1. INTRODUCTION
Since 31 December 2019, over 9 million cases of SARS-CoV-2 have been reported on five continents. During the 2003 SARS outbreak, the reported burden of asymptomatic infections was 7.5% among healthcare workers, and 13% among SARS-CoV positive cases [1]. In the current pandemic (COVID-19), the percentage of asymptomatic carriers is even higher up to 56.5% [2], posing a serious challenge to controlling the spread.
A key factor in the infectious process is the interaction between the viral surface receptor(s) and the host-cell receptor(s). A study using combined molecular docking and structural bioinformatics has demonstrated that the SARS-CoV-2 spike protein can interact with the Glucose Regulating Protein 78 (GRP78) on the surface of human respiratory cells [3]. The same approach could shed light on the possible interactions between SARS-CoV-2 spike protein and GRP78 in different animal species. We hypothesize that some animals that come into close contact with humans might become a reservoir for the disease. To explore this hypothesis, we studied the structure of GRP78 protein in different species and compared it with that in humans. Identifying any potential ecological reservoirs of the SARS-CoV-2 virus could help predict possible future outbreaks.
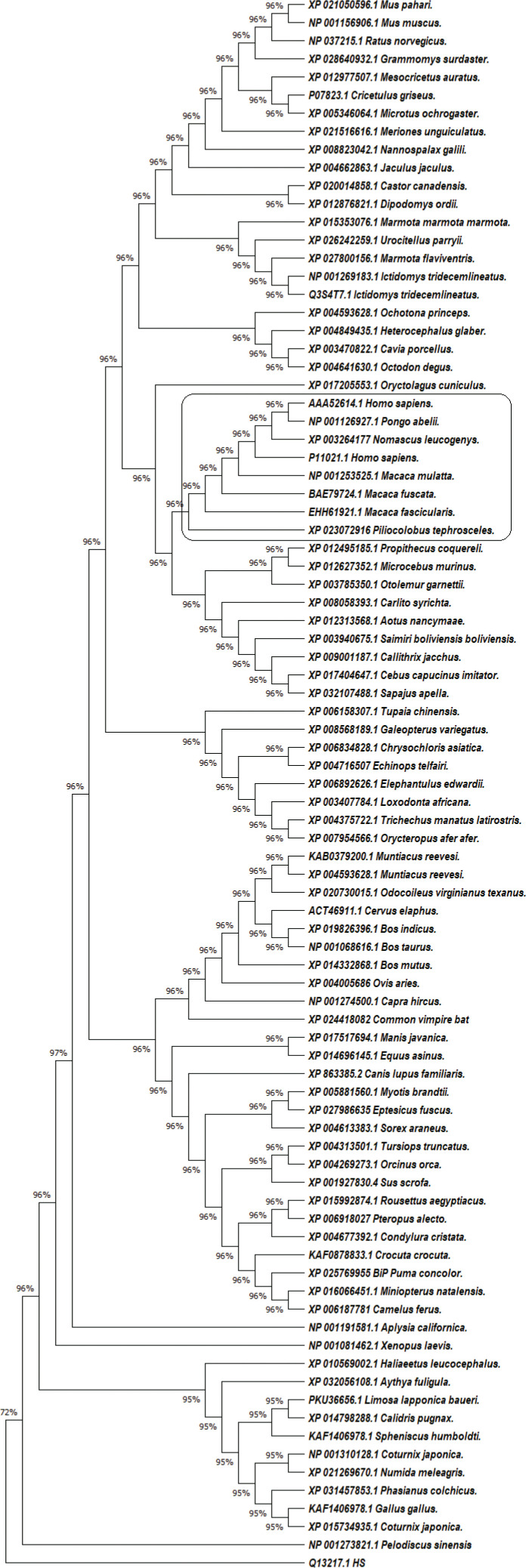
In our research, we retrieved the amino-acid sequences of the GRP78 proteins in 84 mammals, amphibians and birds from the National Center for Biotechnology Information (NCBI) database (Figure 1). For data analysis, we applied statistical Maximum Likelihood (ML) analysis with MEGA X software, Cn3D – including the online version in NCBI, and Phyre2 [4]. In the phylogenetic analysis, we used human DnaJ homolog subfamily C member 3 protein (NCBI accession number Q13217.1) as an outgroup; and the differences between bovine and human GRP78 protein, determined by Yang et al. [5], as an ingroup reference. The human GRP78 deposited by Yang et al. [5] under sequence number P11021.1.
Figure 1.
The evolutionary history of 87 amino acid sequences of GRP78 protein from 84 species was inferred by using the ML statistical method, JTT matrix-based model plus Gamma distribution (+G, parameter = 0.2191) and bootstrap 1000. There were a total of 683 positions in the final dataset. The human GRP78 protein grouped together with GRP78 from Macaca mulatta, Macaca fascicularis, Macaca fuscata, Piliocolobus tephrosceles, Nomascus leucogenys and Pongo abelii.
Multiple sequence alignment and phylogenetic analysis identified several taxa (Figure 1). The species with the highest similarity to the human GRP78 protein were apes from the genera Macaca, Nomascus, Pongo and Piliocolobus. This similarity was also present in the 3D visualization of the proteins.
In humans, GRP78 consists of two domains: a nucleotide-binding domain and a Substrate-binding Domain (SBD). SBD has two subdomains: SBDα, a 10-kDa subdomain, and SBDβ, a 25-kDa subdomain. The active site of the protein consists of amino acid residues located in SBD: L1,2 (I426, T428, V429, V432, T434) and L3,4 (F451, S452, V461, and I463), which interact with the substrate through Van der Waals forces [5]. Ibrahim et al. [3] described the interaction between the spike protein of SARS-CoV-2 and cell-surface GRP78 SBDβ with the corresponding amino-acid residues [3]. While analyzing the structure of GRP78 in the phylogenetically similar species, using the data reported by Ibrahim et al. [3] in their Table 1, we found complete congruence with some of the amino acid residues in human GRP78. Other positions did not have a corresponding match in the human reference sequences of GRP78, e.g.: residues forming hydrogen bonds in positions G489 (P), V453 (T), V490 (A), G489 (P), G454 (A), T456 (D), Q492 (R), T458 (Q) and some hydrophobic residues – V490 (A), Q492 (R), V453 (T), V457 (N) (The letters in brackets indicate the amino acids in the reference molecule P11021.1). We associate the lack of matches in these positions with possible technical errors, due to the immense amount of data analyzed by the authors; else the role of GRP78 receptors would not be as significant as thought.
We found differences in SBD (TASDNQP → TASDTQP) in Macaca fuscata. However, this modification in L3,4 does not have a significant effect on the polypeptide-binding pocket of the protein [6].
Our analysis suggested that domestic animals are not likely to act as an epidemiological factor due to significant differences in the structure of their GRP78 protein. The species that stood out as potential SARS-CoV-2 reservoirs belong to the so-called Old World Мonkey (OWM) family Cercopithecidae, with a broad distribution in Asia (Macaca mulatta), Southeast Asia (Macaca fascicularis), Japan (M. fuscata), and Africa – Tanzania, Uganda, Democratic Republic of Congo, Rwanda, Burundi (Piliocolobus tephrosceles). Potential reservoirs other than OWM are Nomascus leucogenys, found in Vietnam and Laos, and Pongo abelii, from Sumatra (superfamily Hominoidea). The animals from the group of OWM are one of the important reservoirs for Lentiviruses and have played an essential role in the evolution of HIV [7]. In superfamily Hominoidea, there are reports about interspecies transmissions between man and some closely-related species (Pan, Gorilla, Pongo and Hylobates), e.g. evidence of HBV of gibbon and human origin in chimpanzees [8]. Based on these data and our results, we hypothesize that these species could act as potential reservoirs for SARS-CoV-2.
Interestingly, in our analysis, Manis javanica did not demonstrate a significant risk of becoming a reservoir for SARS-CoV-2 despite the reported link between Pangolin-CoV and SARS-CoV-2 [9]. This could be because Li et al. [9] show the probable path of cross-species transmission of the virus from bat or pangolin and its adaptation to the human body. After the virus has adapted to the new host, it can become a single-host pathogen infecting humans only or it can also infect species other than the ones the virus has originated from. An example of the latter evolutionary scenario is the human measles virus, which has resulted from a cattle-to-human host-jump of rinderpest virus [10] or HBV. In our study, we discuss the possibility of transmission of SARS-CoV-2 from humans to primates to humans.
Our findings based on GRP78 protein analysis corroborated with published reports support the hypothesis that some primate species – but not likely domesticated animals – could serve as a reservoir for SARS-CoV-2. Surveillance of those potential reservoirs for SARS-COV-2 can help prevent future outbreaks.
ACKNOWLEDGMENTS
We would like to acknowledge Prof. T. Strateva (Department “Medical Microbiology”, Medical University – Sofia) for her invaluable advice and guidance while writing the manuscript.
CONFLICTS OF INTEREST
The authors declare they have no conflicts of interest.
FUNDING
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
REFERENCES
- [1].Wilder-Smith A, Teleman MD, Heng BH, Earnest A, Ling AE, Leo YS. Asymptomatic SARS coronavirus infection among healthcare workers, Singapore. Emerg Infect Dis. 2005;11:1142–5. doi: 10.3201/eid1107.041165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kimball A, Hatfield KM, Arons M, James A, Taylor J, Spicer K, et al. Asymptomatic and presymptomatic SARS-CoV-2 infections in residents of a long-term care skilled nursing facility — King County, Washington, March 2020. MMWR Morb Mortal Wkly Rep. 2020;69:377–81. doi: 10.15585/mmwr.mm6913e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ibrahim IM, Abdelmalek DH, Elshahat ME, Elfiky AA. COVID-19 spike-host cell receptor GRP78 binding site prediction. J Infect. 2020;80:554–62. doi: 10.1016/j.jinf.2020.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–58. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Yang J, Nune M, Zong Y, Zhou L, Liu Q. Close and allosteric opening of the polypeptide-binding site in a human Hsp70 chaperone BiP. Structure. 2015;23:2191–203. doi: 10.1016/j.str.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yang J, Zong Y, Su J, Li H, Zhu H, Columbus L, et al. Conformation transitions of the polypeptide-binding pocket support an active substrate release from Hsp70s. Nat Commun. 2017;8:1201. doi: 10.1038/s41467-017-01310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].McCarthy KR, Kirmaier A, Autissier P, Johnson WE. Evolutionary and functional analysis of old world primate TRIM5 reveals the ancient emergence of primate lentiviruses and convergent evolution targeting a conserved capsid interface. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005085. e1005085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bonvicino CR, Moreira MA, Soares MA. Hepatitis B virus lineages in mammalian hosts: potential for bidirectional cross-species transmission. World J Gastroenterol. 2014;20:7665–74. doi: 10.3748/wjg.v20.i24.7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li X, Zai J, Zhao Q, Nie Q, Li Y, Foley BT, et al. Evolutionary history, potential intermediate animal host, and cross-species analyses of SARS-CoV-2. J Med Virol. 2020;92:602–11. doi: 10.1002/jmv.25731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Nambulli S, Sharp CR, Acciardo AS, Drexler JF, Duprex WP. Mapping the evolutionary trajectories of morbilliviruses: what, where and whither. Curr Opin Virol. 2016;16:95–105. doi: 10.1016/j.coviro.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]