Abstract
Our recent study demonstrated that the QKI-5 regulated miRNA, miR-196b-5p, and it functions as an onco-microRNA in non-small cell lung cancer (NSCLC) by directly targeting GATA6 and TSPAN12. However, the role of miR-196b-5p in NSCLC progression and metastasis still remains unclear. We found that miR-196b-5p promotes lung cancer cell proliferation and colony formation by directly targeting tumor suppressor, FAS. The expression of FAS was significantly downregulated in NSCLC tissue samples and was negatively correlated with the miR-196b-5p expression. Knocking down FAS activates NFkB signaling and subsequent IL6 secretion, resulting in phosphorylation of signal transducer and activator of transcription 3 (STAT3) to promote lung cancer cell growth. Our findings indicated that miR-196b-5p might exhibit novel oncogenic function by FAS-mediated STAT3 activation in NSCLC, and suggested that targeting the miR-196b-5p/FAS/NFkB/IL6/STAT3 pathway might be a promising therapeutic strategy in treating NSCLC.
Subject terms: Non-small-cell lung cancer, miRNAs
Introduction
Lung cancer is the most commonly diagnosed cancer and the leading cause of cancer-related death worldwide, with ~2.1 million new lung cancer cases and 1.8 million deaths were reported in 20181. Non-small cell lung cancer (NSCLC) accounts for ~80–85% of all lung cancer cases. The main histological subtypes of NSCLC are lung adenocarcinoma (ADC, ~40–50% of all lung cancer cases) and squamous cell carcinoma (SCC, ~20–30% of all lung cancer cases)2. Several targeted therapies were available for patients with NSCLC; however, most of them are only effective in treating cancer patients with certain genetic backgrounds. In addition, the 5-year relative survival rate of NSCLC is only about 20% due to the late onset of clinical symptoms and inadequate screening methods3. Hence, deeply understanding the molecular mechanisms underlying NSCLC tumorigenesis and target molecules for developing novel therapeutic strategies are urgently needed.
microRNAs (miRNAs), a class of small noncoding RNAs, play a key role in cancer pathogenesis by targeting specific mRNAs4,5. miRNA negatively regulates mRNA stability and/or repress mRNA translation by targeting the complementary sequences in the 3′-untranslated region (UTR) of target gene6. Most of miRNAs function as a tumor suppressor or oncogene in a tissue-specific manner5. Accumulating evidence indicated that the dysregulation of miRNAs was closely associated with NSCLC progression and metastasis7. For instance, miR-34 inhibits tumor immune evasion to suppress NSCLC progression by targeting PD-L18. miR-548a-3p has been reported to regulate the Warburg effect and NSCLC growth by inhibiting transcription factor SIX19. let-7, a tumor suppressor in breast cancer, reduces breast tumor-initiating cells and inhibits tumor formation in NOD/SCID mice by silencing multiple targets10. In addition, let-7 regulates the Warburg effect and tumor progression by directly targeting PDK111. Clinical trials using chemically modified antisense oligonucleotides and/or miRNA mimics to targeting miRNAs in cancer have been conducted and demonstrated that potential for developing novel therapeutic methods7.
FAS, also known as CD95, was discovered as an apoptosis-inducing receptor12. Activated FAS recruits FAS-associated protein with death domain (FADD). FADD further recruits caspase-8, caspase-10, and cellular FADD-like interleukin-1-β-converting enzyme-inhibitory protein (c-FLIP) to form the death-inducing signaling complex (DISC)13, which mediates both apoptotic and non-apoptotic signaling pathways14. Accumulated evidence has indicated that FAS plays a key role in cancer progression12. Activated FAS signaling suppresses lung cancer cell growth in a mouse model through increasing TH9 cell differentiation15. In contrast, the FAS pathway facilitates colon cancer growth and metastatic potential by inducing the Erk1/2 pathway16. In addition, it has been reported that the miR-23a/b17 and miR-106a18 could target FAS to promote thymic lymphoma and gastric cancer progression, respectively.
In this study, we found that miR-196b-5p plays oncogenic functions by directly targeting FAS. FAS expression was significantly negatively correlated with the miR-196b-5p expression in NSCLC. Knocking down FAS promoted lung cancer cells growth through NFkB activation-mediated enhanced IL6 secretion and subsequent STAT3 activation. Our results demonstrated that the miR-196b-5p/FAS/NFkB/IL6/STAT3 axis might be a potential therapeutic target in NSCLC.
Materials and methods
Cell culture and reagents
The human lung cancer cell lines (A549, H460, and H292) were purchased from the American Type Culture Collection (ATCC). A549, H460, and H292 cells were cultured in RPMI-1640 medium (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco, Carlsbad, CA, USA) and 1% penicillin–streptomycin (Gibco, Carlsbad, CA, USA). Cell Cycle Regulation Antibody Sampler Kit (9932), anti-p65 antibody (8242), and anti-GAPDH rabbit monoclonal antibody (5174) were purchased from Cell Signaling Technology. Antibodies against p-STAT3 (Y705) (ab76315), STAT3 (ab68153), and Lamin B1 (ab16048) were purchased from Abcam. Anti-FAS/CD95 rabbit polyclonal antibody (13098–1-AP) was obtained from Proteintech, and an anti-vinculin antibody was ordered from Sigma-Aldrich. The miR-196b mimics, siFAS, and siSTAT3 were purchased from Genepharma (Shanghai, China). p65 overexpressing plasmid was kindly provided from Hui-lung Sun (Chicago University).
Transfection
Transfection of siRNAs against FAS, STAT3, and negative control was carried out in Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instruction (Invitrogen). p65 overexpressing vector, control vector, miR-196b mimic, and control mimic were transfected to the cells by using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instruction (Invitrogen). The sequences of siRNAs are shown in Supplementary Table S1.
RNA extraction and quantitative real-time PCR
Total RNAs were extracted by TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instruction. For cDNA synthesis, 1 μg of the total RNA was transcribed using the PrimeScript™ RT reagent Kit with gDNA Eraser (TAKARA, Japan) according to the manufacturer’s instruction. Quantitative real-time PCR was performed by using TB Green Fast qPCR mix (TAKARA, Japan) according to the manufacturer’s instruction. All reactions were conducted in triplicates. RT-PCR primers for IL6, FAS, and GAPDH are shown in Supplementary Table S2.
Cell proliferation assays
Two or three thousand cells in RPMI-1640 medium with 10% FBS were added to each well in a 48-well plate in sextuplicate and cultured at 37 °C for 4 days. The viability of cells was determined by MTT assay. To investigate the FAS knockdown or p65 overexpression-induced IL6 secretion on lung cancer cell proliferation, FAS knockdown or p65 overexpression lung cancer cells were cultured in six-well plates at 5 × 105 cells/well and incubated in RPMI-1640 medium with 2% FBS overnight. The cells were washed with PBS, and 2 ml of serum-free medium were added to each well. After 24 h, conditioned medium were collected and subjected to in vitro cell proliferation assay.
Cell colony-formation assay
After 24 h transfection, 500–1000 cells in RPMI-1640 medium with 10% FBS were added to each six-well plate in triplicate, and cultured at 37 °C for 6–9 days. Cells were fixed with 4% paraformaldehyde in PBS for 15 min, and stained with crystal violet (Beyotime, China). The colony number was calculated by Image J software.
Western blot analysis
The cells were lysed with RIPA buffer (Boster, China) supplemented with protease/phosphatase inhibitor Cocktail (Boster, China). Proteins from the lysate were separated by electrophoresis using 10–12% polyacrylamide gels and electrotransferred to PVDF membranes (Bio-Rad). After blocking with 5% skimmed milk, membranes were incubated with various primary antibodies. Then membranes were incubated with appropriate horseradish peroxidase (HRP)-conjugated secondary antibody. Specific proteins were detected with EZ-ECL Kit (Biological Industries).
Immunohistochemistry
The harvested tumor tissues were fixed with 10% formalin, processed, and embedded in paraffin. In total, 5-µm-thick sections were placed on positively charged slides. The tissue sections were stained by routine immunohistochemical techniques and incubated with primary antibodies against FAS (1:250) overnight at 4 °C. Conjugated secondary antibodies and diaminobenzidine (DAB) were used to detect FAS before hematoxylin staining and a neutral gum sealing. The FAS positive cells were counted and photographed with an ortho-microscope (Laica, Germany). According to the percentage of positive cells in the mean average of five fields, immunohistochemical reactivity for FAS was scored as follows: 0–5% (−), 5–25% (1+), 25–50% (2+), 50–100% (3+). The FAS expression graded as 3+ was defined as strong, 2+ was defined as moderate, and 1+ was defined as weak, respectively.
Flow-cytometry analysis for cell cycle
The cells were fixed with 70% ethanol at −20 °C overnight, then washed with PBS and resuspended in BD Pharmingen™ PI/RNase Staining Buffer (BD Biosciences). The cells were incubated at 37 °C for 40 min, and the cell cycle was detected by FACS Calibur Flow Cytometer (BD Biosciences). The results were processed by FlowJo10 software.
ELISA assay
To investigate IL6 secretion levels in media, miR-196b overexpression, FAS knockdown, or p65 overexpression, lung cancer cells were cultured in six-well plates at 5 × 105 cells/well and incubated in RPMI-1640 medium with 2% FBS overnight. The cells were washed with PBS, and 2 ml of serum-free medium were added to each well. After 24 h, suspensions of medium were collected and subjected to enzyme-linked immunosorbent assay (ELISA). IL6 secretion level in suspensions was detected according to the manufacturer’s instruction of IL6 Human Uncoated ELISA Kit (Invitrogen).
Cytosol and nuclear protein extraction
Cells were harvested and lysed using Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime, China) according to the manufacturer’s instruction. Protein expression of the cytoplasmic and nuclear extractions was determined by western blot analysis. Lamin B was used as a marker of nuclear protein, and GAPDH was used as a marker of cytoplasmic protein.
Patient samples
This study was approved by the Institutional Research Human Ethical Committee of the Wenzhou Medical University for the use of clinical biopsy specimens, and informed consent was obtained from the patients. A total of 30 NSCLC tissues and 30 corresponding NATs were obtained postoperatively from Zhoushan Hospital of Wenzhou Medical University. NSCLC tissues and NATs from patients were immediately placed in liquid nitrogen and then stored at −80 °C until further analysis.
TCGA dataset
The TCGA miRNA-seq and RNA-seq data with clinical information were downloaded on July 31, 2013. Only log-2 transformed level 3 data were used for analysis. For analysis of the TCGA dataset, the Welch t test was conducted to determine if the miR-196b-5p expression is different between NSCLC tissues and NATs. For the correlation analysis between miR-196b-5p expression and FAS expression, Pearson correlation coefficients were calculated.
Statistical analysis
All statistical analyses were performed by GraphPad Prism 7.0 (GraphPad Software, La Jolla, CA, USA). Data are represented as means with standard deviation (SD), and statistical significance was determined with unpaired t tests unless indicated otherwise. P values < 0.05 were considered statistically significant.
Results
Upregulated miR-196b-5p inhibits FAS expression in NSCLC
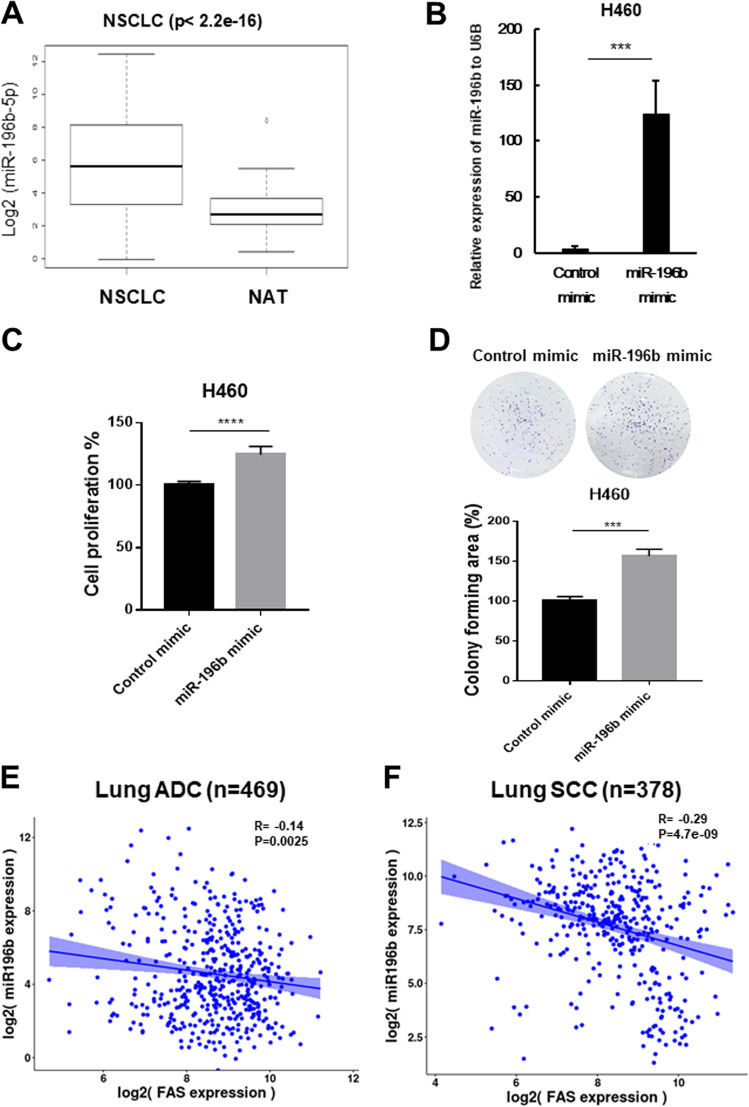
Recently, we found that miR-196b-5p functions as an oncomiRNA in NSCLC by directly targeting GATA6 and TSPAN1219. To further validate the oncogenic function of miR-196b-5p in NSCLC, we analyzed miR-196b-5p expression in NSCLC tissues and normal adjacent tissues (NATs) using The Cancer Genome Atlas (TCGA) dataset. We found that higher miR-196b-5p expression in NSCLC tissues than those in NATs (Fig. 1a). In detail, the average expression level of miR-196b-5p in NSCLC tissues is about ten times higher than those in NATs. Of note, almost 100 times higher miR-196b-5p expression was observed in certain NSCLC tissue samples than the average expression level of miR-196b-5p in NATs. Overexpressing miR-196b-5p in H460 cells (Fig. 1b) promoted cell proliferation (Fig. 1c) and colony formation (Fig. 1d). Similar results were also observed in miR-196b-5p overexpressing H292 cells (Supplementary Fig. 1A–C) and H520 cells (Supplementary Fig. 1D–F). Our recent study suggested that FAS might be a potential target of miR-196b-5p in NSCLC20. Bioinformatics analyses showed that five of seven miRNA target prediction websites indicated that FAS is a direct target of miR-196b-5p (Supplementary Table S3). It has also been reported that FAS might be a target of miR-196b in colorectal cancer20 and mixed-lineage leukemia (MLL)-rearranged leukemia21. Thus, we further examined the correlation between miR-196b-5p and FAS in NSCLC. TCGA lung adenocarcinoma (ADC) (n = 469) samples and lung SCC (n = 378) samples having both miR-196b-5p and FAS expression data available were selected for Pearson correlation analysis. The results showed that miR-196b-5p was negatively correlated with FAS in both lung ADC (R = −0.14, P = 0.0025) and lung SCC (R = −0.29, P = 4.7e-09), suggesting that reduced expression of FAS might be related to the upregulation of miR-196b-5p in NSCLC (Fig. 1e, f).
Fig. 1. miR-196b-5p plays an oncogenic function in lung cancer cells and is negatively correlated with FAS in non-small cell lung cancer (NSCLC).
a miR-196b-5p expression data were obtained from the TCGA miR-seq dataset. Of evaluable 666 NSCLC patients, 86 patients had matched normal adjacent tissues. b qRT-PCR examines miR-196b-5p expression in miR-196b-5p overexpressing H460 cells and control cells. Results are represented as means ± SD (n = 3). c Cell proliferation assay for miR-196b-5p overexpressing H460 cells and control cells. Results are represented as means ± SD (n = 3). d Colony-formation assay for miR-196b-5p overexpressing H460 cells and control cells. Colony-forming areas were measured by Image J software, and relative colony-forming areas were calculated by comparing with corresponding controls. Results are represented as means ± SD (n = 3). e, f FAS expression from TCGA RNA-seq data and miR-196b-5p expression from miR-seq data were used to examine the correlation between miR-196b-5p and FAS expressions in lung ADC dataset (n = 469) (e) and lung squamous cell carcinoma (SCC) dataset (n = 378) (f). ***P < 0.001, ****P < 0.0001.
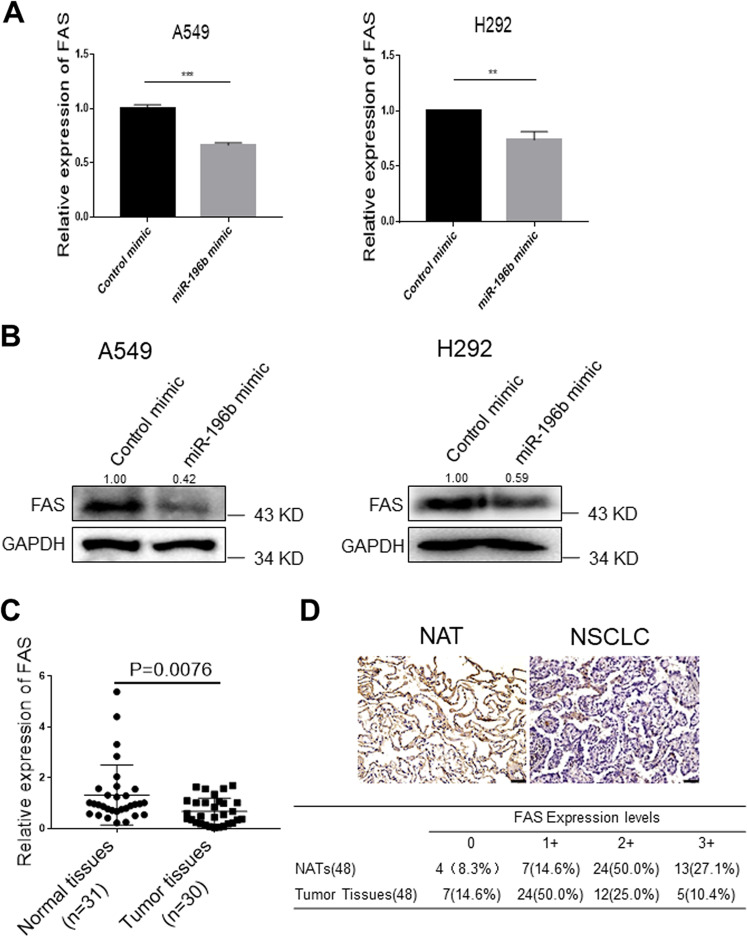
Next, to investigate the effects of miR-196b-5p on the expression of FAS mRNA and protein, we transiently overexpressed miR-196b mimics in A549 and H292 lung cancer cells. Overexpressing miR-196b-5p markedly reduced FAS mRNA (Fig. 2a) and protein (Fig. 2b) levels in both A549 and H292 cells, suggesting that miR-196b-5p might directly target FAS and enhanced expression of miR-196b-5p may reduce FAS expression in NSCLC.
Fig. 2. FAS is the target of miR-196b-5p and is downregulated in non-small cell lung cancer (NSCLC).
a FAS expression in A549 and H292 lung cancer cells after overexpressing miR-196b-5p was determined by qRT-PCR. Results are represented as means ± SD (n = 3). b Western blot analysis of FAS protein in A549 and H292 lung cancer cells after overexpressing miR-196b-5p. The bands were quantified using Image J software, and relative values were obtained by normalizing to the value of each corresponding GAPDH. c Expression level of FAS in 60 paired NSCLC tissues and their matched NATs. The RNA samples were extracted from 30 NSCLC tissues and 30 corresponding NATs. The RNAs were subject to qRT-PCR with a FAS probe, and the expression was normalized by GAPDH. d Upper panel shows representative immunohistochemical staining for FAS in NSCLC tissues and normal adjacent tissues (NATs) from the same patient. Summary of tissue immunohistochemical staining data for FAS in 48 pairs of clinical NSCLC tissues and NATs are shown in the lower panel. **P < 0.01, ***P < 0.001.
FAS expression is downregulated in NSCLC
Several studies have reported that FAS expression was downregulated in various cancers, including lung cancer22–24. Viard-Leveugle et al. has shown that FAS expression was markedly decreased in ~90% of lung cancer tissue samples compared with the normal lung tissue samples, and 24% of lung cancer tissue samples were completely lost FAS expression24. Since miR-196–5p was negatively correlated with FAS expression, we further evaluated FAS expression in NSCLC using TCGA lung ADC and SCC dataset. Analysis of FAS expression in 334 lung ADC patients with 57 matched NATs (Supplementary Fig. 2A) and 349 lung SCC patients with 51 matched NATs (Supplementary Fig. 2B) showed that significantly downregulated FAS expression in both lung ADC (P = 2.72e-14) and SCC (P = 7.92e-28). We further analyzed FAS expression in 30 NSCLC tissues and 31 NATs from Zhoushan Hospital of Wenzhou Medical University. Consistent with TCGA dataset and previous study, we found that FAS expression was downregulated in NSCLC tissues compared with those in NATs (P = 0.0076) (Fig. 2c). In addition, we analyzed FAS protein expression levels in tissue samples from 48 NSCLC patients by immunohistochemical staining. We found that FAS immunoreactivity was markedly reduced in NSCLC tissue samples compared with those in NATs samples. Of note, 77.1% of the NAT specimens showed moderate-to-strong FAS expression; however, only 35.4% of NSCLC tissue specimens presented moderate-to-strong FAS expression (Fig. 2d). Kaplan–Meier survival analysis using 1882 available NSCLC patients from the Kaplan–Meier Plotter showed that high expression of FAS is significantly associated with favorable prognosis of NSCLC patients (HR = 0.77, P = 6.5e-05) (Supplementary Fig. 2C). These results indicate that both mRNA and protein levels of FAS were downregulated in NSCLC tissues, and FAS may play an essential role in NSCLC carcinogenesis. Considering FAS mediates external cell death signaling, we speculate that manipulation of FAS expression without external cell death signal may not impact on the cell death rate of lung cancer cells.
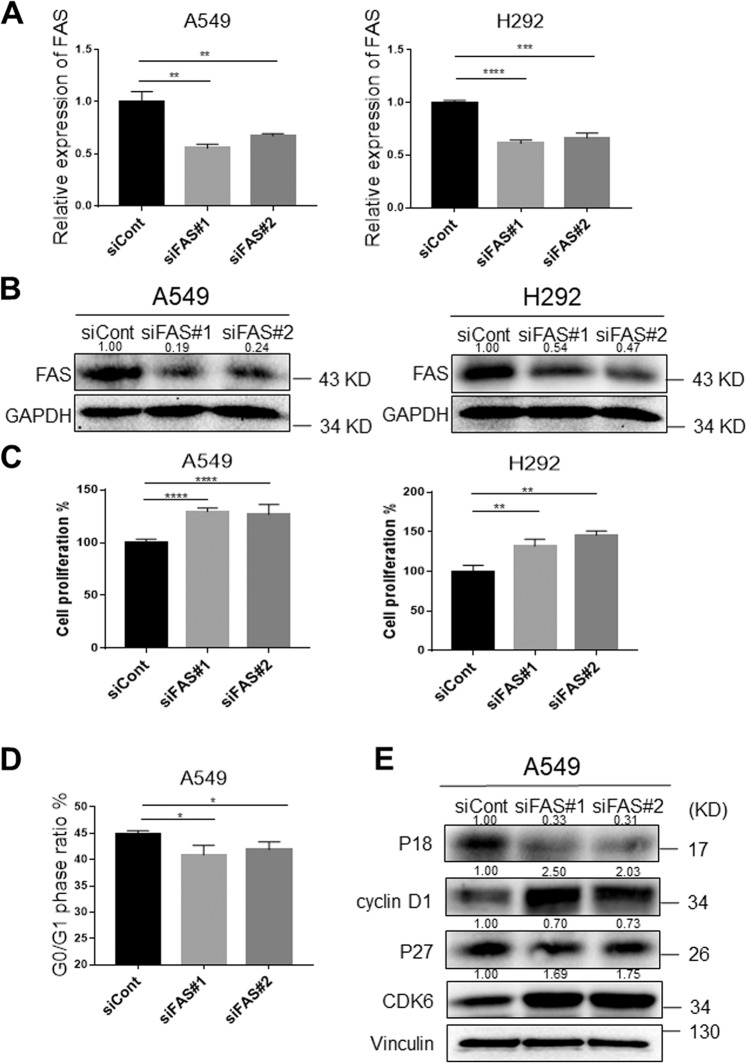
FAS knockdown promotes lung cancer cell proliferation and cell cycle
To further investigate the functions of FAS in lung cancer cells proliferation, colony formation, and cell cycle, we knocking down FAS in A549 and H292 cells by two independent siRNAs since both cell lines have relatively higher FAS expression than other lung cancer cell lines (Supplementary Fig. 3A). The FAS mRNA (Fig. 3a) and protein (Fig. 3b) expression levels after knockdown were confirmed by qRT-PCR analysis and western blot analysis. Knocking down FAS (~50%) significantly promoted cell proliferation (Fig. 3c) and colony formation (Supplementary Fig. 3B, C) in both A549 and H292 cells. We further investigated the effects of FAS knockdown on an SCC cell line, H520 cells. As expected, knocking down FAS enhanced the proliferative and colony-forming ability of H520 cells (Supplementary Fig. 3D, E). In addition, cell cycle analysis showed that knocking down FAS promoted transition of the G1 to S cell cycle in both A549 and H292 cells (Fig. 3d and Supplementary Fig. 4A, B). Accordingly, cell cycle G1 phase-related proteins were consistently changed in both A549 and H292 cells after knocking down FAS, including cyclin D1, CDK6, CDK inhibitor 1B (P27), and 2C (P18) (Fig. 3e and Supplementary Fig. 4C). These findings indicated that increased lung cancer cell proliferative and colony-forming ability by FAS knockdown might be due to accelerated cell cycle.
Fig. 3. FAS knockdown promotes lung cancer cell proliferation and cell cycle.
a qRT-PCR measure FAS expression in FAS knockdown A549 and H292 lung cancer cells. The expression was normalized by GAPDH. Results are represented as means ± SD (n = 3). b Western blot analyses of FAS protein in FAS knockdown A549 and H292 lung cancer cells. The bands were quantified using Image J software, and relative values were obtained by normalizing to the value of each corresponding GAPDH. c Cell proliferation assay for A549 and H292 cells after transfection with siFAS for 4 days. Results are represented as means ± SD (n = 3). d The percentage of cell cycle G1 phase cells in FAS knockdown A549 lung cancer cells and control cells. Results are represented as means ± SD (n = 3). e Western blot analyses for cell cycle G1 phase-related proteins in FAS knockdown A549 lung cancer cells and control cells. The bands were quantified using Image J software, and relative values were obtained by normalizing to the value of each corresponding Vinculin. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
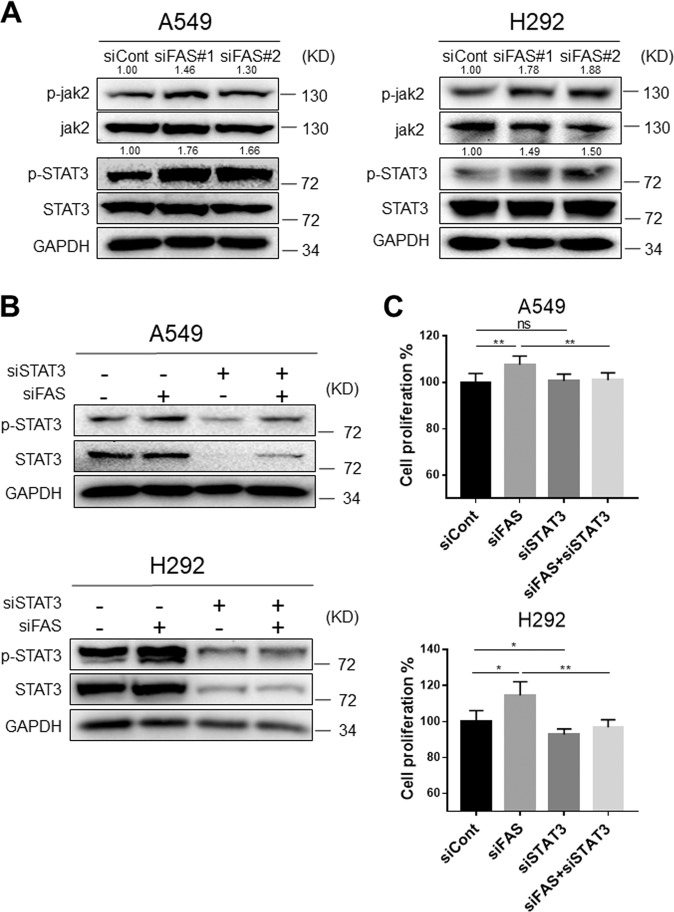
FAS knockdown activates STAT3 signaling
FAS signaling is closely associated with the immunological and inflammatory pathway25. Accumulating evidence suggests that FAS regulates T cells differentiation by in combination with STATs family proteins26. Particularly, STAT3 is highly activated in various cancers, and activated STAT3 promotes cancer progression27. It has been reported that STAT1/3-Mcl-1 signaling regulates FAS-mediated apoptosis in solid cancers28, and anti-FAS antibody-mediated activation of FAS could increase the phosphorylated STAT3 expression level in gastric cancer29. To investigate whether FAS signaling is involved in STAT3 activation in NSCLC, we performed a series of knockdown experiments. Knocking down FAS consistently enhanced phosphorylation levels of jak2 and STAT3 in both A549 and H292 cells (Fig. 4a). Similar results were also observed in FAS knocking down H520 cells (Supplementary Fig. 3F). To further demonstrate the relationship between STAT3 and FAS in NSCLC, we knocking down STAT3, FAS, or both of them in lung cancer cells. As expected, knocking down FAS increased STAT3 phosphorylation, while knocking down STAT3 reduced the levels of total STAT3 and phosphorylated STAT3. Moreover, we found that knocking down both STAT3 and FAS consistently attenuated FAS-knockdown-induced STAT3 phosphorylation (Fig. 4b). Accordingly, increased proliferative (Fig. 4c) and colony-forming abilities (Supplementary Fig. 4D) by FAS knockdown were significantly attenuated by knocking down both STAT3 and FAS. Together, these results suggested that the FAS downregulation promotes lung cancer cell growth by activating the STAT3 signaling pathway.
Fig. 4. FAS knockdown promotes STAT3 phosphorylation.
a Western blot analyses for p-STAT3 (Y705), STAT3, p-Jak2, and Jak2 in FAS knockdown A549 and H292 lung cancer cells. The bands of p-STAT3 and p-Jak2 were quantified using Image J software, and relative values were obtained by normalizing to the value of each corresponding STAT3 and Jak2. b Western blot analysis to detect p-STAT3 (Y705) expression in FAS knockdown A549 and H292 lung cancer cells transfecting with or without STAT3 siRNAs. c Cell proliferation assay in FAS knockdown A549 and H292 lung cancer cells transfecting with or without STAT3 siRNAs. Results are represented as means ± SD (n = 3). *P < 0.05, **P < 0.01, ns no significance.
FAS knockdown facilitates p65 nuclear translocation and promotes IL6 transcription
Our results showed that the FAS knockdown activated STAT3 signaling. Increased IL6 expression has been reported to activate STAT3 signaling, subsequently involving drug resistance and cancer progression30. Accumulating evidence has suggested that FAS facilitates the secretion of proinflammatory cytokines and chemokines, including CXCL1/KC, CXCL2/MIP2, IL6, and IL831–33. Among them, IL6 is a well-characterized cytokine that has been reported to be involved in various cancers progression by stimulating JAK2/STAT3 pathway30. Since both FAS and IL6 were closely associated with cancer pathogenesis and STAT3 activation, we investigated IL6 mRNA and protein levels after FAS knockdown. IL6 mRNA (Fig. 5a) and protein (Fig. 5b) expression was significantly increased in FAS knocking down A549 cells compared to the control cells. As expected, knockdown of FAS in H520 cells significantly enhanced IL6 protein secretion compared to the control cells (Supplementary Fig. 5D). Based on this result, we speculate that FAS knockdown might promote IL6 secretion from NSCLC cells.
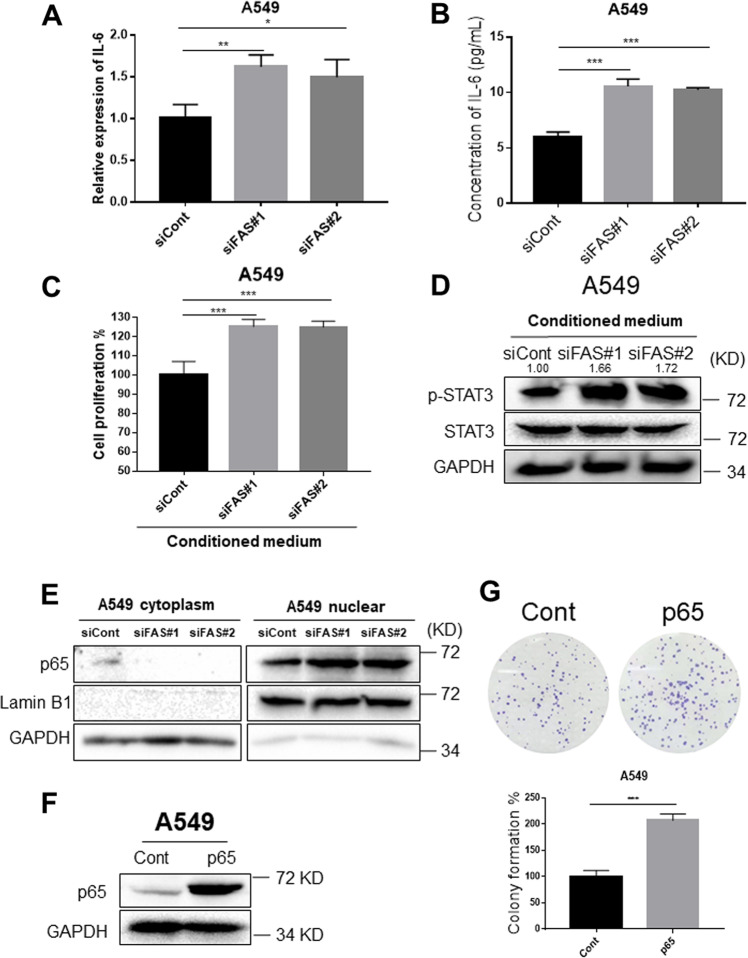
Fig. 5. FAS knockdown enhances IL6 expression.
a qRT-PCR measure IL6 expression in FAS knockdown A549 lung cancer cells. The expression was normalized by GAPDH. Results are represented as means ± SD (n = 3). b ELISA measures secreted IL6 protein levels in conditioned media from FAS knockdown A549 cells and control cells. Results are represented as means ± SD (n = 3). c Cell proliferation assay of A549 cells treated with conditioned mediums from FAS knockdown lung cancer cells or control cells. Results are represented as means ± SD (n = 3). d Western blot analysis for p-STAT3 (Y705) expression in A549 cells treated with conditioned mediums from FAS knockdown lung cancer cells or control cells. The p-STAT3 bands were quantified using Image J software, and relative values were obtained by normalizing to the value of each corresponding STAT3. e Western blot analyses for p65 protein in FAS knockdown A549 cells and control cells. Both nuclear and cytosol proteins were extracted and subjected to western blot analysis. f Western blot analyses of p65 protein in p65 overexpressing A549 cells and control cells. g Colony-formation assay for p65 overexpressing A549 cells and control cells. Colony-forming areas were measured by Image J software, and relative colony-forming areas were calculated by comparing with corresponding controls. Results are represented as means ± SD (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001.
To verify this hypothesis, conditioned medium from FAS knockdown and control cells were subjected to cell proliferation and colony-formation assays (Supplementary Fig. 5A). Conditioned medium from FAS knockdown cells significantly enhanced cell proliferative ability (Fig. 5c) and colony-forming ability compared with those conditioned media from control cells (Supplementary Fig. 5C). Interestingly, conditioned medium from FAS knockdown cells markedly increased STAT3 phosphorylation (Fig. 5d), suggesting that increased IL6 expression by FAS knockdown is responsible for STAT3 activation in lung cancer cells. p65, a member of the NFkB family, is a well-known transcription factor that has been reported to enhance IL6 transcription by binding its promoter region34,35. Hence, we checked whether p65 was activated following FAS knockdown. Indeed, FAS knockdown promoted nuclear translocation of p65 protein compared to those control cells, suggesting that NFkB activation by FAS knockdown is the main cause of increased IL6 expression in lung cancer cells (Fig. 5e).
NF-κB-mediated enhancement of IL6 expression promotes STAT3 activation
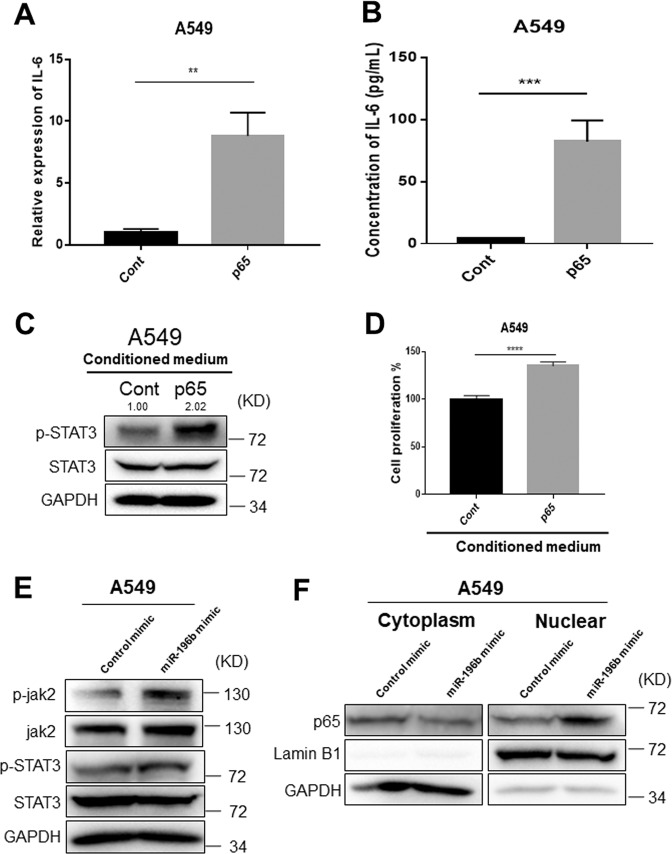
To further demonstrate the relationship between p65, IL6, and STAT3 signaling, we overexpressed p65 in lung cancer cells. Increased p65 protein expression after overexpressing was confirmed by western blot analysis (Fig. 5f). Overexpressing p65 significantly enhanced colony-forming ability of lung cancer cells (Fig. 5g). Since NFkB signaling could regulate IL6 expression in inflammation and cancer35,36, we further investigated IL6 expression in p65 overexpressing lung cancer cells. As expected, p65 overexpression significantly increased IL6 mRNA and protein expressions (Fig. 6a, b). In addition, conditioned medium from p65 overexpressing A549 cells markedly enhanced phosphorylated STAT3 expression, subsequently promoted cell proliferation in A549 cells compared with those conditioned medium from control cells (Fig. 6c, d, and Supplementary Fig. 5B). Pearson correlation analyses using TCGA NSCLC dataset from GEPIA website showed that p65 was positively correlated with both IL6 (P < 2.2e-16, R = 0.2) and STAT3 (P < 2.2e-16, R = 0.41) (Supplementary Fig. 6A, B). A positive correlation between IL6 and STAT3 (P = 1.3e-11, R = 0.2) was also observed (Supplementary Fig. 6C). Considering low R values for correlation between IL6 and P65 or STAT3, further studies need to be done to validate these results. We further investigated the effects of miR-196b-5p overexpression on the expression levels of phospho-STAT3 and nuclear translocation of p65. Overexpression of miR-196b-5p slightly enhanced phospho-STAT3 expression (Fig. 6e); however, consistently increased nuclear translocation of p65 (Fig. 6f). Furthermore, miR-196b-5p significantly upregulated IL6 mRNA expression (Supplementary Fig. 7A); however, no significant difference was observed for secreted IL6 protein levels (Supplementary Fig. 7B). These results suggested that miR-196b-5p-mediated FAS downregulation at least partly contributes to the activation of p65-IL6-STAT3 signaling, subsequently promotes NSCLC progression.
Fig. 6. FAS knockdown-mediated p65 activation promotes IL6 expression.
a qRT-PCR measure IL6 expression in p65 overexpressing A549 cells and control cells. The expression was normalized by GAPDH. Results are represented as means ± SD (n = 3). b ELISA measures secreted IL6 protein levels in conditioned media from p65 overexpressing A549 cells and control cells. c Western blot analysis for p-STAT3 (Y705) expression in A549 cells treated with conditioned mediums from p65 overexpressing lung cancer cells or control cells. The p-STAT3 bands were quantified using Image J software, and relative values were obtained by normalizing to the value of the corresponding STAT3. d Cell proliferation assay of A549 cells treated with conditioned mediums from p65 overexpressing lung cancer cells or control cells. Results are represented as means ± SD (n = 3). e Western blot analyses for p-STAT3 (Y705), STAT3, p-Jak2, and Jak2 in miR-196b-5p overexpressing lung cancer cells and control cells. f Western blot analyses for p65 protein in miR-196b-5p overexpressing A549 cells and control cells. Both nuclear and cytosol proteins were extracted and subjected to western blot analysis. **P < 0.01, ****P < 0.0001.
Discussion
Dysregulated miRNA expression is frequently seen in cancer. In cancer, miRNA may function as an oncogene or tumor suppressor, depending on cancer tissue type37. miR-196b plays dual functions in cancer; however, its role in lung cancer remains controversial. Our recent study showed that QKI-5 negatively regulates miR-196b-5p, and upregulated miR-196b-5p promotes lung cancer cell migration, proliferation, and cell cycle through directly targeting the tumor suppressors, GATA6 and TSPAN12 in NSCLC19. In addition, miR-196b-5p has been reported to function as an oncogene in laryngeal SCC by targeting SOCS238, and in oral cancer through regulating the NME4-JNK-TIMP1-MMP signaling pathway39. miR-196b simultaneously targets both HOXA9 and MEIS1 oncogenes, and FAS tumor suppressor to regulate tumorigenesis in MLL-rearranged leukemia21. In our study, we found that upregulated miR-196b-5p promotes lung cancer cell growth by targeting known tumor suppressor, FAS, in NSCLC. The expression of FAS is negatively correlated with the expression of miR-196b-5p, and is markedly downregulated in NSCLC tissues. Further analysis showed that high expression of FAS is associated with a favorable prognosis of NSCLC. Knocking down FAS promoted lung cancer cell growth by accelerating cell cycle, suggesting that bona fide tumor-suppressive functions of FAS in NSCLC.
The STAT family proteins were discovered as the transcription factors that bind to specific DNA sequences in the promoter region regulating gene transcription40. Persistent STAT3 activation promotes tumor progression and metastasis in various cancer27,41. FAS signaling facilitates apoptotic cell death in lung cancer cells, and advanced lung cancer is associated with decreased expression of FAS in circulating CD8 + T cells42,43. In contrast, it has been reported that FAS signaling promotes gastric cancer progression by activating STAT3 signaling29. Tyrosine phosphorylation of FAS (pY-291-FAS) activates STAT3, and subsequent AKT and MAPK signaling pathway resulting in increased cell proliferation and migration44, suggesting that dual functions of FAS in cancer progression. Nevertheless, the underlying molecular mechanisms of FAS in NSCLC progression, particularly linking between FAS and STAT3 signaling still remain unclear. Here, we found that FAS knockdown consistently activates the JAK2/STAT3 pathway. Since IL6 could promote STAT3 signaling pathway30,35, we further evaluated IL6 expression after knocking down FAS in lung cancer cells. As expected, we observed that increased IL6 expression in FAS knockdown lung cancer cells. Activated NFkB plays a key role in various cancer progression and could enhance IL6 secretion in tumor microenvironment36,45. To further demonstrate underlying molecular mechanisms of FAS knockdown-mediated enhancement of IL6 expression, we investigated p65 protein nuclear translocation in FAS knockdown lung cancer cells. Knocking down FAS indeed increased nuclear p65 expression, indicating that FAS knockdown induced IL6-mediated STAT3 activation might be due to activated NFkB signaling in NSCLC. Furthermore, we confirmed that overexpressing p65 elevated IL6 expression, and subsequent STAT3 activation resulting in enhanced lung cancer cell growth rate. Although a direct link between miR-196b-5p and phospho-STAT3 or IL6 is weak, FAS knockdown-mediated activation of p65-IL6-STAT3 is consistent in our study. We think that a number of downstream targets of miR-196b and their crosstalk might attenuate the effects of miR-196b-5p on the expression of phospho-STAT3 and IL6. Further study needs to validate this hypothesis. Our study revealed for the first time that FAS knockdown facilitates STAT3 activation in lung cancer cells. FAS knockdown accelerated nuclear translocation of p65, which in turn promotes IL6 expression and subsequent STAT3 phosphorylation.
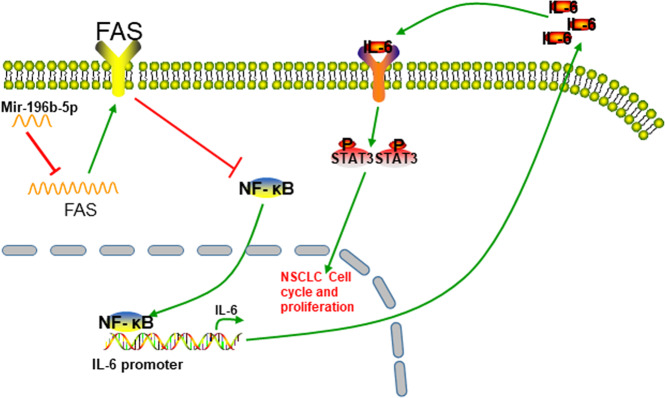
Our current study revealed that miR-196b-5p-mediated downregulation of FAS might involve activation of STAT3 signaling through the NFkB-IL6 axis (Fig. 7). The study provided important insights into understanding the oncogenic functions of miR-196b-5p by targeting FAS, and interaction between FAS and STAT3 in the progression of NSCLC. Our study suggested that targeting miR-196b-5p/FAS/NFkB/IL6/STAT3 pathway might be effective to treat some NSCLCs.
Fig. 7. Schematic illustration of summary of this study.
Proposed dysregulated miR-196b-5p-mediated downregulation of FAS activates STAT3 signaling through the NFkB-IL6 axis in NSCLC.
Supplementary information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81672305 to Ri Cui and 21701194 to Xiaohui Zheng).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by T. Kaufmann
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xiangjie Huang, Sisi Xiao, Xinping Zhu
Contributor Information
Xiaoying Huang, Email: zjwzhxy@126.com.
Ri Cui, Email: wzmucuiri@163.com.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41419-020-02997-7).
References
- 1.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Osmani L, Askin F, Gabrielson E, Li QK. Current WHO guidelines and the critical role of immunohistochemical markers in the subclassification of non-small cell lung carcinoma (NSCLC): moving from targeted therapy to immunotherapy. Semin Cancer Biol. 2018;52:103–109. doi: 10.1016/j.semcancer.2017.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 4.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 8.Cortez, M. A. et al. PDL1 regulation by p53 via miR-34. J. Natl Cancer Inst. 108, djv303 (2016). [DOI] [PMC free article] [PubMed]
- 9.Li L, et al. Transcriptional regulation of the Warburg effect in cancer by SIX1. Cancer Cell. 2018;33:368–385 e367. doi: 10.1016/j.ccell.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 10.Yu F, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 11.Ma X, et al. Lin28/let-7 axis regulates aerobic glycolysis and cancer progression via PDK1. Nat. Commun. 2014;5:5212. doi: 10.1038/ncomms6212. [DOI] [PubMed] [Google Scholar]
- 12.Martin-Villalba A, Llorens-Bobadilla E, Wollny D. CD95 in cancer: tool or target? Trends Mol. Med. 2013;19:329–335. doi: 10.1016/j.molmed.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Kischkel FC, et al. Cytotoxicity-dependent APO-1 (Fas/CD95)associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J. 1995;14:5579–5588. doi: 10.1002/j.1460-2075.1995.tb00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fouque A, Debure L, Legembre P. The CD95/CD95L signaling pathway: a role in carcinogenesis. Biochim. Biophys. Acta. 2014;1846:130–141. doi: 10.1016/j.bbcan.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 15.Shen Y, et al. Fas signaling-mediated TH9 cell differentiation favors bowel inflammation and antitumor functions. Nat. Commun. 2019;10:2924. doi: 10.1038/s41467-019-10889-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, et al. Human and mouse colon cancer utilizes CD95 signaling for local growth and metastatic spread to liver. Gastroenterology. 2009;137:934–944. doi: 10.1053/j.gastro.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B, et al. Up-regulated expression of miR-23a/b targeted the pro-apoptotic Fas in radiation-induced thymic lymphoma. Cell Physiol. Biochem. 2013;32:1729–1740. doi: 10.1159/000356607. [DOI] [PubMed] [Google Scholar]
- 18.Wang Z, et al. miR-106a is frequently upregulated in gastric cancer and inhibits the extrinsic apoptotic pathway by targeting FAS. Mol. Carcinog. 2013;52:634–646. doi: 10.1002/mc.21899. [DOI] [PubMed] [Google Scholar]
- 19.Liang, G. et al. miR-196b-5p mediated downregulation of TSPAN12 and GATA6 promotes tumor progression in non-small cell lung cancer. Proc Natl Acad Sci USA117, 4347–4357 (2020). [DOI] [PMC free article] [PubMed]
- 20.Mo J, et al. MicroRNA 196B regulates FAS-mediated apoptosis in colorectal cancer cells. Oncotarget. 2014;6:2843–2855. doi: 10.18632/oncotarget.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z, et al. miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nat. Commun. 2012;3:688. doi: 10.1038/ncomms1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maas S, et al. Decreased Fas expression in advanced-stage bladder cancer is not related to p53 status. Urology. 2004;63:392–397. doi: 10.1016/j.urology.2003.08.023. [DOI] [PubMed] [Google Scholar]
- 23.Mo JS, et al. MicroRNA 196B regulates FAS-mediated apoptosis in colorectal cancer cells. Oncotarget. 2015;6:2843–2855. doi: 10.18632/oncotarget.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Viard-Leveugle I, Veyrenc S, French LE, Brambilla C, Brambilla E. Frequent loss of Fas expression and function in human lung tumours with overexpression of FasL in small cell lung carcinoma. J. Pathol. 2003;201:268–277. doi: 10.1002/path.1428. [DOI] [PubMed] [Google Scholar]
- 25.Cruz AC, et al. Fas/CD95 prevents autoimmunity independently of lipid raft localization and efficient apoptosis induction. Nat. Commun. 2016;7:13895. doi: 10.1038/ncomms13895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer Zu Horste G, et al. Fas promotes T helper 17 cell differentiation and inhibits T helper 1 cell development by binding and sequestering transcription factor STAT1. Immunity. 2018;48:556–569 e557. doi: 10.1016/j.immuni.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 27.Miklossy G, Hilliard TS, Turkson J. Therapeutic modulators of STAT signalling for human diseases. Nat. Rev. Drug Discov. 2013;12:611–629. doi: 10.1038/nrd4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radhakrishnan H, et al. MACC1 regulates Fas mediated apoptosis through STAT1/3 - Mcl-1 signaling in solid cancers. Cancer Lett. 2017;403:231–245. doi: 10.1016/j.canlet.2017.06.020. [DOI] [PubMed] [Google Scholar]
- 29.Yang Y, et al. Fas signaling promotes gastric cancer metastasis through STAT3-dependent upregulation of fascin. PLoS ONE. 2015;10:e0125132. doi: 10.1371/journal.pone.0125132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson DE, O’Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018;15:234–248. doi: 10.1038/nrclinonc.2018.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altemeier WA, Zhu X, Berrington WR, Harlan JM, Liles WC. Fas (CD95) induces macrophage proinflammatory chemokine production via a MyD88-dependent, caspase-independent pathway. J. Leukoc. Biol. 2007;82:721–728. doi: 10.1189/jlb.1006652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cullen SP, et al. Fas/CD95-induced chemokines can serve as “find-me” signals for apoptotic cells. Mol. Cell. 2013;49:1034–1048. doi: 10.1016/j.molcel.2013.01.025. [DOI] [PubMed] [Google Scholar]
- 33.Shimizu M, et al. Roles of CXC chemokines and macrophages in the recruitment of inflammatory cells and tumor rejection induced by Fas/Apo-1 (CD95) ligand-expressing tumor. Int. J. Cancer. 2005;114:926–935. doi: 10.1002/ijc.20836. [DOI] [PubMed] [Google Scholar]
- 34.Wang X, et al. Long non-coding RNA DILC regulates liver cancer stem cells via IL-6/STAT3 axis. J. Hepatol. 2016;64:1283–1294. doi: 10.1016/j.jhep.2016.01.019. [DOI] [PubMed] [Google Scholar]
- 35.Nan J, Wang Y, YANG J, Stark GR. IRF9 and unphosphorylated STAT2 cooperate with NF-κB to drive IL6 expression. Proc. Natl Acad. Sci. USA. 2018;115:3906–3911. doi: 10.1073/pnas.1714102115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoesel B, Schmid JA. The complexity of NF-κB signaling in inflammation and cancer. Mol. Cancer. 2013;12:86. doi: 10.1186/1476-4598-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu. Rev. Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao X, Zhang W, Ji W. miR-196b is a prognostic factor of human laryngeal squamous cell carcinoma and promotes tumor progression by targeting SOCS2. Biochem. Biophys. Res. Commun. 2018;501:584–592. doi: 10.1016/j.bbrc.2018.05.052. [DOI] [PubMed] [Google Scholar]
- 39.Lu YC, et al. OncomiR-196 promotes an invasive phenotype in oral cancer through the NME4-JNK-TIMP1-MMP signaling pathway. Mol. Cancer. 2014;13:218. doi: 10.1186/1476-4598-13-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Darnell JE., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 41.Frank DA. STAT3 as a central mediator of neoplastic cellular transformation. Cancer Lett. 2007;251:199–210. doi: 10.1016/j.canlet.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 42.Li L, et al. Pigment epithelial-derived factor (PEDF)-triggered lung cancer cell apoptosis relies on p53 protein-driven Fas ligand (Fas-L) up-regulation and Fas protein cell surface translocation. J. Biol. Chem. 2014;289:30785–30799. doi: 10.1074/jbc.M114.590000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu L, et al. Advanced lung cancer is associated with decreased expression of perforin, CD95, CD38 by circulating CD3+CD8+ T lymphocytes. Ann. Clin. Lab. Sci. 2015;45:528. [PubMed] [Google Scholar]
- 44.Ta, N. L., Chakrabandhu, K., Huault, S., & Hueber, A.-O. The tyrosine phosphorylated pro-survival form of Fas intensifies the EGF-induced signal in colorectal cancer cells through the nuclear EGFR/STAT3-mediated pathway. Sci. Rep.8, 1–15 (2018). [DOI] [PMC free article] [PubMed]
- 45.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.