Abstract
Dilated cardiomyopathy (DCM) and ischemic cardiomyopathy (ICM) are two common types of cardiomyopathies leading to heart failure. Accurate diagnostic classification of different types of cardiomyopathies is critical for precision medicine in clinical practice. In this study, we hypothesized that machine learning (ML) can be used as a novel diagnostic approach to analyze cardiac transcriptomic data for classifying clinical cardiomyopathies. RNA-Seq data of human left ventricle tissues were collected from 41 DCM patients, 47 ICM patients, and 49 nonfailure controls (NF) and tested using five ML algorithms: support vector machine with radial kernel (svmRadial), neural networks with principal component analysis (pcaNNet), decision tree (DT), elastic net (ENet), and random forest (RF). Initial ML classifications achieved ~93% accuracy (svmRadial) for NF vs. DCM, ~82% accuracy (RF) for NF vs. ICM, and ~80% accuracy (ENet and svmRadial) for DCM vs. ICM. Next, 50 highly contributing genes (HCGs) for classifying NF and DCM, 68 HCGs for classifying NF and ICM, and 59 HCGs for classifying DCM and ICM were selected for retraining ML models. Impressively, the retrained models achieved ~90% accuracy (RF) for NF vs. DCM, ~90% accuracy (pcaNNet) for NF vs. ICM, and ~85% accuracy (pcaNNet and RF) for DCM vs. ICM. Pathway analyses further confirmed the involvement of those selected HCGs in cardiac dysfunctions such as cardiomyopathies, cardiac hypertrophies, and fibrosis. Overall, our study demonstrates the promising potential of using artificial intelligence via ML modeling as a novel approach to achieve a greater level of precision in diagnosing different types of cardiomyopathies.
Keywords: artificial intelligence, cardiomyopathy, heart failure, machine learning, transcriptome
INTRODUCTION
Heart failure (HF) is one of the leading causes of human mortality (34). Cardiomyopathy is one of the causes of heart failure which has two major clinical presentations, dilated cardiomyopathy (DCM) and ischemic cardiomyopathy (ICM) (15, 30). DCM occurs due to left ventricular systolic dysfunction and dilatation (19), whereas ICM occurs due to continuous imbalance between supply and requirement of oxygen in the heart, which results in the loss of cardiac muscle cells, myocardial scarring, and ventricular failure (7). Since both DCM and ICM are the major risk factors for the development of HF, their timely diagnosis and classification are important. Currently, various clinical methods, such as chest X-ray, echocardiogram, electrocardiogram, cardiac MRI, cardiac CT scan, and blood tests, are available for the diagnosis of the cardiomyopathies (55). However, there is no consistency in the definition and diagnostic classification of these diseases (41). Due to this deficiency, clinicians are unable to intervene in a timely manner to diagnose and treat these clinically different presentations of heart failure.
Machine learning (ML), as a part of artificial intelligence (AI), has been successfully applied in a variety of medical fields for the discovery of new genotype-phenotype associations, disease diagnosis, adverse effects prediction and lowering readmission and death rates (24). Classification of diseases by ML has been applied on several types of individual or combinatorial large-scale data sets such as genomic, transcriptomic, and patient demographic data sets (24). RNA-Seq data have been used for ML classification for detection of diseases such as pneumonia (9), endometriosis (1), and cancer (52). Thus, RNA-Seq data are useful not only for differentiating between various diseases but also may be useful to build ML models for prediction of disease subtypes. Additionally, RNA-Seq data sets are readily available for various cardiomyopathies, which served as the basis for our study, wherein we hypothesized that ML can be used as a novel diagnostic approach to analyze cardiac RNA-Seq data for classifying clinical cardiomyopathies.
Therefore, we evaluated the utility of ML to detect and differentiate transcriptomic signatures from 137 human cardiac RNA-Seq data sets obtained from DCM, ICM, and non-failure (NF) subjects. This was performed in three experiments: first, using five different ML algorithms, we tested their performances to differentiate between NF and DCM. Next, we trained and tested ML models to classify NF and ICM. Finally, we tested the capability of ML models to differentially classify subjects as belonging to either one of the two different types of cardiomyopathies, DCM or ICM. To our knowledge, this is the first study demonstrating the successful application of ML modeling on whole genome transcriptomic data for diagnostic classifications of clinical cardiomyopathies.
METHODS
Data set processing.
Gene Expression Omnibus (GEO) database (8) was queried for RNA-Seq studies performed on human left ventricle tissues collected from NF, DCM, and ICM subjects. Table 1 summarizes the information about seven data sets found in GEO including GSE116250 (43), GSE57344 (28), GSE71613 (40), GSE120852 (37), GSE46224 (51), GSE48166 (47), and GSE108157 (36). A total number of 41 DCM, 47 ICM, and 49 NF samples were identified and downloaded for this study. Raw reads (FASTQ files) were downloaded from the European Nucleotide Archive website (https://www.ebi.ac.uk/ena) and analyzed with FastQC (6) for quality control, Cutadapt (31) for filtering adaptors and low-quality bases (Phred quality score < 10), HISAT2 and SAMtools (21, 26) for aligning the trimmed reads to human reference genome (GRCh38), and HTSeq-count (5) for quantifying gene expression. Only uniquely mapped reads were used for expression quantification. The compute-intensive tasks were performed at the Ohio Supercomputer Center (56).
Table 1.
Summary of data sets found in the GEO database
| Study | Data Set | Platform | Sample Size |
|---|---|---|---|
| Study_1 | GSE116250 (43) | Illumina HiSeq 2500 | 37 DCM, 13 ICM, 14 NF |
| Study_2 | GSE57344 (28) | Illumina Genome Analyzer | 2 DCM, 1 ICM, 3 NF |
| Study_3 | GSE71613 (40) | Illumina HiSeq 2000 | 2 DCM, 4 NF |
| Study_4 | GSE120852 (37) | Illumina HiSeq 2500 | 5 ICM, 5 NF |
| Study_5 | GSE46224 (51) | Illumina HiSeq 2000 | 8 ICM, 8 NF |
| Study_6 | GSE48166 (47) | Illumina Genome Analyzer II | 15 ICM, 15 NF |
| Study_7 | GSE108157 (36) | Illumina HiSeq 2500 | 5 ICM |
GEO, Gene Expression Omnibus; DCM, dilated cardiomyopathy; ICM, ischemic cardiomyopathy; NF, nonfailure controls.
ML training and testing.
The study was conducted in three stages: First, the performance of different ML algorithms was tested for their efficiency to classify RNA-Seq data sets as those from NF or DCM. Next, we trained and tested ML models to classify NF and ICM. Finally, we tested the capability of ML models to differentially classify two different types of cardiomyopathies, DCM and ICM. Read counts, representing quantified gene expression, were used as inputs for ML modeling. The following five ML algorithms were implemented using the caret R package (25): “svmRadial” for support vector machine with radial kernel (svmRadial), “pcaNNet” for neural networks with principal component analysis (pcaNNet), “rpart” for decision tree (DT), “glmnet” for elastic net (ENet), and “rf” for random forest (RF). R packages including kernlab (20), randomForest (27), glmnet (11) and rpart (45) were embedded to be used for implementing the ML algorithms. For the initial step of feature selection, to reduce the dimensionality of the feature space and computational time, features with low variance were filtered. To perform this task, gene-wise variance was calculated for each gene, and the top 1,000 genes with the highest variance across all the samples in each experiment were selected out of 58,884 genes for training the ML models. A 70:30 ratio was used to divide the data into training and testing data sets. In the training phase, each ML model was evaluated by 10-fold cross-validation repeated 10 times. Automatic hyperparameter tuning, by testing 10 different values for each hyperparameter, was performed by caret. The processes of data shuffling, data splitting, training, and testing were independently performed for 50 iterations. In each iteration, the performance parameters, including accuracy, AUC (area under the receiver operating characteristic curve), sensitivity, specificity, precision and F1 score, were computed for the testing data set. Mean value and standard deviation of each performance parameter were calculated from all the values collected through all the iterations.
Highly contributing genes and pathway analysis.
Highly contributing genes (HCGs) to each ML model were identified and proposed as candidate biomarker genes. To perform this task, variable importance scores of the genes were calculated with the “varImp” function from the caret R package (25). A gene with a higher score represents its higher prediction contribution to the ML model. Variable importance calculation was performed for all the models, and the top genes with the highest scores in each model were used to generate the Venn diagram via the tool developed by the Bioinformatics and Evolutionary Genomics group at Ghent University (http://bioinformatics.psb.ugent.be/webtools/Venn/). The genes that were shared among at least two models were included in the final HCGs list. To evaluate how the selected HCGs were able to classify case and control as well as cardiomyopathy subtypes, only selected HCGs were used to train and test the five ML models as described above. The Ingenuity Pathway Analysis (IPA, Qiagen) (23) was performed to test whether the selected HCGs were indicative of any cardiac dysfunction related to cardiomyopathy or HF.
RESULTS
Diagnostic classifications of cardiomyopathies by ML.
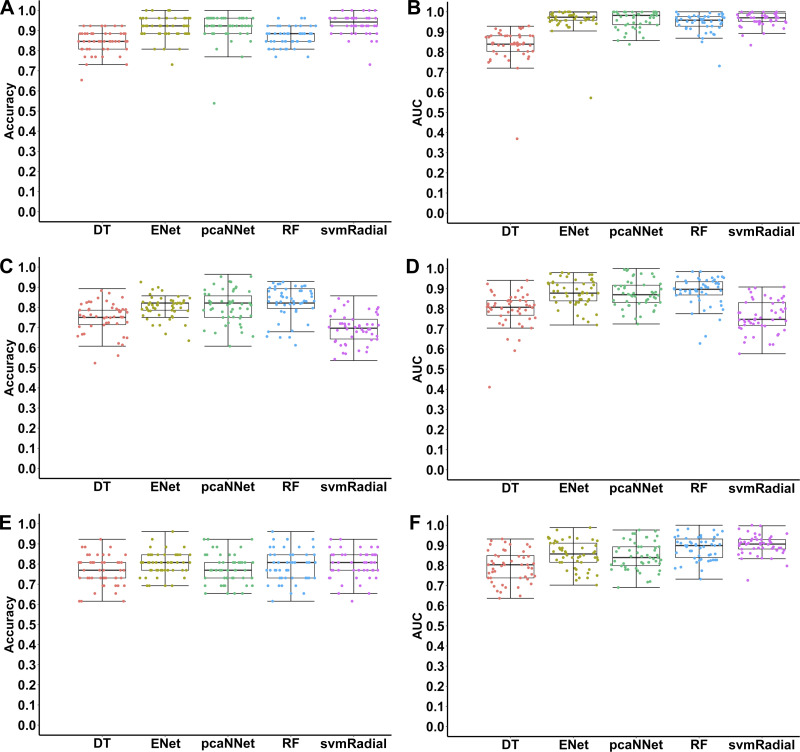
Figure 1 and Table 2 present performance measures of different ML models evaluated on the testing data sets for classifications of different types of cardiomyopathies and subtype recognition using the top 1,000 high-variance gene features. First, we tested the performance of different ML algorithms in differentiating between DCM and NF. Results show that >90% accuracy was achieved by ENet (~92%), pcaNNet (~92%), and svmRadial (~93%) (Fig. 1A, Table 2). In terms of AUC, four algorithms, ENet, pcaNNet, RF, and svmRadial, achieved ≥0.95 AUC in classifying DCM and NF (Fig. 1B, Table 2). If one compares all the performance measures, svmRadial, pcaNNet, and ENet were among the best ML models to classify DCM and NF samples. In terms of variation, no clear difference was observed among different models (Fig. 1, A and B; Table 2). Next, we trained and tested ML models to classify ICM and NF. Results showed that ≥80% accuracy and ≥0.88 AUC were achieved using ENet, pcaNNet, and RF (Fig. 1, C and D; Table 2). It is noteworthy that svmRadial, which was one of the best models in classifying DCM, did not perform well in classifying ICM and had a sharp decrease in all measurements (Fig. 1, Table 2). Another important point is that all models had a slight increase in variation compared with DCM classification (Fig. 1, Table 2). Even so, an impressive ≥80% accuracy was still achieved in diagnosing ICM. Finally, we tested the capability of ML models to classify two different subtypes of cardiomyopathies (DCM vs. ICM). Remarkably, all five algorithms achieved >75% accuracy in classifying DCM vs. ICM, with ENet (~80%) and svmRadial (~80%) as the highest ones (Fig. 1E, Table 2). In terms of AUC, four algorithms including ENet, pcaNNet, RF, and svmRadial achieved ≥0.85 AUC in classifying DCM and ICM (Fig. 1F, Table 2). The performance measures of subtype differentiation decreased compared with DCM-NF classification but were almost similar to ICM-NF classification (Table 2). In terms of variation, all the models had higher variation compared with DCM-NF classification, but compared with ICM-NF, depending on the model and measurement, variation was either similar, higher, or lower (Fig. 1, Table 2).
Fig. 1.
Accuracies and areas under the receiver operating characteristic curve (AUCs) of different machine learning algorithms in classifications of dilated cardiomyopathy (DCM), ischemic cardiomyopathy (ICM), and nonfailure control (NF) samples using the top 1,000 high-variance gene features. Each point represents one iteration (total 50 iterations) using each algorithm in each classification. A, B: NF vs. DCM; C, D: NF vs. ICM; E, F: DCM vs. ICM. DT, decision tree; ENet, elastic net; pcaNNet, neural networks with principal component analysis; RF, random forest; svmRadial, support vector machine with radial kernel.
Table 2.
Performance measures of different machine learning algorithms for classifications of different types of cardiomyopathies and subtype recognition using the top 1,000 high-variance gene features
| Accuracy | AUC | Sensitivity | Specificity | Precision | F1 | |
|---|---|---|---|---|---|---|
| NF vs. DCM | ||||||
| DT | 0.84 ± 0.06 | 0.83 ± 0.08 | 0.76 ± 0.13 | 0.92 ± 0.07 | 0.90 ± 0.08 | 0.81 ± 0.08 |
| ENet | 0.92 ± 0.05 | 0.96 ± 0.06 | 0.93 ± 0.07 | 0.91 ± 0.09 | 0.91 ± 0.09 | 0.91 ± 0.05 |
| pcaNNet | 0.92 ± 0.07 | 0.96 ± 0.04 | 0.91 ± 0.15 | 0.92 ± 0.08 | 0.92 ± 0.08 | 0.92 ± 0.05 |
| RF | 0.87 ± 0.05 | 0.95 ± 0.05 | 0.87 ± 0.09 | 0.87 ± 0.08 | 0.86 ± 0.07 | 0.86 ± 0.05 |
| svmRadial | 0.93 ± 0.05 | 0.96 ± 0.03 | 0.87 ± 0.10 | 0.98 ± 0.04 | 0.98 ± 0.05 | 0.92 ± 0.07 |
| NF vs. ICM | ||||||
| DT | 0.75 ± 0.07 | 0.79 ± 0.09 | 0.76 ± 0.18 | 0.74 ± 0.13 | 0.76 ± 0.10 | 0.74 ± 0.11 |
| ENet | 0.80 ± 0.05 | 0.88 ± 0.07 | 0.79 ± 0.13 | 0.81 ± 0.13 | 0.83 ± 0.10 | 0.80 ± 0.06 |
| pcaNNet | 0.81 ± 0.08 | 0.88 ± 0.07 | 0.83 ± 0.11 | 0.78 ± 0.14 | 0.81 ± 0.10 | 0.81 ± 0.08 |
| RF | 0.82 ± 0.08 | 0.89 ± 0.07 | 0.78 ± 0.14 | 0.87 ± 0.09 | 0.86 ± 0.09 | 0.81 ± 0.10 |
| svmRadial | 0.69 ± 0.07 | 0.76 ± 0.08 | 0.61 ± 0.19 | 0.78 ± 0.17 | 0.76 ± 0.11 | 0.65 ± 0.11 |
| DCM vs. ICM | ||||||
| DT | 0.77 ± 0.07 | 0.79 ± 0.08 | 0.75 ± 0.14 | 0.78 ± 0.13 | 0.76 ± 0.11 | 0.75 ± 0.08 |
| ENet | 0.80 ± 0.06 | 0.85 ± 0.07 | 0.85 ± 0.12 | 0.76 ± 0.11 | 0.76 ± 0.07 | 0.80 ± 0.06 |
| pcaNNet | 0.78 ± 0.08 | 0.85 ± 0.07 | 0.80 ± 0.18 | 0.76 ± 0.13 | 0.76 ± 0.10 | 0.76 ± 0.10 |
| RF | 0.79 ± 0.08 | 0.89 ± 0.06 | 0.88 ± 0.11 | 0.72 ± 0.12 | 0.74 ± 0.09 | 0.80 ± 0.08 |
| svmRadial | 0.80 ± 0.07 | 0.90 ± 0.05 | 0.82 ± 0.11 | 0.78 ± 0.10 | 0.77 ± 0.09 | 0.79 ± 0.08 |
Values are presented as means ± standard deviation (total 50 iterations). AUC, area under the receiver operating characteristic curve; DCM, dilated cardiomyopathy; ICM, ischemic cardiomyopathy; NF, nonfailure controls; DT, decision tree; ENet, elastic net; pcaNNet, neural networks with principal component analysis; RF, random forest; svmRadial, support vector machine with radial kernel.
Identifying HCGs to ML based diagnosis of cardiomyopathies.
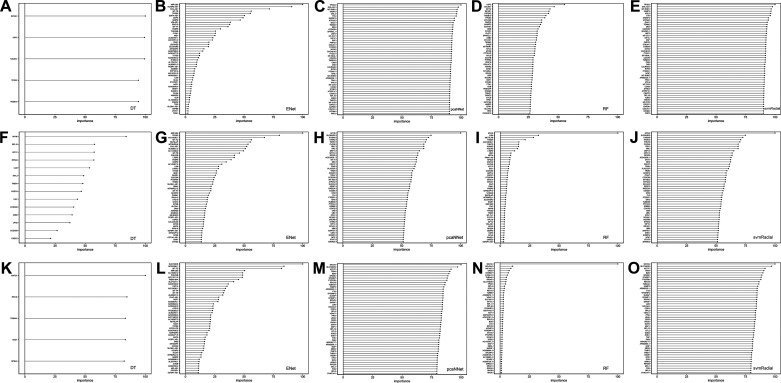
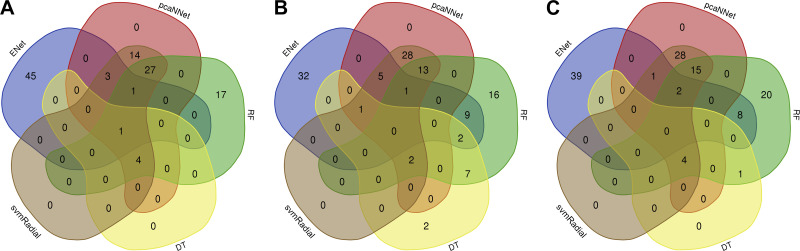
The next step of this study was to further select HCGs, as potential candidate biomarkers, from the top 1,000 high-variance gene features. This step was considered to be the second step of feature selection to further decrease the dimensionality of the feature space. To perform this task, we calculated variable importance scores of the genes, and the top 50 genes with the highest scores were selected for each model. Figure 2 and Supplementary Table S1 (all supplemental material is available at https://doi.org/10.5281/zenodo.3941331) present the top genes and their scores for each model and experiment. Given that the importance scores (ranging from 0 to 100) are relative and specific to each model, only a few genes were scored more than 0 in the DT model, which is why we have a limited number of genes in Fig. 2, A, F, K. HCGs were selected if they were shared among at least two models (Fig. 3). Since the final number of selected HCGs depended on the shared HCGs among different ML models, the number of HCGs selected for each classification experiment is different (Fig. 3). Selected HCGs, including 50 genes for DCM-NF, 68 genes for ICM-NF, and 59 genes for DCM-ICM, are presented in Table 3.
Fig. 2.
Variable importance scores of top gene features highly contributing to classifications of dilated cardiomyopathy (DCM), ischemic cardiomyopathy (ICM), and nonfailure control (NF) in each ML algorithm. A–E: NF vs. DCM, F–J: NF vs. ICM, K–O: DCM vs. ICM. DT, decision tree; ENet, elastic net; pcaNNet, neural networks with principal component analysis; RF, random forest; svmRadial, support vector machine with radial kernel.
Fig. 3.
Venn diagrams of shared highly contributing genes among different machine learning models in each classification. A: nonfailure controls (NF) vs. dilated cardiomyopathy (DCM), B: NF vs. ischemic cardiomyopathy (ICM), C: DCM vs. ICM. DT, decision tree; ENet, elastic net; pcaNNet, neural networks with principal component analysis; RF, random forest; svmRadial, support vector machine with radial kernel.
Table 3.
Selected HCGs for each classification experiment
| Classification | HCGs Shared among at Least Two ML Models |
|---|---|
| NF vs. DCM | TPGS2, HABP2, EIF4G1, TMSB10, USF2, COL6A1, AP005329.1, LDLRAD2, SVIL-AS1, COQ9, DAG1, TENM2, DES, CHCHD10, MT-CO3, MT-RNR1, MYL3, COX7B, PMP22, MT-CO1, MYL9, KIF1C, SH3RF2, FYCO1, MT-ND4L, TPM1, SYNE2, ASB2, CKMT2, COQ8A, DSP, MYH6, CKB, SPARCL1, ATP2A2, COX7A1, NFIC, RPL19, MT-RNR2, S100A9, PTDSS1, PKP2, CCDC69, TNNT2, CKM, TTN, GAPDH, ATP5F1C, RYR2, MT-CYB |
| NF vs. ICM | AC012636.1, CCDC69, KIF1C, MYH6, ANKRD2, STAT3, CNN1, SH3RF2, SYNE2, LUM, THBS4, SRP14, SLC9A3R2, S100A9, CDC37, CLCN6, TRDN, MYH14, TNNT2, KANK2, RPS25, MT-TI, HRC, SERF2, MIR1282, RN7SL4P, LTBP2, RASD1, FKBP5, TPGS2, MTND4P12, PTGFRN, IGFBP7-AS1, PTP4A3, PLA2G2A, NFIC, SLC4A3, DES, MAP7D1, FLII, COX7B, NCAM1, RAD23A, YBX1, EIF4G1, KCNIP2, PTDSS1, ERBB2, ITGA7, TUBB4B, UBC, MT-ND3, UBB, CKM, COQ8A, UQCRB, CKB, TPM2, ATP2A2, IDH2, EPN1, RPL19, MYL3, POSTN, RPS24, CCDC80, CSDC2, CILP |
| DCM vs. ICM | SLC7A6OS, MT-TC, SOD1, TUBA4A, RPS4X, MAT2A, HABP2, C1S, RPL5, MAF1, SPARC, STARD7, SOD2, VDAC1, GOLGA4, B2M, SARAF, TPM4, RHOA, PTGFRN, RPS18, TP53INP2, MTND6P4, AC145207.2, KCNIP2, AC036108.2, AC005726.4, AC008079.1, ATG10, MIF-AS1, TIMP2, EFEMP1, FOS, RPL12, PPIA, COPA, AP002956.1, THBS1, ARHGEF17, LDHA, DYNC1H1, OGDH, SBDS, CFL2, AP005329.1, DCN, MGP, PKM, SLMAP, DSTN, RPS29, CD63, NFIX, LUM, RPL3, ARIH2, TNC, MCL1, RPL38 |
HCG, highly contributing gene; ML, machine learning; DCM, dilated cardiomyopathy; ICM, ischemic cardiomyopathy; NF, nonfailure controls.
Retraining ML models using HCGs.
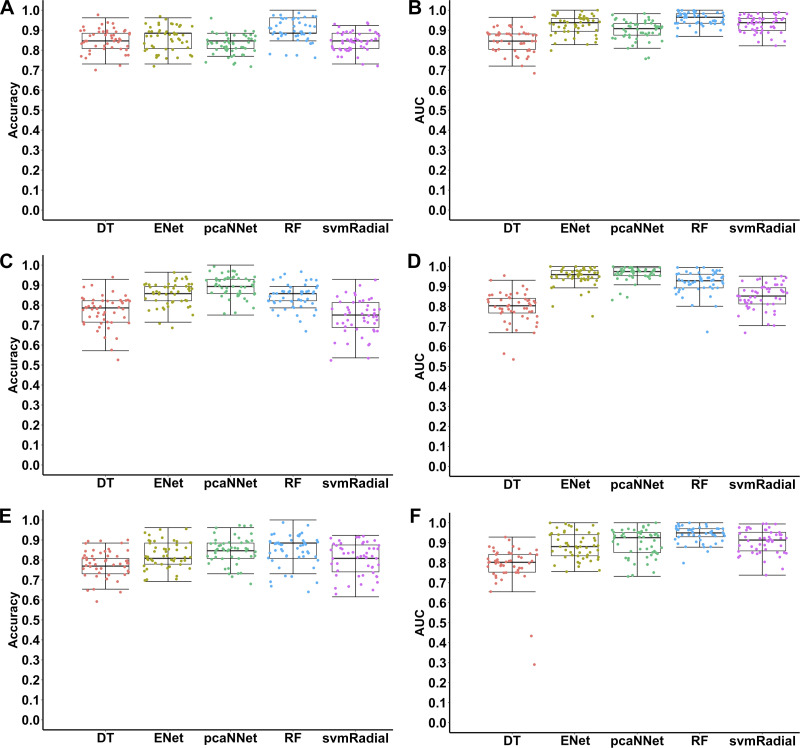
After selecting the HCGs for each classification experiment, we reimplemented five ML algorithms for training and testing using the only selected HCGs in Table 3. Figure 4 and Table 4 present performance measures of different ML models evaluated on the testing data sets for classifications of different types of cardiomyopathies by using HCGs. In the case of classifying DCM and NF, ~90% accuracy was achieved by using RF, and ~85% accuracy was achieved with other models (Fig. 4A, Table 4). In terms of AUC, four algorithms (ENet, pcaNNet, RF, and svmRadial) achieved ≥0.90 AUC in classifying DCM and NF (Fig. 4B, Table 4). RF outperformed other ML models regarding all the performance measures (Table 4). Compared with initial classification using the top 1,000 high-variance genes, an increased prediction accuracy was observed in the RF and DT models using the only selected HCGs (Table 2, Table 4). However, all the measurements in ENet, pcaNNet, and svmRadial decreased with only using the selected HCGs (Table 2, Table 4).
Fig. 4.
Accuracies and areas under the receiver operating characteristic curve (AUCs) of different machine learning algorithms in classifications of dilated cardiomyopathy (DCM), ischemic cardiomyopathy (ICM), and nonfailure control (NF) samples using only selected highly contributing genes. Each point represents one iteration (total 50 iterations) using each algorithm in each classification. A, B: NF vs. DCM; C, D: NF vs. ICM; E, F: DCM vs. ICM. DT, decision tree; ENet, elastic net; pcaNNet, neural networks with principal component analysis; RF, random forest; svmRadial, support vector machine with radial kernel.
Table 4.
Performance measures of different ML algorithms for classifications of different types of cardiomyopathies and subtype recognition using only selected HCGs
| Accuracy | AUC | Sensitivity | Specificity | Precision | F1 | |
|---|---|---|---|---|---|---|
| NF vs. DCM | ||||||
| DT | 0.85 ± 0.06 | 0.85 ± 0.06 | 0.77 ± 0.10 | 0.92 ± 0.06 | 0.90 ± 0.08 | 0.82 ± 0.07 |
| ENet | 0.86 ± 0.06 | 0.93 ± 0.05 | 0.83 ± 0.10 | 0.88 ± 0.07 | 0.86 ± 0.08 | 0.84 ± 0.07 |
| pcaNNet | 0.83 ± 0.05 | 0.90 ± 0.05 | 0.83 ± 0.09 | 0.83 ± 0.09 | 0.82 ± 0.08 | 0.82 ± 0.05 |
| RF | 0.90 ± 0.06 | 0.96 ± 0.03 | 0.87 ± 0.08 | 0.93 ± 0.08 | 0.92 ± 0.08 | 0.89 ± 0.06 |
| svmRadial | 0.84 ± 0.05 | 0.93 ± 0.04 | 0.78 ± 0.09 | 0.89 ± 0.07 | 0.87 ± 0.07 | 0.82 ± 0.06 |
| NF vs. ICM | ||||||
| DT | 0.77 ± 0.08 | 0.79 ± 0.08 | 0.78 ± 0.17 | 0.76 ± 0.13 | 0.78 ± 0.10 | 0.76 ± 0.11 |
| ENet | 0.85 ± 0.06 | 0.95 ± 0.05 | 0.80 ± 0.11 | 0.91 ± 0.07 | 0.91 ± 0.07 | 0.84 ± 0.07 |
| pcaNNet | 0.90 ± 0.06 | 0.97 ± 0.04 | 0.89 ± 0.10 | 0.90 ± 0.09 | 0.91 ± 0.08 | 0.89 ± 0.06 |
| RF | 0.84 ± 0.06 | 0.92 ± 0.06 | 0.82 ± 0.11 | 0.86 ± 0.09 | 0.86 ± 0.08 | 0.84 ± 0.07 |
| svmRadial | 0.74 ± 0.09 | 0.85 ± 0.06 | 0.65 ± 0.18 | 0.84 ± 0.14 | 0.82 ± 0.12 | 0.71 ± 0.12 |
| DCM vs. ICM | ||||||
| DT | 0.78 ± 0.07 | 0.78 ± 0.10 | 0.75 ± 0.15 | 0.81 ± 0.13 | 0.79 ± 0.11 | 0.75 ± 0.09 |
| ENet | 0.82 ± 0.06 | 0.88 ± 0.07 | 0.86 ± 0.11 | 0.79 ± 0.12 | 0.79 ± 0.09 | 0.82 ± 0.07 |
| pcaNNet | 0.85 ± 0.06 | 0.90 ± 0.07 | 0.84 ± 0.10 | 0.85 ± 0.09 | 0.84 ± 0.09 | 0.83 ± 0.07 |
| RF | 0.85 ± 0.07 | 0.94 ± 0.04 | 0.91 ± 0.10 | 0.79 ± 0.14 | 0.80 ± 0.10 | 0.85 ± 0.07 |
| svmRadial | 0.81 ± 0.08 | 0.91 ± 0.06 | 0.79 ± 0.13 | 0.82 ± 0.10 | 0.79 ± 0.10 | 0.79 ± 0.09 |
Values are presented as means ± standard deviation (total 50 iterations). HCG, highly contributing gene; ML, machine learning; DCM, dilated cardiomyopathy; ICM, ischemic cardiomyopathy; NF, nonfailure controls; DT, decision tree; ENet, elastic net; pcaNNet, neural networks with principal component analysis; RF, random forest; svmRadial, support vector machine with radial kernel.
The classification results of ICM vs. NF using HCGs were very promising. Impressively, pcaNNet trained with only HCGs achieved ~90% accuracy (Fig. 4C, Table 4), which was much better than the accuracies of all the ML models trained with the top 1,000 high-variance gene features (Fig. 1C, Table 2). Interestingly, almost all the performance measures trained with HCGs were better than those measures trained with the top 1,000 high-variance genes (Table 2, Table 4). For example, ~9% and ~5% increased accuracies in pcaNNet and ENet were observed, respectively, as well as ~9% increased AUCs in pcaNNet and svmRadial (Table 2, Table 4). The ~90% accuracy (pcaNNet) of classifying ICM and NF with only 68 genes is very promising (Fig. 4C, Table 4). Significant improvements were also represented by >0.90 AUC in ENet, pcaNNet, and RF trained with only 68 HCGs (Fig. 4D, Table 4).
Finally, we tested the capability of five ML models to classify two subtypes of cardiomyopathies (DCM vs. ICM) using only selected HCGs. Remarkably, ENet, pcaNNet, RF, and svmRadial achieved >80% accuracy in classifying DCM and ICM (Fig. 4E, Table 4). In terms of AUC, pcaNNet, RF, and svmRadial achieved ≥0.90 AUC in classifying DCM and ICM (Fig. 4F, Table 4). Compared with the classification results using the top 1,000 high-variance genes (Table 2), almost all the performance measures trained with HCGs were improved (Table 4). In general, in terms of classification results using selected HCGs as potential candidate biomarkers, the classification results of ICM-NF and DCM-ICM were significantly improved compared with DCM-NF. Our results demonstrate that a high prediction accuracy of diagnosing and differentiating clinical cardiomyopathies can be achieved by training various ML models with a small number of gene features.
Pathophysiological pathway analysis.
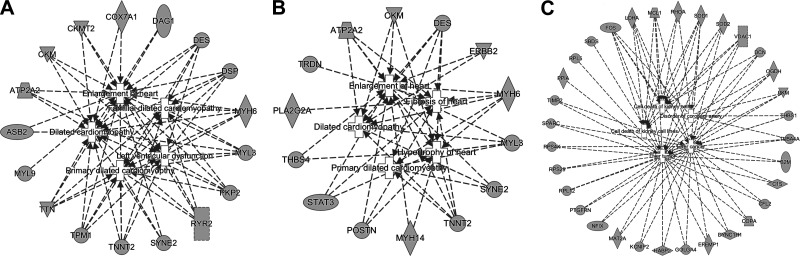
Furthermore, IPA pathway analyses were performed to examine if any selected HCGs, as the potential biomarkers, were enriched in pathophysiological pathways related to HF. Integrated networks of top five diseases and their corresponding genes in each classification experiment are presented in Fig. 5. Table 5 summarizes the top 10 diseases and their corresponding genes in each classification experiment. The selected HCGs from the DCM-NF classification were clearly involved in cardiac dysfunctions, such as DCM, familial DCM, left ventricular dysfunction, primary DCM, and enlargement of heart, and the genes DES, DSP, MYH6, MYL3, PKP2, RYR2, TPM1, and TTN were shared among these top five cardiac dysfunctions (Fig. 5A). Similarly, using the selected HCGs for classifying ICM and NF, fibrosis of heart, hypertrophy of heart, enlargement of heart, primary DCM, and DCM were identified as the top five cardiac dysfunctions, and the genes TNNT2, MYH6, and DES were shared among all these top five cardiac dysfunctions (Fig. 5B). Finally, in the case of subtype differentiation (DCM vs. ICM), the goal was to identify the distinct dysfunctions between the two cardiomyopathies. Interestingly, none of the pathways related to the heart were detected, and the top five dysfunctional indexes were cell death of kidney cells, cell death of kidney cell lines, disorder of coronary artery, liver tumor, and liver cancer (Fig. 5C).
Fig. 5.
Integrated networks of the top 5 pathophysiological dysfunctions and their associated genes. A: NF vs. DCM, B: NF vs. ICM, C: DCM vs. ICM. DCM, dilated cardiomyopathy; ICM, ischemic cardiomyopathy; NF, nonfailure controls.
Table 5.
Top 10 significant pathophysiological dysfunctions in each classification experiment
| Diseases or Functions | P Value | Involved Genes |
|---|---|---|
| NF-DCM | ||
| DCM | 4.28E-16 | ASB2, ATP2A2, CKM, CKMT2, COX7A1, DAG1, DES, DSP, MYH6, MYL3, PKP2, RYR2, SYNE2, TNNT2, TPM1, TTN |
| Familial DCM | 3.10E-14 | DES, DSP, MYH6, MYL3, PKP2, RYR2, TNNT2, TPM1, TTN |
| Left ventricular dysfunction | 4.39E-14 | ATP2A2, CKM, CKMT2, DES, DSP, MYH6, MYL3, PKP2, RYR2, TNNT2, TPM1, TTN |
| Primary DCM | 6.66E-14 | DES, DSP, MYH6, MYL3, PKP2, RYR2, SYNE2, TNNT2, TPM1, TTN |
| Enlargement of heart | 6.97E-13 | ASB2, ATP2A2, CKM, CKMT2, COX7A1, DAG1, DES, DSP, MYH6, MYL3, MYL9, PKP2, RYR2, SYNE2, TNNT2, TPM1, TTN |
| Familial left ventricular noncompaction | 8.86E-13 | MYH6, PKP2, RYR2, TNNT2, TPM1, TTN |
| Arrhythmogenic right ventricular dysplasia familial 9 | 1.72E-11 | DES, MYH6, PKP2, RYR2, TTN |
| Arrhythmogenic right ventricular cardiomyopathy | 2.65E-11 | DES, DSP, MYH6, PKP2, RYR2, TTN |
| DCM 1S | 2.95E-11 | DES, DSP, PKP2, TNNT2, TTN |
| Familial arrhythmia | 3.77E-11 | DES, DSP, MYH6, PKP2, RYR2, TNNT2, TPM1, TTN |
| NF-ICM | ||
| Fibrosis of heart | 5.57E-07 | ATP2A2, DES, MYH6, PLA2G2A, POSTN, STAT3, TNNT2, TRDN |
| Hypertrophy of heart | 8.90E-07 | ATP2A2, CKM, DES, MYH14, MYH6, MYL3, POSTN, STAT3, THBS4, TNNT2 |
| Enlargement of heart | 2.55E-06 | ATP2A2, CKM, DES, ERBB2, MYH14, MYH6, MYL3, POSTN, STAT3, SYNE2, THBS4, TNNT2 |
| Primary DCM | 1.91E-05 | DES, MYH6, MYL3, SYNE2, TNNT2 |
| DCM | 2.51E-05 | ATP2A2, CKM, DES, ERBB2, MYH6, MYL3, SYNE2, TNNT2 |
| Arrhythmia | 2.83E-05 | ATP2A2, DES, KCNIP2, MYH6, TNNT2, TRDN, TUBB4B |
| Left ventricular dysfunction | 3.73E-05 | ATP2A2, CKM, DES, MYH6, MYL3, TNNT2 |
| Familial DCM | 4.52E-05 | DES, MYH6, MYL3, TNNT2 |
| Damage of heart | 6.88E-05 | DES, MYH6, POSTN, STAT3 |
| Enlargement of left ventricle | 1.09E-04 | ATP2A2, CKM, DES, MYL3, TNNT2 |
| DCM-ICM | ||
| Cell death of kidney cells | 1.53E-05 | DCN, FOS, LDHA, MCL1, RHOA, SOD1, SOD2, VDAC1 |
| Cell death of kidney cell lines | 4.23E-05 | FOS, LDHA, MCL1, RHOA, SOD1, SOD2, VDAC1 |
| Disorder of coronary artery | 4.29E-05 | FOS, OGDH, PKM, RHOA, SOD1, THBS1, TUBA4A |
| Liver tumor | 1.29E-04 | B2M, C1S, CFL2, COPA, DCN, DYNC1H1, EFEMP1, FOS, GOLGA4, HABP2, KCNlP2, LDHA, MAT2A, MCL1, NFIX, OGDH, PKM, PPIA, PTGFRN, RPL12, RPL5, RPS29, RPS4X, SBDS, SOD1, SOD2, SPARC, THBS1, TIMP2, TUBA4A, VDAC1 |
| Liver cancer | 1.54E-04 | B2M, C1S, CFL2, COPA, DCN, DYNC1H1, EFEMP1, FOS, GOLGA4, HABP2, KCNlP2, LDHA, MAT2A, MCL1, NFIX, OGDH, PKM, PTGFRN, RPL12, RPS29, RPS4X, SOD1, SOD2, SPARC, THBS1, TIMP2, TUBA4A, VDAC1 |
| Inflammation of heart | 2.05E-04 | ARIH2, B2M, PPIA |
| Coronary artery disease | 4.72E-04 | FOS, OGDH, PKM, RHOA, THBS1 |
| Hepatic adenocarcinoma | 6.78E-04 | MCL1, THBS1, TUBA4A |
| Increased levels of albumin | 1.15E-03 | B2M, THBS1 |
| Apoptosis of cardiomyocytes | 2.11E-03 | MCL1, RHOA, SOD2, TIMP2 |
DCM, dilated cardiomyopathy; ICM, ischemic cardiomyopathy; NF, nonfailure controls.
DISCUSSION
In this study, we demonstrated an accurate and robust diagnostic classification and candidate biomarker selection approach for dilated and ischemic cardiomyopathies by using five different supervised ML algorithms. It should be noted that our study did not adjust for batch effect or normalize gene expression data across different samples as we aimed to test the capacity and adaptability of ML models trained with raw gene read counts to classify and predict new unknown samples without the need for repeated processing of all the previous samples with the new samples. Impressively, initial ML classifications, using the top 1,000 high-variance genes, achieved ~93% accuracy (svmRadial) for NF vs. DCM, ~82% accuracy (RF) for NF vs. ICM, and ~80% accuracy (ENet and svmRadial) for DCM vs. ICM (Table 2). Previous applications of ML classifications of diseases using RNA-Seq data have been reported with an accuracy range of 20–90% for endometriosis (1) and >85% accuracy in the discrimination of hepatobiliary, lung, and pancreatic cancer types (4). High classification accuracy obtained for DCM vs. NF indicates the high potential of ML application for diagnosing DCM by using RNA-Seq data. Although the accuracy for classification of ICM vs. NF and ICM vs. DCM was lower than DCM vs. NF, more than 80% accuracy is still very promising. Due to the fact that DCM often has a genetic pathogenesis (33), the difference in accuracy between DCM vs. NF and ICM vs. NF is expected. In terms of differences among the algorithms, svmRadial had the best performance in DCM vs. NF and also subtype recognition, DCM vs. ICM. However, in ICM vs. NF, RF outperformed other models. In general, initial classification results were very satisfying, and we propose this approach as a promising tool for accurate diagnosis of clinical cardiomyopathies.
After performance evaluation of classification experiments with the top 1,000 high-variance genes, we selected HCGs for each ML model and classification experiment by calculating the variable importance scores of gene features. Feature selection has become a common practice in analyzing high-dimensional data, such as gene expression data in our study, to overcome the large-input dimensionality and relatively small sample sizes (39). In addition, this approach could be implemented for selecting the highly informative features (16). After feature selection, we identified 50 HCGs for classifying NF vs. DCM, 68 HCGs for classifying NF vs. ICM, and 59 HCGs for classifying DCM vs. ICM (Table 3). Among the HCGs identified in the DCM vs. NF classification, several genes, such as HABP2, TMSB10, COL6A1, DES, and KIF1C, have been previously reported to be dysregulated in DCM (3, 10, 48, 54). In addition, genetic mutations in MT-RNR1, MYL3, MT-ND4L, TPM1, TNNT2, and TTN have been reported to be the cause of DCM (2, 12, 14, 17, 18, 53). Impressively, the selected HCGs for classifying DCM and NF were shown to be related to various heart diseases and dysfunctions such as DCM, familial DCM, and left ventricular dysfunction (Fig. 5A, Table 5). In the ICM vs. NF classification, several HCGs have been previously reported to be associated with ICM or other types of cardiac dysfunctions. For example, dysregulation of NCAM1 in ICM (46) and STAT3 in ischemic heart disease (50) has been previously reported. In addition, genetic mutations in TRDN, SLC4A3, MYH6, and TNNT2 have been found to be associated with sudden cardiac death, HF, hypoplastic left heart, and cardiac hypertrophy, respectively (22, 29, 35, 44). Interestingly, the IPA analyses identified fibrosis of heart, hypertrophy of heart, and enlargement of heart as the top diseases, which further confirms that our feature selection procedure successfully identified ICM-related genes (Fig. 5B, Table 5). Furthermore, we identified 59 genes as HCGs for differentiating between DCM and ICM (Table 3). Among these identified HCGs, genetic mutations in TIMP2 and abnormal expression of MGP have been reported to contribute to HF (13, 32). Interestingly, only a few known HF genes were identified as the top genes in this part, and the reason could be the common characteristics between DCM and ICM as cardiovascular diseases. In our study, several known genes for DCM and ICM or other types of heart diseases were identified by using the feature selection method. This shows the capability of the ML approach in identifying the associated genes to cardiomyopathies or other types of cardiac dysfunctions. In addition, several genes discovered from our unique approach were not previously observed in cardiac diseases. These sets of genes could be considered as new diagnostic biomarkers and therapeutic targets for future research and clinical application. Furthermore, these selected genes can be tested in animal models to gain more information about their pathophysiological roles in various types of cardiomyopathies.
ML models were retrained with selected HCGs to test if the prediction accuracy could be further improved through dimensionality reduction of the gene features. Impressively, the retrained classification results using only those selected HCGs achieved ~90% accuracy (RF) for DCM vs. NF, ~90% accuracy (pcaNNet) for ICM vs. NF, and ~85% accuracy (pcaNNet and RF) for DCM vs. ICM (Fig. 4, Table 4). This presents our successful selection of the highest informative features. RF was the only algorithm that had increases in almost all the performance measures in all the classification experiments. Accuracies of ~90, ~84, and ~85% and AUCs of ~0.96, ~0.92, and ~0.94 were achieved by RF in DCM vs. NF, ICM vs. NF, and DCM vs. ICM, respectively. These results present the huge potential of the RF model to differentiate between different types of cardiomyopathies by using RNA-Seq data with only a few genes. Improvements in the performance of the RF model by the feature selection method have been previously reported in classifications of gene expression data from different types of cancer such as colon, prostrate, breast, skin, leukemia, lung, and brain (42, 49). Achieving more than 85% accuracy in differentiating between cardiomyopathies and controls, as well as subtype recognition, could be a turning point in the diagnosis of cardiomyopathies, and it could lead to a more accurate diagnosis. Improvement in performance measures of most of the ML algorithms with different types of samples using only a small set of genes in our study indicates that a robust feature selection is a crucial step for improving the prediction performance as well as reducing the computational complexity.
Overall, our study is the first to demonstrate the promising potential of using large-scale transcriptomic data for training ML models to classify different types of clinical cardiomyopathies toward a greater level of precision medicine. Our findings and approaches indicate that accurate classifications of different cardiomyopathies and subtype differentiation are achievable using ML modeling of whole genome transcriptomic data of cardiac samples collected through clinical cardiac biopsy, and potentially, similar transcriptome-based ML modeling approaches could be applied for diagnosing other diseases by using their related biosamples (e.g., tissue and blood). In addition, we presented a robust approach for identification of potential biomarkers that could be used as an alternative method to common statistical methods. Our feature selection results show that small sets of informative genes could further improve ML performance, and those genes were very closely related to corresponding cardiovascular diseases. Lastly, our study prioritized previously unknown genes and pathways as candidate biomarkers for differentiating between DCM and ICM.
GRANTS
The work was supported by the Dean’s Postdoctoral to Faculty Fellowship from University of Toledo College of Medicine and Life Sciences to X. Cheng. X. Cheng also acknowledges funding support from the P30 Core Center Pilot Grant from the National Institute on Drug Abuse Center of Excellence in Omics, Systems Genetics, and the Addictome. B. Joe acknowledges support from National Heart, Lung, and Blood Institute Grant HL143082. P. B. Munroe acknowledges support from the National Institute of Health Research Cardiovascular Biomedical Research Centre at Barts and Queen Mary University of London.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
X.C. and A.A. conceived and designed research; A.A. and X.C. performed experiments; A.A. and X.C. analyzed data; A.A., I.M., S.A., B.J., and X.C. interpreted results of experiments; A.A. prepared figures; A.A., I.M., and S.A. drafted manuscript; A.A., I.M., S.A., P.B.M., B.J., and X.C. edited and revised manuscript; A.A., I.M., S.A., P.B.M., B.J., and X.C. approved final version of manuscript.
ACKNOWLEDGMENTS
We appreciate the computational support from the Ohio Supercomputer Center.
REFERENCES
- 1.Akter S, Xu D, Nagel SC, Bromfield JJ, Pelch K, Wilshire GB, Joshi T. Machine learning classifiers for endometriosis using Transcriptomics and Methylomics data. Front Genet 10: 766, 2019. doi: 10.3389/fgene.2019.00766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alila-Fersi O, Chamkha I, Majdoub I, Gargouri L, Mkaouar-Rebai E, Tabebi M, Tlili A, Keskes L, Mahfoudh A, Fakhfakh F. Co segregation of the m.1555A>G mutation in the MT-RNR1 gene and mutations in MT-ATP6 gene in a family with dilated mitochondrial cardiomyopathy and hearing loss: A whole mitochondrial genome screening. Biochem Biophys Res Commun 484: 71–78, 2017. doi: 10.1016/j.bbrc.2017.01.070. [DOI] [PubMed] [Google Scholar]
- 3.Alimadadi A, Munroe PB, Joe B, Cheng X. Meta-Analysis of Dilated Cardiomyopathy Using Cardiac RNA-Seq Transcriptomic Datasets. Genes (Basel) 11: 60, 2020. doi: 10.3390/genes11010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alkan CB, Isik Z. Characterization of Cancer Types by Applying Machine Learning Methods on Blood RNA-Sequencing Data. In: 2019 3rd International Symposium on Multidisciplinary Studies and Innovative Technologies (ISMSIT) 2019, p. 1–4. [Google Scholar]
- 5.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169, 2015. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrews S. FastQC: a quality control tool for high throughput sequence data [Online]. Babraham Bioinformatics, Babraham Institute, Cambridge, United Kingdom: https://www.bioinformatics.babraham.ac.uk/projects, 2010. https://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 7.Anversa P, Sonnenblick EH. Ischemic cardiomyopathy: pathophysiologic mechanisms. Prog Cardiovasc Dis 33: 49–70, 1990. doi: 10.1016/0033-0620(90)90039-5. [DOI] [PubMed] [Google Scholar]
- 8.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res 41, D1: D991–D995, 2013. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi Y, Liu TT, Pankratz DG, Colby TV, Barth NM, Lynch DA, Walsh PS, Raghu G, Kennedy GC, Huang J. Identification of usual interstitial pneumonia pattern using RNA-Seq and machine learning: challenges and solutions. BMC Genomics 19, Suppl 2: 101, 2018. doi: 10.1186/s12864-018-4467-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeAguero JL, McKown EN, Zhang L, Keirsey J, Fischer EG, Samedi VG, Canan BD, Kilic A, Janssen PML, Delfín DA. Altered protein levels in the isolated extracellular matrix of failing human hearts with dilated cardiomyopathy. Cardiovasc Pathol 26: 12–20, 2017. doi: 10.1016/j.carpath.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 33: 1–22, 2010. doi: 10.18637/jss.v033.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerull B, Gramlich M, Atherton J, McNabb M, Trombitás K, Sasse-Klaassen S, Seidman JG, Seidman C, Granzier H, Labeit S, Frenneaux M, Thierfelder L. Mutations of TTN, encoding the giant muscle filament titin, cause familial dilated cardiomyopathy. Nat Genet 30: 201–204, 2002. doi: 10.1038/ng815. [DOI] [PubMed] [Google Scholar]
- 13.Givvimani S, Kundu S, Narayanan N, Armaghan F, Qipshidze N, Pushpakumar S, Vacek TP, Tyagi SC. TIMP-2 mutant decreases MMP-2 activity and augments pressure overload induced LV dysfunction and heart failure. Arch Physiol Biochem 119: 65–74, 2013. doi: 10.3109/13813455.2012.755548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Govindaraj P, Rani B, Sundaravadivel P, Vanniarajan A, Indumathi KP, Khan NA, Dhandapany PS, Rani DS, Tamang R, Bahl A, Narasimhan C, Rakshak D, Rathinavel A, Premkumar K, Khullar M, Thangaraj K. Mitochondrial genome variations in idiopathic dilated cardiomyopathy. Mitochondrion 48: 51–59, 2019. doi: 10.1016/j.mito.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Griffin BP, editor. Manual of Cardiovascular Medicine (4th Ed.). Philadelphia, PA: Lippincott Williams & Wilkins, 2013. [Google Scholar]
- 16.Grissa D, Pétéra M, Brandolini M, Napoli A, Comte B, Pujos-Guillot E. Feature selection methods for early predictive biomarker discovery using untargeted metabolomic data. Front Mol Biosci 3: 30, 2016. doi: 10.3389/fmolb.2016.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hershberger RE, Norton N, Morales A, Li D, Siegfried JD, Gonzalez-Quintana J. Coding sequence rare variants identified in MYBPC3, MYH6, TPM1, TNNC1, and TNNI3 from 312 patients with familial or idiopathic dilated cardiomyopathy. Circ Cardiovasc Genet 3: 155–161, 2010. doi: 10.1161/CIRCGENETICS.109.912345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hershberger RE, Pinto JR, Parks SB, Kushner JD, Li D, Ludwigsen S, Cowan J, Morales A, Parvatiyar MS, Potter JD. Clinical and functional characterization of TNNT2 mutations identified in patients with dilated cardiomyopathy. Circ Cardiovasc Genet 2: 306–313, 2009. doi: 10.1161/CIRCGENETICS.108.846733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jefferies JL, Towbin JA. Dilated cardiomyopathy. Lancet 375: 752–762, 2010. doi: 10.1016/S0140-6736(09)62023-7. [DOI] [PubMed] [Google Scholar]
- 20.Karatzoglou A, Smola A, Hornik K, Zeileis A. kernlab-an S4 package for kernel methods in R. J Stat Softw 11: 1–20, 2004. doi: 10.18637/jss.v011.i09. [DOI] [Google Scholar]
- 21.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37: 907–915, 2019. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komamura K, Iwai N, Kokame K, Yasumura Y, Kim J, Yamagishi M, Morisaki T, Kimura A, Tomoike H, Kitakaze M, Miyatake K. The role of a common TNNT2 polymorphism in cardiac hypertrophy. J Hum Genet 49: 129–133, 2004. doi: 10.1007/s10038-003-0121-4. [DOI] [PubMed] [Google Scholar]
- 23.Krämer A, Green J, Pollard J Jr, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 30: 523–530, 2014. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krittanawong C, Zhang H, Wang Z, Aydar M, Kitai T. Artificial intelligence in precision cardiovascular medicine. J Am Coll Cardiol 69: 2657–2664, 2017. doi: 10.1016/j.jacc.2017.03.571. [DOI] [PubMed] [Google Scholar]
- 25.Kuhn M Building predictive models in R using the caret package. J Stat Softw 28: 1–26, 2008. doi: 10.18637/jss.v028.i05.27774042 [DOI] [Google Scholar]
- 26.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup . The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079, 2009. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liaw A, Wiener M. Classification and regression by randomForest. R News 2: 18–22, 2002. [Google Scholar]
- 28.Liu Y, Morley M, Brandimarto J, Hannenhalli S, Hu Y, Ashley E, Tang W, Moravec C, Margulies K, Cappola T, Li M. RNA-Seq identifies novel myocardial gene expression signatures of heart failure [RNA-seq]. GEO Database, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Z, Liu X, Yu H, Pei J, Zhang Y, Gong J, Pu J. Common variants in TRDN and CALM1 are associated with risk of sudden cardiac death in chronic heart failure patients in Chinese Han population. PLoS One 10: e0132459, 2015. doi: 10.1371/journal.pone.0132459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maron BJ, Towbin JA, Thiene G, Antzelevitch C, Corrado D, Arnett D, Moss AJ, Seidman CE, Young JB; American Heart Association; Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; Council on Epidemiology and Prevention . Contemporary definitions and classification of the cardiomyopathies: an American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translation Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation 113: 1807–1816, 2006. doi: 10.1161/CIRCULATIONAHA.106.174287. [DOI] [PubMed] [Google Scholar]
- 31.Martin M Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17: 10–12, 2011. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 32.Mayer O Jr, Seidlerová J, Vaněk J, Karnosová P, Bruthans J, Filipovský J, Wohlfahrt P, Cífková R, Windrichová J, Knapen MHJ, Drummen NE, Vermeer C. The abnormal status of uncarboxylated matrix Gla protein species represents an additional mortality risk in heart failure patients with vascular disease. Int J Cardiol 203: 916–922, 2016. doi: 10.1016/j.ijcard.2015.10.226. [DOI] [PubMed] [Google Scholar]
- 33.McNally EM, Mestroni L. Dilated cardiomyopathy: genetic determinants and mechanisms. Circ Res 121: 731–748, 2017. doi: 10.1161/CIRCRESAHA.116.309396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menasché P, Hagège AA, Scorsin M, Pouzet B, Desnos M, Duboc D, Schwartz K, Vilquin J-T, Marolleau J-P. Myoblast transplantation for heart failure. Lancet 357: 279–280, 2001. doi: 10.1016/S0140-6736(00)03617-5. [DOI] [PubMed] [Google Scholar]
- 35.Al Moamen NJ, Prasad V, Bodi I, Miller ML, Neiman ML, Lasko VM, Alper SL, Wieczorek DF, Lorenz JN, Shull GE. Loss of the AE3 anion exchanger in a hypertrophic cardiomyopathy model causes rapid decompensation and heart failure. J Mol Cell Cardiol 50: 137–146, 2011. doi: 10.1016/j.yjmcc.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pepin M, Crossman D, Barchue J, Pamboukian S, Pogwizd S, Wende A. Genome-Wide DNA Methylation Encodes Cardiac Transcriptional Reprogramming in Human Ischemic Heart Failure [RNA-Seq]. GEO Database, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rau C, Tsai E. Wipi1 is a Genetic Hub that Mediates Right Ventricular Failure. GEO Database, 2018. [Google Scholar]
- 39.Saeys Y, Inza I, Larrañaga P. A review of feature selection techniques in bioinformatics. Bioinformatics 23: 2507–2517, 2007. doi: 10.1093/bioinformatics/btm344. [DOI] [PubMed] [Google Scholar]
- 40.Schiano C, Costa V, Casamassmi A, Aprile M, Rienzo M, Esposito R, Ciccodicola A, Napoli C. RNA-Sequencing shows novel transcriptomic signatures in failing and non-failing human heart. GEO Database, 2015. [Google Scholar]
- 41.Sisakian H Cardiomyopathies: Evolution of pathogenesis concepts and potential for new therapies. World J Cardiol 6: 478–494, 2014. doi: 10.4330/wjc.v6.i6.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Statnikov A, Wang L, Aliferis CF. A comprehensive comparison of random forests and support vector machines for microarray-based cancer classification. BMC Bioinformatics 9: 319, 2008. doi: 10.1186/1471-2105-9-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sweet M, Ambardekar A, Bristow M, Mestroni L, Taylor M. RNA-seq of heart failure in human left ventricles. GEO Database, 2018. [Google Scholar]
- 44.Theis JL, Zimmermann MT, Evans JM, Eckloff BW, Wieben ED, Qureshi MY, O’Leary PW, Olson TM. Recessive MYH6 mutations in hypoplastic left heart with reduced ejection fraction. Circ Cardiovasc Genet 8: 564–571, 2015. doi: 10.1161/CIRCGENETICS.115.001070. [DOI] [PubMed] [Google Scholar]
- 45.Therneau T, Atkinson B, Ripley B, Ripley MB. Package ‘rpart’. cran.ma.ic.ac.uk/web/packages/rpart/rpart. pdf (accessed 20 April 2016).
- 46.Tur MK, Etschmann B, Benz A, Leich E, Waller C, Schuh K, Rosenwald A, Ertl G, Kienitz A, Haaf AT, Bräuninger A, Gattenlöhner S. The 140-kD isoform of CD56 (NCAM1) directs the molecular pathogenesis of ischemic cardiomyopathy. Am J Pathol 182: 1205–1218, 2013. doi: 10.1016/j.ajpath.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 47.Wang L, Hu Y, Pu W. RNA-seq identifies novel transcript elements and transcript processing in the normal and failing hearts. GEO Database, 2013. [Google Scholar]
- 48.Wittchen F, Suckau L, Witt H, Skurk C, Lassner D, Fechner H, Sipo I, Ungethüm U, Ruiz P, Pauschinger M, Tschope C, Rauch U, Kühl U, Schultheiss HP, Poller W. Genomic expression profiling of human inflammatory cardiomyopathy (DCMi) suggests novel therapeutic targets. J Mol Med (Berl) 85: 257–271, 2007. doi: 10.1007/s00109-006-0122-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu XY, Wu ZY, Li K. Identification of differential gene expression for microarray data using recursive random forest. Chin Med J (Engl) 121: 2492–2496, 2008. doi: 10.1097/00029330-200812020-00005. [DOI] [PubMed] [Google Scholar]
- 50.Yamauchi-Takihara K, Kishimoto T. A novel role for STAT3 in cardiac remodeling. Trends Cardiovasc Med 10: 298–303, 2000. doi: 10.1016/S1050-1738(01)00066-4. [DOI] [PubMed] [Google Scholar]
- 51.Yang K, Nerbonne J. Deep RNA Sequencing Reveals Dynamic Regulation of Myocardial Noncoding RNA in Failing Human Heart and Remodeling with Mechanical Circulatory Support. GEO Database, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang Y, Zhang T, Xiao R, Hao X, Zhang H, Qu H, Xie B, Wang T, Fang X. Platform-independent approach for cancer detection from gene expression profiles of peripheral blood cells. Brief Bioinform 21: 1006–1015, 2020. doi: 10.1093/bib/bbz027. [DOI] [PubMed] [Google Scholar]
- 53.Zhao Y, Feng Y, Zhang Y-M, Ding X-X, Song Y-Z, Zhang A-M, Liu L, Zhang H, Ding J-H, Xia X-S. Targeted next-generation sequencing of candidate genes reveals novel mutations in patients with dilated cardiomyopathy. Int J Mol Med 36: 1479–1486, 2015. doi: 10.3892/ijmm.2015.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhuang Y, Gong YJ, Zhong BF, Zhou Y, Gong L. Bioinformatics method identifies potential biomarkers of dilated cardiomyopathy in a human induced pluripotent stem cell-derived cardiomyocyte model. Exp Ther Med 14: 2771–2778, 2017. doi: 10.3892/etm.2017.4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mayo Clinic Staff Cardiomyopathy. https://www.mayoclinic.org/diseases-conditions/ebola-virus/home/ovc-20338671, accessed 10 Apr. 2020.
- 56.Ohio Supercomputer Center Columbus OH. https://www.osc.edu. 1978.