Abstract
Background.
Microbiologists are valued for their time-honed skills in image analysis including identification of pathogens and inflammatory context in Gram stains, ova and parasite preparations, blood smears, and histopathological slides. They also must classify colonial growth on a variety of agar plates for triage and workup. Recent advances in image analysis, in particular application of artificial intelligence (AI), have the potential to automate these processes and support more timely and accurate diagnoses.
Objectives.
To review current artificial intelligence-based image analysis as applied to clinical microbiology and discuss future trends in the field.
Sources.
Material sourced for this review included peer-reviewed literature annotated in the PubMed or Google Scholar databases and preprint articles from bioRxiv. Articles describing use of AI for analysis of images used in infectious disease diagnostics were reviewed.
Content.
We describe application of machine learning towards analysis of different types of microbiological image data. Specifically, we outline progress in smear and plate interpretation and potential for AI diagnostic applications in the clinical microbiology laboratory.
Implications.
Combined with automation, we predict that AI algorithms will be used in the future to pre-screen and pre-classify image data, thereby increasing productivity and enabling more accurate diagnoses through collaboration between the AI and microbiologist. Once developed, image-based AI analysis is inexpensive and amenable to local and remote diagnostic use.
Introduction
Image interpretation is fundamental to clinical microbiology laboratory diagnostics. For example, Gram stains, fecal and blood smears, and histopathological slides all need to be interpreted by highly trained microbiologists and/or pathologists. These specimens provide critical diagnostic information including the presence and categories of microbes, the host inflammatory response, and specimen quality. Taken in clinical context, this information helps establish whether an infection is present and often suggests a differential diagnosis that can direct therapy.
Microbiologists also classify growth of colonies on agar plates. Expertise is required to distinguish potential pathogens from one another and from normal flora. Multiple differential media including chromogenic agar must be correctly interpreted. These discriminatory observations are the basis for the type and extent of microbial workup.
A chronic shortage of medical laboratory scientists provides an additional challenge in the clinical microbiology laboratory [1]. Therefore, automation of the experientially demanding, visual interpretative tasks underlying workflow in the clinical microbiology laboratory has appeal. Until recently, options for automating image-based interpretative tasks were unsatisfactory. However, new artificial intelligence (AI) algorithms that excel in image discrimination now open up the potential for automated clinical microbiology interpretation with associated gains in efficiency and diagnostic accuracy.
AI general concepts
For the purposes of this review, AI is a set of rules or algorithms that allow computers (the artifice) to make decisions with features that simulate human intelligence. Intelligence implies an ability to learn: that is to alter underlying computer code to enhance future task performance. A significant advance in the AI field was the development of complex, multi-layered AI architectures known as deep learning algorithms. A subset of these algorithms, so called convolutional neural networks (CNNs), are highly interconnected networks modeled after the human optical cortex and excel at image classification [2]. For example, these algorithms have been used as aids in diagnostic interpretation of image data to detect markers of cancer in histologcal sections of breast biopsies [3], interpret echocardiograms [4] and identify metastases in brain magnetic resonance imaging datasets (MRI) [5].
By analogy to human learning, interconnected nodes are variably stimulated based on input, for example, images. During learning, connections associated with an enhanced accuracy of image classification are reinforced, while connections that lead to erroneous categorization are weakened [6]. In essence, the wiring of our brains and by analogy computer code in CNNs is optimized based on experience.
During supervised training of CNNs for diagnostic interpretation, a training set, a large number of human classified images, is provided as input. The algorithm classifies each image and compares its accuracy with the human classification. Through a process of trial-and-error, the CNN adjusts its neural network (i.e., its programming) thousands or even millions of times, to optimize its accuracy. At some point, improvements in accuracy level off and the algorithm is said to be trained.
The quality of training material is critical to development of robust interpretative algorithms. Without optimized training material, learning can go off track with surprising results. As an example, if we were to train a CNN to recognize a coffee mug, we would train it on a large number of images of coffee mugs. However, a subset of coffee cups in the training set may be sitting on countertops. The algorithm may then inappropriately associate features of a countertop with a coffee mug classification. This misdirected learning is termed overfitting and is indicative of training with inadequate numbers of images sufficient to allow relevant features in a classified image to be ascertained. Overfitting is diagnosed using a test set, which is an independent set of images not used during training and which should be representative of the type and variety of images likely to be observed in the real world.
Overfitting can be addressed in a variety of ways, one of which is provision of more data during training. In our example, we could supplement our training set with images of coffee mugs in sinks, in a hand, or on a bookshelf. We would also provide many examples of objects that are not coffee mugs including countertops without coffee mugs. Through these strategies, we train the algorithm to focus on the defining features of the classified objects.
Search strategy
With this general introduction, we now turn to practical applications in the clinical microbiology laboratory. We reviewed the literature available in PubMed and Google Scholar through search terms including “artificial intelligence” and “clinical microbiology” on September 21, 2019. Seven articles were identified, only two of which pertained to image analysis (one review and our own publication on use of AI to interpret Gram stains). Therefore, we expanded our search and found that “Machine-learning and bacterial vaginosis,” and “Malaria and machine learning” identified several other AI-based smear analysis citations. We therefore start by discussing smear interpretation and follow with discussion of use of AI for microbial agar plate analysis, where introduction in clinical systems has largely preceded exploration in the literature.
Smear interpretation
In 2018, Smith, Kang and Kirby published a proof of principle study on use of a convolutional neural network (CNN) for automated interpretation of blood culture Gram stains [7]. Several key aspects of future use of AI in clinical microbiology can be abstracted from this study. Here, the authors used a CNN, which, as mentioned previously, excels at image categorization. However, training such a complex network is very computationally intensive, typically requiring specialized infrastructure (https://arxiv.org/abs/1512.00567). By analogy, we liken it to educating a child from primary school through college. However, if this young adult was originally trained to be an economist but needed to change careers, it would not have to start with pre-school again. Instead, its existing education would be used as a basis for training in a new discipline, let’s say as a clinical microbiologist.
Therefore, Smith et al. started with a CNN called Inception 3.0, previously trained by Google to recognize everyday objects (https://arxiv.org/abs/1512.00567), and retrained only the final layers to recognize several common Gram stain morphologies (Gram-negative rods, Gram-positive cocci in clusters and Gram-positive cocci in chains) in positive blood culture smears. In essence, the “college education” was kept intact and used as a foundation for microbiology specialty training. This re-education is referred to in the field as transfer learning and is orders of magnitude less computationally intensive than complete end-to-end training of the CNN. In total, 100,000 classified image crops were fed into the CNN for training and across all Gram stain categories yielded a ~95% crop classification accuracy and a composite 92.5% whole slide classification accuracy.
However, the ultimate goal is to perform interpretation with an accuracy equal to a skilled microbiologist (i.e. >99%) [8]. An important feature of CNNs is that accuracy increases with the size of training sets, which may be comprised of millions of images[9]. It may be difficult or impossible to collect and classify such immense image datasets. Therefore, alternative strategies must be pursued. One of these strategies is data augmentation, a method by which a computer program transforms training images by rotation, inversion, displacement, or distortion to increase the variety of images in a dataset. However, data augmentation itself is computationally intensive, as each image must be transformed individually. With a more powerful computer containing a dedicated graphics processing unit (GPU), Smith and Kirby performed data augmentation, converting 100,000 images crops into 40 million transformed image crops. Training on this augmented data set increased crop accuracy calls to >99% (Smith and Kirby, unpublished data).
We envision that other types of microscope-based clinical microbiology diagnostics would also benefit from application of machine learning. For example, diagnosis of bacterial vaginosis is one potential high volume activity that could be performed inexpensively by training on smears classified by Nugent rules [10, 11]. Song et. al. used segmentation and morphological analysis of individual bacterial cells to classify them based on the Nugent criteria. Using 10 images per slide, they achieved a 79.1% accuracy compared to 90.7% for experienced technologists interpreting the same images. Although not yet equal to a trained technologist, this work provides a proof-of-concept for diagnosis of bacterial vaginosis leveraging CNN capabilities.
In addition to observation of bacterial smears, there is significant potential for use of AI in parasite diagnostics, with almost all published effort thus far devoted to malaria. The gold standard for Plasmodium detection and speciation is microscopic observation of stained thick and thin blood smears. Due to a paucity of trained individuals in endemic areas, several studies have explored alternative AI models to automate image interpretation. Such models routinely achieved sensitivity and specificities of >95% on a per-image basis when data are collected and interpreted in a research setting [12–17]. However, to our knowledge, none have yet been thoroughly evaluated clinically, and therefore, it is unclear how well these methods will perform during diagnostic implementation, especially in resource-limited settings. Nevertheless, these studies provide proof-of-concept that AI can be used for identification of Plasmodium spp.
It is important to note that automated slide interpretation depends on availability of automated microscopy to support high quality, efficient, and targeted acquisition of images. Unfortunately, current automated slide scanners used in anatomic pathology are inadequate to capture the narrow focal plane of microbiology images. Introduction of automated microscopy capabilities in clinical microbiology practice will likely occur in the context of microbiology laboratory automation (MLA), and we expect AI-based slide interpretation to co-evolve with these technologies.
Plate interpretation
The second major type of image-based AI interpretation relevant to clinical microbiology involves analysis of microbial growth on agar plates. In contrast to automated slide imaging, automation of plate inoculation, handling, and imaging has already been incorporated into existing MLA systems. Yet several challenges remain in AI-based interpretation of microbial growth and incorporation within total laboratory automation workflow [18].
For example, individual colonies must be accurately demarcated and different colony types must be distinguished and quantified. This poses a computational challenge because colony morphology varies based on bacterial density and media type [19]. Furthermore, colonies are not always well separated, making it difficult to determine exactly how many morphotypes are present. However, it is technically much simpler to detect presence or absence of colonies, as segmentation and enumeration is not required. Given that many plates in the microbiology laboratory have no growth, reliable AI-based decisions on growth versus no growth would be very useful in limiting the number of negative cultures that need technologist attention.
As such, the first iteration of plate reading AI technology has been applied in two areas: detection of presence or absence of growth in urine cultures and determining the presence of specific target organisms of clinical or epidemiological importance on chromogenic agar plates based on indicator color. Examples of the latter include AI-based screening of specific chromogenic medium for group-A Streptococcus, vancomycin-resistant Enterococcus, and methicillin-resistant Staphylococcus aureus [20–22].
Urine culture workup, including plate interpretation, is likely the first culture type to be nearly or completely automated for several reasons. First, urine can be received in standardized containers, which can be easily processed by MLA systems. Second, pathogens in urine cultures are largely predictable so this limits the need to recognize all possible organisms, and chromogenic agar methods may provide additional cues that improve the ability of the AI to differentiate among these pathogens [23]. Third, mixed, polymicrobial cultures are typically not worked up, meaning that the AI needs to recognize the presence of mixed cultures but not necessarily to distinguish and categorize multiple admixed colonial morphologies.
The future of AI in clinical microbiology
We envision that implementation of AI in the clinical microbiology laboratory will follow from the aforementioned proof-of-concept studies. For example, respiratory Gram stains could be interpreted through automated quantification and discrimination of inflammatory, epithelial, and bacterial cell morphologies. Further, AI may also find a niche in other experientially demanding processes such as readout of fluorescent acid fast bacillus smears, morphological diagnosis of fungal adhesive tape preparations, or stool trichrome smears.
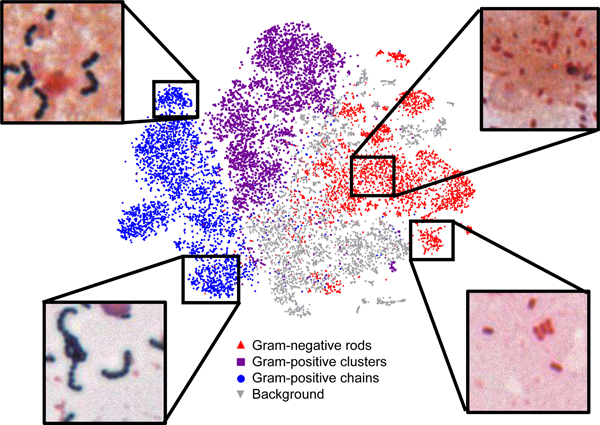
Importantly, AI is capable of generating quantitative and nuanced information in the form of relative probabilities for each potential classification. For example, an image could be interpreted as 90% probability of Gram-positive cocci in clusters and 10% probability of Gram-positive cocci in chains. It is evident from a two-dimensional representation of their classified Gram stain dataset that an even more nuanced readout is possible (Smith and Kirby, Fig. 1, unpublished data). Specifically, subtle distinctions in organism shape and size can be differentiated. This suggests that with further training therapeutically important diagnostic distinctions could likely be made between organisms with similar Gram stain morphologies: for instance, identification of Gram-positive diplococcci suggestive of pneumococci; short Gram-positive rods suggestive of Listeria; or minute Gram-negative coccobacilli suggestive of Brucella or Francisella.
Figure 1. Two-dimensional representation of Gram-stain classifications highlights ability to further subcategorize data.
Here, a t-statistic nearest neighbor embedding (tSNE) algorithm was used to visualize Gram-stain crop classification data in two-dimensions. Clusters of Gram-positive cocci in long (left, lower) or short chains (left, upper) are readily recognizable. Similarly, Gram-negative rods (right, lower) can be distinguished from coccobacilli (right, upper). This information could later be used to provide additional probabilistic subclassification of organisms.
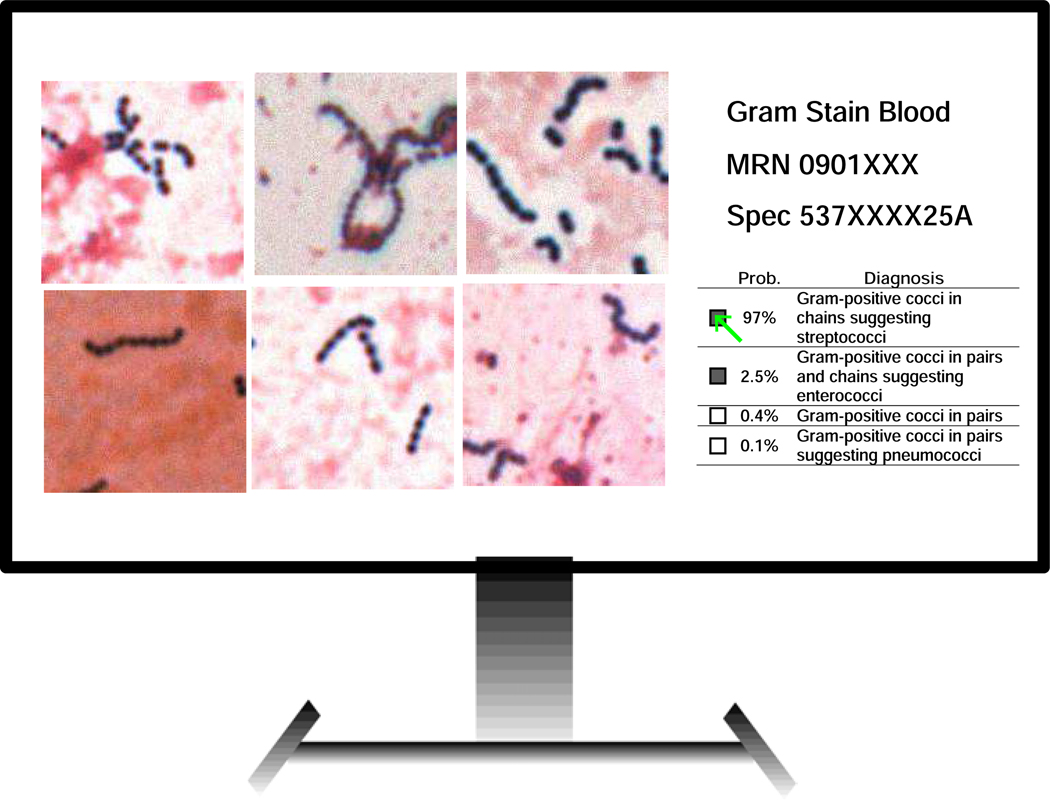
To incorporate this probabilistic output into clinical practice, Smith and Kirby conceived a platform called Technologist Assist (TA) in which a selection of AI-classified image crops are displayed on a computer screen for review along with probabilistic interpretations (See Figs. 2 and 3) [7, 24].
Figure 2. Technologist Assist.
This platform is envisioned as a way for AI and clinical laboratory scientists to collaborate. After analysis of a smear by a trained AI, diagnostic image crops are displayed for technologist review along with a probabilistic differential Gram stain diagnosis. The technologist can then review images and select one or more diagnoses, which would then cross over from the laboratory information system to the patient report. The microbiologist also has the option of reviewing the smear directly in the presumptively rare instances where there is discordance between their assessment of the image crops and the offered AI interpretations or if there were diagnostic uncertainty.
Figure 3. Example of probabilistic AI-assisted Gram stain reports.
The probabilistic report provides a weighted differential diagnosis that can draw attention to infections that require special therapeutic intervention.
Can we afford it?
We close with a final word about cost-effectiveness. Classification of relevant pathogens in Gram stain or other smears by AI could potentially be supplanted by emerging molecular platforms. However, these technologies are expensive on a per assay basis and currently provide only qualitative detection of select pathogens. Direct analysis still provides an immediate understanding of the complexity and context of infection, which may not be otherwise amenable to molecular diagnostic interpretation.
It should be noted that training of AI is labor and computationally intensive.. Training in image-based infectious disease diagnostics relies on image data sets curated with great effort through manual interpretation of images by skilled clinical microbiologists. After training, accuracy and robustness must be extensively verified under real world conditions in comparison again to diagnostic interpretation by trained microbiologists. Acceptable accuracy thresholds for implementation will need to be established based on intended use and risks associated with false positive and false negative results. For example, replacement of human interpretation would require very high diagnostic sensitivity and specificity, whereas implementations that provide diagnostic assistance (for example, prescreening for potential pathogens) with an ultimate skilled human interpretation of curated image data prior to diagnosis would require a high sensitivity with tolerance for a more relaxed specificity.
However, after this resource intensive training, validation, and implementation, AI-based diagnosis is essentially free and image analysis can be performed on lower end computers that already exist in most microbiology laboratories. However, an intrinsic property of AI algorithms is that they can provide predictions only for data sets similar to those used in training. For example, a model trained to interpret colonies on plate media, chromogenic media or slides stained with reagents from a specific manufacturer may fail to function if an alternative manufacturer of media or stain were used. Therefore, it is important to consider the training dataset for any given model and retrain as necessary if new conditions are used.
It is also evident that AI-assisted image analysis can be performed at a distance, only requiring that the remote site have a microscope and a way to transmit images to the AI using the internet or cellular service, i.e., telemicrobiology. Therefore, AI-based image analysis in infectious disease diagnostics will likely find its niche by providing answers in health care systems both large and small where other techniques are not cost effective, immediate, or comprehensive.
Acknowledgements.
This work was conducted with support from Harvard Catalyst | The Harvard Clinical and Translational Science Center (National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health Award UL1 TR001102) and financial contributions from Harvard University and its affiliated academic healthcare centers. K.P.S. was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number F32 AI124590. We thank NVIDIA for supplying a graphic processing unit through an unrestricted grant that enabled data augmentation studies.
Conflict of interest disclosure
Dr. Kirby reports grants from US National Institutes of Health, non-financial support from NVIDIA, during the conduct of the study; In addition, Dr. Kirby has a patent WO2019104003A1 pending. Dr. Smith reports grants from US National Institutes of Health, during the conduct of the study; In addition, Dr. Smith has a patent WO2019104003A1 pending.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Garcia E, Kundu I, Ali A, Soles R, The American Society for Clinical Pathology’s 20162017 Vacancy Survey of Medical Laboratories in the United States, Am J Clin Pathol 149(5) (2018) 387–400. [DOI] [PubMed] [Google Scholar]
- [2].LeCun Y, Bengio Y, Hinton G, Deep learning, Nature 521(7553) (2015) 436–44. [DOI] [PubMed] [Google Scholar]
- [3].Ehteshami Bejnordi B, Mullooly M, Pfeiffer RM, Fan S, Vacek PM, Weaver DL, Herschorn S, Brinton LA, van Ginneken B, Karssemeijer N, Beck AH, Gierach GL, van der Laak J, Sherman ME, Using deep convolutional neural networks to identify and classify tumor-associated stroma in diagnostic breast biopsies, Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc 31(10) (2018) 15021512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Madani A, Arnaout R, Mofrad M, Arnaout R, Fast and accurate view classification of echocardiograms using deep learning, NPJ digital medicine 1 (2018) pii: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Liu Y, Stojadinovic S, Hrycushko B, Wardak Z, Lau S, Lu W, Yan Y, Jiang SB, Zhen X, Timmerman R, Nedzi L, Gu X, A deep convolutional neural network-based automatic delineation strategy for multiple brain metastases stereotactic radiosurgery, PloS one 12(10) (2017) e0185844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kandel ER, Dudai Y, Mayford MR, The molecular and systems biology of memory, Cell 157(1) (2014) 163–86. [DOI] [PubMed] [Google Scholar]
- [7].Smith KP, Kang AD, Kirby JE, Automated Interpretation of Blood Culture Gram Stains by Use of a Deep Convolutional Neural Network, J Clin Microbiol 56(3) (2018) e01521–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Samuel LP, Balada-Llasat JM, Harrington A, Cavagnolo R, Multicenter Assessment of Gram Stain Error Rates, J Clin Microbiol 54(6) (2016) 1442–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Barry-Straume J, Tschannen A, Engels DW, Fine E, An Evaluation of Training Size Impact on Validation Accuracy for Optimized Convolutional Neural Networks, SMU Data Science Review 1(4) (2018) Article 12. [Google Scholar]
- [10].Nugent RP, Krohn MA, Hillier SL, Reliability of diagnosing bacterial vaginosis is improved by a standardized method of gram stain interpretation, J Clin Microbiol 29(2) (1991) 297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Song Y, He L, Zhou F, Chen S, Ni D, Lei B, Wang T, Segmentation, Splitting, and Classification of Overlapping Bacteria in Microscope Images for Automatic Bacterial Vaginosis Diagnosis, IEEE journal of biomedical and health informatics 21(4) (2017) 10951104. [DOI] [PubMed] [Google Scholar]
- [12].Park HS, Rinehart MT, Walzer KA, Chi JT, Wax A, Automated Detection of P falciparum Using Machine Learning Algorithms with Quantitative Phase Images of Unstained Cells, PloS one 11(9) (2016) e0163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Poostchi M, Ersoy I, McMenamin K, Gordon E, Palaniappan N, Pierce S, Maude RJ, Bansal A, Srinivasan P, Miller L, Palaniappan K, Thoma G, Jaeger S, Malaria parasite detection and cell counting for human and mouse using thin blood smear microscopy, Journal of medical imaging (Bellingham, Wash.) 5(4) (2018) 044506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Poostchi M, Silamut K, Maude RJ, Jaeger S, Thoma G, Image analysis and machine learning for detecting malaria, Translational research : the journal of laboratory and clinical medicine 194 (2018) 36–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rajaraman S, Antani SK, Poostchi M, Silamut K, Hossain MA, Maude RJ, Jaeger S, Thoma GR, Pre-trained convolutional neural networks as feature extractors toward improved malaria parasite detection in thin blood smear images, PeerJ 6 (2018) e4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Rajaraman S, Jaeger S, Antani SK, Performance evaluation of deep neural ensembles toward malaria parasite detection in thin-blood smear images, PeerJ 7 (2019) e6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Torres K, Bachman CM, Delahunt CB, Alarcon Baldeon J, Alava F, Gamboa Vilela D, Proux S, Mehanian C, McGuire SK, Thompson CM, Ostbye T, Hu L, Jaiswal MS, Hunt VM, Bell D, Automated microscopy for routine malaria diagnosis: a field comparison on Giemsa-stained blood films in Peru, Malaria journal 17(1) (2018) 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kirn TJ, Automatic Digital Plate Reading for Surveillance Cultures, J Clin Microbiol 54(10) (2016) 2424–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ferrari A, Lombardi S, Signoroni A, Bacterial colony counting with Convolutional Neural Networks in Digital Microbiology Imaging, Pattern Recognition 61 (2017) 629–640. [Google Scholar]
- [20].Faron ML, Buchan BW, Coon C, Liebregts T, van Bree A, Jansz AR, Soucy G, Korver J, Ledeboer NA, Automatic Digital Analysis of Chromogenic Media for Vancomycin-Resistant-Enterococcus Screens Using Copan WASPLab, J Clin Microbiol 54(10) (2016) 2464–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Faron ML, Buchan BW, Vismara C, Lacchini C, Bielli A, Gesu G, Liebregts T, van Bree A, Jansz A, Soucy G, Korver J, Ledeboer NA, Automated Scoring of Chromogenic Media for Detection of Methicillin-Resistant Staphylococcus aureus by Use of WASPLab Image Analysis Software, J Clin Microbiol 54(3) (2016) 620–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Van TT, Mata K, Dien Bard J, Automated Detection of Streptococcus pyogenes Pharyngitis by Use of Colorex Strep A CHROMagar and WASPLab Artificial Intelligence Chromogenic Detection Module Software, J Clin Microbiol 57(11) (2019) e00811–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Faron ML, Buchan BW, Samra H, Ledeboer NA, Evaluation of the WASPLab software to Automatically Read CHROMID CPS Elite Agar for Reporting of Urine Cultures, J Clin Microbiol (2019) e00540–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Smith KP, Kirby JE, Rapid Susceptibility Testing Methods, Clinics in laboratory medicine 39(3) (2019) 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]