acidic pH facilitates fusion of enveloped viruses such as Influenza and Ebola with the endosomal membrane, which is required for the insertion of viral genomes into the cytosol [56]. Endosomal pH also promotes the entry of pathogenic agents such as diphtheria toxin, which enters from sorting endosomes, and anthrax toxin, which first enters multivesicular bodies and then is released from late endosomes [57]. Certain secretory vesicles are acidified by the V-ATPase to facilitate the proteolytic processing of prohormones such as proinsulin [58]. Proteolytic processing also takes place in dendritic cell lysosomes, where internalized antigens are packaged for presentation on major histocompatibility complex class II molecules [59]. The proton gradient or membrane potential imposed by the V-ATPase is also coupled to small molecule transport. Thus, the positive interior membrane potential within synaptic vesicles drives glutamate uptake, while noradrenaline and GABA are loaded into synaptic vesicles by proton/neurotransmitter antiporters [2,60]. In lysosomes, several types of proton/amino acid symporters use the proton gradient to drive amino acid efflux [61].
Because most lysosomal hydrolases require acidic pH for activity, the V-ATPase is essential for proper degradation of macromolecules [62]. Macromolecules are delivered to lysosomes either endocytically, via chaperone-mediated autophagy, or via macroautophagy (hereafter autophagy), a catabolic program for recycling cellular components [63,64]. During autophagy, organelles and other macromolecules are engulfed within double-membraned vesicles called autophagosomes. Autophagosomes eventually fuse with lysosomes to form autolysosomes, in which the lumenal contents are degraded, and the resulting catabolites returned to the cytosol to be reused [65]. During this process, acidification is essential for both autophagosome/lysosome fusion and for subsequent breakdown of lumenal contents [66–68]. In fact, V-ATPase inhibitors such as bafilomycin A1 are such potent blockers of autophagic flux that they are often used as autophagy inhibitors [69]. Autophagy normally operates at low basal levels but can be substantially upregulated during times of energy stress or nutrient deprivation [70]. In addition, autophagy is an important survival mechanism employed by cancer cells (discussed below).
2.2. Function of plasma membrane V-ATPases
V-ATPases are localized to the plasma membrane of certain polarized animal cells where they function to transport protons to the extracellular space. These include osteoclasts, α-intercalated cells of the kidney collecting duct, clear cells of the epididymis, interdental cells of the inner ear and sustentacular cells of the olfactory epithelium [2,5]. In osteoclasts, plasma membrane V-ATPases function in bone resorption, which depends on acidification of the osteoclast/bone interface. Mutations in subunit a3, which targets the V-ATPase to the plasma membrane of osteoclasts, lead to a severe congenital form of osteopetrosis in humans [71,72]. Renal α-intercalated cells possess pools of V-ATPase-rich vesicles poised for fusion with the apical membrane [2,73]. These cells respond to alterations in plasma pH by rapidly adjusting the density of apical V-ATPases, which function to acidify urine, thereby removing excess acid from the blood. Mutations in subunit a4, which targets the V-ATPase to the plasma membrane of intercalated cells, cause recessive distal renal tubular acidosis in humans [74,75]. A similar form of endocytosis/exocytosis of V-ATPase-rich vesicles occurs in the clear cells of the epididymal epithelium. Acidification of the epididymal fluid maintains spermatozoa in a quiescent state, which is necessary for proper maturation and storage [2,76]. While V-ATPase mutations have not been directly linked decreased fertility in humans, male mice engineered to lack plasma membrane V-ATPase expression in the epididymis have elevated epididymal fluid pH, defective sperm, and are infertile [77]. Finally, V-ATPases have been detected at the plasma membrane in numerous types of human cancers, where they appear to aid in tumor cell invasiveness [78]. A detailed discussion of V-ATPases in cancer is presented in the section on V-ATPases in disease.
3. V-ATPase regulation
3.1. Regulated assembly
Because so many essential processes occur within discrete pH ranges, V-ATPase activity is highly regulated. Among the most important forms of V-ATPase regulation is the reversible dissociation of the V-ATPase complex into its component V1 and V0 domains, a process termed regulated assembly (Fig. 2). In the dissociated state, both the ATP-hydrolytic function of V1 and the proton transport function of V0 are inhibited (as discussed in the section on V-ATPase structure and mechanism) [2,79–82]. Since ATP consumption is prevented in the dissociated state, disassembly has been proposed to be a mechanism for conserving energy during times of nutrient limitation. Thus, the first demonstrations of regulated assembly were in insect cells during molting and yeast cells subjected to glucose starvation [83,84].
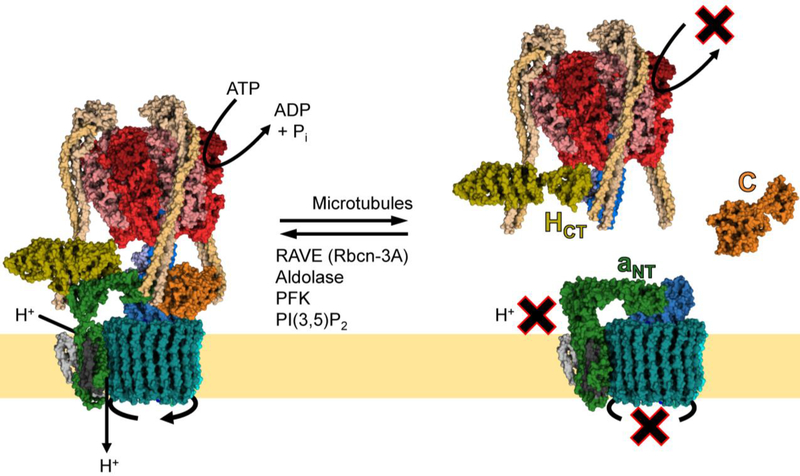
Fig. 2.
Regulated assembly of the V-ATPase.
An important form of V-ATPase regulation is the reversible dissociation of the V1 and V0 domains. In the disassembled state, free V1 cannot hydrolyze ATP, and free V0 cannot conduct protons. In free V1, HCT undergoes a large conformational shift, bringing it into contact with subunit F of the central stalk (PDB ID: 5D80) [45]. In free V0, the N-terminus of Vph1p or Stv1p (aNT) contacts subunit d of the central stalk (PDB ID: 6O7U) [46]. The only subunit that is released from either domain upon disassembly is subunit C. The process of reversible dissociation is rapid, does not require new protein synthesis, and occurs in response to diverse stimuli (see Table 1). In yeast, disassembly requires an intact microtubule network, while reassembly requires the RAVE complex, of which Rav1p is homologous to Rbcn-3A in higher eukaryotes. Other assembly factors common to yeast and higher eukaryotes include the glycolytic enzymes aldolase and phosphofructokinase (PFK). In yeast, PI(3,5)P2 binds to Vph1p and promotes assembly of Vph1p-containing V-ATPase complexes.
Regulated assembly enables rapid and local adjustment of V-ATPase activity. In yeast, for example, glucose starvation causes disassembly of V-ATPases localized to the vacuole [83,85], while Golgi-localized V-ATPases remain assembled [86]. When adequate glucose is present, yeast cells maintain V-ATPase assembly through activation of the Ras/adenylate cyclase/cAMP-dependent protein kinase (PKA) pathway [87]. Interestingly, elevated glucose was found to increase cytosolic pH in yeast, and to promote PKA activity in a V-ATPase-dependent manner, suggesting the existence of a positive feedback loop between PKA and the V-ATPase in yeast [88]. Assembly also responds to alterations in extracellular and vacuolar pH. In yeast grown at elevated pH or treated with chloroquine to neutralize the vacuole, V-ATPase disassembly upon glucose starvation is reduced [89,90].
While PKA controls V-ATPase assembly in response to glucose availability in yeast, it is also involved in controlling assembly in insect cells. In blowfly salivary glands, pharmacological elevation of intracellular cAMP leads to PKA-dependent phosphorylation of subunit C and increased V-ATPase assembly [91–93]. These results suggest that direct phosphorylation of V-ATPase subunits may be involved in regulating assembly, although this has not yet been directly demonstrated. Interestingly, recent FRET experiments in yeast found that glucose starvation leads to dissociation of subunit C from the entire V-ATPase complex, while the rest of V1 remains in close proximity with the membrane [94]. This finding suggests that association of subunit C may be a key determining factor in the stability and activity of V-ATPases in vivo. Another region that appears to be important for controlling assembly is the non-homologous region (NHR) of subunit A, so-named because it is a 100 amino acid insert not present in the otherwise homologous F-ATPase β subunit [1,2,95]. Introduction of specific mutations in this region was found to block disassembly upon glucose starvation without altering catalytic activity [96]. Remarkably, in cells expressing only a NHR-hemagglutinin fusion protein in place of the endogenous A subunit, the NHR was found to associate with the V0 domain in a glucose-dependent manner [90].
Disassembly and reassembly appear to be controlled by different mechanisms. In yeast subjected to glucose starvation, disassembly – but not reassembly – requires an intact microtubule network [97]. Disassembly also requires the V-ATPase to be catalytically active, suggesting the enzyme must be able to enter a particular conformation to undergo dissociation [85,98,99]. Reassembly – but not disassembly – is mediated by a number of proteins referred to as assembly factors. These include the glycolytic enzymes aldolase and phosphofructokinase, which physically interact with the V-ATPase in a glucose-dependent manner and function to promote assembly [100,101]. Another important assembly factor is Regulator of H+-ATPase of vacuoles and endosomes (RAVE), which is required for proper V-ATPase reassembly when cells are returned to normal glucose levels [102,103]. RAVE is believed to act as a chaperone to facilitate reintegration of free C subunits within the V1 subcomplex [104]. Furthermore, RAVE is recruited to vacuolar membranes by Vph1p during glucose sufficiency and is released to the cytosol during starvation, highlighting the dynamic nature of the RAVE/V-ATPase interaction [105]. Interestingly, RAVE displays specificity with respect to V-ATPase isoforms, by promoting assembly of Vph1p-containing, but not Stv1p-containing complexes [106]. In Drosophila and mammals, Rabconnectin-3α (Rbcn-3A) is homologous to Rav1p, and is important for endosomal acidification and Notch signaling [107,108].
Yeast also increase V-ATPase assembly in response to salt stress and this change is mediated by increased levels of PI(3,5)P2, a predominantly vacuolar inositol lipid produced by the phosphatidylinositol 3-phosphate 5-kinase Fab1p (homologous to mammalian PIKfyve) [109,110]. Interestingly, glucose-mediated assembly changes are independent of PI(3,5)P2 levels [109]. PI(3,5)P2 was found to preferentially bind to the N-terminal domain of Vph1p and increase V-ATPase catalytic activity and proton pumping [109,111]. Alternatively, PI(4)P binds to the N-terminal domain of Stv1p, and this interaction appears to be important for proper targeting of Stv1p-containing V-ATPases to the Golgi [112]. Intriguingly, PI(4)P interacts with the N-terminal domain of human a2, suggesting lipid interactions may represent a conserved mechanism for controlling V-ATPase trafficking and possibly activity [112].
Regulated assembly occurs in mammalian cells in response to diverse stimuli. Assembly increases in cells exposed to elevated glucose concentrations in a phosphatidylinositol 4,5-bisphosphate 3-kinase (PI3K)-dependent manner, perhaps as a way to contend with excess glycolytic acid generation [113,114]. Assembly increases in cells infected with Influenza virus in both a PI3K and mitogen-activated protein kinase (MAPK)-dependent pathway in order to acidify endosomes and promote viral fusion [115]. Recently, PI3K, Akt and MAPK1/2 were found to physically associate with subunit E to promote assembly and endosomal acidification in cells infected with rotavirus [116]. Further investigation will be needed to determine whether direct association of these important kinases represents a common mechanism by which assembly is controlled in mammalian cells. PI3K and mTOR complex I (mTORC1) are also important for increased lysosomal assembly in dendritic cells exposed to a maturation stimulus, where enhanced lysosomal acidification promotes antigen processing [117,118]. Assembly also increases in cells exposed to EGF, and V-ATPase inhibition was shown to prevent EGF-stimulated mTORC1 activation [119]. Finally, increasing evidence points to lysosomal V-ATPase assembly functioning in cellular nutrient homeostasis. Glucose starvation rapidly increases V-ATPase assembly and lysosomal acidification in mammalian cells in a PI3K and 5′-AMP-activated protein kinase (AMPK)-dependent manner [120]. Amino acid starvation also increases assembly and lysosomal acidification, and this effect was found to be PI3K-independent [121]. A more detailed discussion of the V-ATPase in nutrient sensing can be found in the section on emerging functions of the V-ATPase.
3.2. Regulated trafficking
Another important form of V-ATPase regulation is the targeting of V-ATPases to different cellular membranes, which is controlled by isoforms of subunit a (also discussed in V-ATPase structure and mechanism). Subunit a is the only V-ATPase subunit with multiple isoforms in yeast, where it is encoded by the genes VPH1 and STV1 [125,126]. Both Vph1p and Stv1p contain targeting information within their N-termini, with Vph1p-containing V-ATPases being targeted to the vacuole, while Stv1p-containing V-ATPases are targeted to the Golgi [127]. In mammals, subunit a exists in four isoforms encoded by the genes ATP6V0A1, ATP6V0A2, TCIRG1 and ATP6V0A4 [128]. a1 and a2 are primarily found within intracellular membranes, although a1 is also targeted to presynaptic plasma membranes of nerve terminals [124,129,130]. a3 is found intracellularly in compartments such as lysosomes, phagosomes and secretory granules, and is also targeted to the plasma membrane of osteoclasts [131–134]. While undifferentiated osteoclast progenitor cells localize a3 to late endosomes and lysosomes, these a3 containing-V-ATPases are trafficked to the plasma membrane upon differentiation into mature osteoclasts [131]. a1-a3 are expressed in a broad range of tissues, whereas a4 appears to be restricted to the kidney, epididymis and inner ear [71,75,135,136]. a4-containing V-ATPases are localized to the apical plasma membrane of renal intercalated cells and epididymal clear cells where they function in urinary acidification and sperm maturation, respectively [75,136].
In addition to isoform composition, which targets V-ATPases to particular membranes, certain polarized cells are able to rapidly alter the density of V-ATPases at the plasma membrane. In both renal intercalated cells and epididymal clear cells, exposure to alkaline pH leads to increased bicarbonate uptake and activation of PKA via a bicarbonate-sensitive adenylate cyclase [5]. cAMP stimulates insertion of V-ATPases into the apical membrane of these cells, and PKA-dependent phosphorylation of subunit A appears to be important for this process [137–139]. Conversely, AMPK was found to block PKA-induced apical accumulation of the V-ATPase in renal and epididymal cells [140]. In kidney cells, this was found to depend on direct phosphorylation of subunit A by AMPK [141]. It should be noted that these studies used localization of V1 subunits as a surrogate for trafficking of the intact enzyme, so it is possible that AMPK and PKA also influence V-ATPase assembly in these cell types.
3.3. Other forms of regulation
Several other forms of V-ATPase regulation have been observed, including disulfide bond formation, changes to coupling efficiency, changes in counterion conductance and regulated expression of V-ATPase subunits. In bovine brain clathrin-coated vesicles, approximately 50% of V-ATPases were found to be inactive, due to disulfide bonding between C254 and C532 in the catalytic A subunit [142,143]. This inhibition can be relieved by disulfide interchange with a third conserved cysteine residue [144].
Another possible mechanism for regulating V-ATPase activity in vivo is by changing the ratio of proton transport to ATP hydrolysis, i.e., the coupling efficiency. Certain point mutations in the non-homologous region of subunit A were shown to increase the coupling efficiency, suggesting that wild-type yeast V-ATPases may not be optimally coupled [96]. Furthermore, coupling is influenced by isoforms of subunit a, with Vph1p-containing V-ATPases being 4–5 times more efficiently coupled than Stv1p-containing V-ATPases [86]. A recent report found that the entrance to the cytosolic hemichannel in subunit a has a negative surface charge in Vph1p, and a positive surface charge in Stv1p [46]. Future studies will be needed to determine whether this property contributes to the observed differences in coupling efficiency between Vph1p- and Stv1p-containing V-ATPases.
Another way that cells may alter V-ATPase activity in particular compartments is through counterion conductance [145]. Chloride channels help to dissipate the interior positive membrane potential generated by the V-ATPase. The H+/Cl− antiporter ClC-5 is present in V-ATPase-rich apical vesicles in renal intercalated cells, and CLC5 mutations cause Dent’s disease, which is characterized by defective endosomal acidification and kidney failure [146]. The major lysosomal H+/Cl− antiporter ClC-7 was found to be important for the ability of primary mouse microglia to degrade amyloid-β (Aβ) – a causative agent of Alzheimer’s disease (discussed below) [147]. Interestingly, plasma membrane ClC7 is also important for osteoclast function, and CLC7 mutations result in osteopetrosis [148].
4. V-ATPases in disease
4.1. Cancer
There is considerable evidence that V-ATPases contribute to the survival and spread of cancer cells through several mechanisms (Fig. 3). One of the ways that V-ATPases have been proposed to promote tumor cell survival is by maintaining a neutral cytosolic pH. Owing to a combination of hypoxia and increased glycolytic metabolism, tumor cells are forced to contend with elevated levels of cytosolic acid [149]. It is believed that cancer cells increase V-ATPase biosynthesis and targeting of V-ATPases to the plasma membrane in order to secrete this acid extracellularly [150]. Plasma membrane V-ATPase localization has been detected in human samples of breast and lung tumors, and numerous cancer cell lines, including Ewing sarcoma, melanoma, breast, liver, pancreatic, prostate and ovarian cancer [151–159]. Indeed, tumors often exhibit a reversed pH gradient, with extracellular pH within the tumor microenvironment being lower than the intracellular pH of the tumor cells [160]. V-ATPase inhibitors have been shown to induce apoptosis in several cancer cell lines, possibly by blocking acid extrusion [161–164].
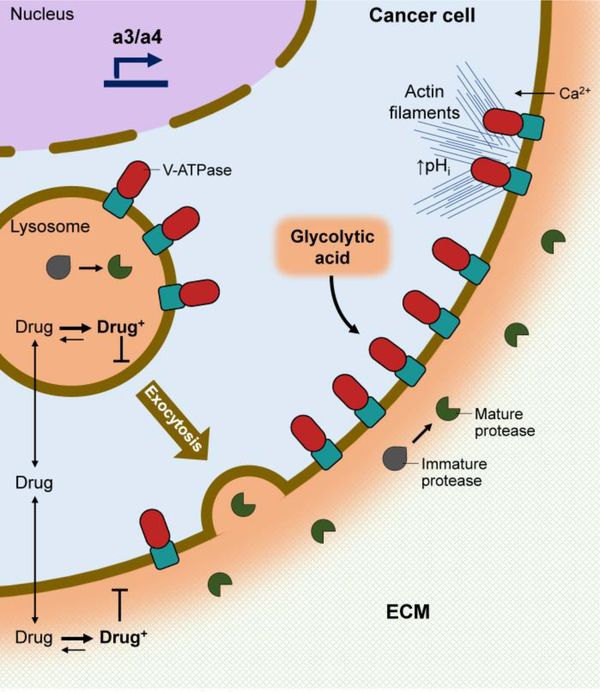
Fig. 3.
Function of V-ATPases in cancer cell survival, migration and invasion.
Cancer cells upregulate a3 and a4, which target V-ATPases to the plasma membrane, where they transport protons extracellularly. An acidic extracellular pH causes protonation of cancer drugs (Drug+), which prevents their diffusion into the cell. Once in the cytosol, cancer drugs diffuse into acidic compartments, where they are retained upon protonation and prevented from reaching their intended targets. Plasma membrane V-ATPases also promote cancer cell survival by removing cytosolic acid produced from glycolysis. Plasma membrane V-ATPases may contribute to cell migration by creating regions of alkaline pH (↑pHi), which promote actin polymerization and branching near the plasma membrane. Alternatively, acidic extracellular pH may contribute to force generation at the leading edge through calcium influx. Finally, plasma membrane V-ATPases are thought to contribute to invasion by activating secreted proteases, which function to cleave extracellular matrix (ECM) components and to activate other secreted proteases such as matrix metalloproteases.
V-ATPases also promote cancer cell survival by preventing cancer drugs from reaching their intended targets. Many chemotherapeutic agents are weak bases that can become protonated in the weakly acidic tumor microenvironment [165]. Protonation in the extracellular environment may prevent drugs from entering cancer cells, and numerous studies have found that raising extracellular pH increases drug uptake, cytosolic retention and tumor cell killing [166–168]. This problem is also enhanced by the fact that drugs in the more alkaline cancer cell cytoplasm are deprotonated and can diffuse out of the cell or partition into acidic compartments [169,170]. These resistance mechanisms may partially explain the enhanced efficacy observed by combining chemotherapeutics with V-ATPase inhibitors [171–174]. Other novel mechanisms promoting cancer cell growth and survival may be tumor type-dependent. A recent study found V-ATPase inhibition stabilized Hypoxia-inducible factor 1-alpha (HIF1α) by preventing its iron-dependent hydroxylation and degradation [175]. Stabilized HIF1α translocates to the nucleus where it represses expression of androgen receptor, which is drives the growth and survival of prostate tumors [175].
In addition to cancer cell survival, a number of studies have suggested that V-ATPases function to promote invasiveness, which is a fundamental property of metastatic cells. While metastasis is the leading cause of death from cancer, current cancer therapies are not specifically designed to prevent metastasis [176]. In order to metastasize, cells must detach from the primary tumor, penetrate basement membrane to enter the vascular or lymphatic systems, and then extravasate to colonize distant sites [177]. Several of these steps require cleavage of extracellular matrix components by secreted proteases such as cathepsins and matrix metalloproteases. Cathepsins have been found to be overexpressed in many human cancers and are correlated with worse prognosis [178]. Encouragingly, pharmacological and genetic inhibition of cathepsins reduced the spread of metastatic breast cancer models in mice [179,180]. Secreted cathepsins are thought to be activated within partially sealed extracellular spaces that are acidified by plasma membrane V-ATPases. Pan-V-ATPase inhibition with membrane-permeant inhibitors is highly effective at reducing in vitro cancer cell migration and/or invasion [151,155,181–187]. While these studies suggest V-ATPases are important for tumor cell invasiveness, they do not differentiate between intracellular and plasma membrane V-ATPases. Moreover, pan-V-ATPase inhibition would likely be too toxic in humans to be therapeutically viable, unless the drug could be selectively delivered to tumor cells.
As described above, V-ATPases have been detected at the plasma membrane of numerous invasive cancer cell lines. Furthermore, plasma membrane V-ATPase localization is significantly higher in invasive MCF10CA1a cells compared to noninvasive parental MCF10a cells [183]. Since plasma membrane targeting is controlled by isoforms of subunit a, it is likely that cancer cells will upregulate particular isoforms in order to increase localization of V-ATPases to the plasma membrane. Invasive MDA-MB-231 and MCF10CA1a cells markedly upregulate a3 relative to non-invasive MCF7 and MCF10a cells, respectively [182,183]. Importantly, RNAi-mediated knockdown of a3 significantly reduces invasion of MDA-MB-231 and MCF10CA1a cells, and overexpression of a3 in MCF10a cells significantly increases their invasion and plasma membrane V-ATPase localization [182,183]. To test whether specific inhibition of plasma membrane V-ATPases is sufficient to reduce invasion, MDA-MB-231 cells were engineered to express the V5 epitope fused to subunit c, such that V5 is oriented extracellularly in plasma membrane V-ATPases. Treatment of c-V5-expressing cells with an anti-V5 antibody inhibited plasma membrane V-ATPase activity and significantly reduced invasion [184]. Similarly, selective inhibition of cell surface V-ATPases using the membrane impermeant reagent biotin-bafilomycin bound to streptavidin inhibited in vitro invasion to the same extent as concanamycin, a membrane permeant inhibitor, arguing that inhibition of plasma membrane V-ATPases is the critical target in reducing tumor cell invasion (157). Furthermore, using isoform-specific antibodies, the invasive lines MDA-MB-231, SUM149 and MCF10CA1a were found to localize more a3-containing V-ATPases to the plasma membrane than non-invasive MCF10a cells [151]. Moreover, examining 42 human breast cancer cDNA samples revealed a3 to be upregulated in all samples, from 2.5 to nearly 50-fold [151]. Finally, immunofluorescence of human tumor samples showed a3 to be upregulated in regions of invasive breast carcinoma relative to adjacent non-invasive tumors and normal tissue [151]. Encouragingly, specific inhibition of a3-containing V-ATPases reduces metastases in the B16 murine melanoma model [154].
In addition to a3 other a isoforms have been observed at the plasma membrane of certain cancer cells. a1 and a3, but not a4 were detected at the plasma membrane of PC-3 prostate cancer cells and siRNA-mediated knockdown of either isoform reduced in vitro invasion [158]. a2 has been detected at the plasma membrane of several different ovarian cancer cell lines [159]. Finally, a recent study using CRISPR/Cas9 to target a-isoform genes found a4 to be critical for in vitro invasion of metastatic murine 4T1 breast cancer cells [185]. In this cell line, only knockout of a4 reduced plasma membrane V-ATPase localization and in vitro invasion [185]. These studies suggest that plasma membrane V-ATPases represent an important and novel target in the development of new therapeutics to reduce cancer metastasis. Orthotopic implantation of tumor cells engineered to reduce plasma membrane V-ATPase activity in mice and the development of inhibitory antibodies directed against extracellular epitopes on the V-ATPase will test the hypothesis that plasma membrane V-ATPases promote breast cancer metastasis in vivo.
It should be noted that many of the studies showing a role for the V-ATPase in promoting cancer cell invasion have also found it to be important for cell motility. While it is still unclear how V-ATPases function in cell migration, one possible mechanism is through actin rearrangement. In normal cells, subunits B and C directly interact with actin microfilaments [188,189]. Moreover, V-ATPases colocalize with F-actin at the leading edge of microvascular endothelial cells and bafilomycin treatment reduced their in vitro migration [190]. A similar pattern of colocalization between V-ATPases and F-actin has been observed at the leading edge of migrating cancer cells [151,182,185,187]. Treatment of HeLa cells with the novel V-ATPase inhibitor iejimalide C, or prostate cancer cells with bafilomycin or concanamycin disrupted actin filament organization, while knockdown of subunit C had a similar effect in breast cancer cells [187,191,192]. It is thought that cell surface V-ATPases contribute to invadopodia formation, and thus cell migration, by creating a pH and/or electrochemical gradient favorable to actin polymerization and branching. Alkaline pH activates the actin binding protein cofilin, which severs actin filaments to create free barbed ends for polymerization and membrane protrusion [149]. Alternatively, certain plasma membrane calcium channels are activated at low extracellular pH, with the resulting calcium influx activating myosin light chain kinase, leading to force-generation at the leading edge [193].
4.2. Neurodegenerative diseases
A pathological hallmark of many neurodegenerative disorders is the toxic buildup of undegraded protein aggregates in the brain. Alzheimer’s disease, for example, is associated with the deposition of extracellular plaques made of insoluble Aβ [194]. Mutations in Presenilin-1 (PS1), a component of the γ-secretase complex responsible for amyloid processing, cause familial Alzheimer’s disease [195]. Defects in PS1 prevent proper a1 trafficking and cause impaired lysosomal acidification, while rescue of lysosomal acidification in PS1 knockout cells reduced Aβ burden [196]. Another major neurodegenerative disorder linked to toxic protein aggregation is Parkinson’s disease, in which α-synuclein aggregates are deposited in neuronal inclusions called Lewy bodies [197]. Mutations in ATP6AP2, which encodes the V-ATPase accessory subunit Ac45, cause X-linked Parkinsonism with spasticity, an early-onset form of Parkinsonism with defective lysosomal acidification [198]. Many other neurodegenerative disorders which are not directly caused by V-ATPase defects nevertheless exhibit lysosomal impairment [199]. Restoration of lysosomal function therefore represents an attractive therapeutic concept that should be investigated further.
4.3. Other genetic disorders
As discussed above, mutations in V-ATPase subunit genes can result in dysfunction of particular cell types. Mutations in a3, which targets the V-ATPase to the plasma membrane of osteoclasts, cause osteopetrosis, which is characterized by dense and brittle bones [72]. In cells expressing a3R444L, which causes a severe form of infantile osteopetrosis, a3 is unglycosylated, retained in the ER and degraded [200]. Mutations in a4 and B1, which are present in plasma membrane V-ATPases in the kidney and cochlea, lead to recessive distal renal tubular acidosis and sensorineural deafness [74,201,202]. In cells expressing the a4R449H mutation, a4 is retained in the ER and degraded [203]. a4 also targets the V-ATPase to the apical membrane of epididymal clear cells, but fertility information is not yet available for patients with a4 mutations [5]. Mutations in a2, which targets the V-ATPase to Golgi and endosomes, cause autosomal recessive cutis laxa type II, a rare connective tissue disorder associated with widespread glycosylation defects [204]. In cells expressing the a2P405L mutation, a2 failed to traffic to the Golgi and was rapidly degraded [203].
5. Emerging functions of the V-ATPase
5.1. Amino acid sensing
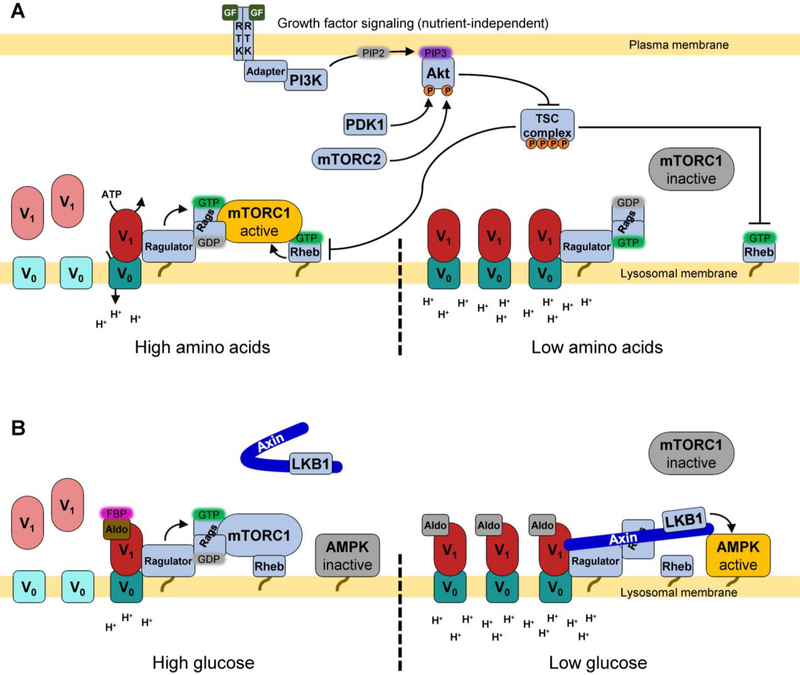
Lysosomal membranes contain a nutrient-sensing supercomplex consisting of the V-ATPase, the Ragulator complex and the Rag GTPases (Fig. 4A). The Rag GTPases respond to amino acid levels by adopting a conformation which promotes binding of mTORC1 (via its Raptor subunit) when amino acids are abundant [205–207]. Upon recruitment to the lysosome, mTORC1 contacts the GTPase Rheb, which in turn is regulated by growth factor signaling [208]. Growth factors activate the PI3K pathway, leading to activation of Akt, which phosphorylates and inhibits Tuberous sclerosis complex 2 (TSC2) [209]. TSC2 is a Rheb GAP, so inhibition of TSC2 enables Rheb to activate mTORC1 when it is present at the lysosome [210]. mTORC1 activation promotes anabolic processes such as translation and ribosome biogenesis, and inhibits catabolic processes such as autophagy [211]. By integrating both amino acid availability and growth factor signaling, mTORC1 promotes growth only when adequate nutrients are present. Interestingly, pharmacological and genetic disruption of the V-ATPase inhibits amino acid-dependent mTORC1 activation [212]. In a separate study, amino acids were found to regulate the V-ATPase, with acute amino acid withdrawal leading to increases in V-ATPase assembly and V-ATPase-dependent lysosomal acidification [121]. While this finding may be suggestive of V-ATPase assembly playing a role in regulating mTORC1, other findings rule out that possibility. First, time-course experiments show that mTORC1 inhibition precedes V-ATPase assembly changes in response to amino acid starvation [121]. Second, removal of particular individual amino acids have differential effects on V-ATPase assembly and mTORC1 activation [121]. Third, while lysosomal neutralization with chloroquine blocks assembly changes in response to amino acid starvation and also inhibits mTORC1, it does not prevent mTORC1 activation upon amino acid readdition [121,212]. These findings suggest that V-ATPase assembly is not involved in transmitting amino acid availability to mTORC1. Instead, enhanced lysosomal acidification upon amino acid starvation represents a mechanism to rapidly increase protein turnover in times of nutrient stress. Interestingly, while other examples of regulated assembly in mammalian cells depend upon PI3K (discussed above), assembly changes in response to amino acid starvation were unaffected by PI3K inhibition [121]. Thus, regulated assembly of the V-ATPase appears to be a novel mechanism involved in amino acid homeostasis. Future studies will focus on identifying the signaling pathways controlling assembly in response to amino acid starvation and measuring the degree to which this process contributes to increased lysosomal acidification and protein degradation in living cells.
Fig. 4.
The V-ATPase participates in nutrient sensing at the lysosomal membrane.
A) The V-ATPase associates with the Ragulator complex, which in turn is bound to the Rag GTPase heterodimer (Rags). When amino acids are abundant, Ragulator GEF activity towards the Rags enables recruitment of mTORC1 via its subunit Raptor (not shown). At the lysosome, mTORC1 associates with Rheb, which, when bound to GTP, stimulates the kinase activity of mTOR. V-ATPase inhibition or knockdown prevents mTORC1 lysosomal recruitment. V-ATPase assembly is also amino acid-dependent, with amino acid depletion leading to increased assembly, possibly as a way to increase protein degradation. mTORC1 activation by also requires growth factor (GF) signaling, which is independent of nutrient status. Growth factors bind receptor tyrosine kinases (RTK), leading to recruitment of PI3K, which phosphorylates PI(4,5)P2 (PIP2) to produce PI(3,4,5)P3 (PIP3). PIP3 recruits Akt to the plasma membrane where it is phosphorylated by 3-phosphoinositide-dependent protein kinase 1 (PDK1), enabling phosphorylation and full activation by mTOR complex II (mTORC2). Once active, Akt phosphorylates and inactivates the TSC complex, thereby relieving inhibition of Rheb. B) When glucose is abundant, the glycolytic enzyme aldolase (Aldo) is occupied by fructose 1,6-bisphosphate (FBP), and promotes Ragulator GEF activity towards the Rags. mTORC1 may then be activated by Rheb if growth factors are present (shown in A). Under these conditions, AMPK present at the lysosome is inactive, because its upstream kinase LKB1 is retained in the cytosol in complex with the scaffolding protein Axin. When glucose is low, Aldolase is no longer occupied by FBP, and Axin-LKB1 is recruited to the lysosome where it activates AMPK. Axin-LKB1 binding also appears to inhibit the Ragulator-Rag complex and displace mTORC1. The V-ATPase is required for glucose dependent recruitment of Axin-LKB1 to the lysosome. V-ATPase assembly is also glucose-dependent, with glucose depletion leading to increased assembly, possibly as a way to increase autophagic flux.
5.2. Glucose Sensing
As with mTORC1, the lysosomal membrane is a platform for AMPK activation. In the canonical model for AMPK activation, AMPK senses cellular AMP:ATP ratios, such that during energy-poor conditions it is able to be activated by liver kinase B1 (LKB1) [213]. Interestingly, another form of AMPK regulation has been discovered, in which AMPK directly responds to glucose levels. During glucose starvation, LKB1 and the scaffolding protein Axin are recruited to the lysosomal membrane, leading to AMPK activation (and mTORC1 displacement) [214] (Fig. 4B). The V-ATPase is required for this process, as RNAi-mediated knockdown of subunit c blocked glucose-dependent translocation of Axin-LKB1 to the lysosome [214]. Interestingly, aldolase appears to be important for this process, as addition of the aldolase glycolytic substrate fructose-1,6-bisphosphate induces dissociation of Axin-LKB1 from the V-ATPase [215]. It was also found that treatment of cells with the V-ATPase inhibitor concanamycin promoted LKB1 recruitment even in unstarved cells [214]. This finding suggests that the V-ATPase inhibitor concanamycin (which prevents conformational changes of the V-ATPase complex) may lock the enzyme in a state that mimics the glucose-starved conformation. While glucose starvation is known to decrease assembly in yeast, a recent study found that acute glucose starvation increases assembly and V-ATPase-dependent lysosomal acidification in mammalian cells [120]. While rapid and reversible, increased assembly upon glucose starvation is preceded by AMPK activation, indicating changes in assembly are not required for glucose-dependent AMPK activation [120]. Furthermore, pharmacological inhibition of either AMPK or PI3K blocked the glucose starvation-dependent increases in V-ATPase assembly and lysosomal acidification, suggesting that these kinases are important for this effect [120]. Future studies will focus on understanding the mechanism by which these signaling pathways modulate V-ATPase activity and testing the hypothesis that cells increase V-ATPase assembly during glucose starvation to promote the utilization of energy sources derived from autophagy.
5.3. Wnt and Notch signaling
The V-ATPase and its accessory subunit PRR appear to be required for full activation of the Wnt signaling pathway. Upon Wnt binding, Frizzled receptors and their coreceptors LDL receptor-related protein 5/6 (LRP5/6) are internalized into signaling endosomes. Here, PRR acts as an adapter between the receptor complex and the V-ATPase. V-ATPase activity appears to be important for LPR5/6 phosphorylation and therefore propagation of the signal [52]. The mechanism by which V-ATPase activity promotes LPR5/6 phosphorylation and activation is still unknown. In Notch signaling, the Notch receptor undergoes a series of cleavage events which release the Notch intracellular domain for translocation to the nucleus. V-ATPase activity appears to be important for cleavage following internalization of the Notch receptor [107]. Interestingly, in Drosophila, Rbcn-3A (homolog of the yeast RAVE subunit Rav1p) was also required for proper Notch trafficking and signal transduction [108].
6. Conclusions and future directions
V-ATPases are essential enzymes with diverse physiological roles, which has led to the evolution of multiple forms of regulation. These include regulated assembly of the peripheral and integral domains, regulated trafficking of assembled complexes and differential properties of complexes based on their isoform composition. A remarkable property of the V-ATPase is the rapidity with which assembly (and thus activity) can be adjusted. Biochemical and genetic studies have begun to elucidate the signaling pathways controlling assembly in response to various stimuli, but many questions still remain. For instance, while protein kinase pathways important for controlling assembly have been identified, the mechanism by which these kinases alter assembly is still unknown. Furthermore, the role of the V-ATPase in nutrient sensing is only beginning to come into focus. It is clear that the V-ATPase is an essential component of a lysosomal signaling hub that functions to balance anabolic and catabolic processes. More studies will be needed to conclusively demonstrate that regulated assembly is a homeostatic mechanism to rapidly adjust lysosomal pH, and thus macromolecule degradation, in response to nutrient availability. A greater understanding of how lysosomal pH is controlled may enable development of novel therapies for diseases characterized by the failure to clear toxic protein aggregates.
A particularly promising area of research is the role of V-ATPases in cancer. There is now considerable evidence that V-ATPases contribute to the survival and invasion of numerous types of human cancer cells. While pan-V-ATPase inhibition with membrane permeant small molecules would likely be too toxic to humans, studies suggest that invasive cell lines and at least some human tumors markedly overexpress a subunit isoforms that target the V-ATPase to the plasma membrane [151,154,182–185]. Furthermore, inhibition of only plasma membrane V-ATPases with either a monoclonal antibody or a membrane-impermeant form of bafilomycin reduced the in vitro invasion of highly invasive breast cancer cells to the same degree as pan-V-ATPase inhibition [184]. Since few normal cell types localize V-ATPases to the plasma membrane, this type of therapy would be inherently tumor-targeted, and likely would have minimal side effects. An added benefit of plasma membrane V-ATPase inhibition would be that such a therapy would likely inhibit osteoclast function and thus bone metastasis. As there are currently no therapies that specifically target the metastatic process, plasma membrane V-ATPases are therefore highly novel targets with the potential to be extremely impactful in the treatment of cancer.
Table 1.
Known regulators of V-ATPase assembly.
| Organism | Stimulus | Assem. change | Signaling pathways | Proposed function | Refs. |
|---|---|---|---|---|---|
| Yeast | |||||
| S. cerevisiae | Glucose starvation | ↓ | Ras/cAMP/PKA | Energy conservation | [83,87] |
| Alkaline cytoplasmic pH | ↑ | ? | PKA activation | [88] | |
| Alkaline extracellular pH | ↑ | ? | pH homeostasis | [89] | |
| Vacuole neutralization | ↑ | ? | Vacuole acidification | [90] | |
| Osmotic stress | ↑ | Fab1p (PIKfyve) | Na+ homeostasis | [109] | |
| Insects | |||||
| M. sexta midgut goblet cells | Molting | ↓ | PKA | Energy conservation | [84,92,93] |
| C. vicina salivary glands | Serotonin | ↑ | PKA | K+ and Cl− efflux | [91–93] |
| A. aegypti malphigian tubules | Blood meal | ↑ | PKA | Solute and water efflux | [122] |
| Mammals | |||||
| MDCK, A549 and HeLa cells | High glucose | ↑ | PI3K | pH homeostasis | [114] |
| MDCK and A549 cells | Influenza infection | ↑ | PI3K/Akt MAPK | Viral entry | [115] |
| MA104 cells | Rotavirus infection | ↑ | PI3K/Akt MAPK | Viral entry | [116] |
| Primary mouse dendritic cells | Maturation | ↑ | PI3K/Akt/mTORC1 | Antigen processing | [117,118] |
| Primary rat hepatocytes | EGF | ↑ | ? (Did not test PI3K pathway inhibitors) | mTORC1 activation | [119] |
| HEK293T and LLCPK cells | Glucose starvation | ↑ | PI3K/Akt AMPK | Autophagic flux | [120] |
| HEK293T cells | Amino acid starvation | ↑ | Not PI3K or mTORC1 | Protein degradation | [121] |
| HEK293T cells | Lysosome neutralization | ↑ | ? | Lysosomal acidification | [121] |
| HL-1 cells | High palmitate | ↓ | ? | Fatty acid uptake | [123] |
| Primary mouse hippocampal neurons | Synaptic vesicle recycling | ↓ | ? | Vesicle exocytosis | [124] |
| ↑ | ? | Vesicle endocytosis | [124] | ||
BBAMEM-20-104 Highlights.
Regulated assembly is a major mechanism for controlling V-ATPase activity in vivo
The V-ATPase is part of a nutrient sensing supercomplex on the lysosomal surface
V-ATPases are upregulated in cancer and promote tumor cell survival and metastasis
Plasma membrane V-ATPases represent promising and novel cancer therapeutic targets
Understanding lysosomal pH control may lead to new treatments for neurodegeneration
Acknowledgements
This work was supported by a Tufts Collaborative Cancer Biology Award to M.P.C., a Breast Cancer Alliance Exceptional Project Award to M.F. and National Institutes of Health Grant GM34478 to M.F.
Abbreviations
- A-ATPase
archaeal-type ATPase
- Ac45
V-type proton ATPase subunit S1
- Akt
cellular homolog of v-Akt, also known as RAC and PKB
- AMPK
5′-AMP-activated protein kinase
- EGF
epidermal growth factor
- F-ATPase
F1FO ATP synthase
- LKB1
liver kinase B1
- MAPK
mitogen-activated protein kinase, also known as ERK
- mTORC1
mTOR complex I
- NHR
non-homologous region
- PI3K
phosphatidylinositol 4,5-bisphosphate 3-kinase
- PKA
cAMP-dependent protein kinase
- PRR
prorenin receptor
- RAVE
regulator of H+-ATPase of vacuoles and endosomes
- Stv1p
V-type proton ATPase subunit a, Golgi isoform
- TSC2
tuberous sclerosis 2 protein, also known as Tuberin
- V-ATPase
vacuolar H+-ATPase
- Voa1
V0 assembly protein 1
- Vph1p
V-type proton ATPase subunit a, vacuolar isoform
Footnotes
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Nishi T, Forgac M, The vacuolar (H+)-ATPases--nature’s most versatile proton pumps, Nat. Rev. Mol. Cell Biol. 3 (2002) 94–103. 10.1038/nrm729. [DOI] [PubMed] [Google Scholar]
- [2].Forgac M, Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology, Nat. Rev. Mol. Cell Biol. 8 (2007) 917–929. 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- [3].Kane PM, The long physiological reach of the yeast vacuolar H+-ATPase, J. Bioenerg. Biomembr. 39 (2007) 415–421. 10.1007/s10863-007-9112-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Saroussi S, Nelson N, Vacuolar H(+)-ATPase-an enzyme for all seasons, Pflugers Arch. 457 (2009) 581–587. 10.1007/s00424-008-0458-9. [DOI] [PubMed] [Google Scholar]
- [5].Breton S, Brown D, Regulation of luminal acidification by the V-ATPase, Physiol. Bethesda Md. 28 (2013) 318–329. 10.1152/physiol.00007.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sun-Wada G-H, Wada Y, Vacuolar-type proton pump ATPases: acidification and pathological relationships, Histol. Histopathol. 28 (2013) 805–815. 10.14670/HH-28.805. [DOI] [PubMed] [Google Scholar]
- [7].Holliday LS, Vacuolar H + -ATPase: An Essential Multitasking Enzyme in Physiology and Pathophysiology, New J. Sci 2014 (2014) 1–21. 10.1155/2014/675430. [DOI] [Google Scholar]
- [8].Marshansky V, Rubinstein JL, Grüber G, Eukaryotic V-ATPase: novel structural findings and functional insights, Biochim. Biophys. Acta. 1837 (2014) 857–879. 10.1016/j.bbabio.2014.01.018. [DOI] [PubMed] [Google Scholar]
- [9].Cotter K, Stransky L, McGuire C, Forgac M, Recent Insights into the Structure, Regulation, and Function of the V-ATPases, Trends Biochem. Sci. 40 (2015) 611–622. 10.1016/j.tibs.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Oot RA, Couoh-Cardel S, Sharma S, Stam NJ, Wilkens S, Breaking up and making up: The secret life of the vacuolar H+ -ATPase, Protein Sci. Publ. Protein Soc. 26 (2017) 896–909. 10.1002/pro.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cross RL, Müller V, The evolution of A-, F-, and V-type ATP synthases and ATPases: reversals in function and changes in the H+/ATP coupling ratio, FEBS Lett. 576 (2004) 1–4. 10.1016/j.febslet.2004.08.065. [DOI] [PubMed] [Google Scholar]
- [12].Grüber G, Manimekalai MSS, Mayer F, Müller V, ATP synthases from archaea: the beauty of a molecular motor, Biochim. Biophys. Acta. 1837 (2014) 940–952. 10.1016/j.bbabio.2014.03.004. [DOI] [PubMed] [Google Scholar]
- [13].Mayer F, Müller V, Adaptations of anaerobic archaea to life under extreme energy limitation, FEMS Microbiol. Rev. 38 (2014) 449–472. 10.1111/1574-6976.12043. [DOI] [PubMed] [Google Scholar]
- [14].Noji H, Yasuda R, Yoshida M, Kinosita K, Direct observation of the rotation of F1-ATPase, Nature. 386 (1997) 299–302. 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- [15].Imamura H, Nakano M, Noji H, Muneyuki E, Ohkuma S, Yoshida M, Yokoyama K, Evidence for rotation of V1-ATPase, Proc. Natl. Acad. Sci. U. S. A. 100 (2003) 2312–2315. 10.1073/pnas.0436796100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hirata T, Iwamoto-Kihara A, Sun-Wada G-H, Okajima T, Wada Y, Futai M, Subunit rotation of vacuolar-type proton pumping ATPase: relative rotation of the G and C subunits, J. Biol. Chem. 278 (2003) 23714–23719. 10.1074/jbc.M302756200. [DOI] [PubMed] [Google Scholar]
- [17].Ludwig J, Kerscher S, Brandt U, Pfeiffer K, Getlawi F, Apps DK, Schägger H, Identification and characterization of a novel 9.2-kDa membrane sector-associated protein of vacuolar proton-ATPase from chromaffin granules, J. Biol. Chem. 273 (1998) 10939–10947. 10.1074/jbc.273.18.10939. [DOI] [PubMed] [Google Scholar]
- [18].Merzendorfer H, Huss M, Schmid R, Harvey WR, Wieczorek H, A novel insect V-ATPase subunit M9.7 is glycosylated extensively, J. Biol. Chem. 274 (1999) 17372–17378. 10.1074/jbc.274.24.17372. [DOI] [PubMed] [Google Scholar]
- [19].Davis-Kaplan SR, Ward DM, Shiflett SL, Kaplan J, Genome-wide analysis of iron-dependent growth reveals a novel yeast gene required for vacuolar acidification, J. Biol. Chem. 279 (2004) 4322–4329. 10.1074/jbc.M310680200. [DOI] [PubMed] [Google Scholar]
- [20].Sambade M, Kane PM, The yeast vacuolar proton-translocating ATPase contains a subunit homologous to the Manduca sexta and bovine e subunits that is essential for function, J. Biol. Chem. 279 (2004) 17361–17365. 10.1074/jbc.M314104200. [DOI] [PubMed] [Google Scholar]
- [21].Arai S, Saijo S, Suzuki K, Mizutani K, Kakinuma Y, Ishizuka-Katsura Y, Ohsawa N, Terada T, Shirouzu M, Yokoyama S, Iwata S, Yamato I, Murata T, Rotation mechanism of Enterococcus hirae V1-ATPase based on asymmetric crystal structures, Nature. 493 (2013) 703–707. 10.1038/nature11778. [DOI] [PubMed] [Google Scholar]
- [22].Zhao J, Benlekbir S, Rubinstein JL, Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase, Nature. 521 (2015) 241–245. 10.1038/nature14365. [DOI] [PubMed] [Google Scholar]
- [23].Suzuki K, Mizutani K, Maruyama S, Shimono K, Imai FL, Muneyuki E, Kakinuma Y, Ishizuka-Katsura Y, Shirouzu M, Yokoyama S, Yamato I, Murata T, Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor, Nat. Commun. 7 (2016) 13235 10.1038/ncomms13235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Vik SB, Antonio BJ, A mechanism of proton translocation by F1F0 ATP synthases suggested by double mutants of the a subunit, J. Biol. Chem. 269 (1994) 30364–30369. [PubMed] [Google Scholar]
- [25].Kawasaki-Nishi S, Nishi T, Forgac M, Interacting helical surfaces of the transmembrane segments of subunits a and c’ of the yeast V-ATPase defined by disulfide-mediated cross-linking, J. Biol. Chem. 278 (2003) 41908–41913. 10.1074/jbc.M308026200. [DOI] [PubMed] [Google Scholar]
- [26].Wang Y, Toei M, Forgac M, Analysis of the membrane topology of transmembrane segments in the C-terminal hydrophobic domain of the yeast vacuolar ATPase subunit a (Vph1p) by chemical modification, J. Biol. Chem. 283 (2008) 20696–20702. 10.1074/jbc.M803258200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mazhab-Jafari MT, Rohou A, Schmidt C, Bueler SA, Benlekbir S, Robinson CV, Rubinstein JL, Atomic model for the membrane-embedded VO motor of a eukaryotic V-ATPase, Nature. 539 (2016) 118–122. 10.1038/nature19828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Roh S-H, Stam NJ, Hryc CF, Couoh-Cardel S, Pintilie G, Chiu W, Wilkens S, The 3.5-Å CryoEM Structure of Nanodisc-Reconstituted Yeast Vacuolar ATPase Vo Proton Channel, Mol. Cell. 69 (2018) 993–1004.e3. 10.1016/j.molcel.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kawasaki-Nishi S, Nishi T, Forgac M, Arg-735 of the 100-kDa subunit a of the yeast V-ATPase is essential for proton translocation, Proc. Natl. Acad. Sci. U. S. A. 98 (2001) 12397–12402. 10.1073/pnas.221291798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wang Y, Inoue T, Forgac M, TM2 but not TM4 of subunit c” interacts with TM7 of subunit a of the yeast V-ATPase as defined by disulfide-mediated cross-linking, J. Biol. Chem. 279 (2004) 44628–44638. 10.1074/jbc.M407345200. [DOI] [PubMed] [Google Scholar]
- [31].Toei M, Toei S, Forgac M, Definition of membrane topology and identification of residues important for transport in subunit a of the vacuolar ATPase, J. Biol. Chem. 286 (2011) 35176–35186. 10.1074/jbc.M111.273409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhang Z, Zheng Y, Mazon H, Milgrom E, Kitagawa N, Kish-Trier E, Heck AJR, Kane PM, Wilkens S, Structure of the yeast vacuolar ATPase, J. Biol. Chem. 283 (2008) 35983–35995. 10.1074/jbc.M805345200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lu M, Vergara S, Zhang L, Holliday LS, Aris J, Gluck SL, The amino-terminal domain of the E subunit of vacuolar H(+)-ATPase (V-ATPase) interacts with the H subunit and is required for V-ATPase function, J. Biol. Chem. 277 (2002) 38409–38415. 10.1074/jbc.M203521200. [DOI] [PubMed] [Google Scholar]
- [34].Wilkens S, Inoue T, Forgac M, Three-dimensional structure of the vacuolar ATPase. Localization of subunit H by difference imaging and chemical cross-linking, J. Biol. Chem. 279 (2004) 41942–41949. 10.1074/jbc.M407821200. [DOI] [PubMed] [Google Scholar]
- [35].Jones RPO, Durose LJ, Findlay JBC, Harrison MA, Defined sites of interaction between subunits E (Vma4p), C (Vma5p), and G (Vma10p) within the stator structure of the vacuolar H+-ATPase, Biochemistry. 44 (2005) 3933–3941. 10.1021/bi048402x. [DOI] [PubMed] [Google Scholar]
- [36].Zhang Z, Inoue T, Forgac M, Wilkens S, Localization of subunit C (Vma5p) in the yeast vacuolar ATPase by immuno electron microscopy, FEBS Lett. 580 (2006) 2006–2010. 10.1016/j.febslet.2006.03.001. [DOI] [PubMed] [Google Scholar]
- [37].Sagermann M, Stevens TH, Matthews BW, Crystal structure of the regulatory subunit H of the V-type ATPase of Saccharomyces cerevisiae, Proc. Natl. Acad. Sci. U. S. A. 98 (2001) 7134–7139. 10.1073/pnas.131192798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Drory O, Frolow F, Nelson N, Crystal structure of yeast V-ATPase subunit C reveals its stator function, EMBO Rep. 5 (2004) 1148–1152. 10.1038/sj.embor.7400294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Oot RA, Wilkens S, Domain characterization and interaction of the yeast vacuolar ATPase subunit C with the peripheral stator stalk subunits E and G, J. Biol. Chem. 285 (2010) 24654–24664. 10.1074/jbc.M110.136960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Oot RA, Wilkens S, Subunit interactions at the V1-Vo interface in yeast vacuolar ATPase, J. Biol. Chem. 287 (2012) 13396–13406. 10.1074/jbc.M112.343962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Couoh-Cardel S, Milgrom E, Wilkens S, Affinity Purification and Structural Features of the Yeast Vacuolar ATPase Vo Membrane Sector, J. Biol. Chem. 290 (2015) 27959–27971. 10.1074/jbc.M115.662494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Parra KJ, Keenan KL, Kane PM, The H subunit (Vma13p) of the yeast V-ATPase inhibits the ATPase activity of cytosolic V1 complexes, J. Biol. Chem. 275 (2000) 21761–21767. 10.1074/jbc.M002305200. [DOI] [PubMed] [Google Scholar]
- [43].Jefferies KC, Forgac M, Subunit H of the vacuolar (H+) ATPase inhibits ATP hydrolysis by the free V1 domain by interaction with the rotary subunit F, J. Biol. Chem. 283 (2008) 4512–4519. 10.1074/jbc.M707144200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Diab H, Ohira M, Liu M, Cobb E, Kane PM, Subunit interactions and requirements for inhibition of the yeast V1-ATPase, J. Biol. Chem. 284 (2009) 13316–13325. 10.1074/jbc.M900475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Oot RA, Kane PM, Berry EA, Wilkens S, Crystal structure of yeast V1-ATPase in the autoinhibited state, EMBO J. 35 (2016) 1694–1706. 10.15252/embj.201593447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Vasanthakumar T, Bueler SA, Wu D, Beilsten-Edmands V, Robinson CV, Rubinstein JL, Structural comparison of the vacuolar and Golgi V-ATPases from Saccharomyces cerevisiae, Proc. Natl. Acad. Sci. U. S. A. 116 (2019) 7272–7277. 10.1073/pnas.1814818116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ryan M, Graham LA, Stevens TH, Voa1p functions in V-ATPase assembly in the yeast endoplasmic reticulum, Mol. Biol. Cell. 19 (2008) 5131–5142. 10.1091/mbc.e08-06-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Jansen EJR, Timal S, Ryan M, Ashikov A, van Scherpenzeel M, Graham LA, Mandel H, Hoischen A, Iancu TC, aymond K, Steenbergen G, Gilissen C, Huijben K, van Bakel NHM, Maeda Y, Rodenburg RJ, Adamowicz M, Crushell E, Koenen H, Adams D, Vodopiutz J, Greber-Platzer S, Müller T, Dueckers G, Morava E, Sykut-Cegielska J, Martens GJM, Wevers RA, Niehues T, Huynen MA, Veltman JA, Stevens TH, Lefeber DJ, ATP6AP1 deficiency causes an immunodeficiency with hepatopathy, cognitive impairment and abnormal protein glycosylation, Nat. Commun. 7 (2016) 11600 10.1038/ncomms11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Guida MC, Hermle T, Graham LA, Hauser V, Ryan M, Stevens TH, Simons M, ATP6AP2 functions as a V-ATPase assembly factor in the endoplasmic reticulum, Mol. Biol. Cell. 29 (2018) 2156–2164. 10.1091/mbc.E18-04-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Supek F, Supekova L, Mandiyan S, Pan YC, Nelson H, Nelson N, A novel accessory subunit for vacuolar H(+)-ATPase from chromaffin granules, J. Biol. Chem. 269 (1994) 24102–24106. [PubMed] [Google Scholar]
- [51].Feng H, Cheng T, Pavlos NJ, Yip KHM, Carrello A, Seeber R, Eidne K, Zheng MH, Xu J, Cytoplasmic terminus of vacuolar type proton pump accessory subunit Ac45 is required for proper interaction with V(0) domain subunits and efficient osteoclastic bone resorption, J. Biol. Chem. 283 (2008) 13194–13204. 10.1074/jbc.M709712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Cruciat C-M, Ohkawara B, Acebron SP, Karaulanov E, Reinhard C, Ingelfinger D, Boutros M, Niehrs C, Requirement of prorenin receptor and vacuolar H+-ATPase-mediated acidification for Wnt signaling, Science. 327 (2010) 459–463. 10.1126/science.1179802. [DOI] [PubMed] [Google Scholar]
- [53].Maxfield FR, McGraw TE, Endocytic recycling, Nat. Rev. Mol. Cell Biol. 5 (2004) 121–132. 10.1038/nrm1315. [DOI] [PubMed] [Google Scholar]
- [54].Ghosh P, Dahms NM, Kornfeld S, Mannose 6-phosphate receptors: new twists in the tale, Nat. Rev. Mol. Cell Biol. 4 (2003) 202–212. 10.1038/nrm1050. [DOI] [PubMed] [Google Scholar]
- [55].Gu F, Gruenberg J, ARF1 regulates pH-dependent COP functions in the early endocytic pathway, J. Biol. Chem. 275 (2000) 8154–8160. 10.1074/jbc.275.11.8154. [DOI] [PubMed] [Google Scholar]
- [56].Grove J, Marsh M, The cell biology of receptor-mediated virus entry, J. Cell Biol. 195 (2011) 1071–1082. 10.1083/jcb.201108131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Gruenberg J, van der Goot FG, Mechanisms of pathogen entry through the endosomal compartments, Nat. Rev. Mol. Cell Biol. 7 (2006) 495–504. 10.1038/nrm1959. [DOI] [PubMed] [Google Scholar]
- [58].Rhodes CJ, Lucas CA, Mutkoski RL, Orci L, Halban PA, Stimulation by ATP of proinsulin to insulin conversion in isolated rat pancreatic islet secretory granules. Association with the ATP-dependent proton pump, J. Biol. Chem. 262 (1987) 10712–10717. [PubMed] [Google Scholar]
- [59].Trombetta ES, Mellman I, Cell biology of antigen processing in vitro and in vivo, Annu. Rev. Immunol. 23 (2005) 975–1028. 10.1146/annurev.immunol.22.012703.104538. [DOI] [PubMed] [Google Scholar]
- [60].Farsi Z, Preobraschenski J, van den Bogaart G, Riedel D, Jahn R, Woehler A, Single-vesicle imaging reveals different transport mechanisms between glutamatergic and GABAergic vesicles, Science. 351 (2016) 981–984. 10.1126/science.aad8142. [DOI] [PubMed] [Google Scholar]
- [61].Xu H, Ren D, Lysosomal physiology, Annu. Rev. Physiol. 77 (2015) 57–80. 10.1146/annurev-physiol-021014-071649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Mindell JA, Lysosomal acidification mechanisms, Annu. Rev. Physiol. 74 (2012) 69–86. 10.1146/annurev-physiol-012110-142317. [DOI] [PubMed] [Google Scholar]
- [63].Dice JF, Chaperone-mediated autophagy, Autophagy. 3 (2007) 295–299. 10.4161/auto.4144. [DOI] [PubMed] [Google Scholar]
- [64].Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y, Dynamics and diversity in autophagy mechanisms: lessons from yeast, Nat. Rev. Mol. Cell Biol. 10 (2009) 458–467. 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- [65].Feng Y, He D, Yao Z, Klionsky DJ, The machinery of macroautophagy, Cell Res. 24 (2014) 24–41. 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Yoshimori T, Yamamoto A, Moriyama Y, Futai M, Tashiro Y, Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells, J. Biol. Chem. 266 (1991) 17707–17712. [PubMed] [Google Scholar]
- [67].Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y, Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells, Cell Struct. Funct. 23 (1998) 33–42. 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- [68].Kawai A, Uchiyama H, Takano S, Nakamura N, Ohkuma S, Autophagosome-lysosome fusion depends on the pH in acidic compartments in CHO cells, Autophagy. 3 (2007) 154–157. 10.4161/auto.3634. [DOI] [PubMed] [Google Scholar]
- [69].Galluzzi L, Bravo-San Pedro JM, Levine B, Green DR, Kroemer G, Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles, Nat. Rev. Drug Discov. 16 (2017) 487–511. 10.1038/nrd.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Mizushima N, Komatsu M, Autophagy: renovation of cells and tissues, Cell. 147 (2011) 728–741. 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- [71].Toyomura T, Oka T, Yamaguchi C, Wada Y, Futai M, Three subunit a isoforms of mouse vacuolar H(+)-ATPase. Preferential expression of the a3 isoform during osteoclast differentiation, J. Biol. Chem. 275 (2000) 8760–8765. 10.1074/jbc.275.12.8760. [DOI] [PubMed] [Google Scholar]
- [72].Frattini A, Orchard PJ, Sobacchi C, Giliani S, Abinun M, Mattsson JP, Keeling DJ, Andersson AK, Wallbrandt P, Zecca L, Notarangelo LD, Vezzoni P, Villa A, Defects in TCIRG1 subunit of the vacuolar proton pump are responsible for a subset of human autosomal recessive osteopetrosis, Nat. Genet. 25 (2000) 343–346. 10.1038/77131. [DOI] [PubMed] [Google Scholar]
- [73].Brown D, Paunescu TG, Breton S, Marshansky V, Regulation of the V-ATPase in kidney epithelial cells: dual role in acid-base homeostasis and vesicle trafficking, J. Exp. Biol. 212 (2009) 1762–1772. 10.1242/jeb.028803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Smith AN, Skaug J, Choate KA, Nayir A, Bakkaloglu A, Ozen S, Hulton SA, Sanjad SA, Al-Sabban EA, Lifton RP, Scherer SW, Karet FE, Mutations in ATP6N1B, encoding a new kidney vacuolar proton pump 116-kD subunit, cause recessive distal renal tubular acidosis with preserved hearing, Nat. Genet. 26 (2000) 71–75. 10.1038/79208. [DOI] [PubMed] [Google Scholar]
- [75].Oka T, Murata Y, Namba M, Yoshimizu T, Toyomura T, Yamamoto A, Sun-Wada GH, Hamasaki N, Wada Y, Futai M, a4, a unique kidney-specific isoform of mouse vacuolar H+-ATPase subunit a, J. Biol. Chem. 276 (2001) 40050–40054. 10.1074/jbc.M106488200. [DOI] [PubMed] [Google Scholar]
- [76].Shum WWC, Da Silva N, Brown D, Breton S, Regulation of luminal acidification in the male reproductive tract via cell-cell crosstalk, J. Exp. Biol. 212 (2009) 1753–1761. 10.1242/jeb.027284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Blomqvist SR, Vidarsson H, Söder O, Enerbäck S, Epididymal expression of the forkhead transcription factor Foxi1 is required for male fertility, EMBO J. 25 (2006) 4131–4141. 10.1038/sj.emboj.7601272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Stransky L, Cotter K, Forgac M, The Function of V-ATPases in Cancer, Physiol. Rev. 96 (2016) 1071–1091. 10.1152/physrev.00035.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Puopolo K, Forgac M, Functional reassembly of the coated vesicle proton pump, J. Biol. Chem. 265 (1990) 14836–14841. [PubMed] [Google Scholar]
- [80].Zhang J, Myers M, Forgac M, Characterization of the V0 domain of the coated vesicle (H+)-ATPase, J. Biol. Chem. 267 (1992) 9773–9778. [PubMed] [Google Scholar]
- [81].Parra KJ, Kane PM, Wild-type and mutant vacuolar membranes support pH-dependent reassembly of the yeast vacuolar H+-ATPase in vitro, J. Biol. Chem. 271 (1996) 19592–19598. 10.1074/jbc.271.32.19592. [DOI] [PubMed] [Google Scholar]
- [82].Kane PM, Targeting reversible disassembly as a mechanism of controlling V-ATPase activity, Curr. Protein Pept. Sci. 13 (2012) 117–123. 10.2174/138920312800493142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Kane PM, Disassembly and reassembly of the yeast vacuolar H(+)-ATPase in vivo, J. Biol. Chem. 270 (1995) 17025–17032. [PubMed] [Google Scholar]
- [84].Sumner JP, Dow JA, Earley FG, Klein U, Jäger D, Wieczorek H, Regulation of plasma membrane V-ATPase activity by dissociation of peripheral subunits, J. Biol. Chem. 270 (1995) 5649–5653. 10.1074/jbc.270.10.5649. [DOI] [PubMed] [Google Scholar]
- [85].Parra KJ, Kane PM, Reversible association between the V1 and V0 domains of yeast vacuolar H+-ATPase is an unconventional glucose-induced effect, Mol. Cell. Biol. 18 (1998) 7064–7074. 10.1128/mcb.18.12.7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Kawasaki-Nishi S, Nishi T, Forgac M, Yeast V-ATPase complexes containing different isoforms of the 100-kDa a-subunit differ in coupling efficiency and in vivo dissociation, J. Biol. Chem. 276 (2001) 17941–17948. 10.1074/jbc.M010790200. [DOI] [PubMed] [Google Scholar]
- [87].Bond S, Forgac M, The Ras/cAMP/protein kinase A pathway regulates glucose-dependent assembly of the vacuolar (H+)-ATPase in yeast, J. Biol. Chem. 283 (2008) 36513–36521. 10.1074/jbc.M805232200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Dechant R, Binda M, Lee SS, Pelet S, Winderickx J, Peter M, Cytosolic pH is a second messenger for glucose and regulates the PKA pathway through V-ATPase, EMBO J. 29 (2010) 2515–2526. 10.1038/emboj.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Diakov TT, Kane PM, Regulation of vacuolar proton-translocating ATPase activity and assembly by extracellular pH, J. Biol. Chem. 285 (2010) 23771–23778. 10.1074/jbc.M110.110122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Shao E, Forgac M, Involvement of the nonhomologous region of subunit A of the yeast V-ATPase in coupling and in vivo dissociation, J. Biol. Chem. 279 (2004) 48663–48670. 10.1074/jbc.M408278200. [DOI] [PubMed] [Google Scholar]
- [91].Dames P, Zimmermann B, Schmidt R, Rein J, Voss M, Schewe B, Walz B, Baumann O, cAMP regulates plasma membrane vacuolar-type H+-ATPase assembly and activity in blowfly salivary glands, Proc. Natl. Acad. Sci. U. S. A. 103 (2006) 3926–3931. 10.1073/pnas.0600011103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Voss M, Vitavska O, Walz B, Wieczorek H, Baumann O, Stimulus-induced phosphorylation of vacuolar H(+)-ATPase by protein kinase A, J. Biol. Chem. 282 (2007) 33735–33742. 10.1074/jbc.M703368200. [DOI] [PubMed] [Google Scholar]
- [93].Rein J, Voss M, Blenau W, Walz B, Baumann O, Hormone-induced assembly and activation of V-ATPase in blowfly salivary glands is mediated by protein kinase A, Am. J. Physiol. Cell Physiol. 294 (2008) C56–65. 10.1152/ajpcell.00041.2007. [DOI] [PubMed] [Google Scholar]
- [94].Tabke K, Albertmelcher A, Vitavska O, Huss M, Schmitz H-P, Wieczorek H, Reversible disassembly of the yeast V-ATPase revisited under in vivo conditions, Biochem. J. 462 (2014) 185–197. 10.1042/BJ20131293. [DOI] [PubMed] [Google Scholar]
- [95].Zimniak L, Dittrich P, Gogarten JP, Kibak H, Taiz L, The cDNA sequence of the 69-kDa subunit of the carrot vacuolar H+-ATPase. Homology to the beta-chain of F0F1-ATPases, J. Biol. Chem. 263 (1988) 9102–9112. [PubMed] [Google Scholar]
- [96].Shao E, Nishi T, Kawasaki-Nishi S, Forgac M, Mutational analysis of the non-homologous region of subunit A of the yeast V-ATPase, J. Biol. Chem. 278 (2003) 12985–12991. 10.1074/jbc.M212096200. [DOI] [PubMed] [Google Scholar]
- [97].Xu T, Forgac M, Microtubules are involved in glucose-dependent dissociation of the yeast vacuolar [H+]-ATPase in vivo, J. Biol. Chem. 276 (2001) 24855–24861. 10.1074/jbc.M100637200. [DOI] [PubMed] [Google Scholar]
- [98].Liu J, Kane PM, Mutational analysis of the catalytic subunit of the yeast vacuolar proton-translocating ATPase, Biochemistry. 35 (1996) 10938–10948. 10.1021/bi9608065. [DOI] [PubMed] [Google Scholar]
- [99].MacLeod KJ, Vasilyeva E, Merdek K, Vogel PD, Forgac M, Photoaffinity labeling of wild-type and mutant forms of the yeast V-ATPase A subunit by 2-azido-[(32)P]ADP, J. Biol. Chem. 274 (1999) 32869–32874. 10.1074/jbc.274.46.32869. [DOI] [PubMed] [Google Scholar]
- [100].Lu M, Ammar D, Ives H, Albrecht F, Gluck SL, Physical interaction between aldolase and vacuolar H+-ATPase is essential for the assembly and activity of the proton pump, J. Biol. Chem. 282 (2007) 24495–24503. 10.1074/jbc.M702598200. [DOI] [PubMed] [Google Scholar]
- [101].Chan C-Y, Parra KJ, Yeast phosphofructokinase-1 subunit Pfk2p is necessary for pH homeostasis and glucose-dependent vacuolar ATPase reassembly, J. Biol. Chem. 289 (2014) 19448–19457. 10.1074/jbc.M114.569855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Seol JH, Shevchenko A, Shevchenko A, Deshaies RJ, Skp1 forms multiple protein complexes, including RAVE, a regulator of V-ATPase assembly, Nat. Cell Biol. 3 (2001) 384–391. 10.1038/35070067. [DOI] [PubMed] [Google Scholar]
- [103].Smardon AM, Tarsio M, Kane PM, The RAVE complex is essential for stable assembly of the yeast V-ATPase, J. Biol. Chem. 277 (2002) 13831–13839. 10.1074/jbc.M200682200. [DOI] [PubMed] [Google Scholar]
- [104].Smardon AM, Kane PM, RAVE is essential for the efficient assembly of the C subunit with the vacuolar H(+)-ATPase, J. Biol. Chem. 282 (2007) 26185–26194. 10.1074/jbc.M703627200. [DOI] [PubMed] [Google Scholar]
- [105].Smardon AM, Nasab ND, Tarsio M, Diakov TT, Kane PM, Molecular Interactions and Cellular Itinerary of the Yeast RAVE (Regulator of the H+-ATPase of Vacuolar and Endosomal Membranes) Complex, J. Biol. Chem. 290 (2015) 27511–27523. 10.1074/jbc.M115.667634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Smardon AM, Diab HI, Tarsio M, Diakov TT, Nasab ND, West RW, Kane PM, The RAVE complex is an isoform-specific V-ATPase assembly factor in yeast, Mol. Biol. Cell. 25 (2014) 356–367. 10.1091/mbc.E13-05-0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Yan Y, Denef N, Schüpbach T, The vacuolar proton pump, V-ATPase, is required for notch signaling and endosomal trafficking in Drosophila, Dev. Cell. 17 (2009) 387–402. 10.1016/j.devcel.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Sethi N, Yan Y, Quek D, Schupbach T, Kang Y, Rabconnectin-3 is a functional regulator of mammalian Notch signaling, J. Biol. Chem. 285 (2010) 34757–34764. 10.1074/jbc.M110.158634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Li SC, Diakov TT, Xu T, Tarsio M, Zhu W, Couoh-Cardel S, Weisman LS, Kane PM, The signaling lipid PI(3,5)P₂ stabilizes V₁ -V(o) sector interactions and activates the V-ATPase, Mol. Biol. Cell. 25 (2014) 1251–1262. 10.1091/mbc.E13-10-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Kane PM, Proton Transport and pH Control in Fungi, Adv. Exp. Med. Biol. 892 (2016) 33–68. 10.1007/978-3-319-25304-6_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Banerjee S, Clapp K, Tarsio M, Kane PM, Interaction of the late endo-lysosomal lipid PI(3,5)P2 with the Vph1 isoform of yeast V-ATPase increases its activity and cellular stress tolerance, J. Biol. Chem. 294 (2019) 9161–9171. 10.1074/jbc.RA119.008552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Banerjee S, Kane PM, Direct interaction of the Golgi V-ATPase a-subunit isoform with PI(4)P drives localization of Golgi V-ATPases in yeast, Mol. Biol. Cell. 28 (2017) 2518–2530. 10.1091/mbc.E17-05-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Nakamura S, Glucose activates H(+)-ATPase in kidney epithelial cells, Am. J. Physiol. Cell Physiol. 287 (2004) C97–105. 10.1152/ajpcell.00469.2003. [DOI] [PubMed] [Google Scholar]
- [114].Sautin YY, Lu M, Gaugler A, Zhang L, Gluck SL, Phosphatidylinositol 3-kinase-mediated effects of glucose on vacuolar H+-ATPase assembly, translocation, and acidification of intracellular compartments in renal epithelial cells, Mol. Cell. Biol. 25 (2005) 575–589. 10.1128/MCB.25.2.575-589.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Marjuki H, Gornitzky A, Marathe BM, Ilyushina NA, Aldridge JR, Desai G, Webby RJ, Webster RG, Influenza A virus-induced early activation of ERK and PI3K mediates V-ATPase-dependent intracellular pH change required for fusion, Cell. Microbiol. 13 (2011) 587–601. 10.1111/j.1462-5822.2010.01556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Soliman M, Seo J-Y, Kim D-S, Kim J-Y, Park J-G, Alfajaro MM, Baek Y-B, Cho E-H, Kwon J, Choi J-S, Kang M-I, Park S-I, Cho K-O, Activation of PI3K, Akt, and ERK during early rotavirus infection leads to V-ATPase-dependent endosomal acidification required for uncoating, PLoS Pathog. 14 (2018) e1006820 10.1371/journal.ppat.1006820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Trombetta ES, Ebersold M, Garrett W, Pypaert M, Mellman I, Activation of lysosomal function during dendritic cell maturation, Science. 299 (2003) 1400–1403. 10.1126/science.1080106. [DOI] [PubMed] [Google Scholar]
- [118].Liberman R, Bond S, Shainheit MG, Stadecker MJ, Forgac M, Regulated assembly of vacuolar ATPase is increased during cluster disruption-induced maturation of dendritic cells through a phosphatidylinositol 3-kinase/mTOR-dependent pathway, J. Biol. Chem. 289 (2014) 1355–1363. 10.1074/jbc.M113.524561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Xu Y, Parmar A, Roux E, Balbis A, Dumas V, Chevalier S, Posner BI, Epidermal growth factor-induced vacuolar (H+)-atpase assembly: a role in signaling via mTORC1 activation, J. Biol. Chem. 287 (2012) 26409–26422. 10.1074/jbc.M112.352229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].McGuire CM, Forgac M, Glucose starvation increases V-ATPase assembly and activity in mammalian cells through AMP kinase and phosphatidylinositide 3-kinase/Akt signaling, J. Biol. Chem. 293 (2018) 9113–9123. 10.1074/jbc.RA117.001327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Stransky LA, Forgac M, Amino Acid Availability Modulates Vacuolar H+-ATPase Assembly, J. Biol. Chem. 290 (2015) 27360–27369. 10.1074/jbc.M115.659128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Tiburcy F, Beyenbach KW, Wieczorek H, Protein kinase A-dependent and -independent activation of the V-ATPase in Malpighian tubules of Aedes aegypti, J. Exp. Biol. 216 (2013) 881–891. 10.1242/jeb.078360. [DOI] [PubMed] [Google Scholar]
- [123].Liu Y, Steinbusch LKM, Nabben M, Kapsokalyvas D, van Zandvoort M, Schönleitner P, Antoons G, Simons PJ, Coumans WA, Geomini A, Chanda D, Glatz JFC, Neumann D, Luiken JJFP, Palmitate-Induced Vacuolar-Type H+-ATPase Inhibition Feeds Forward Into Insulin Resistance and Contractile Dysfunction, Diabetes. 66 (2017) 1521–1534. 10.2337/db16-0727. [DOI] [PubMed] [Google Scholar]
- [124].Bodzęta A, Kahms M, Klingauf J, The Presynaptic v-ATPase Reversibly Disassembles and Thereby Modulates Exocytosis but Is Not Part of the Fusion Machinery, Cell Rep. 20 (2017) 1348–1359. 10.1016/j.celrep.2017.07.040. [DOI] [PubMed] [Google Scholar]
- [125].Manolson MF, Proteau D, Preston RA, Stenbit A, Roberts BT, Hoyt MA, Preuss D, Mulholland J, Botstein D, Jones EW, The VPH1 gene encodes a 95-kDa integral membrane polypeptide required for in vivo assembly and activity of the yeast vacuolar H(+)-ATPase, J. Biol. Chem. 267 (1992) 14294–14303. [PubMed] [Google Scholar]
- [126].Manolson MF, Wu B, Proteau D, Taillon BE, Roberts BT, Hoyt MA, Jones EW, STV1 gene encodes functional homologue of 95-kDa yeast vacuolar H(+)-ATPase subunit Vph1p, J. Biol. Chem. 269 (1994) 14064–14074. [PubMed] [Google Scholar]
- [127].Kawasaki-Nishi S, Bowers K, Nishi T, Forgac M, Stevens TH, The amino-terminal domain of the vacuolar proton-translocating ATPase a subunit controls targeting and in vivo dissociation, and the carboxyl-terminal domain affects coupling of proton transport and ATP hydrolysis, J. Biol. Chem. 276 (2001) 47411–47420. 10.1074/jbc.M108310200. [DOI] [PubMed] [Google Scholar]
- [128].Miranda KC, Karet FE, Brown D, An extended nomenclature for mammalian V-ATPase subunit genes and splice variants, PloS One. 5 (2010) e9531 10.1371/journal.pone.0009531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Hurtado-Lorenzo A, Skinner M, El Annan J, Futai M, Sun-Wada G-H, Bourgoin S, Casanova J, Wildeman A, Bechoua S, Ausiello DA, Brown D, Marshansky V, V-ATPase interacts with ARNO and Arf6 in early endosomes and regulates the protein degradative pathway, Nat. Cell Biol. 8 (2006) 124–136. 10.1038/ncb1348. [DOI] [PubMed] [Google Scholar]
- [130].Morel N, Dedieu J-C, Philippe J-M, Specific sorting of the a1 isoform of the V-H+ATPase a subunit to nerve terminals where it associates with both synaptic vesicles and the presynaptic plasma membrane, J. Cell Sci. 116 (2003) 4751–4762. 10.1242/jcs.00791. [DOI] [PubMed] [Google Scholar]
- [131].Toyomura T, Murata Y, Yamamoto A, Oka T, Sun-Wada G-H, Wada Y, Futai M, From lysosomes to the plasma membrane: localization of vacuolar-type H+ -ATPase with the a3 isoform during osteoclast differentiation, J. Biol. Chem. 278 (2003) 22023–22030. 10.1074/jbc.M302436200. [DOI] [PubMed] [Google Scholar]
- [132].Matsumoto N, Sekiya M, Tohyama K, Ishiyama-Matsuura E, Sun-Wada G-H, Wada Y, Futai M, Nakanishi-Matsui M, Essential Role of the a3 Isoform of V-ATPase in Secretory Lysosome Trafficking via Rab7 Recruitment, Sci. Rep 8 (2018) 6701 10.1038/s41598-018-24918-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Saw NMN, Kang S-YA, Parsaud L, Han GA, Jiang T, Grzegorczyk K, Surkont M, Sun-Wada G-H, Wada Y, Li L, Sugita S, Vacuolar H(+)-ATPase subunits Voa1 and Voa2 cooperatively regulate secretory vesicle acidification, transmitter uptake, and storage, Mol. Biol. Cell. 22 (2011) 3394–3409. 10.1091/mbc.E11-02-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Sun-Wada G-H, Tabata H, Kawamura N, Aoyama M, Wada Y, Direct recruitment of H+-ATPase from lysosomes for phagosomal acidification, J. Cell Sci. 122 (2009) 2504–2513. 10.1242/jcs.050443. [DOI] [PubMed] [Google Scholar]
- [135].Nishi T, Forgac M, Molecular cloning and expression of three isoforms of the 100-kDa a subunit of the mouse vacuolar proton-translocating ATPase, J. Biol. Chem. 275 (2000) 6824–6830. 10.1074/jbc.275.10.6824. [DOI] [PubMed] [Google Scholar]
- [136].Pietrement C, Sun-Wada G-H, Silva ND, McKee M, Marshansky V, Brown D, Futai M, Breton S, Distinct expression patterns of different subunit isoforms of the V-ATPase in the rat epididymis, Biol. Reprod. 74 (2006) 185–194. 10.1095/biolreprod.105.043752. [DOI] [PubMed] [Google Scholar]
- [137].Pastor-Soler NM, Hallows KR, Smolak C, Gong F, Brown D, Breton S, Alkaline pH- and cAMP-induced V-ATPase membrane accumulation is mediated by protein kinase A in epididymal clear cells, Am. J. Physiol. Cell Physiol. 294 (2008) C488–494. 10.1152/ajpcell.00537.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Alzamora R, Thali RF, Gong F, Smolak C, Li H, Baty CJ, Bertrand CA, Auchli Y, Brunisholz RA, Neumann D, Hallows KR, Pastor-Soler NM, PKA regulates vacuolar H+-ATPase localization and activity via direct phosphorylation of the a subunit in kidney cells, J. Biol. Chem. 285 (2010) 24676–24685. 10.1074/jbc.M110.106278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Păunescu TG, Ljubojevic M, Russo LM, Winter C, McLaughlin MM, Wagner CA, Breton S, Brown D, cAMP stimulates apical V-ATPase accumulation, microvillar elongation, and proton extrusion in kidney collecting duct A-intercalated cells, Am. J. Physiol. Renal Physiol 298 (2010) F643–654. 10.1152/ajprenal.00584.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Hallows KR, Alzamora R, Li H, Gong F, Smolak C, Neumann D, Pastor-Soler NM, AMP-activated protein kinase inhibits alkaline pH- and PKA-induced apical vacuolar H+-ATPase accumulation in epididymal clear cells, Am. J. Physiol. Cell Physiol. 296 (2009) C672–681. 10.1152/ajpcell.00004.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Alzamora R, Al-Bataineh MM, Liu W, Gong F, Li H, Thali RF, Joho-Auchli Y, Brunisholz RA, Satlin LM, Neumann D, Hallows KR, Pastor-Soler NM, AMP-activated protein kinase regulates the vacuolar H+-ATPase via direct phosphorylation of the A subunit (ATP6V1A) in the kidney, Am. J. Physiol. Renal Physiol. 305 (2013) F943–956. 10.1152/ajprenal.00303.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Feng Y, Forgac M, Cysteine 254 of the 73-kDa A subunit is responsible for inhibition of the coated vesicle (H+)-ATPase upon modification by sulfhydryl reagents, J. Biol. Chem. 267 (1992) 5817–5822. [PubMed] [Google Scholar]
- [143].Feng Y, Forgac M, Inhibition of vacuolar H(+)-ATPase by disulfide bond formation between cysteine 254 and cysteine 532 in subunit A, J. Biol. Chem. 269 (1994) 13224–13230. [PubMed] [Google Scholar]
- [144].Feng Y, Forgac M, A novel mechanism for regulation of vacuolar acidification, J. Biol. Chem. 267 (1992) 19769–19772. [PubMed] [Google Scholar]
- [145].Jefferies KC, Cipriano DJ, Forgac M, Function, structure and regulation of the vacuolar (H+)-ATPases, Arch. Biochem. Biophys. 476 (2008) 33–42. 10.1016/j.abb.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Günther W, Lüchow A, Cluzeaud F, Vandewalle A, Jentsch TJ, ClC-5, the chloride channel mutated in Dent’s disease, colocalizes with the proton pump in endocytotically active kidney cells, Proc. Natl. Acad. Sci. U. S. A. 95 (1998) 8075–8080. 10.1073/pnas.95.14.8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Majumdar A, Capetillo-Zarate E, Cruz D, Gouras GK, Maxfield FR, Degradation of Alzheimer’s amyloid fibrils by microglia requires delivery of ClC-7 to lysosomes, Mol. Biol. Cell. 22 (2011) 1664–1676. 10.1091/mbc.E10-09-0745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Kornak U, Kasper D, Bösl MR, Kaiser E, Schweizer M, Schulz A, Friedrich W, Delling G, Jentsch TJ, Loss of the ClC-7 chloride channel leads to osteopetrosis in mice and man, Cell. 104 (2001) 205–215. 10.1016/s00928674(01)00206-9. [DOI] [PubMed] [Google Scholar]
- [149].Webb BA, Chimenti M, Jacobson MP, Barber DL, Dysregulated pH: a perfect storm for cancer progression, Nat. Rev. Cancer. 11 (2011) 671–677. 10.1038/nrc3110. [DOI] [PubMed] [Google Scholar]
- [150].Collins MP, Forgac M, Regulation of V-ATPase Assembly in Nutrient Sensing and Function of V-ATPases in Breast Cancer Metastasis, Front. Physiol 9 (2018) 902 10.3389/fphys.2018.00902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Cotter K, Liberman R, Sun-Wada G, Wada Y, Sgroi D, Naber S, Brown D, Breton S, Forgac M, The a3 isoform of subunit a of the vacuolar ATPase localizes to the plasma membrane of invasive breast tumor cells and is overexpressed in human breast cancer, Oncotarget. 7 (2016) 46142–46157. 10.18632/oncotarget.10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Lu Q, Lu S, Huang L, Wang T, Wan Y, Zhou CX, Zhang C, Zhang Z, Li X, The expression of V-ATPase is associated with drug resistance and pathology of non-small-cell lung cancer, Diagn. Pathol 8 (2013) 145 10.1186/1746-1596-8-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Avnet S, Di Pompo G, Lemma S, Salerno M, Perut F, Bonuccelli G, Granchi D, Zini N, Baldini N, V-ATPase is a candidate therapeutic target for Ewing sarcoma, Biochim. Biophys. Acta. 1832 (2013) 1105–1116. 10.1016/j.bbadis.2013.04.003. [DOI] [PubMed] [Google Scholar]
- [154].Nishisho T, Hata K, Nakanishi M, Morita Y, Sun-Wada G-H, Wada Y, Yasui N, Yoneda T, The a3 isoform vacuolar type H+-ATPase promotes distant metastasis in the mouse B16 melanoma cells, Mol. Cancer Res. MCR 9 (2011) 845–855. 10.1158/1541-7786.MCR-10-0449. [DOI] [PubMed] [Google Scholar]
- [155].Sennoune SR, Bakunts K, Martínez GM, Chua-Tuan JL, Kebir Y, Attaya MN, Martínez-Zaguilán R, Vacuolar H+-ATPase in human breast cancer cells with distinct metastatic potential: distribution and functional activity, Am. J. Physiol. Cell Physiol. 286 (2004) C1443–1452. 10.1152/ajpcell.00407.2003. [DOI] [PubMed] [Google Scholar]
- [156].Xu J, Xie R, Liu X, Wen G, Jin H, Yu Z, Jiang Y, Zhao Z, Yang Y, Ji B, Dong H, Tuo B, Expression and functional role of vacuolar H(+)-ATPase in human hepatocellular carcinoma, Carcinogenesis. 33 (2012) 2432–2440. 10.1093/carcin/bgs277. [DOI] [PubMed] [Google Scholar]
- [157].Chung C, Mader CC, Schmitz JC, Atladottir J, Fitchev P, Cornwell ML, Koleske AJ, Crawford SE, Gorelick F, The vacuolar-ATPase modulates matrix metalloproteinase isoforms in human pancreatic cancer, Lab. Investig. J. Tech. Methods Pathol 91 (2011) 732–743. 10.1038/labinvest.2011.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Smith GA, Howell GJ, Phillips C, Muench SP, Ponnambalam S, Harrison MA, Extracellular and Luminal pH Regulation by Vacuolar H+-ATPase Isoform Expression and Targeting to the Plasma Membrane and Endosomes, J. Biol. Chem. 291 (2016) 8500–8515. 10.1074/jbc.M116.723395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Kulshrestha A, Katara GK, Ibrahim S, Pamarthy S, Jaiswal MK, Gilman Sachs A, Beaman KD, Vacuolar ATPase “a2” isoform exhibits distinct cell surface accumulation and modulates matrix metalloproteinase activity in ovarian cancer, Oncotarget. 6 (2015) 3797–3810. 10.18632/oncotarget.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Gerweck LE, Seetharaman K, Cellular pH gradient in tumor versus normal tissue: potential exploitation for the treatment of cancer, Cancer Res. 56 (1996) 1194–1198. [PubMed] [Google Scholar]
- [161].von Schwarzenberg K, Wiedmann RM, Oak P, Schulz S, Zischka H, Wanner G, Efferth T, Trauner D, Vollmar AM, Mode of cell death induction by pharmacological vacuolar H+-ATPase (V-ATPase) inhibition, J. Biol. Chem. 288 (2013) 1385–1396. 10.1074/jbc.M112.412007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Schempp CM, von Schwarzenberg K, Schreiner L, Kubisch R, Müller R, Wagner E, Vollmar AM, V-ATPase inhibition regulates anoikis resistance and metastasis of cancer cells, Mol. Cancer Ther. 13 (2014) 926–937. 10.1158/1535-7163.MCT-13-0484. [DOI] [PubMed] [Google Scholar]
- [163].Lu X, Chen L, Chen Y, Shao Q, Qin W, Bafilomycin A1 inhibits the growth and metastatic potential of the BEL-7402 liver cancer and HO-8910 ovarian cancer cell lines and induces alterations in their microRNA expression, Exp. Ther. Med 10 (2015) 1829–1834. 10.3892/etm.2015.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Zhang S, Schneider LS, Vick B, Grunert M, Jeremias I, Menche D, Müller R, Vollmar AM, Liebl J, Anti-leukemic effects of the V-ATPase inhibitor Archazolid A, Oncotarget. 6 (2015) 43508–43528. 10.18632/oncotarget.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Wojtkowiak JW, Verduzco D, Schramm KJ, Gillies RJ, Drug resistance and cellular adaptation to tumor acidic pH microenvironment, Mol. Pharm. 8 (2011) 2032–2038. 10.1021/mp200292c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Vukovic V, Tannock IF, Influence of low pH on cytotoxicity of paclitaxel, mitoxantrone and topotecan, Br. J. Cancer. 75 (1997) 1167–1172. 10.1038/bjc.1997.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Raghunand N, He X, van Sluis R, Mahoney B, Baggett B, Taylor CW, Paine-Murrieta G, Roe D, Bhujwalla ZM, Gillies RJ, Enhancement of chemotherapy by manipulation of tumour pH, Br. J. Cancer. 80 (1999) 1005–1011. 10.1038/sj.bjc.6690455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Mahoney BP, Raghunand N, Baggett B, Gillies RJ, Tumor acidity, ion trapping and chemotherapeutics. I. Acid pH affects the distribution of chemotherapeutic agents in vitro, Biochem. Pharmacol. 66 (2003) 1207–1218. 10.1016/s0006-2952(03)00467-2. [DOI] [PubMed] [Google Scholar]
- [169].Martínez-Zaguilán R, Raghunand N, Lynch RM, Bellamy W, Martinez GM, Rojas B, Smith D, Dalton WS, Gillies RJ, pH and drug resistance. I. Functional expression of plasmalemmal V-type H+-ATPase in drug-resistant human breast carcinoma cell lines, Biochem. Pharmacol. 57 (1999) 1037–1046. 10.1016/s0006-2952(99)00022-2. [DOI] [PubMed] [Google Scholar]
- [170].Raghunand N, Martínez-Zaguilán R, Wright SH, Gillies RJ, pH and drug resistance. II. Turnover of acidic vesicles and resistance to weakly basic chemotherapeutic drugs, Biochem. Pharmacol. 57 (1999) 1047–1058. 10.1016/s0006-2952(99)00021-0. [DOI] [PubMed] [Google Scholar]
- [171].Marquardt D, Center MS, Involvement of vacuolar H(+)-adenosine triphosphatase activity in multidrug resistance in HL60 cells, J. Natl. Cancer Inst. 83 (1991) 1098–1102. 10.1093/jnci/83.15.1098. [DOI] [PubMed] [Google Scholar]
- [172].Ouar Z, Bens M, Vignes C, Paulais M, Pringel C, Fleury J, Cluzeaud F, Lacave R, Vandewalle A, Inhibitors of vacuolar H+-ATPase impair the preferential accumulation of daunomycin in lysosomes and reverse the resistance to anthracyclines in drug-resistant renal epithelial cells, Biochem. J. 370 (2003) 185–193. 10.1042/BJ20021411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Luciani F, Spada M, De Milito A, Molinari A, Rivoltini L, Montinaro A, Marra M, Lugini L, Logozzi M, Lozupone F, Federici C, Iessi E, Parmiani G, Arancia G, Belardelli F, Fais S, Effect of proton pump inhibitor pretreatment on resistance of solid tumors to cytotoxic drugs, J. Natl. Cancer Inst. 96 (2004) 1702–1713. 10.1093/jnci/djh305. [DOI] [PubMed] [Google Scholar]
- [174].Kulshrestha A, Katara GK, Ibrahim SA, Riehl V, Sahoo M, Dolan J, Meinke KW, Pins MR, Beaman KD, Targeting V-ATPase Isoform Restores Cisplatin Activity in Resistant Ovarian Cancer: Inhibition of Autophagy, Endosome Function, and ERK/MEK Pathway, J. Oncol 2019 (2019) 2343876 10.1155/2019/2343876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Licon-Munoz Y, Fordyce CA, Hayek SR, Parra KJ, V-ATPase-dependent repression of androgen receptor in prostate cancer cells, Oncotarget. 9 (2018) 28921–28934. 10.18632/oncotarget.25641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Steeg PS, Targeting metastasis, Nat. Rev. Cancer. 16 (2016) 201–218. 10.1038/nrc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Lambert AW, Pattabiraman DR, Weinberg RA, Emerging Biological Principles of Metastasis, Cell. 168 (2017) 670–691. 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Mohamed MM, Sloane BF, Cysteine cathepsins: multifunctional enzymes in cancer, Nat. Rev. Cancer. 6 (2006) 764–775. 10.1038/nrc1949. [DOI] [PubMed] [Google Scholar]
- [179].Vasiljeva O, Papazoglou A, Krüger A, Brodoefel H, Korovin M, Deussing J, Augustin N, Nielsen BS, Almholt K, Bogyo M, Peters C, Reinheckel T, Tumor cell-derived and macrophage-derived cathepsin B promotes progression and lung metastasis of mammary cancer, Cancer Res. 66 (2006) 5242–5250. 10.1158/0008-5472.CAN-05-4463. [DOI] [PubMed] [Google Scholar]
- [180].Withana NP, Blum G, Sameni M, Slaney C, Anbalagan A, Olive MB, Bidwell BN, Edgington L, Wang L, Moin K, Sloane BF, Anderson RL, Bogyo MS, Parker BS, Cathepsin B inhibition limits bone metastasis in breast cancer, Cancer Res. 72 (2012) 1199–1209. 10.1158/0008-5472.CAN-11-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Martinez-Zaguilan R, Lynch RM, Martinez GM, Gillies RJ, Vacuolar-type H(+)-ATPases are functionally expressed in plasma membranes of human tumor cells, Am. J. Physiol. 265 (1993) C1015–1029. 10.1152/ajpcell.1993.265.4.C1015. [DOI] [PubMed] [Google Scholar]
- [182].Hinton A, Sennoune SR, Bond S, Fang M, Reuveni M, Sahagian GG, Jay D, Martinez-Zaguilan R, Forgac M, Function of a subunit isoforms of the V-ATPase in pH homeostasis and in vitro invasion of MDA-MB231 human breast cancer cells, J. Biol. Chem. 284 (2009) 16400–16408. 10.1074/jbc.M901201200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Capecci J, Forgac M, The function of vacuolar ATPase (V-ATPase) a subunit isoforms in invasiveness of MCF10a and MCF10CA1a human breast cancer cells, J. Biol. Chem. 288 (2013) 32731–32741. 10.1074/jbc.M113.503771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Cotter K, Capecci J, Sennoune S, Huss M, Maier M, Martinez-Zaguilan R, Forgac M, Activity of plasma membrane V-ATPases is critical for the invasion of MDA-MB231 breast cancer cells, J. Biol. Chem. 290 (2015) 3680–3692. 10.1074/jbc.M114.611210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].McGuire CM, Collins MP, Sun-Wada G, Wada Y, Forgac M, Isoform-specific gene disruptions reveal a role for the V-ATPase subunit a4 isoform in the invasiveness of 4T1–12B breast cancer cells, J. Biol. Chem. 294 (2019) 11248–11258. 10.1074/jbc.RA119.007713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Wiedmann RM, von Schwarzenberg K, Palamidessi A, Schreiner L, Kubisch R, Liebl J, Schempp C, Trauner D, Vereb G, Zahler S, Wagner E, Müller R, Scita G, Vollmar AM, The V-ATPase-inhibitor archazolid abrogates tumor metastasis via inhibition of endocytic activation of the Rho-GTPase Rac1, Cancer Res. 72 (2012) 5976–5987. 10.1158/0008-5472.CAN-12-1772. [DOI] [PubMed] [Google Scholar]
- [187].Licon-Munoz Y, Michel V, Fordyce CA, Parra KJ, F-actin reorganization by V-ATPase inhibition in prostate cancer, Biol. Open 6 (2017) 1734–1744. 10.1242/bio.028837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Holliday LS, Lu M, Lee BS, Nelson RD, Solivan S, Zhang L, Gluck SL, The amino-terminal domain of the B subunit of vacuolar H+-ATPase contains a filamentous actin binding site, J. Biol. Chem. 275 (2000) 32331–32337. 10.1074/jbc.M004795200. [DOI] [PubMed] [Google Scholar]
- [189].Vitavska O, Wieczorek H, Merzendorfer H, A novel role for subunit C in mediating binding of the H+-V-ATPase to the actin cytoskeleton, J. Biol. Chem. 278 (2003) 18499–18505. 10.1074/jbc.M212844200. [DOI] [PubMed] [Google Scholar]
- [190].Rojas JD, Sennoune SR, Maiti D, Bakunts K, Reuveni M, Sanka SC, Martinez GM, Seftor EA, Meininger CJ, Wu G, Wesson DE, Hendrix MJC, Martínez-Zaguilán R, Vacuolar-type H+-ATPases at the plasma membrane regulate pH and cell migration in microvascular endothelial cells, Am. J. Physiol. Heart Circ. Physiol. 291 (2006) H1147–1157. 10.1152/ajpheart.00166.2006. [DOI] [PubMed] [Google Scholar]
- [191].Kazami S, Takaine M, Itoh H, Kubota T, Kobayashi J, Usui T, Iejimalide C is a potent V-ATPase inhibitor, and induces actin disorganization, Biol. Pharm. Bull. 37 (2014) 1944–1947. 10.1248/bpb.b14-00548. [DOI] [PubMed] [Google Scholar]
- [192].Cai M, Liu P, Wei L, Wang J, Qi J, Feng S, Deng L, Atp6v1c1 may regulate filament actin arrangement in breast cancer cells, PloS One. 9 (2014) e84833 10.1371/journal.pone.0084833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Stock C, Schwab A, Protons make tumor cells move like clockwork, Pflugers Arch. 458 (2009) 981–992. 10.1007/s00424-009-0677-8. [DOI] [PubMed] [Google Scholar]
- [194].Masters CL, Bateman R, Blennow K, Rowe CC, Sperling RA, Cummings JL, Alzheimer’s disease, Nat. Rev. Dis. Primer. 1 (2015) 15056 10.1038/nrdp.2015.56. [DOI] [PubMed] [Google Scholar]
- [195].Lee J-H, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, Wolfe DM, Martinez-Vicente M, Massey AC, Sovak G, Uchiyama Y, Westaway D, Cuervo AM, Nixon RA, Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations, Cell. 141 (2010) 1146–1158. 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Lee J-H, McBrayer MK, Wolfe DM, Haslett LJ, Kumar A, Sato Y, Lie PPY, Mohan P, Coffey EE, Kompella U, Mitchell CH, Lloyd-Evans E, Nixon RA, Presenilin 1 Maintains Lysosomal Ca(2+) Homeostasis via TRPML1 by Regulating vATPase-Mediated Lysosome Acidification, Cell Rep. 12 (2015) 1430–1444. 10.1016/j.celrep.2015.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Poewe W, Seppi K, Tanner CM, Halliday GM, Brundin P, Volkmann J, Schrag A-E, Lang AE, Parkinson disease, Nat. Rev. Dis. Primer. 3 (2017) 17013 10.1038/nrdp.2017.13. [DOI] [PubMed] [Google Scholar]
- [198].Korvatska O, Strand NS, Berndt JD, Strovas T, Chen D-H, Leverenz JB, Kiianitsa K, Mata IF, Karakoc E, Greenup JL, Bonkowski E, Chuang J, Moon RT, Eichler EE, Nickerson DA, Zabetian CP, Kraemer BC, Bird TD, Raskind WH, Altered splicing of ATP6AP2 causes X-linked parkinsonism with spasticity (XPDS), Hum. Mol. Genet. 22 (2013) 3259–3268. 10.1093/hmg/ddt180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Colacurcio DJ, Nixon RA, Disorders of lysosomal acidification-The emerging role of v-ATPase in aging and neurodegenerative disease, Ageing Res. Rev. 32 (2016) 75–88. 10.1016/j.arr.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Bhargava A, Voronov I, Wang Y, Glogauer M, Kartner N, Manolson MF, Osteopetrosis mutation R444L causes endoplasmic reticulum retention and misprocessing of vacuolar H+-ATPase a3 subunit, J. Biol. Chem. 287 (2012) 26829–26839. 10.1074/jbc.M112.345702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Karet FE, Finberg KE, Nelson RD, Nayir A, Mocan H, Sanjad SA, Rodriguez-Soriano J, Santos F, Cremers CW, Di Pietro A, Hoffbrand BI, Winiarski J, Bakkaloglu A, Ozen S, Dusunsel R, Goodyer P, Hulton SA, Wu DK, Skvorak AB, Morton CC, Cunningham MJ, Jha V, Lifton RP, Mutations in the gene encoding B1 subunit of H+-ATPase cause renal tubular acidosis with sensorineural deafness, Nat. Genet. 21 (1999) 84–90. 10.1038/5022. [DOI] [PubMed] [Google Scholar]
- [202].Stover EH, Borthwick KJ, Bavalia C, Eady N, Fritz DM, Rungroj N, Giersch ABS, Morton CC, Axon PR, Akil I, Al-Sabban EA, Baguley DM, Bianca S, Bakkaloglu A, Bircan Z, Chauveau D, Clermont M-J, Guala A, Hulton SA, Kroes H, Li Volti G, Mir S, Mocan H, Nayir A, Ozen S, Rodriguez Soriano J, Sanjad SA, Tasic V, Taylor CM, Topaloglu R, Smith AN, Karet FE, Novel ATP6V1B1 and ATP6V0A4 mutations in autosomal recessive distal renal tubular acidosis with new evidence for hearing loss, J. Med. Genet. 39 (2002) 796–803. 10.1136/jmg.39.11.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Esmail S, Kartner N, Yao Y, Kim JW, Reithmeier RAF, Manolson MF, Molecular mechanisms of cutis laxa- and distal renal tubular acidosis-causing mutations in V-ATPase a subunits, ATP6V0A2 and ATP6V0A4, J. Biol. Chem. 293 (2018) 2787–2800. 10.1074/jbc.M117.818872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Kornak U, Reynders E, Dimopoulou A, van Reeuwijk J, Fischer B, Rajab A, Budde B, Nürnberg P, Foulquier F, ARCL Debré-type Study Group, Lefeber D, Urban Z, Gruenewald S, Annaert W, Brunner HG, van Bokhoven H, Wevers R, Morava E, Matthijs G, Van Maldergem L, Mundlos S, Impaired glycosylation and cutis laxa caused by mutations in the vesicular H+-ATPase subunit ATP6V0A2, Nat. Genet. 40 (2008) 32–34. 10.1038/ng.2007.45. [DOI] [PubMed] [Google Scholar]
- [205].Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM, The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1, Science. 320 (2008) 1496–1501. 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM, Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids, Cell. 141 (2010) 290–303. 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM, Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1, Cell. 150 (2012) 1196–1208. 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Manning BD, Toker A, AKT/PKB Signaling: Navigating the Network, Cell. 169 (2017) 381–405. 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Inoki K, Li Y, Zhu T, Wu J, Guan K-L, TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling, Nat. Cell Biol. 4 (2002) 648–657. 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- [210].Zhang Y, Gao X, Saucedo LJ, Ru B, Edgar BA, Pan D, Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins, Nat. Cell Biol. 5 (2003) 578–581. 10.1038/ncb999. [DOI] [PubMed] [Google Scholar]
- [211].Dibble CC, Manning BD, Signal integration by mTORC1 coordinates nutrient input with biosynthetic output, Nat. Cell Biol. 15 (2013) 555–564. 10.1038/ncb2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM, mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase, Science. 334 (2011) 678–683. 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Lin S-C, Hardie DG, AMPK: Sensing Glucose as well as Cellular Energy Status, Cell Metab. 27 (2018) 299–313. 10.1016/j.cmet.2017.10.009. [DOI] [PubMed] [Google Scholar]
- [214].Zhang C-S, Jiang B, Li M, Zhu M, Peng Y, Zhang Y-L, Wu Y-Q, Li TY, Liang Y, Lu Z, Lian G, Liu Q, Guo H, Yin Z, Ye Z, Han J, Wu J-W, Yin H, Lin S-Y, Lin S-C, The lysosomal v-ATPase-Ragulator complex is a common activator for AMPK and mTORC1, acting as a switch between catabolism and anabolism, Cell Metab. 20 (2014) 526–540. 10.1016/j.cmet.2014.06.014. [DOI] [PubMed] [Google Scholar]
- [215].Zhang C-S, Hawley SA, Zong Y, Li M, Wang Z, Gray A, Ma T, Cui J, Feng J-W, Zhu M, Wu Y-Q, Li TY, Ye Z, Lin S-Y, Yin H, Piao H-L, Hardie DG, Lin S-C, Fructose-1,6-bisphosphate and aldolase mediate glucose sensing by AMPK, Nature. 548 (2017) 112–116. 10.1038/nature23275. [DOI] [PMC free article] [PubMed] [Google Scholar]