Abstract
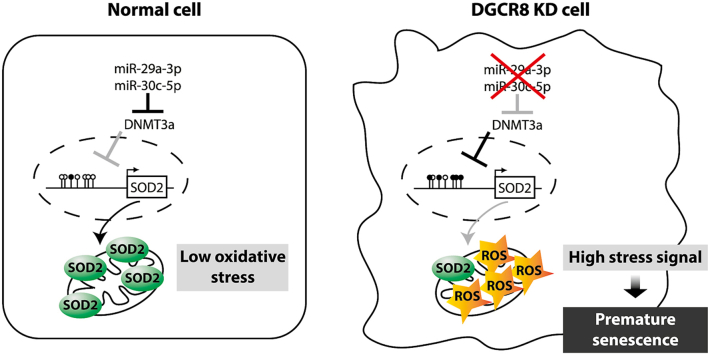
The use of human mesenchymal stem cells (hMSCs) in clinical applications requires large-scale cell expansion prior to administration. However, the prolonged culture of hMSCs results in cellular senescence, impairing their proliferation and therapeutic potentials. To understand the role of microRNAs (miRNAs) in regulating cellular senescence in hMSCs, we globally depleted miRNAs by silencing the DiGeorge syndrome critical region 8 (DGCR8) gene, an essential component of miRNA biogenesis. DGCR8 knockdown hMSCs exhibited severe proliferation defects and senescence-associated alterations, including increased levels of reactive oxygen species (ROS). Transcriptomic analysis revealed that the antioxidant gene superoxide dismutase 2 (SOD2) was significantly downregulated in DGCR8 knockdown hMSCs. Moreover, we found that DGCR8 silencing in hMSCs resulted in hypermethylation in CpG islands upstream of SOD2. 5-aza-2′-deoxycytidine treatment restored SOD2 expression and ROS levels. We also found that these effects were dependent on the epigenetic regulator DNA methyltransferase 3 alpha (DNMT3A). Using computational and experimental approaches, we demonstrated that DNMT3A expression was regulated by miR-29a-3p and miR-30c-5p. Overexpression of miR-29a-3p and/or miR-30c-5p reduced ROS levels in DGCR8 knockdown hMSCs and rescued proliferation defects, mitochondrial dysfunction, and premature senescence. Our findings provide novel insights into hMSCs senescence regulation by the miR-29a-3p/miR-30c-5p/DNMT3A/SOD2 axis.
Keywords: Cellular senescence, Mitochondrial oxidative stress, microRNAs, DNMT3A, SOD2, Human mesenchymal stem cells
Graphical abstract
Highlights
-
•
The global loss of miRNAs in hMSCs by DGCR8 gene silencing induces mitochondrial dysfunction and premature senescence.
-
•
Superoxide dismutase 2 (SOD2) is misregulated in DGCR8 knockdown hMSCs.
-
•
SOD2 deregulation is in part responsible for oxidative stress observed in DGCR8 knockdown hMSCs.
-
•
miR-29a-3p and miR-30c-5p regulate SOD2 expression and oxidative homeostasis in hMSCs by directly suppressing DNMT3A.
1. Introduction
The use of mesenchymal stem cells (MSCs) in novel therapeutic strategies is auspicious due to their tissue repair and regeneration capabilities [1]. MSCs can be readily isolated from the stroma of virtually all tissues, including the bone marrow, adipose tissue, and umbilical cord [2,3]. Numerous studies in animal models demonstrated the therapeutic potentials of the transplantation of ex vivo expanded MSCs. Numerous ongoing clinical trials are assessing the clinical efficacy of human MSCs (hMSCs) in various diseases [4]. To obtain a sufficient number of cells for cell therapy, a reproducible and efficient ex vivo expansion method is required. However, similar to other primary human cells, hMSCs can only divide a limited number of times under standard culture conditions [5]. Unlimited cell division is hindered by cellular senescence, a permanent state of cell-cycle arrest that is triggered by various external or internal stimuli [6]. Cellular senescence is characterised by morphological alterations, induction of tumour suppressor networks, increased senescence-associated β-galactosidase (SA-β-gal) activity, and a loss of proliferative capacity [7]. Furthermore, senescent MSCs exhibit reduced multilineage differentiation potential, as well as impaired migration, homing, and immunomodulatory abilities, limiting the therapeutic efficacy of hMSCs [8,9].
Several cellular senescence types have been characterised depending on the stimuli. Reactive oxygen species (ROS)-mediated oxidative stress is one of the major causes of cellular senescence [10,11]. Mitochondrial dysfunction is another common cause of senescence with discrete secretory phenotypes [12]. Due to the heterogeneity of senescent cells and their associated phenotypes, the elucidation of the mechanisms underlying cellular senescence is challenging. Nevertheless, various mechanisms regulating cellular senescence have been reported [6]. For example, cyclin-dependent kinase (CDK) inhibitors are involved in the regulation of cell cycle arrest. The CDK inhibitors CKDN2A (p16) and CDKN1A (p21), together with retinoblastoma (RB) and p53, are essential regulators of senescence [13]. Both p16 and p21 are regulated at the genetic and epigenetic level, as several mechanisms influence their transcription and post-translational modification [6].
MicroRNAs (miRNAs) are small (~22 nucleotides long) non-coding RNAs; they can be found in all plant and animal species, playing crucial roles in the regulation of gene expression [14,15]. Once primary miRNAs (pri-miRNAs) are produced by RNA polymerase II, they are processed by a complex consisting of the RNase III enzyme Drosha and the RNA-binding protein DiGeorge Syndrome Critical Region Gene 8 (DGCR8) in the cell nucleus. The resulting precursor miRNAs are then exported into the cytoplasm, where they are cleaved by the RNase III enzyme Dicer [14]. The functions of numerous miRNAs have been determined in vitro using DGCR8-depleted embryonic stem cells (ESCs) followed by rescue with miRNA mimics [16,17]. Such approaches revealed that certain miRNAs are important regulators of cell cycle and pluripotency [[18], [19], [20]]. Moreover, using DGCR8-depleted mouse glial progenitor cells (GPCs), miR-125 and let-7 have been identified as essential regulators of GPC differentiation into astrocytes. However, the global effects of miRNA loss have not been systematically studied in hMSCs.
Global expression profiling and functional analyses have been used to study the role of miRNAs in cellular senescence [21]. Frequently, miRNAs directly bind and repress key signalling pathway components that modulate senescence. For instance, miR-106b family members, miR-130b, miR-302a/b/c/d, miR-512–3p, and miR-515–3p suppress oncogene-induced senescence by negatively regulating p21 expression [22]. Additionally, depletion of Dicer and DGCR8 induces cellular senescence in mouse and human fibroblasts partly through p21 downregulation [23,24], suggesting an important role of miRNAs in senescence.
To gain further insight into the role of miRNAs in regulating cellular senescence in hMSCs, we depleted miRNAs globally by silencing DGCR8. DGCR8 depletion in hMSCs resulted in severe proliferation defects and senescence-associated alterations, including enlarged and flattened morphology, activation of tumour suppressor networks, and enhanced senescence-associated β-galactosidase (SA-β-gal) activity. Furthermore, ROS levels were markedly increased in DGCR8-depleted hMSCs. Transcriptome profiling analysis revealed that an antioxidant gene, superoxide dismutase 2 (SOD2) was significantly downregulated in DGCR8-depleted cells. Overexpression of SOD2 reduced ROS levels and rescued mitochondrial dysfunction. We demonstrated that CpG islands in the upstream regulatory region of the SOD2 gene were hypermethylated in DGCR8-depleted hMSCs; 5-aza-2′- deoxycytidine (decitabine; 5-aza-dC) restored SOD2 protein expression and ROS levels in a DNA methyltransferase 3 alpha (DNMT3A)-dependent manner. We further demonstrated that miR-29a-3p and miR-30c-5p directly target DNMT3A, regulating ROS generation, cell proliferation, mitochondrial dysfunction, and early onset of cellular senescence. Our findings provide novel insight into the role of miR-29a-3p and miR-30c-5p on SOD2 expression and oxidative homeostasis in hMSCs by regulating DNMT3A expression.
2. Materials and methods
2.1. Cell culture
This study was performed according to the guidelines of the Institutional Review Boards of Asan Medical Center and Soonchunhyang University (IRB no. 1040875-201708-BM-034). hMSCs were isolated from the adipose tissues of three donors (male or female, 33–46 years old) as previously described [25,26]. hMSCs were cultured in Dulbecco's Modified Eagle's Medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (Gibco), 100 units/mL penicillin, and 100 μg/mL streptomycin (Gibco). Cells were maintained in a humidified atmosphere containing 5% CO2 at 37 °C. Cell culture medium was replaced every three days, and cells were used at passage 3 to 5.
2.2. Transfection
For transfection with small interfering RNAs (siRNAs), hMSCs were seeded at 50%–60% confluence in complete cell growth medium (day 0). On day 1, the cells were transfected with 32 nM DGCR8-targeting siRNAs (siDGCR8) or GFP control (siGFP; ST Pharm) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The siRNA sequences were previously described [27,28]: siGFP (sense, 5′-GUU CAG CGU GUC CGG CGA GT TdTdT-3'; antisense, 5′-CUC GCC GGA CAC GCU GAA CT TdTdT-3′) and siDGCR8 (sense, 5′-CAU CGG ACA AGA GUG UGA UdTdT-3'; antisense, 5′-AUC ACA CUC U UG UCC GAU GdTdT-3′). For transfection with miRNA mimics, 48 h after siDGCR8 transfection, 20 nM miR-20a-3p and miR-30c-5p mimics (Qiagen) were transfected using DharmaFECT1 (Thermo Fisher Scientific) following the manufacturer's protocol. For SOD2 gene overexpression, 72 h after siRNAs transfection, cells were transfected with a total of 1 μg of SOD2 pCMV-SPORT6 construct (NM_001024465) using Fugene 6 (Promega) in accordance with the manufacturer's protocol.
2.3. Cell proliferation and viability assays
To determine the growth rate of hMSCs, we seeded 3.5 × 104 cells in complete cell growth medium in 6-well plates (day 0). The next day, the cells were transfected with 32 nM siRNAs. At days 3, 5, 7, and 10, the total cell numbers were counted. To assess cell proliferation, we used the CellTiter 96 Aqueous One Solution Cell Proliferation Assay (Promega) in accordance with the manufacturer's instructions. Briefly, 3 × 103 cells per well were seeded in 96-well plates (day 0). After transfection with siRNAs on day 1, cells were analysed 3, 5, 7, and 10 days after transfection. At the indicated time points, 20 μL of CellTiter 96 Aqueous One Solution containing 3-(4,5-dimethyl-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) (MTS; Promega) was added to each well. Optical absorbance at 490 nm was measured using a multilabel plate reader (PerkinElmer). Each assay was performed with at least six replicate wells for each condition.
2.4. Terminal deoxynucleotidyl transferase biotin-dUTP nick end-labelling (TUNEL) assay
TUNEL assays were performed using a TUNEL Apoptosis Detection Kit (Millipore) according to the manufacturer's instructions and as previously described [29]. Briefly, cells were fixed in 4% paraformaldehyde for 15 min at room temperature (RT) and then permeabilised with 0.5% Tween-20 and 0.2% BSA in PBS buffer for 15 min at RT. After washing, cells were incubated with TdT end-labelling cocktail for 1 h at RT. Subsequently, samples were incubated with avidin-FITC in the dark for 30 min at RT. Images were acquired using an EVOS fluorescence inverted microscope (Thermo Fisher Scientific).
2.5. 5-Bromo-2′-deoxyuridine (BrdU) assay
hMSCs were seeded in 96-well plates (3 × 103 cells/well) in complete cell growth medium. To detect cells in the S phase of the cell cycle, we used the Cell Proliferation ELISA/BrdU Kit (Roche) according to the manufacturer's instructions. Optical absorbance was measured at 370 and 492 nm using a multilabel plate reader (PerkinElmer). Each assay was performed with at least three replicate wells for each condition.
2.6. Quantitative real-time PCR
Total RNA was isolated from cultured cells using a mirVana miRNA isolation kit (Life Technologies) according to the manufacturer's protocol. Total RNA (200 ng) was reverse-transcribed using random hexamers and SuperScript III reverse transcriptase (Life Technologies). Quantitative real-time PCR (qRT-PCR) was performed using a Power SYBR Green PCR Master Mix (Applied Biosystems) on a 7900 Real-Time PCR system (Applied Biosystems). The primers used for qRT-PCR are shown in Supplementary Table S1. GAPDH was used as an endogenous control for normalisation. To assess the expression levels of miRNAs, we used 400 ng of total RNA, which was reverse-transcribed using a miScript II RT kit (Qiagen). qRT-PCR reactions were prepared using a miScript SYBR Green PCR kit (Qiagen) according to the manufacturer's instructions. SNORD68 was used as an endogenous control for normalisation. Each reaction was performed with at least three biological replicates and three replicate wells for each condition.
2.7. Western blot analysis
Cells were collected and lysed in RIPA buffer (Biosesang) containing Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific). After incubating for 30 min on ice with vortexing every 10 min, cellular debris was removed by centrifugation at 12,000 rpm for 20 min at 4 °C. Proteins were separated by 4–20% gradient gel (Bio-Rad) or 8–12% SDS-polyacrylamide gel electrophoresis. After electrophoresis, proteins were transferred onto nitrocellulose membranes (GE Healthcare) and incubated overnight at 4 °C with appropriate primary antibodies: H-Ras rabbit polyclonal antibody (sc-520, Santa Cruz; 1:200), MEK1/2 (D1A5) rabbit monoclonal antibody (8727; Cell Signalling; 1:1000), phospho-Erk1/2 mouse monoclonal antibody (9106, Cell Signalling; 1:1000), Erk1/2 rabbit polyclonal antibody (9102, Cell Signalling; 1:1000), phospho-p53 (Ser15) rabbit antibody (9284, Cell Signalling; 1:500), p53 rabbit polyclonal antibody (sc-6243, Santa Cruz; 1:1000), p21 rabbit monoclonal antibody (2947, Cell Signalling; 1:1000), SOD1 rabbit polyclonal antibody (sc-11407, Santa Cruz; 1:1000), SOD2 mouse monoclonal antibody (611,580, BD Biosciences; 1:1000), SOD3 mouse monoclonal antibody (ab28442, Abcam; 1:2000), DNMT3A rabbit polyclonal antibody (2160, Cell Signalling; 1:1000), alpha-tubulin rabbit antibody (SAB3501071, Sigma Aldrich; 1:5000), and alpha-tubulin mouse antibody (T9026, Sigma Aldrich; 1:5000). Alpha-tubulin was used as a loading control. Membranes were washed with 1 × TBST (Tris-Buffered Saline, 0.1% Tween 20 Detergent) for 30 min, followed by incubation with the corresponding horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h at RT. The following secondary antibodies were used: anti-mouse IgG, HRP-linked antibody (7076, Cell Signalling Technology; 1:3000) and anti-rabbit IgG, HRP-linked antibody (7074, Cell Signalling Technology; 1:3000). Antibody incubations were performed in 1 × TBST containing 5% BSA or non-fat dry milk. After washing with 1 × TBST for 30 min, proteins were visualised using ImmobilonTM Western ECL solution (Millipore, WBKLO500). Images were acquired using a chemiluminescent image analyser (GE Healthcare, LAS-4000).
2.8. SA-β-gal
Cells were seeded in 24-well plates (1 × 104 cells/well) in complete cell growth medium and transfected with siRNAs on the next day. Cells were fixed in 4% paraformaldehyde for 15 min at 25 °C 7 days post-transfection. After washing with Dulbecco's phosphate-buffered saline (DPBS), cells were stained with SA-β-gal staining solution (1 mg/mL X-gal, 0.04 M citric acid, 5 mM potassium ferrocyanide, 4.8 mM potassium ferrocyanide, 0.15 M NaCl, 2 mM MgCl2; pH 6.0) overnight at 37 °C. The number of SA-β-gal-positive cells was determined under a phase-contrast microscope.
2.9. Measurement of intracellular ROS levels
Intracellular ROS levels were determined using the oxidation-sensitive fluorescent probe 2′,7′-dichlorofluorescin diacetate (H2DCFDA; Thermo Fisher Scientific). Four days after transfection, cells were incubated with or without 10 mM N-acetyl-l-cysteine (NAC) (Sigma) for 48 h and then washed with PBS. Six days after siRNA transfection, cells were stained with 25 μM H2DCFDA for 30 min and washed with PBS. Fluorescence images were acquired using an inverted EVOS fluorescence microscope (Thermo Fisher Scientific). Fluorescence (490 nm excitation, 520 nm emission) was measured using a fluorescence microplate reader (PerkinElmer).
Measurement of hydrogen peroxide (H2O2), specifically released from cells, was performed by Amplex red assay. The staining mixture contained 50 μM Amplex Red reagent (Invitrogen Molecular Probes) and 0.1 U/mL HRP in KRPG buffer (145 mM NaCl, 5.7 mM sodium phosphate, 4.86 mM KCL, 0.54 mM CaCl2, 1.22 mM MgSO4, 5.5 mM glucose; pH 7.35). The reaction mixture was pre-warmed at 37 °C for 10 min. Cells were washed with HBSS (Hanks’ Balanced Salt Solution) and then suspended in 20 μL of KRPG buffer. Pre-warmed reaction mixture (100 μL) was added, and cells were incubated at 37 °C in a CO2 incubator for 90 min. Fluorescence (530 nm excitation, 590 nm emission) was measured using a fluorescence microplate reader (PerkinElmer). KRPG buffer (20 μL) without cells was measured as a negative control, while 3.0% H2O2 in KRPG buffer served as a positive control. Each experiment was performed in triplicate. The ROS levels were expressed as fold change relative to the siGFP transfection control.
The mitochondrial superoxide anion levels were measured in live cells using the indicator MitoSOX Red (Thermo Fisher Scientific). Transfected cells in black 96-well plates were washed twice with HBSS, and incubated with 5 μM MitoSOX in HBSS for 20 min. After removing staining solution, cells were washed again with HBSS, and then the fluorescence intensity was measured using a Victor X3 multiplate reader (PerkinElmer) with excitation and emission wavelengths of 510 and 580 nm, respectively. Images of the cells labelled with MitoSOX Red were acquired using an Eclipse Ts2 fluorescence microscope (Nikon) and fluorescence intensity was quantified using ImageJ software (Wayne Rasband at the National Institutes of Health; http://rsbweb.nih.gov/ij/).
2.10. Treatments
Three days after siDGCR8 transfection, cells were treated with 2 μM 5-aza-2′-deoxycytidine (decitabine, 5-aza-dC; Sigma); fresh serum-free medium was refreshed every day. Additionally, cells were treated with 5 μg/mL recombinant human SOD2 (Sigma) in complete cell culture medium three days after transfection.
2.11. Global gene expression profiling
hMSCs from three donors were subjected to global gene expression profiling using the Illumina Human HT-12 v4.0 Expression BeadChip (Illumina) (GEO database with accession number GSE149171). Total RNA was isolated from early-passage hMSCs (passage 4–5) at population doubling level (PDL) 3–5, late-passage hMSCs (passage 22–25) at PDL15–39, and siGFP or siDGCR8-transfected hMSCs using Trizol (Invitrogen). Extracted total RNA was hybridised to the arrays according to the manufacturer's protocol. The arrays were scanned using an Illumina bead array reader, and the scanned images were imported into BeadStudio software for data extraction and quality control. We identified all genes showing differential expression (P < 0.05) between three individual sets of either late and early-passage hMSCs or DGCR8-depleted and control hMSCs. Data were validated across all samples using real-time RT-PCR. Differentially expressed genes were analysed using Ingenuity Pathway Analysis (IPA; Qiagen) software to predict the core canonical pathways and functional networks significantly affected by DGCR8 depletion and replicative senescence.
2.12. JC-1 staining
Mitochondrial dysfunction in DGCR8-depleted cells was assessed using 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanide iodide (JC-1) probes (Thermo Fisher Scientific) according to the manufacturer's instructions. Cells (3 × 104 or 1 × 104) were seeded in imaging dishes (μ-Dish 35 mm, high or μ-Slide 8 Well; Ibidi) in complete cell growth medium. siDGCR8 transfected cells with or without SOD2 overexpression were treated with 1 μM Hoechst (Thermo Fisher) and 10 μM JC-1 probes dissolved in dimethyl sulfoxide (DMSO) for 10 min at 37 °C. Fluorescence images were acquired using either an EVOS fluorescence inverted microscope (Thermo Fisher Scientific) or confocal microscopes (LSM-780 or LSM-810, Zeiss).
2.13. Bisulphite sequencing
Genomic DNA was isolated from cultured cells using a DNeasy Blood & Tissue kit (Qiagen) according to the manufacturer's protocol. Genomic DNA (500 ng) was bisulphate-modified using the Imprint DNA Modification kit (Sigma) per the manufacturer's instructions. Primers specific to bisulphate-modified DNA were designed using the Meth primer program (http://www.urogene.org/methprimer). The upstream element region of the SOD2 gene was amplified using the forward primer 5′-AAA AAT AAG AGT ATT TAT AAT TTG GTT TTA -3′ and the reverse primer 5′-AAA CAA CAA AAA ATT CTT TCC TAC -3′. The core promoter region was amplified using the forward primer 5′-GTA ATT AAA ATT TAG GGG TAG G -3′ and reverse primer 5′-AAA AAA AAC TAC AAA CTA ACC TC -3′. PCR products were purified using a gel extraction kit (Qiagen). Purified PCR products were cloned into the TOPO TA Cloning Vector (Invitrogen) according to the manufacturer's instructions. Selected clones (10–12) from each sample were sequenced externally (Macrogen Inc., Seoul, Korea). BioEdit was used for sequence alignment.
2.14. Reporter vectors and luciferase reporter assay
To construct a luciferase reporter for the wild-type DNMT3A 3′-UTR, we used genomic DNA from hMSCs to PCR-amplify a partial fragment (662–868) of the DNMT3A 3′ UTR containing the miR-29a and miR-30c binding sites. PCR products were cloned into the psiCHECK-2 vector (Promega). A mutant DNMT3A 3′ UTR was generated by site-directed mutagenesis using Phusin High-Fidelity DNA Polymerase (Thermo Fisher Scientific) as previously described [30]. The sequences of the primers for cloning and site-directed mutagenesis are listed in Supplementary Table S2. For transfection, 293T-cells were seeded in 24-well plates (3 × 104 cells/well) and grown to 60% confluence. Subsequently, cells were transfected with mixtures containing 100 ng of the reporter constructs and miRNA mimic using DharmaFECT1 (Thermo Fisher Scientific) per the manufacturer's instructions. After 48 h of transfection, the cells were washed with DPBS and lysed in reporter lysis buffer (RLB). Luciferase activity was determined using a dual-luciferase reporter assay system (Promega) and a microplate reader (PerkinElmer).
2.15. Enzyme-linked immunosorbent assays (ELISAs)
For quantitative measurement of IL-8 chemokine levels, the medium from hMSCs was harvested after transfection. The levels of chemokines were determined using a Human IL-8/CXCL8 DuoSet ELISA kit (R&D Systems) according to the manufacturer's protocol. Optical absorbance at 450 nm (A450) was measured using a microplate spectrophotometer (BioTek).
2.16. Oxygen consumption rate (OCR) measurement
To measure the mitochondrial respiratory activity in real time, the oxygen consumption rate (OCR) was monitored using a Seahorse XFp Analyzer (Agilent). Cells were plated at 90% confluence in XFp 96-well plates in complete medium. The assay medium was Seahorse XF Base Medium (Agilent) supplemented with 25 mM glucose (Agilent), 10 mM pyruvate (Agilent), and 2 mM glutamine (Agilent), pH 7.4. Assays consisted of mixing for 3 min, a 2-min waiting period, followed by 3 min of measurement. Following measurement of basal respiration, oligomycin (1.5 μM) (Sigma) was added, followed by FCCP (1.0 μM) (Sigma) and finally by rotenone (Sigma) and antimycin (Sigma) mixture (1.5 μM). The cells were then collected for BCA assay for normalisation to determine the protein content (Pierce).
3. Results
3.1. DGCR8 loss in hMSCs leads to proliferation defects and induction of senescent phenotypes
To determine the effect of the global loss of miRNAs during ex vivo expansion of hMSCs, we depleted DGCR8, an essential component of miRNA biogenesis, in early-passage hMSCs (passage 3–5). Quantitative RT-PCR (qRT-PCR) showed a 60%–77% reduction in DGCR8 mRNA levels up to 10 days after siRNA transfection (Supplementary Fig. S1A) [27]. Immunoblot analysis confirmed the reduced DGCR8 protein levels in siDGCR8-transfected hMSCs (Supplementary Fig. S1B).
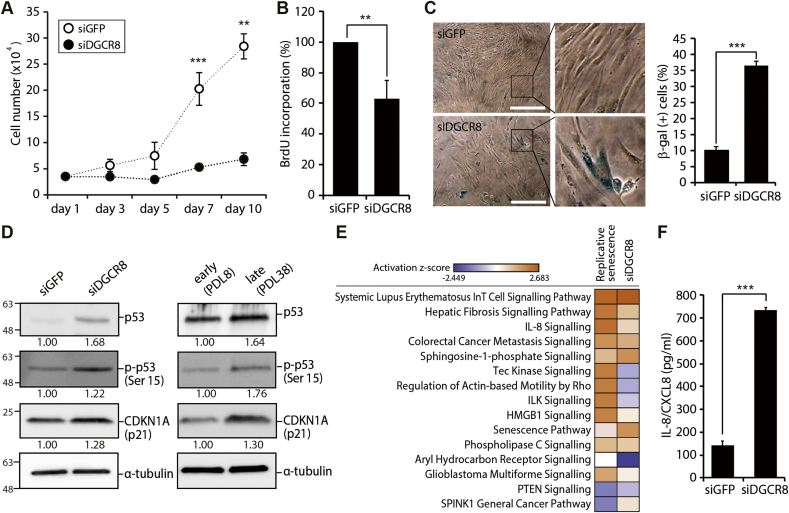
We then investigated the effects of the global loss of miRNAs on cellular growth. siDGCR8-transfected cells exhibited cell proliferation impairment with an extended doubling time compared with siGFP-transfected control cells (Fig. 1A). Cell viability was assessed using the MTS assay; siDGCR8-transfected hMSCs showed severe cell proliferation defects (Supplementary Fig. S1C). Quantification of BrdU incorporation confirmed the significant reduction of the proliferation rate in DGCR8 knockdown cells compared to control cells (Fig. 1B). Notably, the growth suppression observed after DGCR8 silencing was not a result of increased apoptosis (Supplementary Fig. S2).
Fig. 1.
DGCR8 silencing in hMSCs induces severe proliferation defects and cellular senescence. (A) Growth curves of hMSCs transfected with either siGFP (white dots) or siDGCR8 (black dots). Error bars indicate the standard error of the mean of at least three independent experiments (**P < 0.01, ***P < 0.001). (B) BrdU incorporation was quantified 5 days after siRNA transfection. Error bars indicate the standard error of the mean of three independent experiments (**P < 0.01). (C) Senescence-associated-β-galactosidase (SA-β-gal) staining. Compared with the siGFP-transfected group, siDGCR8-transfected hMSCs showed increased numbers of SA-β-gal-positive cells and enlarged size (left). The percentage of SA-β-gal-positive cells was significantly higher in siDGCR8-transfected hMSCs (right; ***P < 0.001). Magnification, 40 × ; scale bar, 150 μm. (D) Western blot showing the protein levels of tumour suppressors and senescence markers, including p21, p53, and p-p53 (ser15). (E) A heatmap of the canonical pathways enriched in replicatively senescent or DGCR8-depleted cells. Pathways were sorted by the activation z-score, which was calculated by total −log (P-value) from Fisher's Exact test across compared observations (observation 1, early-vs. late-passage; observation 2, siGFP- vs. siDGCR8-transfected cells). Orange (positive z-score) or blue (negative z-score) colour codes represent the activation or inhibition of the given pathway. (F) The levels of IL-8 secretion upon DGCR8 knockdown were quantified by enzyme-linked immunosorbent assay (ELISA). Error bars indicate the standard error of the mean of three independent experiments (***P < 0.001).
Proliferation arrest is the hallmark of cellular senescence. Thus, we next investigated the effects of DGCR8 knockdown on cellular senescence in hMSCs by measuring SA-β-gal activity. Interestingly, SA-β-gal activity was markedly higher in siDGCR8-transfected cells than in control cells (Fig. 1C, left, enlarged images on the right). The percentage of SA-β-gal-positive cells was also considerably higher in DGCR8 knockdown hMSCs (Fig. 1C, right). Furthermore, DGCR8-depleted hMSCs underwent morphological alterations resembling senescent cells, including the acquisition of flat, large, and irregular shapes (Supplementary Fig. S1D).
Because the tumour suppressor networks are commonly activated during cellular senescence, we next examined the expression of key tumour suppressor genes. DGCR8 knockdown cells exhibited increased levels of p53, p-p53 (Ser15), and CDKN1A (p21) at both the mRNA (Supplementary Fig. S1E) and protein levels (Fig. 1D, left), comparable to the replicatively senescent cells at PDL38 (Fig. 1D, right). Additionally, the mRNA levels of p53 target genes, including SESN1 (antioxidative stress protein Sestrin1) and GADD45 (the growth arrest and DNA damage-induced gene 45) were significantly increased after DGCR8 depletion (Supplementary Fig. S1E).
To identify shared features of the transcriptomes of replicatively senescent and DGCR8 knockdown cells, we determined the mRNA expression profiles of early- and late-passage hMSCs and siGFP- or siDGCR8-transfected cells. As expected, both replicatively senescent and DGCR8-depleted hMSCs showed marked alterations in gene expression profiles (Supplementary Fig. S3). In the senescent hSMCs, 1,414 probes showed upregulation and 1,740 probes showed downregulation compared to early-passage controls (P < 0.05, Fisher's exact test). Meanwhile among the 47,231 probes of the array, 1,247 probes were upregulated, and 1,136 probes were downregulated in DGCR8 knockdown hMSCs compared with control hMSCs (P < 0.05, Fisher's exact test). We next performed extensive bioinformatics analysis using IPA software. Specifically, we compared the canonical pathways of replicative senescence (late vs. early passage) and premature senescence upon DGCR8 loss (siDGCR8- vs. siGFP-transfected cells). The results indicated that the canonical pathways, i.e. IL-8 signalling, senescence pathways, and phospholipase C signalling, were all increased in both replicatively senescent and DGCR8-depleted hMSCs (Fig. 1E, activation z-score from 0.626 to 2.236). Consistent with the bioinformatics and phenotypic results, the levels of interleukin-8 (IL-8/CXCL8), a classic component of the senescence-associated secretory phenotype (SASP), were 5.2-fold higher in siDGCR8-transfected cells than in siGFP-transfected cells (Fig. 1F). Taken together, these results further support the induction of premature senescence after global miRNA loss.
3.2. Knockdown of DGCR8 results in increased ROS levels and Ras signalling induction
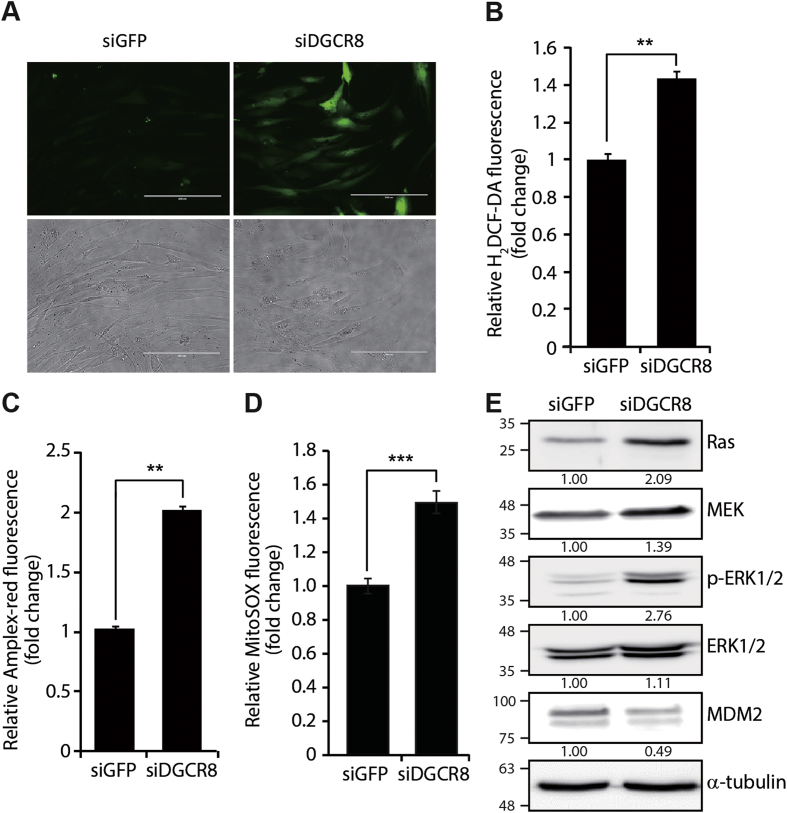
ROS production and oxidative stress is a major cause of cellular senescence. Hence, we investigated the effects of global miRNA loss on ROS levels. H2DCF-DA analysis revealed that ROS production was greatly increased in siDGCR8-transfected hMSCs compared with control cells (Fig. 2A). We confirmed that the increase in ROS generation was specifically inhibited by treatment with the free radical scavenger N-acetyl-l-cysteine (NAC) (Supplementary Fig. S4). The ROS production was further confirmed by quantitative measurements of total ROS and H2O2 which showed a 1.5-fold increase in H2DCF-DA (Fig. 2B) and a 2-fold increase in H2O2 by Amplex red assay in DGCR8 knockdown cells (Fig. 2C). Measurement of the intracellular level of mitochondrial superoxide anion (O2−) showed a significant 1.5-fold increase in siDGCR8-transfected cells (Fig. 2D). In addition to high levels of ROS, activation of oncogenes, such as Ras, is another major trigger of premature cellular senescence [31]. This process is known as oncogene-induced senescence (OIS) and can affect the levels of intracellular ROS [32]. To assess the potential involvement of Ras signalling in premature hMSC senescence after DGCR8 depletion, we investigated the protein levels of key components of the pathway. The levels of RAS, MEK, and p-ERK1/2 were significantly increased in DGCR8 knockdown hMSCs, indicating Ras signalling pathway activation (Fig. 2E). In addition, the expression of MDM2, a p53-specific E3 ubiquitin ligase, was decreased in DGCR8-depleted cells, which may have contributed to the induction of p53 shown in Fig. 1D. This is consistent with the well understood p53-MDM2 autoregulatory loop [33]. Taken together, these findings suggest that global loss of miRNAs after DGCR8 depletion induces premature senescent-associated alterations in hMSCs, including increased ROS levels and induction of oncogenic pathways.
Fig. 2.
DGCR8 knockdown leads to increased ROS levels and Ras signalling pathway activation. (A) Representative fluorescence images of 2′,7′-dichlorofluorescein diacetate (H2DCF-DA)-stained cells. DGCR8 knockdown in hMSCs significantly increased ROS generation 5 days after siRNA transfection. Scale bar, 200 μm. (B) Quantitative measurement of global ROS production by H2DCF-DA. Error bars denote the standard error of the mean of three independent experiments (**P < 0.01) (C) Quantitative measurement of hydrogen peroxide (H2O2) in siGFP- and siDGCR8-transfected cells using Amplex-red assay. Error bars denote the standard error of the mean of three independent experiments (**P < 0.01). (D) Quantitative measurement of mitochondrial superoxide anion (O2−) by MitoSOX staining. Error bars denote the standard error of the mean of three independent experiments (***P < 0.001). (E) Western blot showing increased Ras, MEK, and p-ERK1/2 protein levels in DGCR8 knockdown hMSCs. Images are representative of three independent experiments.
3.3. Depletion of DGCR8 induces mitochondrial dysfunction and SOD2 downregulation
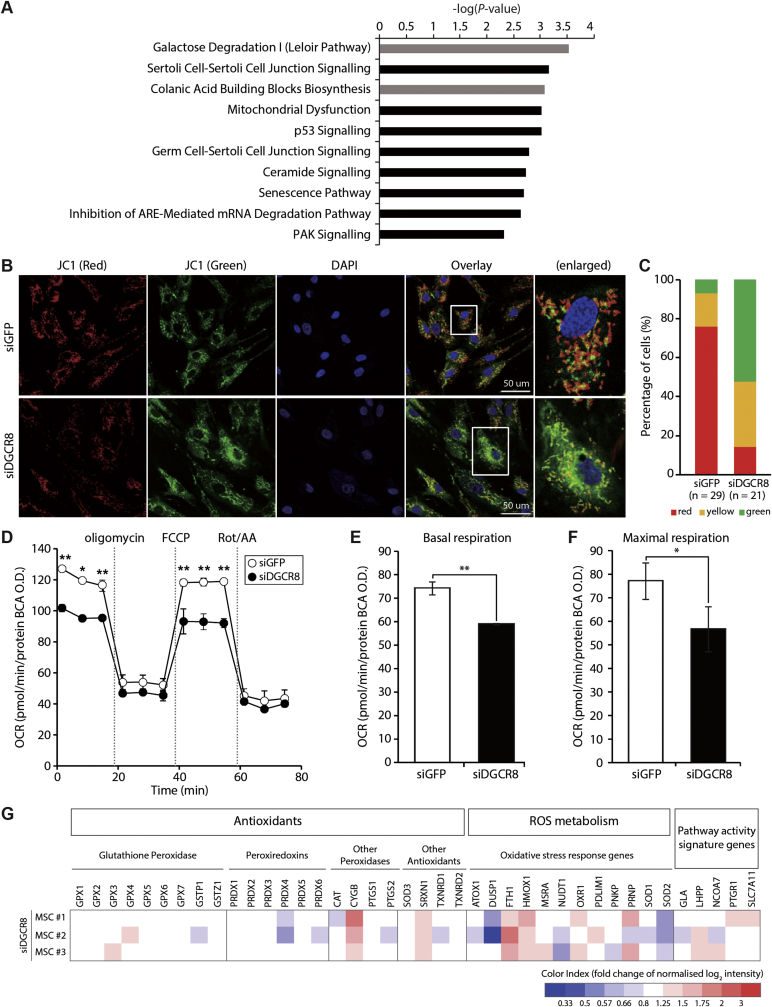
To systematically assess the underlying mechanisms of premature senescence in DGCR8 knockdown hMSCs, we compared the mRNA expression profiles of siDGCR8-transfected and siGFP control cells obtained from three donors. As previously described, DGCR8 knockdown in hMSCs resulted in dramatic alterations in gene expression profiles (Supplementary Fig. S3B; Supplementary Table S3 and S4). We performed canonical pathway analysis of all differentially expressed transcripts in DGCR8-depleted hMSCs. IPA indicated that the top 10 canonical pathways including metabolic pathways and signalling pathways were enriched in DGCR8 knockdown hMSCs (Fig. 3A, grey and black bars, respectively). Interestingly, eight of these signalling pathways were functionally linked to cellular growth, proliferation, cell cycle regulation, cellular stress/injury, mitochondrial dysfunction, and senescence, consistent with the senescence phenotypes and proliferation defects observed in siDGCR8-transfected cells.
Fig. 3.
DGCR8 silencing induces mitochondrial dysfunction and deregulation of ROS metabolism-related genes. (A) Differentially expressed genes (DEGs) were subjected to Ingenuity Pathway Analysis (IPA), and the top 10 canonical pathways enriched in DGCR8-depleted hMSCs are shown. Metabolic pathways are shown in grey, while signalling pathways are indicated in black. (B) Mitochondrial membrane potential (MMP) in DGCR8-depleted hMSCs was assessed using the fluorescent dye JC-1. Representative confocal microscopy images showing high MMP (red fluorescence aggregates) and low MMP (green fluorescence monomers). Scale bars, 50 μm. (C) Quantification of JC-1 ratios. (D) Kinetic responses of oxygen consumption rate (OCR) in DGCR8-depleted hMSCs upon sequential addition of ATP synthase inhibitor oligomycin (1.5 μM), electron chain uncoupler carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP) (1.0 μM), and complex I and III inhibitors rotenone and antimycin (1.5 μM each). The OCR values were normalised relative to the BCA protein assay. Error bars indicate the standard error of the mean of three independent experiments (*P < 0.05, **P < 0.01). (E) Basal mitochondrial respiration rates (basal OCR measurement minus rotenone/antimycin A response) and (F) maximal respiration rates (FCCP response minus rotenone/antimycin A response) were quantified in DGCR8 knockdown cells (*P < 0.05, **P < 0.01). (G) Heatmap showing the expression patterns of 40 oxidative stress-related genes upon DGCR8 depletion (n = 3 independent biological samples).
To verify the significance of enriched pathways in DGCR8 knockdown cells at the cellular level, we evaluated the effect of DGCR8 loss on mitochondrial membrane potential using the fluorescent cationic dye JC-1. The mitochondria inner membrane is impermeable to most small molecules and ions, maintaining a gradient of protons and ions [34]. However, oxidative stress and ROS accumulation often disrupt the inner membrane polarisation by affecting the influx of protons and ions [35]. Confocal microscopy revealed a profound colour shift from red to green, suggesting mitochondrial depolarisation in DGCR8 knockdown hMSCs (Fig. 3B). Quantitative analysis revealed a significant decrease in the number of red cells upon DGCR8 silencing while the number of green cells increased, confirming mitochondrial dysfunction (Fig. 3C). Next, mitochondrial function was examined by measuring the OCR using a Seahorse XFp instrument. Consistent with the results of genomics and cellular analyses, DGCR8-depleted cells showed significant reductions of the basal and maximal respiration rates compared to siGFP-transfected cells indicating mitochondrial dysfunction (Fig. 3D–F).
We next focused on the expression patterns of 40 genes involved in oxidative stress. Peroxidases, including glutathione peroxidase (GPX) and peroxiredoxins (PRDX), are classic antioxidant genes. Oxidative stress response genes and genes involved in superoxide metabolism, such as SOD genes, are essential for ROS metabolism. Among these genes, SOD2 was the only gene that was significantly downregulated upon DGCR8 knockdown in all three independent biological replicates (Fig. 3G). The SOD family is an important antioxidant defence system found in nearly all living cells exposed to oxygen; it includes enzymes that mediate the detoxification of superoxide by converting it into oxygen and H2O2 [36]. Consistently, SOD2 exhibited the strongest downregulation among the 26 genes associated with mitochondrial dysfunction (Supplementary Fig. S5A). siRNA-mediated SOD2 knockdown induced mitochondrial depolarisation defects similar to those observed in siDGCR8-transfected hMSCs (Supplementary Fig. S5B). These findings suggested a potential role of SOD2 downregulation in the enhanced ROS production and premature senescence observed in DGCR8 knockdown cells.
3.4. Expression of recombinant SOD2 protein in siDGCR8 restores ROS levels and mitochondrial function
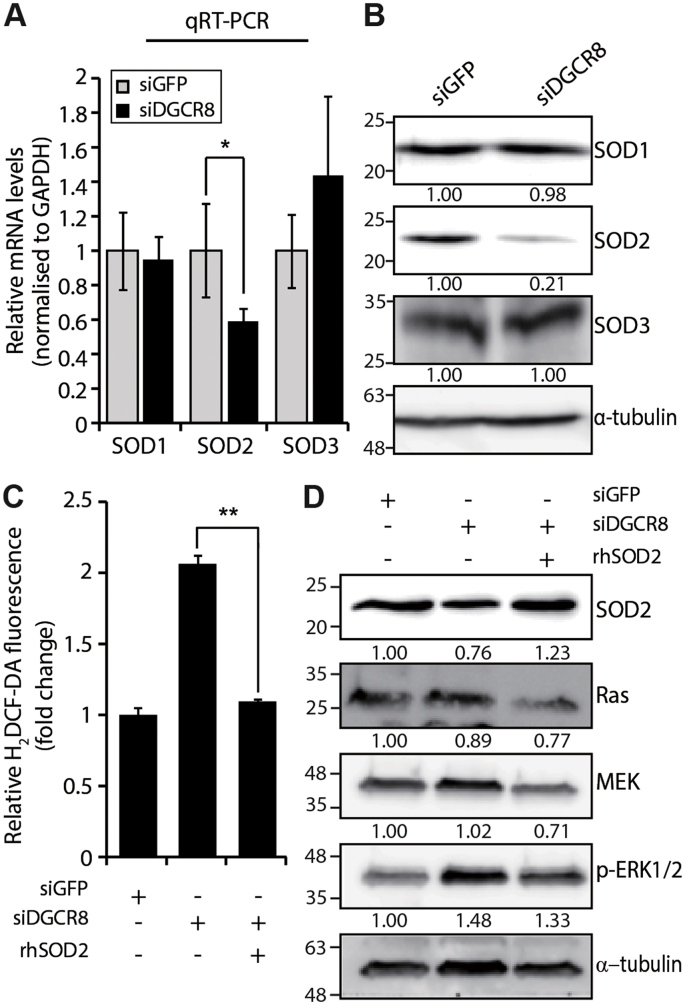
There are three major SOD isoforms; SOD1 is localised to the intracellular cytoplasmic compartment while SOD3 is the secreted extracellular isoform. SOD2 is localized to mitochondria of aerobic cells and uses manganese (Mn) as a cofactor [36]. Consistent with the microarray data, qRT-PCR analysis revealed that unlike other SODs, SOD2 mRNA levels were significantly reduced in siDGCR8-transfected hMSCs compared with control cells (Fig. 4A and Supplementary Fig. S6A). This was also confirmed at the protein level (Fig. 4B).
Fig. 4.
Overexpression of human recombinant SOD2 (rhSOD2) restores ROS levels and mitochondrial function in DGCR8 knockdown hMSCs. (A) The mRNA levels of SOD genes were analysed by quantitative real-time PCR. Data were normalised to GAPDH mRNA levels. Error bars indicate the standard error of the mean of three independent experiments (*P < 0.05). (B) Immunoblot analysis of SOD proteins in DGCR8 knockdown cells. (C) Total ROS levels were measured in siDGCR8-transfected cells after rhSOD2 overexpression (5 μg/mL), using H2DCF-DA. Error bars indicate the standard error of the mean of three independent experiments (**P < 0.01). (D) Western blot showing the protein levels of Ras signalling components in rhSOD2-overexpressing DGCR8 knockdown hMSCs.
To assess the relevance of SOD2 downregulation in the increase in ROS levels upon DGCR8 knockdown, we forced the expression of a recombinant human SOD2 protein (rhSOD2) in siDGCR8-transfected hMSCs. Overexpression of SOD2 in DGCR8 knockdown hMSCs decreased ROS levels to levels similar to those observed in siGFP-transfected control cells (Fig. 4C). Similarly, overexpression of SOD2 in DGCR8 knockdown hMSCs reduced the protein levels of key Ras signalling components, including Ras, MEK, and p-ERK1/2 (Fig. 4D). We further confirmed the functional recovery of DGCR8-depleted hMSCs by transient expression of the SOD2 gene under the control of the CMV promoter. When SOD2 was overexpressed in siDGCR8-transfected cells (Supplementary Fig. S6B), we observed a significant reduction in mitochondrial superoxide anion levels (Supplementary Fig. S6C) and partial recovery of the mitochondrial membrane potential (Supplementary Fig. S6D). More importantly, the cellular OCR in siDGCR8-transfected cells overexpressing the SOD2 gene was functionally rescued (Supplementary Fig. S6E). These results suggest that downregulation of SOD2 proteins contributes to premature senescence in DGCR8 knockdown hMSCs by deregulating ROS production and mitochondrial oxidative stress.
3.5. DNMT3A is an intermediate regulator of SOD2 expression
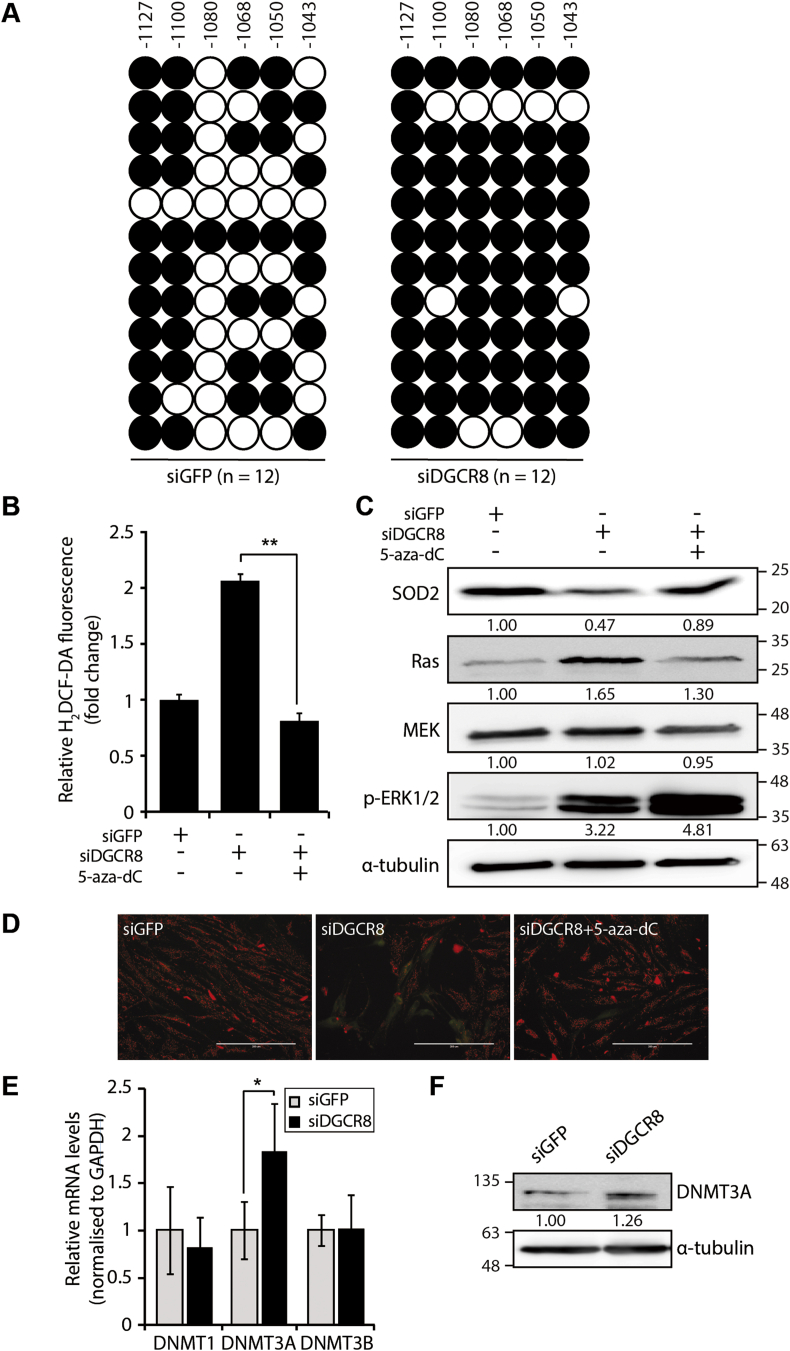
Since miRNAs typically repress the expression of target mRNAs [37], we hypothesised that an intermediate regulator is responsible for the SOD2 downregulation observed in DGCR8 knockdown hMSCs. The expression of SOD2 is tightly regulated by various epigenetic mechanisms [38]. Hence, using Promoter 2.0 Prediction (www.cbs.dtu.dk) and TRANFACv8.0 software, we identified the core promoter and upstream regulatory region of the SOD2 gene (Supplementary Fig. S7A), which have been previously shown to regulate SOD2 expression [38,39]. CpG islands in these regulatory regions are potential targets for DNA methylation. Methylation analysis of these CpG islands using bisulphite sequencing revealed that the methylation of six CpG sites within the upstream regulatory region (−1127 to −1043 from the transcription start site) was increased by 30% upon DGCR8 loss (12 independent clones; Fig. 5A). By contrast, the core promoter was unmethylated in both siGFP- and siDGCR8-transfected hMSCs (Supplementary Fig. S7B).
Fig. 5.
Hypermethylation of the SOD2 upstream regulatory region is dependent on DNMT3A. (A) Location and methylation status of six CpG sites in the SOD2 upstream regulatory region analysed by bisulphite sequencing (BSP) in control and DGCR8 knockdown hMSCs (n = 12 individual clones). Each circle represents a single sequencing reaction of each CpG site (white circles, unmethylated CpG sites; black circles, methylated CpGs). (B) Total ROS levels are measured by H2DCF-DA after 5-aza-dC treatment (2 μM) in DGCR8 knockdown hMSCs. Error bars denote the standard error of the mean of three independent experiments (**P < 0.01). (C) Immunoblot showing the levels of SOD2 and Ras signalling components after 5-aza-dC treatment (2 μM) in DGCR8 knockdown hMSCs. (D) Mitochondrial membrane potential (MMP) was assessed by JC-1 staining in DGCR8 knockdown hMSCs. Representative fluorescence microscopy images of siGFP- or siDGCR8-transfected cells, as well as 5-aza-dC-treated DGCR8 knockdown cells. Scale bars, 200 μm. (E) The mRNA levels of DNMTs were measured by quantitative real-time PCR. Data were normalised to GAPDH mRNA levels. Error bars indicate the standard error of the mean of three independent experiments (*P < 0.05). (F) Western blot analysis of DNMT3A protein levels in DGCR8 knockdown hMSCs.
To confirm the relevance of SOD2 promoter hypermethylation after DGCR8 loss, we treated cells with the DNA methylation inhibitor 5-aza-2′-deoxycytidine (5-aza-dC); DNA methylation inhibition restored SOD2 protein levels in DGCR8 knockdown cells (Fig. 5C, top row). This result was consistent with a previous report demonstrating the dose-dependent increases in SOD2 mRNA and protein levels upon 5-aza-dC treatment in fawn-hooded rat (FHR) pulmonary artery smooth muscle cells (PASMCs) [40]. More importantly, 5-aza-dC treatment in siDGCR8-transfected hMSCs reduced the levels of ROS (Fig. 5B) and the mitochondrial superoxide anion to physiological levels (Supplementary Fig. S8), as well as restored the mitochondrial membrane potential (Fig. 5D). Moreover, RAS and MEK protein levels were significantly decreased after 5-aza-dC treatment (Fig. 5C, bottom rows). Interestingly, treatment of DGCR8-depleted hMSCs with 5-aza-dC increased ERK1/2 phosphorylation. As a genome-wide demethylation drug, 5-aza-dC can affect diverse signalling pathways. Indeed, it has been reported that 5-aza-dC indirectly activates ERK through CCL2 and CCR-2 in human monocytic leukaemia cells [41].
DNA methylation is one of the most common epigenetic modifications in higher eukaryotes and is mediated by highly conserved enzymes called DNA methyltransferases (DNMTs) [42]. In mammals, DNMT1, DNMT3A, and DNMT3B are the three major DNMTs. We found that DNMT3A was upregulated in siDGCR8-transfected cells both at the mRNA (Fig. 5E) and protein levels (Fig. 5F). These results suggest that DNMT3A is an intermediate regulator of SOD2 expression and that DNMT3A upregulation in DGCR8 knockdown cells might contribute to SOD2 hypermethylation and increased ROS generation.
3.6. miR-29a-3p and miR-30c-5p directly repress DNMT3A and rescue the oxidative stress and premature senescence induced by DGCR8 knockdown in hMSCs
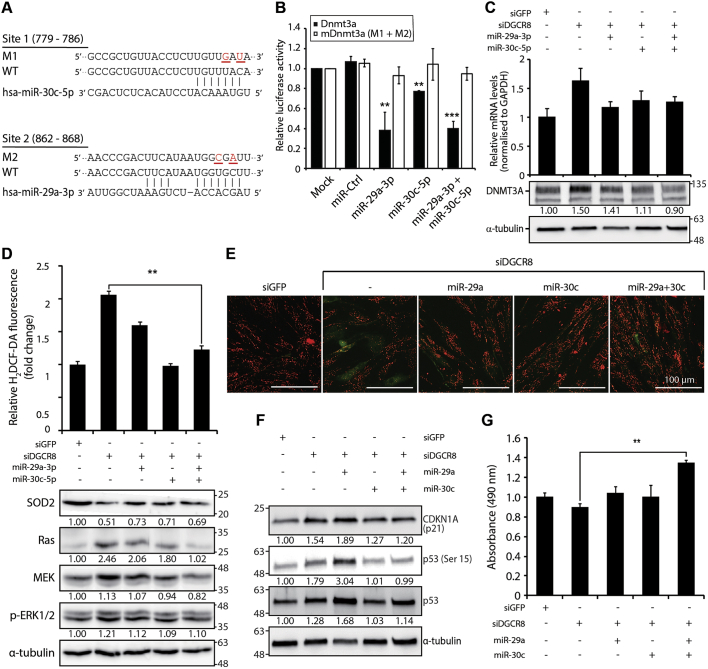
DNMT3A is a crucial epigenetic modifier; hence, it is tightly regulated by diverse mechanisms, including miRNAs. Analysis using TargetScan software (release 7.2) predicted that 54 miRNA families conserved among vertebrates could directly repress DNMT3A expression. Notably, family members of miR-29 (29a, 29b, and 29c) and miR-30 (30a, 30b, 30c, 30d, and 30e) shared high sequence homology and represented the top predicted DNMT3A repressors based on the cumulative weighted context++ score or the aggregate PCT score [43,44]. Consistently, both miRNA families had the highest mirSVR scores in the miRanda database (Supplementary Fig. S9). To determine whether DNMT3A is a direct target of the miR-29 and miR-30 families, we cloned the partial 3′ UTR of DNMT3A (662–868) downstream of the Renilla luciferase reporter gene and co-transfected the reporter construct with miRNA mimics into 293T cells (Fig. 6A). Compared with mock and miR-control co-transfected cells, luciferase activity was significantly decreased by ~60% by miR-29a-3p and by 20% by miR-30c-5p (Fig. 6B, black bars). Co-transfection with miR-29a-3p and miR-30c-5p mimics also significantly decreased luciferase activity. Moreover, mutagenesis of the putative miRNA binding sites increased luciferase activity (Fig. 6B, white bars), suggesting that miR-29a-3p and miR-30c-5p exert their inhibitory functions by directly binding to the DNMT3A 3′ UTR.
Fig. 6.
miR-29a-3p and miR-30c-5p directly repress DNMT3A and rescue oxidative stress and premature senescence induced by DGCR8 knockdown. (A) Schematic illustration of miR-30c-5p (site 1) and miR-29a-3p (site 2) binding sites within the human DNMT3A 3′ UTR (662–868) region, which was cloned downstream of the Renilla luciferase reporter gene in the psiCHECK-2 vector. Two predicted targeting sites and the corresponding mutations are listed. The mutagenised nucleotides are underlined and highlighted in red. The construct for the mutant 3′ UTR contained both mutagenised sites (M1+M2). Data were normalised to firefly luciferase. (B) Luciferase reporter assay. 293T cells were co-transfected with luciferase reporters carrying either the wild-type DNMT3A 3′ UTR (Dnmt3a) or the mutagenised DNMT3A 3′ UTR (mDnmt3a), as well as 50 nM negative control mimic (miR-Control) or miR-29a-3p or/and miR-30c-5p mimics. Error bars indicate the standard error of the mean of three experiments. (**P < 0.01 and ***P < 0.001 compared to the miR-control). (C) The mRNA and protein expression levels of DNMT3A in DGCR8 knockdown hMSCs with or without miR-29a-3p or/and miR-30c-5p overexpression. The mRNA levels of DNMT3A were normalised to GAPDH. Error bars denote the standard error of the mean of three independent experiments (top). Immunoblot showing DNMT3A protein levels in DGCR8 knockdown cells with or without miR-29a-3p or/and miR-30c-5p forced expression (bottom). (D) Total ROS levels in DGCR8 knockdown hMSCs after miR-29a-3p or/and miR-30c-5p overexpression were measured using H2DCF-DA. Error bars denote the standard error of the mean of three independent experiments (**P < 0.01). Immunoblot showing the protein levels of SOD2 and Ras signalling components (bottom). (E) Mitochondrial membrane potential (MMP) in DGCR8 knockdown hMSCs after miR-29a-3p or/and miR-30c-5p overexpression was determined using JC-1. Representative fluorescence microscopy images in siGFP, siDGCR8, and mimic-transfected siDGCR8 hMSCs. Scale bar, 100 μm. (F) Western blot showing the protein levels of tumour suppressors and senescence markers, including p21, p53, and p-p53 (ser15), after miR-29a-3p or/and miR-30c-5p overexpression in DGCR8 knockdown hMSCs. (G) Mean growth rate of siGFP and siDGCR8 hMSCs co-transfected with miR-29a-3p or/and miR-30c-5p mimics, as determined by MTS. Error bars denote the standard error of the mean of three independent experiments (**P < 0.01).
To determine the relevance of miR-29a-3p and miR-30c-5p in DGCR8 knockdown-mediated premature senescence, we forced their expression in siDGCR8-transfected cells. Overexpression of miR-29a-3p and/or miR-30c-5p significantly downregulated DNMT3A expression both at the mRNA and protein levels (Fig. 6C). Furthermore, forced expression of miR-29a-3p or miR-30c-5p significantly reduced ROS levels (Fig. 6D, top bar graphs) and protein levels of Ras signalling components, as well as restored SOD2 protein levels (Fig. 6D, bottom blots); these effects were more potent when miR-29a-3p and miR-30c-5p mimics were co-transfected. Intriguingly, overexpression of either miRNA or their combination restored the mitochondrial polarisation (Fig. 6E). We further confirmed the reduced levels of mitochondrial superoxide anion upon miR-29a-3p and/or miR-30c-5p overexpression as determined by MitoSOX staining (Supplementary Fig. S10A). These results suggest that miR-29a-3p and miR-30c-5p modulate mitochondrial oxidative stress in hMSCs by directly repressing DNMT3A expression and thereby regulating the expression of SOD2 and Ras signalling components.
We next examined the expression of senescence markers after transfection with miR-29a-3p and/or miR-30c-5p mimics in DGCR8 knockdown hMSCs. Interestingly, the levels of p21, p53, and p-p53 (Ser 15) were significantly reduced after transfection with miR-30c-5p or the combination of miR-29a-3p and miR-30c-5p (Fig. 6F). Oxidative stress and the aberrant accumulation of ROS could affect not only the early onset of senescence but also cell proliferation and survival [45]. Consistently, transfection of DGCR8 knockdown hMSCs with miR-29a-3p or miR-30c-5p mimics significantly decreased the production of IL-8, an SASP factor (Supplementary Fig. S10B), and increased the cell proliferation rate, and the combination of both had the most potent effect (**P < 0.01; Fig. 6G). These findings suggest that miR-29a-3p and miR-30c-5p can restore the abnormal senescence phenotypes observed in DGCR8 knockdown hMSCs.
4. Discussion
The use of hMSCs in clinical applications requires large numbers of cells. These cells can be produced by in vitro expansion; however, prolonged in vitro maintenance often compromises their quality [46]. Similarly to other primary cell types, long-term culture of hMSCs can result in cellular senescence, triggered by various internal or external stimuli, including accumulation of DNA damage, activation of oncogenes, oxidative stress, and mitochondrial dysfunction [6,7]. Frequently, multiple signals promote cellular senescence, and downstream effector pathways often overlap. Therefore, a better understanding of the regulatory mechanisms underlying cellular senescence is essential for the widespread application of stem cells in clinical applications.
miRNAs are critical regulators of various biological processes, including cellular senescence [21]. The elucidation of the biological functions of miRNAs can be extremely challenging, because each miRNA can regulate the expression of hundreds of genes [47] and different miRNAs can have redundant functions. Hence, to explore the importance of miRNAs during in vitro expansion of hMSCs, we silenced miRNAs globally by silencing DGCR8. We found that DGCR8 knockdown in early-passage hMSCs induced premature senescence with enhanced ROS production and mitochondrial impairment. Consistently, transcriptomics and bioinformatics analyses in DGCR8 knockdown hMSCs indicated a functional enrichment in mitochondrial dysfunction-related circuits. Similar approaches were used to address the role of miRNAs in replicative senescence in fibroblasts; DGCR8 depletion in mouse or human fibroblasts promoted cellular senescence by inducing the expression of specific miRNAs, such as the p21-targeting miR-93 [24].
Oxidative stress modulates mitochondrial function and induces cellular senescence directly or indirectly [48]; the role of miRNAs in these processes is also crucial. For example, under oxidative stress conditions, miR-210 promoted survival and suppressed apoptosis in cancer cells by reducing ROS levels [49]. Consistently, overexpression of miR-210 in rat MSCs increased the survival of MSCs during oxidative stress [50]. The survival of hMSCs under hypoxic conditions was enhanced by miR-210 through a positive feedback loop with hypoxia-inducible factor-1 (HIF-1) [51], suggesting a critical role of miR-210 in response to hypoxia [52]. Furthermore, miR-302 has been shown to prevent oxidant-induced cell death in human adipose-derived MSCs by targeting CCL5 [53]. Several other miRNAs have been shown to regulate the expression of antioxidant enzymes. For instance, miR-335 and miR-34a promoted senescence in rat kidney cells by downregulating the expression of mitochondrial antioxidant genes, such as SOD2 and thioredoxin reductase 2 (Txnrd2) [54]. By contrast, in this study, we demonstrated that miR-29a and miR-30c directly suppressed the expression of DNMT3A, thereby restoring SOD2 expression and promoting hMSC survival. The net effect of SOD2 loss is controversial. Archer et al. reported that SOD2 knockdown decreased H2O2 production [40]. In contrast, life-long reduction of SOD2 induced DNA damage and cancer risk [55]. In our prematurely senescent siDGCR8-transfected hMSCs, we observed misregulation of another antioxidant gene, peroxiredoxin 4 (PRDX4), encoding an enzyme responsible for reducing H2O2 levels. The mRNA levels of PRDX4 were decreased by about 21% in DGCR8-depleted cells, which may have been partially responsible for accumulation of H2O2 even under conditions of SOD2 repression upon DGCR8 loss.
Epigenetic modifications are also important regulators of cellular senescence, replicative senescence, and organismal ageing [56]. Replicative senescence in human fibroblasts was accompanied by DNA hypomethylation and focal hypermethylation [57]. More recently, Koch et al. [58] identified six CpG sites as epigenetic senescence biomarkers. Considering the vital role of DNMTs in DNA methylation [59], we analysed their relevance in the regulation of cellular senescence by miR-29a and miR-34c; we found that some of the effects of miR-29a and miR-34c are dependent on DNMT3A downregulation. The role of miR-29 family members in regulating DNA methylation by targeting DNMT3A has been demonstrated in various cancer types [60]. In most cases, deregulation of miR-29 expression alters the epigenetic landscape, promoting malignant transformation [[61], [62], [63], [64], [65], [66]] and cell invasion [67,68]. Furthermore, miR-29a has been shown to maintain self-renewal in mouse hematopoietic stem cells by targeting DNMT3A [69]. Therefore, several lines of evidence support the role of miR-29 family members in regulating key epigenetic mechanisms.
In conclusion, we demonstrated the role of DNMT3A in negatively regulating SOD2 expression. We also identified the critical components of this regulatory axis, which comprises miRNAs, DNA methyltransferases, and antioxidant enzymes. miR-29a and miR-30c repress DNMT3A expression, subsequently restoring SOD2 expression and alleviating oxidative stress in hMSCs. Based on these findings, we proposed a comprehensive regulatory circuit for SOD2 composed of two consecutive inhibitory steps. Although in this study, we identified a novel axis regulating cellular senescence in hMSCs, the implementation of these findings to develop miRNA-based therapeutic interventions could be challenging. Recently, DGCR8-independent stable miRNA expression (DISME) has emerged as a novel approach to stably express miRNAs, making the analysis of the long-term effects of specific miRNAs feasible [70]. miRNA-based therapies are considered to be the next-generation biopharmaceuticals for various diseases, including cancer and viral infections [71,72]; hence, the long-term in vitro and in vivo functions of miR-29a-3p and miR-30c-5p in cellular senescence and organismal ageing merit further investigation.
Accession number
All microarray data are available at the Gene Expression Omnibus (GEO) database with accession number GSE149171.
Declaration of competing interest
The authors declare no competing financial interests.
Acknowledgements
We thank all the members of Suh laboratory for their helpful and constructive comments. This work was supported by the Soon Chun Hyang University Research Fund and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute funded by the Ministry of Health & Welfare, Republic of Korea [HI18C0283].
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101716.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary data associated with this article can be found in the online version at Redox Biology's website.
References
- 1.Bianco P., Cao X., Frenette P.S., Mao J.J., Robey P.G., Simmons P.J. The meaning, the sense and the significance: translating the science of mesenchymal stem cells into medicine. Nat. Med. 2013;19(1):35–42. doi: 10.1038/nm.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.da Silva Meirelles L., Chagastelles P.C., Nardi N.B. Mesenchymal stem cells reside in virtually all post-natal organs and tissues. J. Cell Sci. 2006;119(Pt 11):2204–2213. doi: 10.1242/jcs.02932. [DOI] [PubMed] [Google Scholar]
- 3.Ullah I., Subbarao R.B., Rho G.J. Human mesenchymal stem cells - current trends and future prospective. Biosci. Rep. 2015;35(2) doi: 10.1042/BSR20150025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daley G.Q. The promise and perils of stem cell therapeutics. Cell Stem Cell. 2012;10(6):740–749. doi: 10.1016/j.stem.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayflick L., Moorhead P.S. The serial cultivation of human diploid cell strains. Exp. Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 6.Hernandez-Segura A., Nehme J., Demaria M. Hallmarks of cellular senescence. Trends Cell Biol. 2018;28(6):436–453. doi: 10.1016/j.tcb.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Kuilman T., Michaloglou C., Mooi W.J., Peeper D.S. The essence of senescence. Genes Dev. 2010;24(22):2463–2479. doi: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Witte S.F.H., Lambert E.E., Merino A., Strini T., Douben H., O'Flynn L. Aging of bone marrow- and umbilical cord-derived mesenchymal stromal cells during expansion. Cytotherapy. 2017;19(7):798–807. doi: 10.1016/j.jcyt.2017.03.071. [DOI] [PubMed] [Google Scholar]
- 9.De Becker A., Riet I.V. Homing and migration of mesenchymal stromal cells: how to improve the efficacy of cell therapy? World J. Stem Cell. 2016;8(3):73–87. doi: 10.4252/wjsc.v8.i3.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu T., Finkel T. Free radicals and senescence. Exp. Cell Res. 2008;314(9):1918–1922. doi: 10.1016/j.yexcr.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernandez-Segura A., de Jong T.V., Melov S., Guryev V., Campisi J., Demaria M. Unmasking transcriptional heterogeneity in senescent cells. Curr. Biol. 2017;27(17):2652–26560 e4. doi: 10.1016/j.cub.2017.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiley C.D., Velarde M.C., Lecot P., Liu S., Sarnoski E.A., Freund A. Mitochondrial dysfunction induces senescence with a distinct secretory phenotype. Cell Metabol. 2016;23(2):303–314. doi: 10.1016/j.cmet.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lowe S.W., Cepero E., Evan G. Intrinsic tumour suppression. Nature. 2004;432(7015):307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 14.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 15.Bartel D.P. Metazoan MicroRNAs. Cell. 2018;173(1):20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suh N., Blelloch R. Small RNAs in early mammalian development: from gametes to gastrulation. Development. 2011;138(9):1653–1661. doi: 10.1242/dev.056234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo W.T., Wang Y. Dgcr8 knockout approaches to understand microRNA functions in vitro and in vivo. Cell. Mol. Life Sci. 2019;76(9):1697–1711. doi: 10.1007/s00018-019-03020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y., Medvid R., Melton C., Jaenisch R., Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 2007;39(3):380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y., Baskerville S., Shenoy A., Babiarz J.E., Baehner L., Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat. Genet. 2008;40(12):1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu K.L., Zhang Q., Yan Y., Li T.T., Duan F.F., Hao J. Pluripotency-associated miR-290/302 family of microRNAs promote the dismantling of naive pluripotency. Cell Res. 2016;26(3):350–366. doi: 10.1038/cr.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suh N. MicroRNA controls of cellular senescence. BMB Rep. 2018;51(10):493–499. doi: 10.5483/BMBRep.2018.51.10.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borgdorff V., Lleonart M.E., Bishop C.L., Fessart D., Bergin A.H., Overhoff M.G. Multiple microRNAs rescue from Ras-induced senescence by inhibiting p21(Waf1/Cip1) Oncogene. 2010;29(15):2262–2271. doi: 10.1038/onc.2009.497. [DOI] [PubMed] [Google Scholar]
- 23.Mudhasani R., Zhu Z., Hutvagner G., Eischen C.M., Lyle S., Hall L.L. Loss of miRNA biogenesis induces p19Arf-p53 signaling and senescence in primary cells. J. Cell Biol. 2008;181(7):1055–1063. doi: 10.1083/jcb.200802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gomez-Cabello D., Adrados I., Gamarra D., Kobayashi H., Takatsu Y., Takatsu K. DGCR8-mediated disruption of miRNA biogenesis induces cellular senescence in primary fibroblasts. Aging Cell. 2013;12(5):923–931. doi: 10.1111/acel.12117. [DOI] [PubMed] [Google Scholar]
- 25.Zuk P.A., Zhu M., Mizuno H., Huang J., Futrell J.W., Katz A.J. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng. 2001;7(2):211–228. doi: 10.1089/107632701300062859. [DOI] [PubMed] [Google Scholar]
- 26.Zuk P.A., Zhu M., Ashjian P., De Ugarte D.A., Huang J.I., Mizuno H. Human adipose tissue is a source of multipotent stem cells. Mol. Biol. Cell. 2002;13(12):4279–4295. doi: 10.1091/mbc.E02-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park S.K., Lee J.S., Choi E.K., You D., Kim C.S., Suh N. Global knockdown of microRNAs affects the expression of growth factors and cytokines in human adipose-derived mesenchymal stem cells. BMB Rep. 2014;47(8):469–474. doi: 10.5483/BMBRep.2014.47.8.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han J., Pedersen J.S., Kwon S.C., Belair C.D., Kim Y.K., Yeom K.H. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136(1):75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J., Tian Z., Gao B., Kunos G. Dose-dependent activation of antiapoptotic and proapoptotic pathways by ethanol treatment in human vascular endothelial cells: differential involvement of adenosine. J. Biol. Chem. 2002;277(23):20927–20933. doi: 10.1074/jbc.M110712200. [DOI] [PubMed] [Google Scholar]
- 30.Hwang S., Park S.K., Lee H.Y., Kim S.W., Lee J.S., Choi E.K. miR-140-5p suppresses BMP2-mediated osteogenesis in undifferentiated human mesenchymal stem cells. FEBS Lett. 2014;588(17):2957–2963. doi: 10.1016/j.febslet.2014.05.048. [DOI] [PubMed] [Google Scholar]
- 31.Dimauro T., David G. Ras-induced senescence and its physiological relevance in cancer. Curr. Cancer Drug Targets. 2010;10(8):869–876. doi: 10.2174/156800910793357998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Courtois-Cox S., Jones S.L., Cichowski K. Many roads lead to oncogene-induced senescence. Oncogene. 2008;27(20):2801–2809. doi: 10.1038/sj.onc.1210950. [DOI] [PubMed] [Google Scholar]
- 33.Moll U.M., Petrenko O. The MDM2-p53 interaction. Mol. Canc. Res. : MCR. 2003;1(14):1001–1008. [PubMed] [Google Scholar]
- 34.Kuhlbrandt W. Structure and function of mitochondrial membrane protein complexes. BMC Biol. 2015;13:89. doi: 10.1186/s12915-015-0201-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jezek J., Cooper K.F., Strich R. Reactive oxygen species and mitochondrial dynamics: the yin and yang of mitochondrial dysfunction and cancer progression. Antioxidants. 2018;7(1) doi: 10.3390/antiox7010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fukai T., Ushio-Fukai M. Superoxide dismutases: role in redox signaling, vascular function, and diseases. Antioxidants Redox Signal. 2011;15(6):1583–1606. doi: 10.1089/ars.2011.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huntzinger E., Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011;12(2):99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 38.Cyr A.R., Hitchler M.J., Domann F.E. Regulation of SOD2 in cancer by histone modifications and CpG methylation: closing the loop between redox biology and epigenetics. Antioxidants Redox Signal. 2013;18(15):1946–1955. doi: 10.1089/ars.2012.4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim H.P., Roe J.H., Chock P.B., Yim M.B. Transcriptional activation of the human manganese superoxide dismutase gene mediated by tetradecanoylphorbol acetate. J. Biol. Chem. 1999;274(52):37455–37460. doi: 10.1074/jbc.274.52.37455. [DOI] [PubMed] [Google Scholar]
- 40.Archer S.L., Marsboom G., Kim G.H., Zhang H.J., Toth P.T., Svensson E.C. Epigenetic attenuation of mitochondrial superoxide dismutase 2 in pulmonary arterial hypertension: a basis for excessive cell proliferation and a new therapeutic target. Circulation. 2010;121(24):2661–2671. doi: 10.1161/CIRCULATIONAHA.109.916098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao X., Xu Q., Sun Y., Lu Z., Li R., Wang X. 5aza2'deoxycytidine promotes migration of acute monocytic leukemia cells via activation of CCL2CCR2ERK signaling pathway. Mol. Med. Rep. 2017;16(2):1417–1424. doi: 10.3892/mmr.2017.6737. [DOI] [PubMed] [Google Scholar]
- 42.Greenberg M.V.C., Bourc'his D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019;20(10):590–607. doi: 10.1038/s41580-019-0159-6. [DOI] [PubMed] [Google Scholar]
- 43.Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Friedman R.C., Farh K.K., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dimozi A., Mavrogonatou E., Sklirou A., Kletsas D. Oxidative stress inhibits the proliferation, induces premature senescence and promotes a catabolic phenotype in human nucleus pulposus intervertebral disc cells. Eur. Cell. Mater. 2015;30:89–102. doi: 10.22203/ecm.v030a07. discussion 3. [DOI] [PubMed] [Google Scholar]
- 46.Wagner W., Horn P., Castoldi M., Diehlmann A., Bork S., Saffrich R. Replicative senescence of mesenchymal stem cells: a continuous and organized process. PloS One. 2008;3(5) doi: 10.1371/journal.pone.0002213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Subramanyam D., Blelloch R. From microRNAs to targets: pathway discovery in cell fate transitions. Curr. Opin. Genet. Dev. 2011;21(4):498–503. doi: 10.1016/j.gde.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vono R., Jover Garcia E., Spinetti G., Madeddu P. Oxidative stress in mesenchymal stem cell senescence: regulation by coding and noncoding RNAs. Antioxidants Redox Signal. 2018;29(9):864–879. doi: 10.1089/ars.2017.7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Favaro E., Ramachandran A., McCormick R., Gee H., Blancher C., Crosby M. MicroRNA-210 regulates mitochondrial free radical response to hypoxia and krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU. PloS One. 2010;5(4) doi: 10.1371/journal.pone.0010345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu J., Huang Z., Lin L., Fu M., Gao Y., Shen Y. miR-210 over-expression enhances mesenchymal stem cell survival in an oxidative stress environment through antioxidation and c-Met pathway activation. Sci. China Life Sci. 2014;57(10):989–997. doi: 10.1007/s11427-014-4725-z. [DOI] [PubMed] [Google Scholar]
- 51.Chang W., Lee C.Y., Park J.H., Park M.S., Maeng L.S., Yoon C.S. Survival of hypoxic human mesenchymal stem cells is enhanced by a positive feedback loop involving miR-210 and hypoxia-inducible factor 1. J. Vet. Sci. 2013;14(1):69–76. doi: 10.4142/jvs.2013.14.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bavelloni A., Ramazzotti G., Poli A., Piazzi M., Focaccia E., Blalock W. MiRNA-210: a current overview. Anticancer Res. 2017;37(12):6511–6521. doi: 10.21873/anticanres.12107. [DOI] [PubMed] [Google Scholar]
- 53.Kim J.Y., Shin K.K., Lee A.L., Kim Y.S., Park H.J., Park Y.K. MicroRNA-302 induces proliferation and inhibits oxidant-induced cell death in human adipose tissue-derived mesenchymal stem cells. Cell Death Dis. 2014;5:e1385. doi: 10.1038/cddis.2014.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bai X.Y., Ma Y., Ding R., Fu B., Shi S., Chen X.M. miR-335 and miR-34a Promote renal senescence by suppressing mitochondrial antioxidative enzymes. J. Am. Soc. Nephrol. 2011;22(7):1252–1261. doi: 10.1681/ASN.2010040367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Remmen H., Ikeno Y., Hamilton M., Pahlavani M., Wolf N., Thorpe S.R. Life-long reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging. Physiol. Genom. 2003;16(1):29–37. doi: 10.1152/physiolgenomics.00122.2003. [DOI] [PubMed] [Google Scholar]
- 56.Wagner W. The link between epigenetic clocks for aging and senescence. Front. Genet. 2019;10:303. doi: 10.3389/fgene.2019.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cruickshanks H.A., McBryan T., Nelson D.M., Vanderkraats N.D., Shah P.P., van Tuyn J. Senescent cells harbour features of the cancer epigenome. Nat. Cell Biol. 2013;15(12):1495–1506. doi: 10.1038/ncb2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koch C.M., Joussen S., Schellenberg A., Lin Q., Zenke M., Wagner W. Monitoring of cellular senescence by DNA-methylation at specific CpG sites. Aging Cell. 2012;11(2):366–369. doi: 10.1111/j.1474-9726.2011.00784.x. [DOI] [PubMed] [Google Scholar]
- 59.Liao J., Karnik R., Gu H., Ziller M.J., Clement K., Tsankov A.M. Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat. Genet. 2015;47(5):469–478. doi: 10.1038/ng.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fabbri M., Garzon R., Cimmino A., Liu Z., Zanesi N., Callegari E. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. U. S. A. 2007;104(40):15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oliveira L.H., Schiavinato J.L., Fraguas M.S., Lucena-Araujo A.R., Haddad R., Araujo A.G. Potential roles of microRNA-29a in the molecular pathophysiology of T-cell acute lymphoblastic leukemia. Canc. Sci. 2015;106(10):1264–1277. doi: 10.1111/cas.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gasque Schoof C.R., Izzotti A., Jasiulionis M.G., Vasques Ldos R. The roles of miR-26, miR-29, and miR-203 in the silencing of the epigenetic machinery during melanocyte transformation. BioMed Res. Int. 2015;2015:634749. doi: 10.1155/2015/634749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robaina M.C., Mazzoccoli L., Arruda V.O., Reis F.R., Apa A.G., de Rezende L.M. Deregulation of DNMT1, DNMT3B and miR-29s in Burkitt lymphoma suggests novel contribution for disease pathogenesis. Exp. Mol. Pathol. 2015;98(2):200–207. doi: 10.1016/j.yexmp.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 64.Xu H., Sun J., Shi C., Sun C., Yu L., Wen Y. miR-29s inhibit the malignant behavior of U87MG glioblastoma cell line by targeting DNMT3A and 3B. Neurosci. Lett. 2015;590:40–46. doi: 10.1016/j.neulet.2015.01.060. [DOI] [PubMed] [Google Scholar]
- 65.Parpart S., Roessler S., Dong F., Rao V., Takai A., Ji J. Modulation of miR-29 expression by alpha-fetoprotein is linked to the hepatocellular carcinoma epigenome. Hepatology. 2014;60(3):872–883. doi: 10.1002/hep.27200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nguyen T., Kuo C., Nicholl M.B., Sim M.S., Turner R.R., Morton D.L. Downregulation of microRNA-29c is associated with hypermethylation of tumor-related genes and disease outcome in cutaneous melanoma. Epigenetics. 2011;6(3):388–394. doi: 10.4161/epi.6.3.14056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Palmbos P.L., Wang L., Yang H., Wang Y., Leflein J., Ahmet M.L. ATDC/TRIM29 drives invasive bladder cancer formation through miRNA-mediated and epigenetic mechanisms. Canc. Res. 2015;75(23):5155–5166. doi: 10.1158/0008-5472.CAN-15-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cui H., Wang L., Gong P., Zhao C., Zhang S., Zhang K. Deregulation between miR-29b/c and DNMT3A is associated with epigenetic silencing of the CDH1 gene, affecting cell migration and invasion in gastric cancer. PloS One. 2015;10(4) doi: 10.1371/journal.pone.0123926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu W., Dooley J., Chung S.S., Chandramohan D., Cimmino L., Mukherjee S. miR-29a maintains mouse hematopoietic stem cell self-renewal by regulating Dnmt3a. Blood. 2015;125(14):2206–2216. doi: 10.1182/blood-2014-06-585273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang X.W., Hao J., Guo W.T., Liao L.Q., Huang S.Y., Guo X. A DGCR8-independent stable MicroRNA expression strategy reveals important functions of miR-290 and miR-183-182 families in mouse embryonic stem cells. Stem Cell Reports. 2017;9(5):1618–1629. doi: 10.1016/j.stemcr.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Christopher A.F., Kaur R.P., Kaur G., Kaur A., Gupta V., Bansal P. MicroRNA therapeutics: discovering novel targets and developing specific therapy. Perspect Clin Res. 2016;7(2):68–74. doi: 10.4103/2229-3485.179431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chakraborty C., Sharma A.R., Sharma G., Doss C.G.P., Lee S.S. Therapeutic miRNA and siRNA: moving from bench to clinic as next generation medicine. Mol. Ther. Nucleic Acids. 2017;8:132–143. doi: 10.1016/j.omtn.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.