Abstract
Over the past five years, technological advances have made it possible to image the translation of single mRNA in the natural context of living cells. With these advances, researchers are beginning to shed light on when, where, and to what degree mRNA are translated with single-molecule precision. These works provide insight into the heterogeneity of translation amongst single transcripts, behavior that is averaged out in complementary bulk assays. In this review, we discuss the rapidly maturing field of live-cell, single-mRNA imaging of translation, beginning with a brief overview of recent technological advances. The remainder of the review focuses on the new biological insights gained from these technologies. We conclude with a discussion of the future of this technology.
Keywords: Translational gene regulation, Single Molecule Imaging, live-cell fluorescence Microscopy
Introduction
Translation is the process by which a ribosome decodes an mRNA codon by codon to synthesize a nascent polypeptide chain. The process has traditionally been studied in cells by detecting expression levels via radiolabeling, fluorescence or bioluminescence (Reviewed in [1]). More recently, ribosome profiling has also been used, in this case, to see where translating ribosomes are distributed across the whole transcriptome[2,3]. While powerful, a drawback of these assays is their reliance on population averaging, which limits their temporal resolution and averages out any heterogeneity in translation that might exist from one mRNA to another [4]. Complementary techniques are therefore required to study translation at the single-mRNA level with higher temporal resolution, preferably in the natural context of living cells.
In an eventful few months in 2016, five publications developed technology to visualize translation in living cells at the single-molecule level [5–9]. The technology, which we refer to collectively as Nascent Chain Tracking (NCT) for simplicity here, uses complementary probes to label mRNA and nascent polypeptide chains in translation sites with different colored fluorophores (Figure 1). mRNA are labeled with the MS2 or PP7 systems (Reviewed in [10]). Similarly, nascent chains are labeled by fluorescent antibody-based probes, either single-chain variable fragments (scFv) [11] or fragmented antibodies (Fab) [5,12]. These probes bind short, repeated epitopes fused to the N-terminus of a protein of interest.
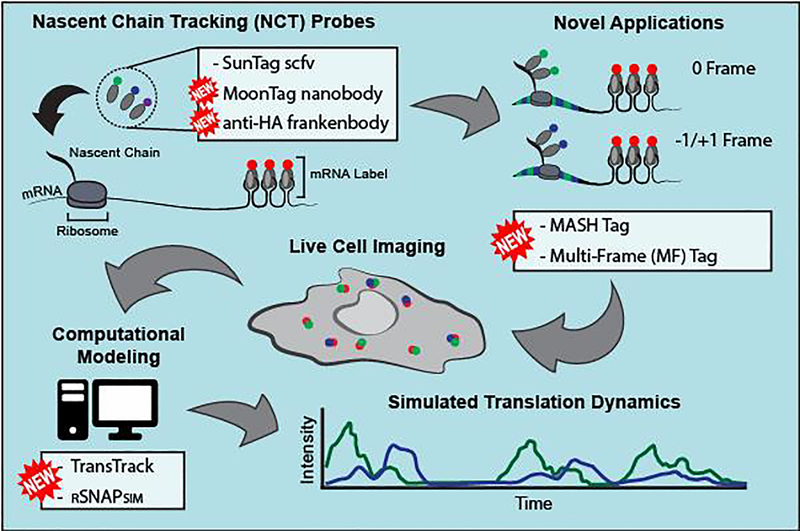
Figure 1. Advances in single-mRNA translation imaging technology.
In the Nascent Chain Tracking (NCT) assay, mRNA (red) and nascent polypeptide chains (green or blue) in translation sites co-move in live cells. New probes, including the MoonTag nanobody and anti-HA frankenbody, can be used to label translation sites in different colors. Together with new reporter tags, including the MoonTag and SunTag Hybrid (MASH) tag and the multi-frame tag, translation sites in live cells can be lit up in different colors depending on what frame is being translated. Translation intensity signals over time can be modeled using new software, including TransTrack or RNA Sequence to NAscent Protein SIMulator (RSNAPSIM).
Unlike more traditional fluorescent fusion tags such as GFP, which take minutes to fold and fluoresce, the major advantage of using fluorescent Fab or scFv is they can light up a nascent polypeptide chain co-translationally, within seconds of its emergence from the exit tunnel [13]. With NCT, it is now possible to visualize where the translating mRNA are in cells, the heterogeneity in translation dynamics between single mRNA, and the specific kinetics of translation initiation and elongation.
This sudden burst of papers from several different labs signifies the need and power of NCT. Since NCT has been cross-verified across research labs, several groups are now starting to use it to extend what we know about translation. In what follows, we highlight the most recent advances of NCT and discuss how the technology is being used to dissect the kinetics of translational gene regulation with unprecedented spatiotemporal resolution.
New Advances in Nascent Chain Tracking
Since 2016, a couple of significant advances have made NCT more versatile and user-friendly, as summarized in Figure 1. First, considerable effort has been made to make more genetically encoded antibody-based probes. To complement the popular SunTag scFv [11], the anti-MoonTag probe was derived from a camelid nanobody that binds the HIV gp41 epitope (KNEQELLELDKWASL, referred to as the MoonTag) [14]. Likewise, to complement the HA spaghetti monster tag [16], the anti-HA frankenbody was developed to bind the classic HA epitope (YPYDVPDYA) [17]. Since the SunTag scFv, MoonTag nanobody, and frankenbody can be encoded in plasmids, they are cheap, easy to use, and can be further genetically engineered to bind their target epitopes with higher affinity or bind new epitopes entirely. Concurrent with the development of these probes, new repeat epitope tags were designed to simultaneously visualize translation in different reading frames [14,18]. In the MoonTag and SunTag Hybrid (MASH) tag [14] and the multi-frame tag [18], epitopes of one type are interspersed between epitopes of another type encoded in a different frame. In this way, as translation progresses, nascent chains are bound and lit up by different fluorescent probes depending on which frame is being translated [14,18].
Another significant advance in NCT has been computational in nature. Translation is inherently a stochastic process that requires sophisticated modeling to accurately fit and predict single-mRNA dynamics. To make this easier, several labs have now published their modeling software alongside methods papers [19–21]. RSNAPSIM, for example, includes an interface that allows users to design translation imaging plasmids and simulate common experiments such as FRAP (fluorescence recovery after photobleaching), FCS (fluorescence correlation spectroscopy), and translation inhibitor experiments [21].
New Applications of Nascent Chain Tracking
Recently, NCT has been applied to answer numerous biological questions, not only about how translation is activated, but also how cells turn off translation and regulate aberrant mRNA. Here we summarize recent NCT applications to investigate translation control.
mRNA Structure During Translation
During canonical translation, interactions between initiation factors bound to the 5’ Cap and poly-A tail have led researchers to believe mRNA exist in a stable closed-loop conformation [22]. Recent single-molecule experiments are beginning to question this model. Specifically, Adivarahan et al. and Khong et al. used smFISH ([23,24]; Reviewed in [25]) to precisely measure distances from the 5’ to 3’ end of single mRNA in untreated and translation inhibited cells (Figure 2a; [26,27]). These studies revealed mRNA ends are further apart in translating versus non-translating mRNA species. Furthermore, by using NCT, these studies in fixed cells and more recently Koch et al. in live cells showed that mRNA become more spread out as more ribosomes load [26–28], indicating the translation machinery is responsible for the spreading out. These data suggest either mRNA is not in a closed loop during translation or the closed-loop conformation is a transient event[29].
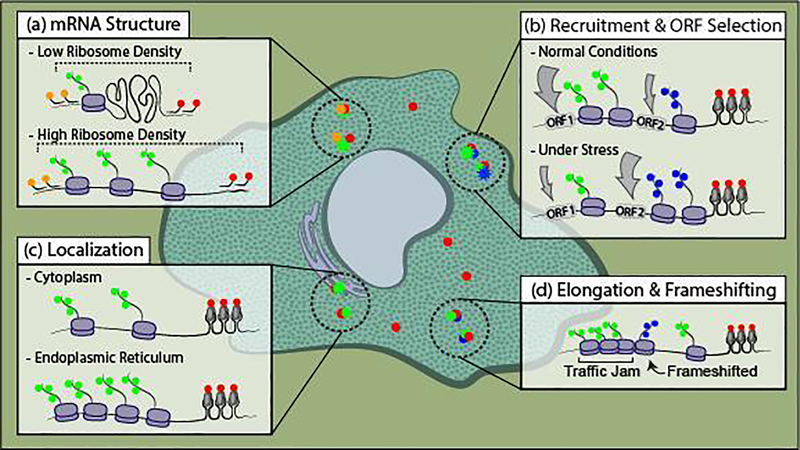
Figure 2. Recent applications of NCT to study active translation dynamics.
(a) NCT was combined with Single-Molecule Fluorescence In Situ Hybridization (smFISH) in fixed cells to measure the distance between the ends of the mRNA (red and orange). This was combined with antibody staining of nascent polypeptide chains (green) to count the translating ribosomes. By measuring the distance between 3’ (red) and 5’ (orange) mRNA probes and counting the number of translating ribosomes, 2D mRNA structures were quantified. (b) Two NCT systems were used to measure ribosome recruitment kinetics to a canonical (green) and non-canonical (blue) Open Reading Frame (ORF) and how ribosomes pick a start site once recruited to the mRNA. These assays were also conducted under stress conditions and differences in ribosome recruitment and ORF selection were measured. (c) By combining endoplasmic reticulum staining with NCT, mRNA localization and ribosomal content were determined. (d) NCT was used to investigate the heterogeneity of ribosomal elongation kinetics, including stalling at specific pause sites or frameshifting at a frameshift sequence. Both of which were associated with ribosomal traffic jams.
Ribosome Recruitment Mechanisms
Different mechanisms have evolved to recruit ribosomes to mRNA and facilitate translation. The canonical mechanism employed by most eukaryotic transcripts relies on the Cap Binding Complex (CBC) to recruit the first round of ribosomes after nuclear export [30]. To investigate this at the single-molecule level, Hoek et al. used NCT to image translation in wildtype cells and cells with eIF4E inactivated by ha4EBP1 over-expression (leaving only CBC for ribosome recruitment)[31]. According to this work, CBC can recruit multiple ribosomes in bursts during early rounds of translation before being replaced by eIF4E. This work highlights how NCT can be combined with inactivation/knock-down assays to dissect the contribution of a specific factor to translational gene regulation.
In addition to Cap-dependent ribosome recruitment, other non-canonical mechanisms have evolved to recruit ribosomes. One mechanism used by a wide range of viruses to hijack host ribosomes is an Internal Ribosomal Entry Site (IRES), an mRNA sequence that attracts ribosomal initiation factors [32,33]. Through the development of an NCT bicistronic reporter, Koch et al. uncovered the translation kinetics of Cap-dependent versus IRES-mediated translation (Figure 2b; [28]). According to this work, the IRES recruits two to three times fewer ribosomes than the Cap in normal conditions, but the balance shifts dramatically in favor of the IRES during stress.
Open Reading Frame Selection
Once the ribosome is recruited to an mRNA, it must scan the 5’ untranslated region (UTR) to find a start codon and thereby choose an open reading frame (ORF). Dr. Marilyn Kozak showed in classic bulk studies that the eukaryotic ribosome typically recognizes an AUG start codon within specific nucleotide contexts when choosing an ORF [34,35]. However, some transcripts have multiple ORFs and/or multiple AUGs, so the ribosome must decide where to initiate translation. NCT is now being used to investigate the stochasticity and heterogeneity of this choice. In one study, NCT was used to examine the regulation of the stress-response gene ATF4, the transcript of which contains two upstream ORFs [8]. According to this work, the third ORF, which codes for the ATF4 protein, was rarely chosen by ribosomes under normal conditions but was dramatically upregulated in a short-lived burst upon stress. NCT revealed these bursts were initiated in a matter of seconds and lasted for minutes at a time.
More recently, Boersma et al. used the MASH tag to investigate ribosomal start-site selection (Figure 2b; [14]). In their MASH reporter, nascent chains were labeled in different colors depending on the start site chosen. This revealed a surprising degree of heterogeneity amongst mRNA, with multiple start sites used intermittently and to varying degrees. Different 5’ UTRs resulted in different start site preferences, directly demonstrating a role for 5’ UTRs in dictating which start site is chosen. In addition, computational fits suggested ribosomes often reinitiate after upstream ORF translation.
Subcellular Localization and Translation
To regulate genes in a spatiotemporal manner, mRNAs are localized to specific cellular compartments for translation. Multiple groups have now used NCT to study translation dynamics in neurons ([7,19,36,37]; Reviewed in [38]). In particular, Wu et al. showed that translation is not necessarily repressed during active mRNA transport in neurons [7]. More recently, Cioni & Lin et al. showed that endosome-associated mRNA are actively translated [36]. Further, blocking the maturation of endosomes with a drug mutating an important endosomal protein inhibited translation without disrupting the mRNA association with endosomes.
Translation can also be targeted to specific subcellular locations. mRNAs encoding membrane and secreted proteins are translated mainly in the endoplasmic reticulum (ER), whereas mRNAs encoding cytosolic proteins are translated mainly in the cytosol. To further study this, Voigt et al. combined ER staining with NCT to track mRNA encoding cytosolic proteins (Figure 2c; [39]). Surprisingly, they found that a subset of these mRNA were localized to the ER during translation. Furthermore, they showed that mRNA localized to the ER were translated by more ribosomes on average, directly demonstrating subcellular localization can alter translation efficiency.
Heterogeneity in Ribosome Elongation and Frameshifting
The regulation of ribosomal elongation rates is an important form of translation control. For example, ribosomes could pause or stall at specific nucleotide sequences to modulate the folding of nascent chains (Reviewed in [40]). Pausing can also lead to ribosomal traffic jams that are known to trigger quality control and the unfolded protein response [41]. NCT is now being used to investigate ribosome elongation dynamics in the context of specific mRNA sequences. First, Yan et al. demonstrated that ribosomes pause for extended times at the XBP1 pause site (which is known to induce ribosome pausing [42]), after which they exit together in bursts (Figure 2d; [9]). More recently, Lyon et al. used the multi-frame tag to visualize frameshifting dynamics at the HIV-1 frameshift sequence (Figure 2d; [18]). This sequence contains a ribosomal pause site and a slippery sequence that causes ribosomes to occasionally slip from the 0 frame to the −1 frame. NCT revealed frameshifting occurs in bursts and is associated with long pauses at the frameshift sequence that induce ribosomal traffic jams.
Nonsense-Mediated mRNA Decay
Cells govern which mRNA should be translated by using mechanisms called mRNA surveillance to “survey” and eliminate wrong or mismanufactured mRNA. One type of mRNA surveillance called Nonsense-Mediated Decay (NMD) targets and destroys mRNA containing a Premature Termination Codon (PTC) [43]. To study how ribosomes impact NMD, Hoek et al. used NCT to detect ribosomes at reporter mRNA targeted for NMD by a PTC (Figure 3, Above; [31]). They used variations of mRNA reporters to test the impact of PTC location and context. Strikingly, NMD-triggered mRNA cleavage was found to be induced with equal probability by each translating ribosome. Also, some mRNA (10–20%, depending on the mRNA reporter) were completely NMD-resistant, suggesting heterogeneity in mRNA sensitivity to NMD.
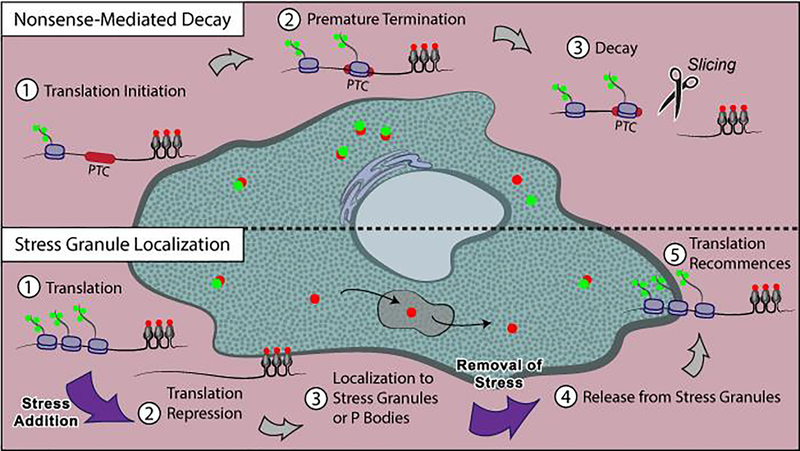
Figure 3. Recent applications of NCT to study translation repression dynamics.
Above: When a ribosome initiates translation, a green signal appears at a red-labeled mRNA (1). Once translating ribosomes reach a Premature Termination Codon (PTC), they can trigger Nonsense-Mediated Decay (NMD)(2). NMD slices an mRNA in half, which physically separates the green translating ribosome signal on the upstream half of the mRNA from the red mRNA tag signal (3). The count and dwell time of ribosomes and the mRNA slicing event can be determined from the green signal location and intensity. Below: After adding acute cell stress, translation is repressed, seen in the cell as loss of green translation signal (1–2). Translationally-silent, red signal-only mRNA can enter stress granules or P bodies (3). After the stress is removed, stress granules dissolve, releasing the mRNA (4). Then, translation recommences, as seen in the cell by green-labeled nascent peptide colocalizing with red mRNA (5).
siRNA-mediated mRNA Surveillance
Another type of mRNA surveillance is the small-interfering RNA (siRNA) pathway. To silence translation, siRNA locates a binding site on a target mRNA and slices it in half, promoting rapid mRNA degradation [44,45]. Horvathova et al. first demonstrated that siRNA-mediated mRNA silencing could be seen in real-time in a live-cell, single-mRNA assay called 3′-RNA End Accumulation during Turnover (TREAT), gaining insight into the timing and subcellular localization of these events [46]. More recently, Ruijtenberg et al. used NCT to show that siRNA silenced an mRNA most efficiently after a ribosome translated across the siRNA binding site [47]. Further, they showed that this was most likely due to translating ribosomes disrupting weak, intra-mRNA interactions, thereby changing the mRNA structure. These structural changes exposed binding sites for siRNA-enzyme complexes, promoting siRNA silencing.
Integrated Stress Response
To conserve resources during stress, cells repress translation and target mRNA to membraneless organelles called stress granules or P bodies (Reviewed in [48]). Moon & Morisaki et al. and Wilbertz et al. used NCT to study the dynamics of this process (Figure 3, Below; [49,50]). They showed translationally active mRNA can transiently interact with stress granules and P bodies, but only for a few seconds at a time. In contrast, non-translating mRNA can stably enter stress granules in a “dock and lock” model [49,51]. During stress, mRNA decay was inhibited, and mRNA immobilized within granules rarely transferred to other granules [46,50]. After stress, stress granules dissolved and released mRNA back into the cytoplasm, after which translation returned to near pre-stress levels [49,50]. These data suggest mRNA in stress granules can reinitiate translation.
Future direction
As the breadth of recent applications indicates, NCT is a rapidly maturing technology that is now beginning to shed light on traditionally hard-to-see aspects of translational gene regulation. The ability of NCT to resolve precisely when, where, and to what degree single mRNA are translated in living cells makes it ideally suited to address questions of translational heterogeneity and stochasticity. In the future we anticipate NCT will be used to clarify how various post-transcriptional mRNA modifications--chemical marks [52,53], subcellular and higher-order structure [54–57], and bound regulatory factors [58,59]--all impact translational control.
While the versatility of NCT has certainly improved with the additions of the MoonTag nanobody [14], frankenbody [17], MASH tag [14] and multi-frame tag [18], there is still much room for improvement. To facilitate multiplexed imaging, additional probes binding complementary epitopes are still needed. Since the frankenbody is modular, loop grafting can be used to generate new probes quickly [17]. Likewise, new nanobodies continue to be developed to bind epitopes in vivo, such as the BC2 [60] and ALFA nanobodies [61]. These may have future applications as NCT probes. As well, additional RNA tags could enhance multicolor applications. For this, riboglow [62,63] and fluorogenic Mango arrays [64] show great promise. Finally, as the probe and tag sets for NCT continue to expand, software will also need to improve to assist with experimental design and interpretation.
To fully exploit NCT and better decipher translational regulatory mechanisms, it will be important to combine it with other technologies. Since NCT marks translation sites, specific translational regulatory factors can be tagged in separate colors or depleted/enriched to directly visualize their impact on translation dynamics. This strategy was used, for example, to distinguish the contribution of the CBC from eIF4E during translation initiation [31]. Alternatively, in the future NCT could be combined with tethering assays [65] to force a factor to a translation site and directly visualize its perturbative effect. Tethering could even be made inducible by chemicals [66] or light [67] to perturb translation in a controlled, reversible fashion.
Ultimately, the final frontier for NCT is to move closer to imaging native translation in vivo. Antibodies against endogenous epitopes could light up native translation. A proof-of-concept of this approach was used to image the translation of endogenous PCNT protein in fixed cells [68]. With a full suite of genetically encoded probes and tags at hand, NCT in tissue or model organisms is also possible by genome engineering. However, advanced light-sheet microscopy [69] with adaptive optics [70] will likely be necessary to focus deep within specimen while maintaining good signal-to-noise and spatiotemporal resolution with minimal phototoxicity.
Regardless of how NCT precisely evolves or which biological questions it eventually helps tackle, its unique ability to light up single mRNA translation in living cells means its future will undoubtedly be bright.
Acknowledgements
A.L.K. and T.J.S. were supported by the National Institutes of Health (NIH) grant R35GM119728. C.A.C. was supported by the National Science Foundation NRT award DGE-1450032. G.G. was supported by the National Science Foundation CAREER grant MCB-1845761 to T.J.S.
Footnotes
Declarations of interest
None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
** of greatest interest
* of special interest
- 1.Tahmasebi S, Sonenberg N, Hershey JWB, Mathews MB: Protein Synthesis and Translational Control: A Historical Perspective. Cold Spring Harb Perspect Biol 2019, 11:a035584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ingolia NT: Ribosome Footprint Profiling of Translation throughout the Genome. Cell 2016, 165:22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ingolia NT, Hussmann JA, Weissman JS: Ribosome Profiling: Global Views of Translation. Cold Spring Harb Perspect Biol 2019, doi: 10.1101/cshperspect.a032698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chao JA, Yoon YJ, Singer RH: Imaging Translation in Single Cells Using Fluorescent Microscopy. Cold Spring Harb Perspect Biol 2012, 4:a012310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morisaki T, Lyon K, DeLuca KF, DeLuca JG, English BP, Zhang Z, Lavis LD, Grimm JB, Viswanathan S, Looger LL, et al. : Real-time quantification of single RNA translation dynamics in living cells. Science 2016, 352:1425–1429. [DOI] [PubMed] [Google Scholar]
- 6.Pichon X, Bastide A, Safieddine A, Chouaib R, Samacoits A, Basyuk E, Peter M, Mueller F, Bertrand E: Visualization of single endogenous polysomes reveals the dynamics of translation in live human cells. J Cell Biol 2016, 214:769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu B, Eliscovich C, Yoon YJ, Singer RH: Translation dynamics of single mRNAs in live cells and neurons. Science 2016, 352:1430–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang C, Han B, Zhou R, Zhuang X: Real-Time Imaging of Translation on Single mRNA Transcripts in Live Cells. Cell 2016, 165:990–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan X, Hoek TA, Vale RD, Tanenbaum ME: Dynamics of Translation of Single mRNA Molecules In Vivo. Cell 2016, 165:976–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coulon A, Chow CC, Singer RH, Larson DR: Eukaryotic transcriptional dynamics: from single molecules to cell populations. Nat Rev Genet 2013, 14:572–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD: A Protein-Tagging System for Signal Amplification in Gene Expression and Fluorescence Imaging. Cell 2014, 159:635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayashi-Takanaka Y, Yamagata K, Wakayama T, Stasevich TJ, Kainuma T, Tsurimoto T, Tachibana M, Shinkai Y, Kurumizaka H, Nozaki N, et al. : Tracking epigenetic histone modifications in single cells using Fab-based live endogenous modification labeling. Nucleic Acids Res 2011, 39:6475–6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morisaki T, Stasevich TJ: Quantifying Single mRNA Translation Kinetics in Living Cells. Cold Spring Harb Perspect Biol 2018, 10:a032078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boersma S, Khuperkar D, Verhagen BMP, Sonneveld S, Grimm JB, Lavis LD, Tanenbaum ME: Multi-Color Single-Molecule Imaging Uncovers Extensive Heterogeneity in mRNA Decoding. Cell 2019, 178:458–472.* To study translation start site selection, this work first created a new translation labeling system called the MoonTag. With the SunTag and the MoonTag systems in alternate reading frames, they discovered that alternative start site selection happens frequently. Different 5’ UTRs resulted in different start site preferences, directly demonstrating a role for 5’ UTRs in dictating which start site is chosen. In addition, computational fits suggested ribosomes often reinitiate after upstream ORF translation.
- 15.Hulsik DL, Liu Y, Strokappe NM, Battella S, Khattabi ME, McCoy LE, Sabin C, Hinz A,Hock M, Macheboeuf P, et al. : A gp41 MPER-specific Llama VHH Requires a Hydrophobic CDR3 for Neutralization but not for Antigen Recognition. PLOS Pathog 2013, 9:e1003202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Viswanathan S, Williams ME, Bloss EB, Stasevich TJ, Speer CM, Nern A, Pfeiffer BD, Hooks BM, Li W-P, English BP, et al. : High-performance probes for light and electron microscopy. Nat Methods 2015, 12:568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao N, Kamijo K, Fox PD, Oda H, Morisaki T, Sato Y, Kimura H, Stasevich TJ: A genetically encoded probe for imaging nascent and mature HA-tagged proteins in vivo. Nat Commun 2019, 10:1–16.** Here, a new genetically encodable scFv probe called the anti-HA frankenbody was developed. This probe can be expressed in cells and was used for imaging and tracking HA-tagged proteins in vivo and for performing NCT with the HA-spaghetti monster tag.
- 18.Lyon K, Aguilera LU, Morisaki T, Munsky B, Stasevich TJ: Live-Cell Single RNA Imaging Reveals Bursts of Translational Frameshifting. Mol Cell 2019, 75:172–183.e9.** To study ribosomal frameshitting, this work created an NCT multi-frame tag with a frameshift sequence from HIV. A small number of mRNA frameshifted for extended times in bursts, and frameshifting was associated with ribosomal traffic jams and mRNA-mRNA dimers.
- 19.Fonkeu Y, Kraynyukova N, Hafner A-S, Kochen L, Sartori F, Schuman EM,Tchumatchenko T: How mRNA Localization and Protein Synthesis Sites Influence Dendritic Protein Distribution and Dynamics. Neuron 2019, 103:1109–1122.e7. [DOI] [PubMed] [Google Scholar]
- 20.Voigt F, Gerbracht JV, Boehm V, Horvathova I, Eglinger J, Chao JA, Gehring NH: Detection and quantification of RNA decay intermediates using XRN1-resistant reporter transcripts. Nat Protoc 2019, 14:1603–1633.* (ER localization) Here, the authors used NCT to investigate translation at different subcellular locations. They revealed that cytoplasmic proteins can localize and translate at the endoplasmic reticulum and those translation sites were loaded by more ribosomes than the cytoplasmic mRNA.
- 21.Aguilera LU, Raymond W, Fox ZR, May M, Djokic E, Morisaki T, Stasevich TJ, Munsky B:Computational design and interpretation of single-RNA translation experiments. PLOS Comput Biol 2019, 15:e1007425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Svitkin YV, Evdokimova VM, Brasey A, Pestova TV, Fantus D, Yanagiya A, Imataka H, Skabkin MA, Ovchinnikov LP, Merrick WC, et al. : General RNA-binding proteins have a function in poly(A)-binding protein-dependent translation. EMBO J 2009, 28:58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Femino AM, Fay FS, Fogarty K, Singer RH: Visualization of Single RNA Transcripts in Situ. Science 1998, 280:585–590. [DOI] [PubMed] [Google Scholar]
- 24.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S: Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods 2008, 5:877–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taliaferro JM: Classical and emerging techniques to identify and quantify localized RNAs. WIREs RNA 2019, 10:e1542. [DOI] [PubMed] [Google Scholar]
- 26.Khong A, Parker R: mRNP architecture in translating and stress conditions reveals an ordered pathway of mRNP compactionTranslating and nontranslating mRNP architecture. J Cell Biol 2018, 217:4124–4140.* Here, the distances between the ends of mRNA were measured using fluorescent probes. They determined that mRNA take on an open conformation when translated. Their results corroborate [27].
- 27.Adivarahan S, Livingston N, Nicholson B, Rahman S, Wu B, Rissland OS, Zenklusen D: Spatial Organization of Single mRNPs at Different Stages of the Gene Expression Pathway. Mol Cell 2018, 72:727–738.e5.* Here, the distances between the ends of mRNA were measured using smFISH. They discovered that mRNA are stretched out while being translated. Further, more translating ribosomes on an mRNA correlated with the mRNA ends being further apart. Their results corroborate [26].
- 28.Koch A, Aguilera L, Morisaki T, Munsky B, Stasevich TJ: Quantifying the spatiotemporal dynamics of IRES versus Cap translation with single-molecule resolution in living cells. bioRxiv 2020, doi: 10.1101/2020.01.09.900829.* Here, a bicistronic NCT biosensor was developed to light up in different colors depending on whether or not translation occurred in a Cap-dependent or IRES-mediated manner. This revealed the EMCV IRES recruits fewer ribosomes than the Cap in normal conditions, but that balance shifts to favor the IRES during stress. Also, the biosensor was used to demonstrate that actively translating mRNA become more stretched out as more ribosomes load (Currently a preprint).
- 29.Vicens Q, Kieft JS, Rissland OS: Revisiting the closed loop model and the nature of mRNA 5′–3′ communication. Mol Cell 2018, 72:805–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gonatopoulos-Pournatzis T, Cowling VH: Cap-binding complex (CBC). Biochem J 2014, 457:231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoek TA, Khuperkar D, Lindeboom RGH, Sonneveld S, Verhagen BMP, Boersma S, Vermeulen M, Tanenbaum ME: Single-Molecule Imaging Uncovers Rules Governing Nonsense-Mediated mRNA Decay. Mol Cell 2019, 75:324–339.e11. )** NCT was used to visualize Nonsense Mediated Decay of mRNA. Decay was triggered when ribosomes reached a premature stop codon in a reporter mRNA.
- 32.Filbin ME, Kieft JS: Toward a structural understanding of IRES RNA function. Curr Opin Struct Biol 2009, 19:267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gebauer F, Hentze MW: IRES unplugged. Science 2016, 351:228–228. [DOI] [PubMed] [Google Scholar]
- 34.Kozak M: Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 1986, 44:283–292. [DOI] [PubMed] [Google Scholar]
- 35.Kozak M: At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J Mol Biol 1987, 196:947–950. [DOI] [PubMed] [Google Scholar]
- 36.Cioni J-M, Lin JQ, Holtermann AV, Koppers M, Jakobs MAH, Azizi A, Turner-Bridger B, Shigeoka T, Franze K, Harris WA, et al. : Late Endosomes Act as mRNA Translation Platforms and Sustain Mitochondria in Axons. Cell 2019, 176:56–72.e15.* Here, the authors studied endosomal trafficking to mitochondria in neurons. Among other results, they used NCT to discover that endosome maturation drove translation of specific mRNA docked to endosomes.
- 37.Langille JJ, Ginzberg K, Sossin WS: Polysomes identified by live imaging of nascent peptides are stalled in hippocampal and cortical neurites. Learn Mem 2019, 26:351–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Latallo MJ, Livingston NM, Wu B: Translation imaging of single mRNAs in established cell lines and primary cultured neurons. Methods 2019, 162–163:12–22. [DOI] [PubMed] [Google Scholar]
- 39.Voigt F, Zhang H, Cui XA, Triebold D, Liu AX, Eglinger J, Lee ES, Chao JA, Palazzo AF: Single-Molecule Quantification of Translation-Dependent Association of mRNAs with the Endoplasmic Reticulum. Cell Rep 2017, 21:3740–3753.* (ER localization) Here, the authors used NCT to investigate translation at different subcellular locations. They revealed that cytoplasmic proteins can localize and translate at the endoplasmic reticulum and those translation sites were loaded by more ribosomes than the cytoplasmic mRNA.
- 40.Collart MA, Weiss B: Ribosome pausing, a dangerous necessity for co-translational events. Nucleic Acids Res [date unknown], doi: 10.1093/nar/gkz763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Juszkiewicz S, Chandrasekaran V, Lin Z, Kraatz S, Ramakrishnan V, Hegde RS: ZNF598 Is a Quality Control Sensor of Collided Ribosomes. Mol Cell 2018, 72:469–481.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ingolia NT, Lareau LF, Weissman JS: Ribosome Profiling of Mouse Embryonic Stem Cells Reveals the Complexity and Dynamics of Mammalian Proteomes. Cell 2011, 147:789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shoemaker CJ, Green R: Translation drives mRNA quality control. Nat Struct Mol Biol 2012, 19:594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC: Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391:806–811. [DOI] [PubMed] [Google Scholar]
- 45.Hamilton AJ, Baulcombe DC: A Species of Small Antisense RNA in Posttranscriptional Gene Silencing in Plants. Science 1999, 286:950–952. [DOI] [PubMed] [Google Scholar]
- 46.Horvathova I, Voigt F, Kotrys AV, Zhan Y, Stadler MB, Giorgetti L, Chao JA: The Dynamics of mRNA Turnover Revealed by Single-Molecule Imaging in Single Cells. Mol Cell 2017, 68:615–625.e9.* This work developed a single-mRNA biosensor for mRNA decay via fluorescence imaging. They used the biosensor to visualize various types of decay in live human cells, including siRNA-mediated mRNA silencing.
- 47.Ruijtenberg S, Sonneveld S, Cui TJ, Logister I, de Steenwinkel D, Xiao Y, MacRae IJ, Joo C, Tanenbaum ME: mRNA structural dynamics shape Argonaute-target interactions. bioRxiv 2019, doi: 10.1101/822452.* Using NCT, this work studied the impact of translating ribosomes on siRNA/Argonaute-mediated mRNA decay. They found that translation increased the probability of Argonaute targeting to an mRNA, thus increasing mRNA decay (Currently a preprint).
- 48.Treeck BV, Parker R: Principles of Stress Granules Revealed by Imaging Approaches. Cold Spring Harb Perspect Biol 2019, 11:a033068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moon SL, Morisaki T, Khong A, Lyon K, Parker R, Stasevich TJ: Multicolor single-molecule tracking of mRNA interactions with RNP granules. Nat Cell Biol 2019, 21:162–168.** Using NCT, the authors visualized translating mRNA in cells undergoing stress. They saw translation shutoff before mRNA localize to stress granules, and some mRNA form stable docking interactions with the granules. After stress relief, the stress granules dissolved then translation resumed. Their results corroborate [50].
- 50.Wilbertz JH, Voigt F, Horvathova I, Roth G, Zhan Y, Chao JA: Single-Molecule Imaging of mRNA Localization and Regulation during the Integrated Stress Response. Mol Cell 2019, 73:946–958.e7.** This work saw that translation shut down at NCT reporter mRNA soon after cell stress was induced. Stress granules stored mRNA during stress but did not impact mRNA decay. Their results corroborate [49].
- 51.Pitchiaya S, Mourao MDA, Jalihal AP, Xiao L, Jiang X, Chinnaiyan AM, Schnell S, Walter NG: Dynamic Recruitment of Single RNAs to Processing Bodies Depends on RNA Functionality. Mol Cell 2019, 74:521–533.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simms CL, Hudson BH, Mosior JW, Rangwala AS, Zaher HS: An Active Role for the Ribosome in Determining the Fate of Oxidized mRNA. Cell Rep 2014, 9:1256–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoernes TP, Hüttenhofer A, Erlacher MD: mRNA modifications: Dynamic regulators of gene expression? RNA Biol 2016, 13:760–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Babendure JR, Babendure JL, Ding J-H, Tsien RY: Control of mammalian translation by mRNA structure near caps. RNA 2006, 12:851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wen J-D, Lancaster L, Hodges C, Zeri A-C, Yoshimura SH, Noller HF, Bustamante C, Tinoco I: Following translation by single ribosomes one codon at a time. Nature 2008, 452:598–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jung H, Gkogkas CG, Sonenberg N, Holt CE: Remote Control of Gene Function by Local Translation. Cell 2014, 157:26–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buxbaum AR, Haimovich G, Singer RH: In the right place at the right time: visualizing and understanding mRNA localization. Nat Rev Mol Cell Biol 2015, 16:95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu B, Buxbaum AR, Katz ZB, Yoon YJ, Singer RH: Quantifying Protein-mRNA Interactions in Single Live Cells. Cell 2015, 162:211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simsek D, Barna M: An emerging role for the ribosome as a nexus for posttranslational modifications. Curr Opin Cell Biol 2017, 45:92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Braun MB, Traenkle B, Koch PA, Emele F, Weiss F, Poetz O, Stehle T, Rothbauer U: Peptides in headlock – a novel high-affinity and versatile peptide-binding nanobody for proteomics and microscopy. Sci Rep 2016, 6:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Götzke H, Kilisch M, Martínez-Carranza M, Sograte-Idrissi S, Rajavel A, Schlichthaerle T, Engels N, Jungmann R, Stenmark P, Opazo F, et al. : The ALFA-tag is a highly versatile tool for nanobody-based bioscience applications. Nat Commun 2019, 10:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Braselmann E, Wierzba AJ, Polaski JT, Chromiński M, Holmes ZE, Hung S-T, Batan D, Wheeler JR, Parker R, Jimenez R, et al. : A multicolor riboswitch-based platform for imaging of RNA in live mammalian cells. Nat Chem Biol 2018, 14:964–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Braselmann E, Stasevich TJ, Lyon K, Batey RT, Palmer AE: Detection and quantification of single mRNA dynamics with the Riboglow fluorescent RNA tag. bioRxiv 2019, doi: 10.1101/701649. [DOI] [Google Scholar]
- 64.Cawte AD, Unrau PJ, Rueda DS: Live Cell Imaging of Single RNA Molecules with Fluorogenic Mango II Arrays. bioRxiv 2019, doi: 10.1101/840082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baron-Benhamou J, Gehring NH, Kulozik AE, Hentze MW: Using the lambdaN peptide to tether proteins to RNAs. Methods Mol Biol Clifton NJ 2004, 257:135–154. [DOI] [PubMed] [Google Scholar]
- 66.Stanton BZ, Chory EJ, Crabtree GR: Chemically induced proximity in biology and medicine. Science 2018, 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spiltoir JI, Tucker CL: Photodimerization systems for regulating protein–protein interactions with light. Curr Opin Struct Biol 2019, 57:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sepulveda G, Antkowiak M, Brust-Mascher I, Mahe K, Ou T, Castro NM, Christensen LN, Cheung L, Jiang X, Yoon D, et al. : Co-translational protein targeting facilitates centrosomal recruitment of PCNT during centrosome maturation in vertebrates. eLife 2018, 7:e34959.* During mitosis, the translation of localized mRNA causes a local enrichment of the cytoplasmic PCNT protein at centrosomes. This work used antibodies to label both ends of endogenous PCNT proteins to study native translation.
- 69.Power RM, Huisken J: A guide to light-sheet fluorescence microscopy for multiscale imaging. Nat Methods 2017, 14:360–373. [DOI] [PubMed] [Google Scholar]
- 70.Ji N: Adaptive optical fluorescence microscopy. Nat Methods 2017, 14:374–380. [DOI] [PubMed] [Google Scholar]