Abstract
Autophagy and apoptosis play crucial roles in tumorigenesis. Recent studies have shown that autophagy and apoptosis have a cross-talk relationship in anti-tumor therapy. It is well established that apoptosis is one of the main pathways of tumor cell death. While autophagy can occurs in tumors with opposite function: protective autophagy and lethal autophagy. Protective autophagy can inhibit tumor apoptosis induced by anticancer drugs, while lethal autophagy can induce tumor cell apoptosis in cooperation with anticancer drugs. Hence, autophagy and apoptosis have synergistic and antagonistic effects in tumor. Colorectal cancer is a common malignant tumor with high morbidity and mortality. In recent years, colorectal carcinoma has achieved improved clinical efficacy with drug treatment. Nonetheless, increasing drug-resistance limit the treatment efficacy, highlighting the urgency of exploring the molecular events that drive drug resistance. Researchers have found that autophagy is one of the major factors leading to drug resistance in colon cancer. Therefore, elucidating the interaction between autophagy and apoptosis is helpful to improve the efficacy of anticancer drugs in clinical treatment of colorectal cancer. This review attaches great importance to the relationship between autophagy and apoptosis and related factors in colorectal cancer.
Keywords: Colorectal carcinoma (CRC), Autophagy, Apoptosis, Interaction, Balance
Introduction
Colorectal Carcinoma is one of the most common types of cancer and ranks third in morbidity and mortality among all tumors [1]. The pathogenesis of colorectal cancer is a complicated multifactorial process involving mutations in specific oncogenes and tumor suppressor genes. Recent detailed studies have defined three major genetic pathways that lead to the occurrence of CRC: the chromosomal instability pathway (CIN), serrated pathway (CPG island methylator phenotype pathway-CIMP), and microsatellite instability pathway (MSI) [2]. In recent years, the incidence of colorectal cancer has been continually increasing and tends to be younger, but its mortality has declined [3]. This has to do with changes in personal eating habits and lifestyles as well as the improvement of medical levels through economic development, making socio-economic development important factors affecting the incidence and mortality of colorectal cancer [4,5]. In a word, the situation of diagnosis and treatment of colorectal cancer is not optimistic. Surgery remains the primary treatment in cases diagnosed early, but it is no longer effective in advanced cancer cases [6]. In these patients, the efficacy of neoadjuvant and cytotoxic therapy has been inhibited by rapidly evolving drug resistance and cancer recurrence [7]. Therefore, early prevention, diagnosis and treatment of colorectal cancer is essential to reduce the incidence and improve the survival rate of patients [8]. At present, a variety of targeted therapeutic drugs for colorectal cancer has been developed clinically. There are three main classes of drugs: monoclonal antibodies against EGFR (cetuximab and panitumumab), monoclonal antibodies against VEGF-A (bevacizumab), fusion proteins against various angiogenic growth factors (e.g., aflibercept), and small-molecule multi-kinase inhibitors (e.g., regorafenib) [9]. With the development of new targeted drug therapies and the in-depth study of drug resistance mechanisms, it seems that the survival rate of colorectal cancer patients can be greatly improved.
Autophagy is a dynamic, multi-step process of degradation of cytoplasmic proteins and organelles by lysosomal pathway, which is closely related to highly conserved genes, namely autophagy-related genes (ATG), and can be subdivided into five stages: initiation, nucleation of the autophagosome, expansion and elongation of the autophagosome membrane, closure and fusion with the lysosome, and the degradation of intravesicular products [10,11]. Based on the pathways that transport cytoplasm to lysosomal lumens, there are three different types of autophagy: macroautophagy, microautophagy and chaperone-mediated autophagy. Autophagy is generally referred to as macro-autophagy [12,13], which is mainly mediated through the PI3K/Akt/mTOR and AMPK/mTOR signaling pathways [14], the molecular mechanism by which mTOR kinase is an important regulatory molecule that induces autophagy. mTOR can be activated by PI3K/AKT/mTOR pathway to inhibit autophagy, while the AMPK/mTOR pathway negatively regulates mTOR to promote autophagy [15]. Autophagy occurs frequently during tumorigenesis and chemotherapy. In general, autophagy protects cancer cells from apoptosis during chemotherapy, leading to drug resistance and refractory cancers [16]. However, other researchers have found that autophagy can also cause cell death and inhibit cell growth or have no effect [17], which depends on tissue type, stage of tumor development, and degree of autophagy activity [18]. Thus, autophagy is a double-edged sword in tumors. Nowadays, enhancing enhance the anti-tumor effect through autophagy induction has become a hot topic in medical biology.
Apoptosis, also known as type I programmed cell death (PCD I), is an autonomic regulated physiological process leading to cell death characterized by cell shrinkage, membrane blebbing, DNA fragmentation and the formation of apoptotic bodies. There are two main apoptotic pathways: exogenous or death receptor pathway and intrinsic or mitochondrial pathway, as well as a perforin/granzyme pathway. These three pathways converge on the same terminal, or execution pathway, which is initiated by the cleavage of caspase-3 [19,20]. Once caspases-3 are initially activated, cell death seems irreversible. Activation of caspase-3 occurs through the upstream signaling molecule Caspase-9 or caspase-8, and then the activated Caspase-3 splices PARP, inactivating PARP and subsequently leading to apoptosis. Thus, the regulation of cell death by apoptosis is unidirectional [21,22], and caspases-3 are central to the mechanism of apoptosis. Meanwhile, the balance of pro-apoptotic and anti-apoptotic protein regulators is the key to determination whether apoptosis occurs [23]. Cancer is often found to over express anti-apoptotic proteins to resist apoptosis [24], helping cancer cells survive, proliferate, and resist drugs [25]. Hence, deregulation in apoptotic cell death machinery is a hallmark of cancer [26]. To sum up, apoptosis is a promising target in the treatment of tumor.
The main way of crosstalk between autophagy and apoptosis in CRC
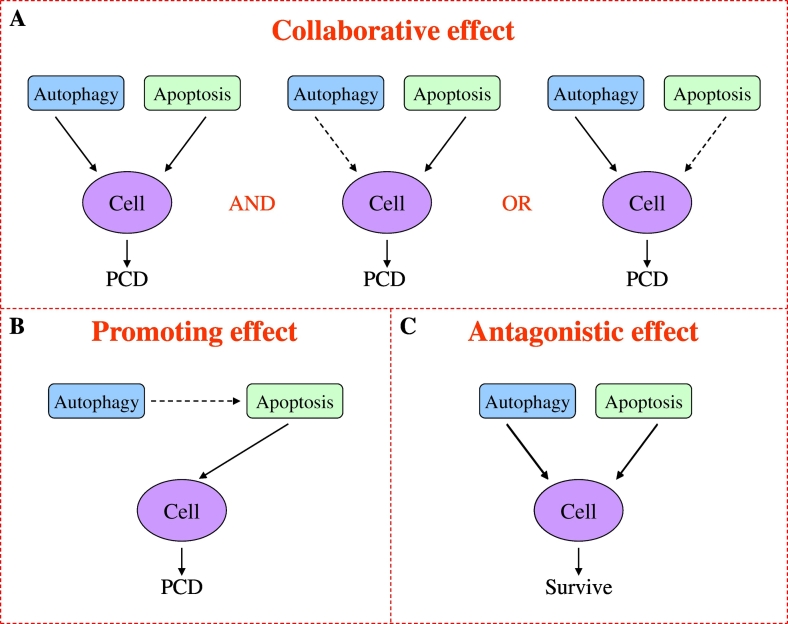
Autophagy and apoptosis are both typical types of programmed cell death, and dysfunction of their pathways can lead to the development of tumors. Nevertheless, relevant studies have shown that autophagy has a dual role in tumors, which can also help tumor cells survival under stress conditions such as hypoxia or low nutrition [27]. Autophagy and apoptosis usually occur within the same cell, and on the sequence of autophagy is in advance of apoptosis [28]. At the same time, whether autophagy induces or inhibits apoptosis depending on the type of cell, nature, and duration of the stimulus/stress [29]. Therefore, the relationship between autophagy and apoptosis is complex in CRC, involving a variety of signal transduction pathways and regulators. According to the different interaction of autophagy and apoptosis in tumor, it can be roughly divided into synergistic effect, promoting effect and antagonistic effect [27,28,30,31] (Fig. 1).
Fig. 1.
Interaction patterns between autophagy and apoptosis in the regulation of cell death in colorectal cancer. (A) Autophagy and apoptosis can induce cell death by the form of synchronous or one main and auxiliary. (B) Autophagy can induce cell death by promoting apoptosis. (C) Autophagy antagonizes apoptosis during cell death.
Collaborative effect
Autophagy, also known as type II programmed cell death, like apoptosis, can lead to tumor cell death by chemotherapeutic drugs in tumor-dependent autophagy [32]. It depends on the tissue type, stage of tumor development, and degree of autophagy activity [18]. For example, excessive or prolonged autophagy can lead to cell death [27]. At present, many studies have found that autophagy and apoptosis play a collaborative role in the treatment of CRC, but the precise mechanism of their collaboration is unclear. According to the report, the activation of Galectin-1 induced by shikonin potently activates apoptosis in colorectal carcinoma cells and autophagic cell death both in vitro and in vivo [33]. The accumulation of ROS induced by shikonin promotes the formation of Galectin-1 dimerization, which induces autophagy and apoptosis of downstream cells by activating the JNK signaling pathway, eventually leading to cell death together. In addition, M4IDP induces apoptosis through the inhibition of Akt phosphorylation and the increase of Bad protein expression, and induces lethal autophagy through the prolonged inhibition of Akt/mTOR/p70S6K pathway [32]. This result shows that the PI3K/AKT/mTOR pathway plays different roles in autophagy and apoptosis of colorectal cancer. Because of PI3K/AKT/mTOR pathway can not only inhibit autophagy through the activation of mTOR but also phosphorylate caspase-3, Caspase-9, and Bad through the activation of AKT, to prevent their activation and further inhibit the apoptosis of cells [[34], [35], [36]]. It has been reported recently, As4O6 induced apoptosis of SW620 cells by up-regulating the exogenous pathway of caspase-8 activation by DR5 and the endogenous pathway mediated by mitochondria, while it also induce beclin-1 independent autophagic cell death [37]. Thus, these findings suggest that autophagy and apoptosis can comparably induce cancer cell death in CRC. However, another form of cooperation between autophagy and apoptosis has been reported, RCE induces HT-29 cell apoptosis through the caspase-7-dependent pathway, but it does not constitute the main mechanism of cell death, which mainly occurs through PCD-II probably as result of excessive autophagy [38]. PCDH17 combined with 5-FU in the treatment of colorectal cancer has also been found to induce apoptosis and autophagic cell death. In addition, autophagy played a leading role in cell death induced by PCDH17, as an autophagy inhibitor blocked cell death to better than the pan-caspase inhibitor Z-VAD-FMK [39]. However, compared with apoptosis, autophagy in promoting cell death is not always a dominant position. For example, Hsuan S et al. found that the main mechanism of WE induced cytotoxicity is the direct effect of apoptosis and the indirect effect of autophagy. Therefore, apoptosis plays a leading role in the death of colorectal cancer cells induced by WE [40]. These results indicated that the collaborative effect of autophagy and apoptosis also exists in the form of one main and one auxiliary in CRC. To sum up, the synergistic effect of autophagy and apoptosis in colon cancer mainly exists in two forms: equivalent, one main and auxiliary.
Promoting effect
In the study of inhibiting radiation-induced autophagy to sensitize malignant glioma cells, it found that autophagy could enhance cell apoptosis, while inhibiting autophagy did not affect the function of cell apoptosis, indicating that autophagy may not directly lead to cell death, but only played a promoting role [41]. This relationship between autophagy and apoptosis has also been found in studies of colorectal cancer [[42], [43], [44]]. Petroni G et al. found that Cla induced and blocked autophagy by regulating both the PI3K/Akt pathway and the herg1-dependent regulation of P53 in colorectal cancer. The former is related to the reduction of the phosphorylation levels of Akt and erk1/2, while the latter is related to the inhibition of autophagy flux. Late damage of autophagy-mediated degradation triggers apoptotic cell death, which is caused by activation of P53 and caspases [42]. The stereoselective assembly of Multifunctional Spirocyclohexene pyrazolone also induced autophagy-dependent apoptosis in colorectal cancer cells [43]. These may be related to autophagy activation of apoptosis-related factors, and the sequence of autophagy is usually before apoptosis [28]. For instance, Emodin induces autophagy, which leads to apoptosis in colon cancer cells in a ROS-dependent manner [44]. In a word, these results suggest that autophagy can trigger cell apoptosis in colorectal cancer. However, there are few studies on the role of apoptosis signal in the regulation of autophagy in colorectal cancer, so its specific mechanism of action remains unclear.
Antagonistic effect
Protective autophagy has been found in many diseases, especially cancer. It is easy to induce protective autophagy during tumor chemotherapy, which helps tumor cells survive under stress conditions such as hypoxia or low nutrition [27], leading to drug resistance and refractory tumors [16]. For example, Zhao Z et al. found that the expression of VEGF-A protein in colorectal cancer tissues was up-regulated. Anti-VEGF (bevacizumab) inhibited cell activity and induced apoptosis. In addition, bevacizumab induced autophagy through increased HIF-1α expression. Inhibition of autophagy by chloroquine or small interfering RNA promoted apoptosis and proliferation inhibition induced by bevacizumab [45], indicating that bevacizumab induced protective autophagy. Recent studies have shown that LncRNA plays a regulatory role between autophagy and apoptosis [46,47], LncRNA (HAGLROS) are highly expressed in CRC tissues and cells which is associated with shorter survival time in CRC patients. Down-regulation of HAGLROS in HCT116 cells can induce apoptosis and inhibit autophagy by regulating the mir-100 /ATG5 axis and the PI3K/AKT/mTOR pathway [46]. Moreover, LncRNA (Malat1) activated autophagy and promoted cell proliferation, yet inhibited apoptosis by sponging miR-101 in colorectal cancer cells [47]. Thus, LncRNA-induced autophagy plays an important role in inhibiting the apoptosis of colorectal cancer cells. All in all, the antagonistic effects of autophagy and apoptosis are prevalent in colorectal cancer, which contributes to the formation and development of CRC in an adverse environment such as low oxygen and low nutrition [27].
The main regulatory factors of the interaction between autophagy and apoptosis in CRC
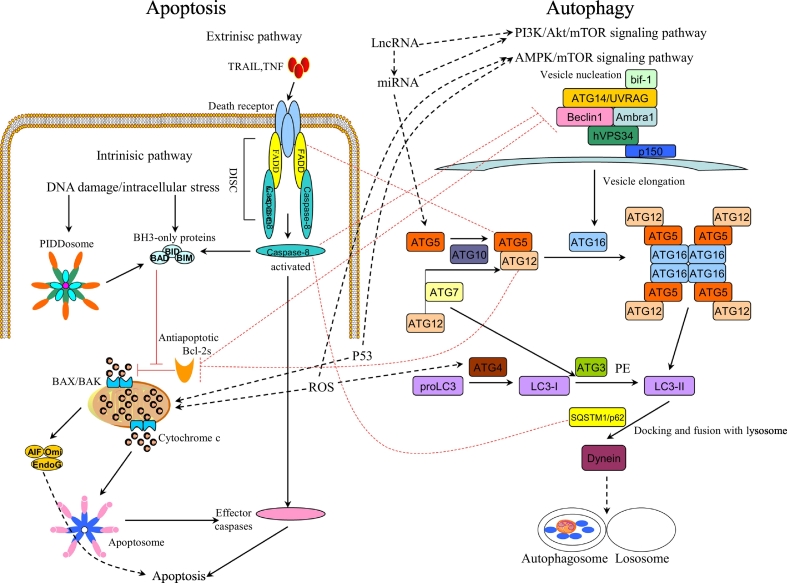
The interaction between autophagy and apoptosis in colorectal cancer is mainly regulated by related signal transduction pathways and factors [[48], [49], [50]] (Fig. 2). At present, relevant literature has reported related factors are involved in regulating the interaction between autophagic and cell apoptosis in other tumors, such as Beclin-1, P62, caspase, Bcl-2 and so on [51,52]. It was also found that these related regulatory factors were involved in the interaction between autophagic and apoptosis in colorectal cancer [48,53] (Fig. 2), the following key describes several major factors of interaction between autophagy and apoptosis: Fig. 2. The interaction between autophagy and apoptosis in colorectal cancer. The interaction between autophagy and apoptosis in colorectal cancer is mainly regulated by related signal transduction pathways and factors, such as the PI3K/Akt/mTOR signaling pathway, AMPK/mTOR signaling pathway, LncRNA, miRNA.
Fig. 2.
Signal network of autophagy and apoptosis.
LC3
Microtubule-associated protein 1A/1b Light chain 3 (LC3), which is the mammalian homologue of yeast ATG8 [54]. During autophagy, the cytoplasmic form of LC3 (LC3-I) is conjured with phosphatidylethanolamine (PE) to form the LC3-phosphatidylethanolamine conjugation (LC3-II). When autophagosomes fuse with lysosomes to form autolysosomes, intra-autophagosomal lc3-II degrades with the contents [55]. Therefore, LC3 is a hallmark of autophagy [56]. At the same time, LC3 has also reported playing an important role in the interaction between autophagy and apoptosis, such as Tao R et al. found that Cantharidin sodium activates Caspase-3 through the LC3 autophagy pathway to induce HepG2 cell apoptosis [57]. Furthermore, Spred2 co-localized and interacted with LC3 through the LC3-interacting region (LIR) motif-in its SPR domain to induce caspase-independent but autophagy-dependent cell death in human cervical cancer HeLa and lung cancer A549 cells [54]. Similarly, anti-tumor drugs can also induce cell apoptosis through the LC3-autophagy pathway in colorectal cancer. Selumetinib (AZD6244), a potent, orally bioavailable, allosteric MEK1/2 inhibitor, while MEK1/2 is one of the critical molecules in the RAS/RAF/MEK/ERK pathway [58]. Moreover, the existence of an intense crosstalk between RAS/RAF/MEK and PI3K/AKT/mTOR pathways has been demonstrated [58]. Therefore, Selumetinib (AZD6244) can induce apoptosis of colorectal cancer cells through the LC3-autophagy pathway [59]. The main mechanism of Selumetinib is to promote autophagy in the early stage of autophagy and inhibit autophagy in the final stage, leading to the accumulation of autophagosomes and the arrest of autophagy, thus inducing apoptosis. To sum up, LC3 can be a potential target for targeted drug therapy for colorectal cancer.
Beclin-1
Beclin-1 is an important component of autophagy signal transduction pathway. It affects every major step in autophagic pathways, from autophagosome formation to the maturation of autophagosome/endosome [60]. The initiation of autophagy requires dissociation of Beclin-1 from Bcl-2 and subsequent binding with PIK3C3/ VPS34 (PtdIns3K), whereas the BH3 domain of Beclin-1 is inhibited by Bcl-2 or Bcl-XL [60,61]. Previous studies have confirmed that ROCK1 is one of the upstream regulators of Beclin-1 mediated autophagy and controls the homeostasis between autophagy and apoptosis [58]. Lin P et al. found that ARV S1133 caused autophagy and apoptosis in Vero and DF1 cells, which was accompanied by the activation of the Beclin-1 promoter in the early to middle stages, and induction of caspase-3 expression in the middle to late stages of infection. Therefore, RhoA/ROCK1 signaling pathway plays an important role in the process of cell autophagy to apoptosis [62]. Furthermore, Zhou et al. showed a correlation between the under-expression of Beclin-1 and the induction of apoptosis in colorectal cancer cells by Icaritine [63]. Interestingly, Icaritin activates the AMPM/mTOR signaling pathway. Activation of mTORC1 will inhibit autophagy, while AMPK-induced inactivation of mTORC1 could provoke autophagy. Previous studies have demonstrated that Icaritine induces apoptosis by inhibiting autophagy in CRC cells [64]. In another study, the transition from autophagy to apoptosis after cleavage of Beclin-1 was observed in steroidal saponin-treated HCT-116 cells [65]. These results indicated that autophagy-related protein Blin -1 was also involved in the regulation of apoptosis. Therefore, Beclin-1 is one of the important factors involved in regulating the interaction between autophagy and apoptosis in colorectal cancer.
SQSTM1\P62
SQSTM1\P62 is an autophagic adaptive protein that bind to ubiquitinated protein aggregates and delivers them to autophagosomes [66]. At the same time, P62 is also an important molecular for tumor genesis and tumor treatment [53,67], autophagy can suppress tumorigenesis through elimination of P62. Li et al. found that combination of rapamycin and SAMC induces HCT-116 cells apoptosis through autophagy/P62/Nrf2 pathway, and P62 played a negative-regulatory role between Nrf2 and autophagy [53]. Previous studies have shown that autophagy can inhibit tumor development by eliminating P62. So, rapamycin can down-regulate p62 expression by promoting autophagy. Also, the recent report that Nrf2- Keap1 binding competed p62 for autophagy degradation [68]. Therefore, the Nrf2-Keap1 system activated by rapamycin and SAMC co-treatment could be related to autophagy through p62. In addition, studies have shown that P62 can regulate ATM/ H2AX pathway mediated escin-induced DNA damage and apoptosis in colorectal cancer [69]. In another study, P62 blocks apoptosis by inhibiting Fas/Cav-1 complex formation. To sum up, P62 plays a regulatory role in the interaction between autophagy and apoptosis in CRC.
Atg5-Atg12
In the autophagy signal transduction pathway, the Atg5-Atg12 conjugate has been shown to promote Atg8-PE formation in a manner analogous to the function of E3 enzymes during classical ubiquitin conjugation reactions [70]. However, in CRC cells with high microsatellite instability (such as HCT116 cells), Atg5 and Atg12 mutations may contribute to the tumor progression by deregulating the autophagy process [71]. Won S et al. found that JA-induced formation of Atg5-Atg12 contributed to the promotion of HT-29 cells apoptosis by JA-induced autophagy [72]. In another study, interaction of Atg5-Atg-12 with FADD can induce autophagic death of HT29 cells [73]. These studies indicate that the ATG5-ATG12 conjugate may be involved in regulating the cascade between autophagy and apoptosis in colorectal cancer. Thus, the Atg5-Atg12 conjugate plays an important role in balancing autophagy and apoptosis in colorectal cancer.
Bcl-2 family proteins
Bcl-2 family proteins include both pro- and anti- apoptotic proteins and are the major regulators of the apoptotic process. Bax and Bad are pro-apoptotic proteins, while Bcl-2 and Bcl-x are anti-apoptotic proteins [74]. Bax permeabilizes the mitochondrial outer membrane, releasing pro-apoptotic factors that activate caspases [75], which can then active the nuclear protein PARP [76]. While Bcl-2 can bind to and sequester Bax to prevent activation of the downstream mitochondrial death cascade [77]. Therefore, the Bcl-2/Bax ratio is very important in regulating caspase-dependent apoptosis. Cokergurkan A et al. found that Long-term use of low-dose purvalanol in the treatment of HCT 116 cells can trigger caspase-dependent apoptotic cell death by regulating bcl-2 family members [78]. Other studies also found that non-phosphorylated Bad promotes apoptosis by binding to and inactivating the anti-apoptotic proteins of Bcl-2 or Bcl-XL [32]. In addition, there is increasing evidence that apoptosis and autophagy were regulated by the Bcl-2 protein family [79]. Huang S et al. showed Celecoxib can induce apoptosis and autophagy in human colorectal cancer cells, both of which can be negatively regulated by Bcl-2/Bcl-XL. At the same time, ABT-737 can enhance celecoxib-mediated apoptosis and autophagy, and play a synergistic cytotoxic effect [80]. Celecoxib-induced apoptosis and autophagy may be related to its known ability to trigger ER stress [81], while ABT-737 combined with celecoxib treatment produced synergistic cytotoxic effects that were due primarily to caspase-dependent apoptosis. In a word, in CRC, Bcl-2 family proteins are involved not only in the regulation of apoptosis, but also in the regulation of autophagy.
ROS
Reactive oxygen species (ROS), generated during cellular metabolism through leakage of electrons by mitochondrial electron transport as well as the endoplasmic reticulum(ER) [82,83], is one of the important mechanisms of mitochondrial dysfunction and cell damage [84,85]. Research has shown that ROS have an important role in the crosstalk between autophagy and apoptosis [86]. ROS has a negative feedback effect in autophagy. On the one hand, ROS can promote the autophagy process. On the other hand, autophagy reduces ROS by removing damaged mitochondria and ER as well as other substances that contribute to ROS production [87,88]. Meanwhile, ROS is a mediator of cell apoptosis [82]. Kim K et al. showed intracellular ROS production played a critical role during ECZ-induced apoptosis and autophagy of colon cancer cells and that increasing ROS generation by the inhibition of autophagy results in enhanced apoptosis [89]. Furthermore, studies have found that Shikonin trigger the dimerization of Galectin-1 by inducing the accumulation of ROS, which eventually lead to autophagy and apoptosis of colorectal cancer cells [33]. It is well known that ROS over-production can trigger ER stress. Peng Y et al. found that M4IDP triggers ER stress by promoting ROS production, subsequently leading to apoptosis and autophagy in colorectal cancer cells [32]. Therefore, ROS from mitochondria and endoplasmic reticulum can be involved in regulating the cascade reaction between autophagy and apoptosis in colorectal cancer. Overall, ROS can be considered as the target molecules for chemotherapy drugs for colorectal cancer.
P53
P53 functions as the most efficient suppressor of cancer development and progression [50], which plays essential roles in cellular protection mechanisms against a variety of stress stimuli, such as DNA damages, metabolic changes and oncogene activation [90,91]. The activation of p53 induces apoptosis or autophagy in certain cancer cells [90]. At the same time, p53 has been shown to simultaneously control both apoptosis and autophagy by up regulating apoptotic genes and regulating multiple levels of the AMP-activated protein kinase-mTOR axis, respectively [92]. For example, An Y et al. found that digiferrol activated p53-dependent gene expression by phosphorylating the p53 protein at ser-15 residue, inducing cell cycle arrest, apoptosis, and autophagy in colon cancer cells [50]. In another study, long-term treatment of colorectal cancer cells by Cla leads to the blocking of autophagy, activation of p53 and caspases, and induction of cell apoptosis [42]. In addition, Garufi A showed in normal/high glucose condition a mutual unbalance between p53-dependent apoptosis (PUMA) and autophagy (DRAM) gene occurred, modifying the ADR-induced cancer cell death in HG both in vitro and in vivo [93]. It is well known that PUMA and DRAM are two target genes of p53. PUMA is mainly involved in p53-induced apoptosis. DRAM interacts with other downstream factors of p53 to induce autophagy. Interestingly, DRAM has also involved in p53 induced cell death [93,94]. Therefore, p53 can participate in the regulation of autophagy and apoptosis through the interaction between the regulatory factors PUMA and DRAM. All in all, p53 plays a regulatory role in the interaction between autophagy and apoptosis in CRC.
LncRNA
Long noncoding RNAs (LncRNAs) are widely defined as transcriptional RNA molecules with a length of more than 200 nt and no protein-coding ability [95]. Bioinformatics analysis showed that LncRNA is the center of normal development and tumorigenesis at the transcriptional level or post-transcriptional level [96], which is actively involved in the regulation of malignant phenotypes, such as apoptosis, autophagy, cell cycle, cell migration, et al. [97,98]. It is reported that inhibition of autophagy by 3-MA can not only alleviate Malat1-induced cell proliferation, but also promote Malat1-induced cell apoptosis. Therefore, Malat1 can activate autophagy and promote cell proliferation. In addition, LncRNA Malat1, as an endogenous sponge, can directly adsorb miR-101, thereby inhibiting the effect of miR-101 on the apoptosis of colorectal cancer cells [47]. It follows that LncRNA can be used as ceRNAs to regulate gene expression through sponge miRNA and play a key role in tumors [99,100]. Song F et al. showed downregulation of LncRNA UCA1 leads to increased phosphorylation of mTOR, PI3K and AKT, and significantly decreased levels of LC3 and autophagy-related gene 5 (ATG5) protein, leading to increased P62 expression. Instead, autophagy activator RAPA reversed the effect. Therefore, down-regulation of UCA1 induces autophagy inhibition by activating the AKT/mTOR signaling pathway, thereby inhibiting cell proliferation and promoting apoptosis of caco2 cells [101]. Mounting evidence suggests that LncRNA is a key factor in autophagy [102]. Moreover, through the negative regulation of miR100/ATG5 axis, the down-regulation of LncRNA HAGLROS can induce the apoptosis of CRC cells and inhibit autophagy [46]. Thus, LncRNA plays an important role in balancing autophagy and apoptosis.
At present, with the in-depth study on the interaction between autophagy and apoptosis in colorectal cancer, an increasing number of factors have been found to be involved in regulating the interaction between autophagy and apoptosis, such as miRNA [46,103]. MicroRNAs (miRNAs) are a class of small, endogenous, non-coding, single-stranded RNAs of 18–25 nucleotides in length that regulated the expression of their target genes through mRNA degradation or translational inhibition. At present, miRNAs have been found to regulate many cellular biological processes including apoptosis, cell proliferation, and autophagy [104]. It has been reported that Mir-218 inhibits the protective autophagy of cells through the expression of YEATS4, making HCT-116/L-OHP cells sensitive to L-OHP-induced apoptosis [103]. However, it is not the only type of miRNA involved in regulating the interaction between autophagy and apoptosis in colorectal cancer. Studies have found that more and more different types of miRNAs are involved in regulating various stages of autophagy and apoptosis interactions in CRC [46,47,104].
Conclusions
In summary, autophagy and apoptosis play an important role in the development and progression of colorectal cancer. However, the relationship between autophagy and apoptosis is anfractuous. On one hand, autophagic proteins can be involved in regulating the process of apoptosis. Moreover, whether autophagy induces or inhibits apoptosis depends on the type of cell, nature, and duration of the stimulus/stress [29]. On the other hand, regulators of apoptosis also function as regulators of autophagic activation [105]. Therefore, the identification of the interaction pattern between autophagy and apoptosis, as well as the regulatory factors and signal transduction pathways involved, plays a crucial role in the treatment of colorectal cancer. As the research further develops, the interaction mechanism between autophagy and apoptosis will be further elucidated. Autophagy or apoptosis can be used as a new target to provide a basis for the development of targeted tumor drugs for colorectal cancer.
Author contributions statement
Xuefeng Li, Qingqiang Xie, and Yuan Liu contributed to the design and implementation of the research, to the analysis of the results and to the writing of the manuscript.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
This work was supported by National Natural Science Foundation of China (81972204 and 81702327), Natural Science Foundation of Guangdong Province (2019A1515011097), Innovation Program of Shenzhen (Grant No. JCYJ20180508165208399), Science and Technology Planning Project of Guangzhou (201904010089), China Postdoctoral Science Foundation (2018M640834 and 2019T120756), the grant from the State Key Lab of Respiratory Disease, Guangzhou Medical University (SKLRD-Z-202002), and the 111 Project (D18010) from the Ministry of Education of China.
Author contributions
Qingqiang Xie: Writing- Original draft preparation, Software. Yuan Liu: Writing- Original Draft preparation, Data Curation. Xuefeng Li: Conceptualization, Writing- Reviewing and Editing, Supervision, Funding acquisition.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Mundade R., Imperiale T.F., Prabhu L., Loehrer P.J., Lu T. Genetic pathways, prevention, and treatment of sporadic colorectal cancer. Oncoscience. 2014;1:400–406. doi: 10.18632/oncoscience.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel R.L., Miller K.D., Sauer A.G., Fedewa S.A., Butterly L., Anderson J.C., Cercek A., Smith R.A., Jemal A. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020;70:145–164. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 4.Chen W., Sun K., Zheng R., Zeng H., Zhang S., Xia C., Yang Z., Li H., Zou X., He J. Cancer incidence and mortality in China, 2014. Chin. J. Cancer Res. 2018;30:1–12. doi: 10.21147/j.issn.1000-9604.2018.01.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bray F.I., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 6.Kekelidze M., Derrico L., Pansini M., Tyndall A., Hohmann J. Colorectal cancer: current imaging methods and future perspectives for the diagnosis, staging and therapeutic response evaluation. World J. Gastroenterol. 2013;19:8502–8514. doi: 10.3748/wjg.v19.i46.8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colussi D., Brandi G., Bazzoli F., Ricciardiello L. Molecular pathways involved in colorectal cancer: implications for disease behavior and prevention. Int. J. Mol. Sci. 2013;14:16365–16385. doi: 10.3390/ijms140816365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haggar F., Boushey R.P. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clinics in Colon and Rectal Surgery. 2009;22:191–197. doi: 10.1055/s-0029-1242458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dekker E., Tanis P.J., Vleugels J.L.A., Kasi P.M., Wallace M.B. Colorectal cancer. Lancet (London, England) 2019;394:1467–1480. doi: 10.1016/S0140-6736(19)32319-0. [DOI] [PubMed] [Google Scholar]
- 10.Jmm L., Towers C.G., Thorburn A. Targeting autophagy in cancer. Nat. Rev. Cancer. 2017;17:528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klionsky D.J., Abeliovich H., Agostinis P., Agrawal D.K., Aliev G., Askew D.S., Baba M., Baehrecke E.H., Bahr B.A., Ballabio A. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eskelinen E., Saftig P. Autophagy: a lysosomal degradation pathway with a central role in health and disease. Biochim. Biophys. Acta. 2009;1793:664–673. doi: 10.1016/j.bbamcr.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 13.Devenport S.N., Shah Y.M. Functions and implications of autophagy in colon cancer. Cells. 2019;8:1349. doi: 10.3390/cells8111349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manning B.D., Toker A. AKT/PKB signaling: navigating the network. Cell. 2017;169:381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardie D.G. AMPK: positive and negative regulation, and its role in whole-body energy homeostasis. Curr. Opin. Cell Biol. 2015;33:1–7. doi: 10.1016/j.ceb.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 16.Li Y., Lei Y., Yao N., Wang C., Hu N., Ye W., Zhang D., Chen Z. Autophagy and multidrug resistance in cancer. Chin J Cancer. 2017;36:52. doi: 10.1186/s40880-017-0219-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gewirtz D.A. The four faces of autophagy: implications for cancer therapy. Cancer Res. 2014;74:647–651. doi: 10.1158/0008-5472.CAN-13-2966. [DOI] [PubMed] [Google Scholar]
- 18.Kaluzki I., Hailemariamjahn T., Doll M., Kaufmann R., Balermpas P., Zoller N., Kippenberger S., Meissner M. Dimethylfumarate inhibits colorectal carcinoma cell proliferation: evidence for cell cycle arrest, apoptosis and autophagy. Cells. 2019;8:1329. doi: 10.3390/cells8111329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elmore S.A. Apoptosis: a review of programmed cell death. Toxicol. Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pistritto G., Trisciuoglio D., Ceci C., Garufi A., D'Orazi G. Apoptosis as anticancer mechanism: function and dysfunction of its modulators and targeted therapeutic strategies. Aging. 2016;8:603–619. doi: 10.18632/aging.100934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoeppner D.J., Hengartner M.O., Schnabel R. Engulfment genes cooperate with ced-3 to promote cell death in Caenorhabditis elegans. Nature. 2001;412:202–206. doi: 10.1038/35084103. [DOI] [PubMed] [Google Scholar]
- 22.Damelio M., Cavallucci V., Cecconi F. Neuronal caspase-3 signaling: not only cell death. Cell Death Differ. 2010;17:1104–1114. doi: 10.1038/cdd.2009.180. [DOI] [PubMed] [Google Scholar]
- 23.Plati J., Bucur O., Khosravifar R. Dysregulation of apoptotic signaling in cancer: molecular mechanisms and therapeutic opportunities. J. Cell. Biochem. 2008;104:1124–1149. doi: 10.1002/jcb.21707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korsmeyer S.J., Shutter D.J., Veis D.E., Merry Z.N. Oltvai, Bcl-2/Bax: a rheostat that regulates an anti-oxidant pathway and cell death. Semin. Cancer Biol. 1993;4:327–332. [PubMed] [Google Scholar]
- 25.Fulda S. Tumor resistance to apoptosis. Int. J. Cancer. 2009;124:511–515. doi: 10.1002/ijc.24064. [DOI] [PubMed] [Google Scholar]
- 26.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 27.Su M., Mei Y., Sinha S.C. Role of the crosstalk between autophagy and apoptosis in cancer. Journal of Oncology. 2013;2013:102735. doi: 10.1155/2013/102735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marino G., Nisosantano M., Baehrecke E.H., Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2014;15:81–94. doi: 10.1038/nrm3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Athamneh K., Alneyadi A., Alsamri H., Alrashedi A., Palakott A., El-Tarabily K.A., Eid A.H., Al Dhaheri Y., Iratni R. Origanum majorana essential oil triggers p38 MAPK-mediated protective autophagy, apoptosis, and caspase-dependent cleavage of P70S6K in colorectal cancer cells. Biomolecules. 2020;10 doi: 10.3390/biom10030412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gump J.M., Thorburn A. Autophagy and apoptosis: what is the connection? Trends Cell Biol. 2011;21:387–392. doi: 10.1016/j.tcb.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryter S.W., Mizumura K., Choi A.M.K. The impact of autophagy on cell death modalities. International Journal of Cell Biology. 2014;2014:502676. doi: 10.1155/2014/502676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng Y., Qiu L., Xu D., Zhang L., Yu H., Ding Y., Deng L., Lin J. M4IDP, a zoledronic acid derivative, induces G1 arrest, apoptosis and autophagy in HCT116 colon carcinoma cells via blocking PI3K/Akt/mTOR pathway. Life Sci. 2017;185:63–72. doi: 10.1016/j.lfs.2017.07.024. [DOI] [PubMed] [Google Scholar]
- 33.Zhang N., Peng F., Wang Y., Yang L., Wu F., Wang X., Ye C., Han B., He G. Shikonin induces colorectal carcinoma cells apoptosis and autophagy by targeting galectin-1/JNK signaling axis. Int. J. Biol. Sci. 2020;16:147–161. doi: 10.7150/ijbs.36955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brech A., Ahlquist T.C., Lothe R.A., Stenmark H.A. Autophagy in tumour suppression and promotion. Mol. Oncol. 2009;3:366–375. doi: 10.1016/j.molonc.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henshall D.C., Araki T., Schindler C.K., Lan J.Q., Tiekoter K.L., Taki W., Simon R.P. Activation of Bcl-2-associated death protein and counter-response of Akt within cell populations during seizure-induced neuronal death. J. Neurosci. 2002;22:8458–8465. doi: 10.1523/JNEUROSCI.22-19-08458.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xin M., Deng X. Nicotine inactivation of the proapoptotic function of Bax through phosphorylation. J. Biol. Chem. 2005;280:10781–10789. doi: 10.1074/jbc.M500084200. [DOI] [PubMed] [Google Scholar]
- 37.Nagappan A., Lee W.S., Yun J.W., Lu J.N., Chang S., Jeong J., Kim G.S., Jung J., Hong S. Tetraarsenic hexoxide induces G2/M arrest, apoptosis, and autophagy via PI3K/Akt suppression and p38 MAPK activation in SW620 human colon cancer cells. PLoS One. 2017;12:0174591. doi: 10.1371/journal.pone.0174591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Athamneh K., Hasasna H.E., Samri H.A., Attoub S., Arafat K., Benhalilou N., Rashedi A.A., Dhaheri Y.A., Abuqamar S., Eid A.H. Rhus coriaria increases protein ubiquitination, proteasomal degradation and triggers non-canonical Beclin-1-independent autophagy and apoptotic cell death in colon cancer cells. Sci. Rep. 2017;7:11633. doi: 10.1038/s41598-017-11202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu S., Lin H., Wang D., Li Q., Luo H., Li G., Chen X., Li Y., Chen P., Zhai B. PCDH17 increases the sensitivity of colorectal cancer to 5-fluorouracil treatment by inducing apoptosis and autophagic cell death. Signal Transduction and Targeted Therapy. 2019;4:1–10. doi: 10.1038/s41392-019-0087-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsuan S., Chyau C., Hung H., Chen J., Chou F. The induction of apoptosis and autophagy by Wasabia japonica extract in colon cancer. Eur. J. Nutr. 2016;55:491–503. doi: 10.1007/s00394-015-0866-5. [DOI] [PubMed] [Google Scholar]
- 41.Ito H., Daido S., Kanzawa T., Kondo S., Kondo Y. Radiation-induced autophagy is associated with LC3 and its inhibition sensitizes malignant glioma cells. Int. J. Oncol. 2005;26:1401–1410. [PubMed] [Google Scholar]
- 42.Petroni G., Bagni G., Iorio J., Duranti C., Lottini T., Stefanini M., Kragol G., Becchetti A., Arcangeli A. Clarithromycin inhibits autophagy in colorectal cancer by regulating the hERG1 potassium channel interaction with PI3K. Cell Death Dis. 2020;11:161. doi: 10.1038/s41419-020-2349-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li X., Chen F., Kang J., Zhou J., Peng C., Huang W., Zhou M., He G., Han B. Stereoselective assembly of multifunctional spirocyclohexene pyrazolones that induce autophagy-dependent apoptosis in colorectal cancer cells. J. Organomet. Chem. 2019;84:9138–9150. doi: 10.1021/acs.joc.9b01098. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y., Luo Q., He X., Wei H., Wang T., Shao J., Jiang X. Emodin induces apoptosis of colon cancer cells via induction of autophagy in a ROS-dependent manner. Oncol. Res. 2017;26:889–899. doi: 10.3727/096504017X15009419625178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao Z., Xia G., Li N., Su R., Chen X., Zhong L. Autophagy inhibition promotes bevacizumab-induced apoptosis and proliferation inhibition in colorectal cancer cells. J. Cancer. 2018;9:3407–3416. doi: 10.7150/jca.24201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng Y., Tan K., Huang H. Long noncoding RNA HAGLROS regulates apoptosis and autophagy in colorectal cancer cells via sponging miR-100 to target ATG5 expression. J. Cell. Biochem. 2019;120:3922–3933. doi: 10.1002/jcb.27676. [DOI] [PubMed] [Google Scholar]
- 47.Si Y., Yang Z., Ge Q., Yu L., Yao M., Sun X., Ren Z., Ding C. Long non-coding RNA Malat1 activated autophagy, hence promoting cell proliferation and inhibiting apoptosis by sponging miR-101 in colorectal cancer. Cell. Mol. Biol. Lett. 2019;24:50. doi: 10.1186/s11658-019-0175-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maranhao S.S., Moura A.F., Oliveira A.C.A., Lima D.J.B., Barrosnepomuceno F.W.A., Paier C.R.K., Pinheiro A.C., Nogueira T.C.M., De Souza M.V.N., Pessoa C. Synthesis of PJOV56, a new quinoxalinyl-hydrazone derivative able to induce autophagy and apoptosis in colorectal cancer cells, and related compounds. Bioorg. Med. Chem. Lett. 2020;30:126851. doi: 10.1016/j.bmcl.2019.126851. [DOI] [PubMed] [Google Scholar]
- 49.Tan H., Li X., Yang W.H., Kang Y. A flavone, Wogonin from Scutellaria baicalensis inhibits the proliferation of human colorectal cancer cells by inducing of autophagy, apoptosis and G2/M cell cycle arrest via modulating the PI3K/AKT and STAT3 signalling pathways. Journal of BUON : official journal of the Balkan Union of Oncology. 2019;24:1143–1149. [PubMed] [Google Scholar]
- 50.An Y., Quan K.T., Gwak J., Ju B.G., Na M., Oh S. Activation of the p53 pathway with digiferrol isolated from Rubia philippinensis induces cell cycle arrest, apoptosis, and autophagy in colon cancer cells. Food Chem. Toxicol. 2018;118:514–522. doi: 10.1016/j.fct.2018.05.054. [DOI] [PubMed] [Google Scholar]
- 51.Noguchi M., Hirata N., Tanaka T., Suizu F., Nakajima H., Chiorini J.A. Autophagy as a modulator of cell death machinery. Cell. Death. Dis. 2020;11:517. doi: 10.1038/s41419-020-2724-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fu L.L., Cheng Y., Liu B. Beclin-1: autophagic regulator and therapeutic target in cancer. Int. J. Biochem. Cell Biol. 2013;45:921–924. doi: 10.1016/j.biocel.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 53.Li S., Yang G., Zhu X., Cheng L., Sun Y., Zhao Z. Combination of rapamycin and garlic-derived S-allylmercaptocysteine induces colon cancer cell apoptosis and suppresses tumor growth in xenograft nude mice through autophagy/p62/Nrf2 pathway. Oncol. Rep. 2017;38:1637–1644. doi: 10.3892/or.2017.5849. [DOI] [PubMed] [Google Scholar]
- 54.Jiang K., Liu M., Lin G., Mao B., Cheng W., Liu H., Gal J., Zhu H., Yuan Z., Deng W. Tumor suppressor Spred2 interaction with LC3 promotes autophagosome maturation and induces autophagy-dependent cell death. Oncotarget. 2016;7:25652–25667. doi: 10.18632/oncotarget.8357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tanida I., Ueno T., Kominami E. LC3 and autophagy. Methods Mol. Biol. 2008;445:77–88. doi: 10.1007/978-1-59745-157-4_4. [DOI] [PubMed] [Google Scholar]
- 56.Kang J.H., Lee S., Cheong H., Lee C.H., Kim S. Transglutaminase 2 promotes autophagy by LC3 induction through p53 depletion in cancer cell. Biomol. Ther. 2019;27:34–40. doi: 10.4062/biomolther.2018.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tao R., Sun W., Yu D., Qiu W., Yan W., Ding Y., Wang G., Li H. Sodium cantharidinate induces HepG2 cell apoptosis through LC3 autophagy pathway. Oncol. Rep. 2017;38:1233–1239. doi: 10.3892/or.2017.5779. [DOI] [PubMed] [Google Scholar]
- 58.Chappell W.H., Steelman L.S., Long J.M., Kempf R.C., Abrams S.L., Franklin R.A., Bäsecke J., Stivala F., Donia M., Fagone P., Malaponte G., Mazzarino M.C., Nicoletti F., Libra M., Maksimovic-Ivanic D., Mijatovic S., Montalto G., Cervello M., Laidler P., Milella M., Tafuri A., Bonati A., Evangelisti C., Cocco L., Martelli A.M., McCubrey J.A. Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR inhibitors: rationale and importance to inhibiting these pathways in human health. Oncotarget. 2011;2:135–164. doi: 10.18632/oncotarget.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grasso S., Pereira G.J.S., Palmeiradossantos C., Calgarotto A.K., Martinezlacaci I., Ferragut J.A., Smaili S.S., Bincoletto C. Autophagy regulates Selumetinib (AZD6244) induced-apoptosis in colorectal cancer cells. Eur. J. Med. Chem. 2016;122:611–618. doi: 10.1016/j.ejmech.2016.06.043. [DOI] [PubMed] [Google Scholar]
- 60.Kang R., Zeh H.J., Lotze M.T., Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Green D.R., Levine B. To be or not to be? How selective autophagy and cell death govern cell fate. Cell. 2014;157:65–75. doi: 10.1016/j.cell.2014.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin P., Chang C., Chen Y., Shih W. RhoA/ROCK1 regulates Avian Reovirus S1133-induced switch from autophagy to apoptosis. BMC Vet. Res. 2015;11:103. doi: 10.1186/s12917-015-0417-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou C., Gu J., Zhang G., Dong D., Yang Q., Chen M., Xu D. AMPK-autophagy inhibition sensitizes icaritin-induced anti-colorectal cancer cell activity. Oncotarget. 2017;8:14736–14747. doi: 10.18632/oncotarget.14718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou C., Chen Z., Lu X., Wu H., Yang Q., Xu D. Icaritin activates JNK-dependent mPTP necrosis pathway in colorectal cancer cells. Tumor Biol. 2016;37:3135–3144. doi: 10.1007/s13277-015-4134-3. [DOI] [PubMed] [Google Scholar]
- 65.Pedro J.M.B., Wei Y., Sica V., Maiuri M.C., Zou Z., Kroemer G., Levine B. BAX and BAK1 are dispensable for ABT-737- induced dissociation of the BCL2-BECN1 complex and autophagy. Autophagy. 2015;11:452–459. doi: 10.1080/15548627.2015.1017191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiang T., Harder B., La Vega M.R.D., Wong P.K., Chapman E., Zhang D.D. p62 links autophagy and Nrf2 signaling. Free Radic. Biol. Med. 2015;88:199–204. doi: 10.1016/j.freeradbiomed.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shi X., Zhang H., Jin W., Liu W., Yin H., Li Y., Dong H. Metronomic photodynamic therapy with 5-aminolevulinic acid induces apoptosis and autophagy in human SW837 colorectal cancer cells. Journal of Photochemistry and Photobiology B-biology. 2019;198:111586. doi: 10.1016/j.jphotobiol.2019.111586. [DOI] [PubMed] [Google Scholar]
- 68.Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y., Ueno I., Sakamoto A., Tong K.I. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 69.Wang Z., Chen Q., Li B., Xie J., Yang X., Zhao K., Wu Y., Ye Z., Chen Z., Qin Z. Escin-induced DNA damage promotes escin-induced apoptosis in human colorectal cancer cells via p62 regulation of the ATM/γH2AX pathway. Acta Pharmacol. Sin. 2018;39:1645–1660. doi: 10.1038/aps.2017.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Romanov J., Walczak M., Ibiricu I., Schuchner S., Ogris E., Kraft C., Martens S. Mechanism and functions of membrane binding by the Atg5–Atg12/Atg16 complex during autophagosome formation. EMBO J. 2012;31:4304–4317. doi: 10.1038/emboj.2012.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kang M.R., Kim M.S., Oh J.E., Kim Y.R., Song S.Y., Kim S.S., Ahn C.H., Yoo N.J., Lee S.H. Frameshift mutations of autophagy-related genes ATG2B, ATG5, ATG9B and ATG12 in gastric and colorectal cancers with microsatellite instability. J. Pathol. 2009;217:702–706. doi: 10.1002/path.2509. [DOI] [PubMed] [Google Scholar]
- 72.Won S.J., Yen C.H., Liu H.S., Wu S.Y., Lan S.H., Jiang-Shieh Y.F., Lin C.N., Su C.L. Justicidin A-induced autophagy flux enhances apoptosis of human colorectal cancer cells via class III PI3K and Atg5 pathway. J. Cell. Physiol. 2015;230:930–946. doi: 10.1002/jcp.24825. [DOI] [PubMed] [Google Scholar]
- 73.Codogno P., Meijer A.J. Atg5: more than an autophagy factor. Nat. Cell Biol. 2006;8:1045–1047. doi: 10.1038/ncb1006-1045. [DOI] [PubMed] [Google Scholar]
- 74.Gross A. BCL-2 family proteins as regulators of mitochondria metabolism. Biochim. Biophys. Acta. 2016;1857:1243–1246. doi: 10.1016/j.bbabio.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 75.Martinou J., Green D.R. Breaking the mitochondrial barrier. Nat. Rev. Mol. Cell Biol. 2001;2:63–67. doi: 10.1038/35048069. [DOI] [PubMed] [Google Scholar]
- 76.Chaitanya G.V., Alexander J.S., Babu P.P. PARP-1 cleavage fragments: signatures of cell-death proteases in neurodegeneration. Cell Communication and Signaling. 2010;8:31. doi: 10.1186/1478-811X-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lin C.H., Wu M.R., Li C.H., Cheng H.W., Huang S.H., Tsai C.H., Lin F.L., Der Ho J., Kang J.J., Hsiao G. Editor's highlight: periodic exposure to smartphone-mimic low-luminance blue light induces retina damage through Bcl-2/BAX-dependent apoptosis. Toxicol. Sci. 2017;157:196–210. doi: 10.1093/toxsci/kfx030. [DOI] [PubMed] [Google Scholar]
- 78.Cokergurkan A., Arisan E.D., Obakan P., Akalin K., Ozbey U., Palavanunsal N. Purvalanol induces endoplasmic reticulum stress-mediated apoptosis and autophagy in a time-dependent manner in HCT116 colon cancer cells. Oncol. Rep. 2015;33:2761–2770. doi: 10.3892/or.2015.3918. [DOI] [PubMed] [Google Scholar]
- 79.Levine B., Sinha S.C., Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4:600–606. doi: 10.4161/auto.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang S., Sinicrope F.A. Celecoxib-induced apoptosis is enhanced by ABT-737 and by inhibition of autophagy in human colorectal cancer cells. Autophagy. 2010;6:256–269. doi: 10.4161/auto.6.2.11124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tsutsumi S., Namba T., Tanaka K.I., Arai Y., Ishihara T., Aburaya M., Mima S., Hoshino T., Mizushima T. Celecoxib upregulates endoplasmic reticulum chaperones that inhibit celecoxib-induced apoptosis in human gastric cells. Oncogene. 2006;25:1018–1029. doi: 10.1038/sj.onc.1209139. [DOI] [PubMed] [Google Scholar]
- 82.Sharma N., Kumar A., Sharma P., Qayum A., Singh S.K., Dutt P., Paul S., Gupta V.K., Verma M., Satti N.K. A new clerodane Furano diterpene glycoside from Tinospora cordifolia triggers autophagy and apoptosis in HCT-116 colon cancer cells. J. Ethnopharmacol. 2018;211:295–310. doi: 10.1016/j.jep.2017.09.034. [DOI] [PubMed] [Google Scholar]
- 83.Kuwabara W.M.T., Zhang L., Schuiki I., Curi R., Volchuk A., Albaloureiro T.C. NADPH oxidase-dependent production of reactive oxygen species induces endoplasmatic reticulum stress in neutrophil-like HL60 cells. PLoS One. 2015;10:1–15. doi: 10.1371/journal.pone.0116410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Scherzshouval R., Elazar Z. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol. 2007;17:422–427. doi: 10.1016/j.tcb.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 85.Fandy T.E., Jiemjit A., Thakar M., Rhoden P., Suarez L., Gore S.D. Decitabine induces delayed reactive oxygen species (ROS) accumulation in leukemia cells and induces the expression of ROS generating enzymes. Clin. Cancer Res. 2014;20:1249–1258. doi: 10.1158/1078-0432.CCR-13-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang Y., Luo H., Hui K., Ci Y., Shi K., Chen G., Shi L., Xu C. Selenite-induced autophagy antagonizes apoptosis in colorectal cancer cells in vitro and in vivo. Oncol. Rep. 2016;35:1255–1264. doi: 10.3892/or.2015.4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Navarroyepesjuliana Burnsmichaela, Anandhanannadurai Khalimonchukoleh, D.R. Maria Quintanillavegabetzabet, Pappaaglaia I., Panayiotidismihalis Francorodrigo. Oxidative stress, redox signaling, and autophagy: cell death versus survival. Antioxid. Redox Signal. 2014;21:66–85. doi: 10.1089/ars.2014.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee J., Giordano S., Zhang J. Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling. Biochem. J. 2012;441:523–540. doi: 10.1042/BJ20111451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim K., Oh T., Yang H., Kim Y.W., Ma J., Park K. Ethanol extract of Chrysanthemum zawadskii Herbich induces autophagy and apoptosis in mouse colon cancer cells through the regulation of reactive oxygen species. BMC Complement. Altern. Med. 2019;19:274. doi: 10.1186/s12906-019-2688-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Son Y., An Y., Jung J., Shin S., Park I., Gwak J., Ju B.G., Chung Y., Na M., Oh S. Protopine isolated from Nandina domestica induces apoptosis and autophagy in colon cancer cells by stabilizing p53. Phytother. Res. 2019;33:1689–1696. doi: 10.1002/ptr.6357. [DOI] [PubMed] [Google Scholar]
- 91.Altman B.J., Rathmell J.C. Metabolic Stress in autophagy and cell death pathways. Cold Spring Harb. Perspect. Biol. 2012;4 doi: 10.1101/cshperspect.a008763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee H., Chung K., Hwang I.H., Gwak J., Park S., Ju B.G., Yun E., Kim D., Chung Y., Na M. Activation of p53 with Ilimaquinone and Ethylsmenoquinone. Marine Sponge Metabolites, Induces Apoptosis and Autophagy in Colon Cancer Cells, Marine Drugs. 2015;13:543–557. doi: 10.3390/md13010543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Garufi A., Pistritto G., Baldari S., Toietta G., Cirone M., Dorazi G. p53-dependent PUMA to DRAM antagonistic interplay as a key molecular switch in cell-fate decision in normal/high glucose conditions. J. Exp. Clin. Cancer Res. 2017;36:126. doi: 10.1186/s13046-017-0596-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhu B., Xing C., Lin F., Fan X., Zhao K., Qin Z. Blocking NF-κB nuclear translocation leads to p53-related autophagy activation and cell apoptosis. World J. Gastroenterol. 2011;17:478–487. doi: 10.3748/wjg.v17.i4.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fan Y., Shen B., Tan M., Mu X., Qin Y., Zhang F., Liu Y. Long non-coding RNA UCA1 increases chemoresistance of bladder cancer cells by regulating Wnt signaling. FEBS J. 2014;281:1750–1758. doi: 10.1111/febs.12737. [DOI] [PubMed] [Google Scholar]
- 96.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 97.Ishikawa T., Nishida T., Ono M., Takarada T., Nguyen H.T., Kurihara S., Furumatsu T., Murase Y., Takigawa M., Oohashi T. Physiological role of urothelial cancer-associated one long noncoding RNA in human skeletogenic cell differentiation. J. Cell. Physiol. 2018;233:4825–4840. doi: 10.1002/jcp.26285. [DOI] [PubMed] [Google Scholar]
- 98.Ruhle F., Stoll M. Long non-coding RNA databases in cardiovascular research. Genomics, Proteomics & Bioinformatics. 2016;14:191–199. doi: 10.1016/j.gpb.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jiao C., Song Z., Chen J., Zhong J., Cai W., Tian S., Chen S., Yi Y., Xiao Y. lncRNA-UCA1 enhances cell proliferation through functioning as a ceRNA of Sox4 in esophageal cancer. Oncol. Rep. 2016;36:2960–2966. doi: 10.3892/or.2016.5121. [DOI] [PubMed] [Google Scholar]
- 100.Zhang X., Ye Y., Zhao S. LncRNA Gas5 acts as a ceRNA to regulate PTEN expression by sponging miR-222-3p in papillary thyroid carcinoma. Oncotarget. 2018;9:3519–3530. doi: 10.18632/oncotarget.23336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Song F., Li L., Liang D., Zhuo Y., Wang X., Dai H. Knockdown of long noncoding RNA urothelial carcinoma associated 1 inhibits colorectal cancer cell proliferation and promotes apoptosis via modulating autophagy. J. Cell. Physiol. 2019;234:7420–7434. doi: 10.1002/jcp.27500. [DOI] [PubMed] [Google Scholar]
- 102.Lu Y., Li Y., Chai X., Kang Q., Zhao P., Xiong J., Wang J. Long noncoding RNA HULC promotes cell proliferation by regulating PI3K/AKT signaling pathway in chronic myeloid leukemia. Gene. 2017;607:41–46. doi: 10.1016/j.gene.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 103.Fu Q., Cheng J., Zhang J., Zhang Y., Chen X., Xie J., Luo S. Downregulation of YEATS4 by miR-218 sensitizes colorectal cancer cells to L-OHP-induced cell apoptosis by inhibiting cytoprotective autophagy. Oncol. Rep. 2016;36:3682–3690. doi: 10.3892/or.2016.5195. [DOI] [PubMed] [Google Scholar]
- 104.Fesler A., Liu H., Wu N., Liu F., Ling P., Ju J. Autophagy regulated by miRNAs in colorectal cancer progression and resistance. Cancer Translational Medicine. 2017;3:96. doi: 10.4103/ctm.ctm_64_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fimia G.M., Piacentini M. Regulation of autophagy in mammals and its interplay with apoptosis. Cell. Mol. Life Sci. 2010;67:1581–1588. doi: 10.1007/s00018-010-0284-z. [DOI] [PMC free article] [PubMed] [Google Scholar]