Graphical abstract
Keywords: non-coding RNA, Promoter, Sequence length effect, Ensemble learning
Highlights
-
•
A computational method for identifying non-coding promoters was proposed for the first time.
-
•
A high-quality dataset was built to train and test the models for identifying non-coding promoters.
-
•
A user-friendly web server was developed to recognize non-coding promoters.
Abstract
The promoter is located near the transcription start sites and regulates transcription initiation of the gene. Accurate identification of promoters is essential for understanding the mechanism of gene regulation. Since experimental methods are costly and ineffective, developing efficient and accurate computational tools to identify promoters are necessary. Although a series of methods have been proposed for identifying promoters, none of them is able to identify the promoters of non-coding RNA (ncRNA). In the present work, a new method called ncPro-ML was proposed to identify the promoter of ncRNA in Homo sapiens and Mus musculus, in which different kinds of sequence encoding schemes were used to convert DNA sequences into feature vectors. To test the length effect, for each species, datasets including sequences with different lengths were built. The results demonstrated that ncPro-ML achieved the best performance based on the dataset with the sequence length of 221 nucleotides for human and mouse. The performances of ncPro-ML were also satisfying from both independent dataset test and cross-species test. The results indicate that the proposed predictor can server as a powerful tool for the discovery of ncRNA promoters. In addition, a web-server for ncPro-ML was developed, which can be freely accessed at http://www.bio-bigdata.cn/ncPro-ML/.
1. Introduction
Non-coding RNA (ncRNA) is a kind of transcripts that lack clear potential to encode proteins or peptides [1]. A large portion of the human genome is transcribed into ncRNA with many different forms, namely long-noncoding RNA (lncRNA), micro RNA (miRNA), circular RNA (circRNA), etc. [1], [2], [3]. Although ncRNAs lack potential to encode proteins, numerous investigations have shown that they play critical roles in many important biological processes including cell cycle, differentiation, development, metabolism, and so on [1], [2], [4], [5], [6]. Moreover, accumulated evidences have demonstrated that ncRNAs exhibit complex interactions with a broad spectrum of human diseases [1], [2], [7], [8]. Deep sequencing of size-fractionated RNAs has become a primary technique for discovering ncRNAs, which generated a myriad of ncRNA candidates. However, the mechanisms of ncRNA are obscure or controversial in some biological process [9], [10]. Therefore, in order to accurately understand their functions, the genomic annotations of the identified ncRNAs are necessary.
The first step of functional genomic annotation is promoter identification. The promoter is an important functional element in non-coding region, which immediately locates near and upstream of the transcription start site (TSS) and is mainly in charge of the gene transcription initiation. Due to their extensive roles in gene transcription, the accurate prediction of promoters becomes an essential step for understanding gene expression and the function of genetic regulatory networks. There were two main kinds of biological experiments for identification of promoters such as mutational analysis and immunoprecipitation assays [11], [12], [13]. Given that these methods were both expensive and time-consuming, computational methods have been proposed to identify promoters. In the past several years, several classifiers have been proposed to identify promoters in multiple species [14], [15], [16]. All these works concerned on the identification of promoters for coding genes. To the best of our knowledge, there exist no computational methods able to identify promoters of non-coding RNA (ncRNA) genes.
Keeping this in mind, we proposed a support vector machine (SVM) based method, called ncPro-ML, to identify promoters of ncRNA. In order to comprehensively extract the sequence based information, eight kinds of feature representation schemes (binary and k-mer frequency [BKF], dinucleotide binary profile and frequency [DBPF], dinucleotide physical-chemical properties [DPCP], trinucleotide physical-chemical properties [TPCP], electron-ion interaction pseudopotentials of trinucleotide [triEIIP], ring-function-hydrogen-chemical properties [RFHCP], pseudo dinucleotide composition [PseDNC] and multivariate mutual information [MMI]) were used to convert DNA sequences into numerical vectors. To obtain a robust model, the feature selection process was utilized to select optimal feature subsets from the candidate feature list for each feature representation scheme. Based on multiple optimal subsets, we trained different models and integrated them by setting the weights according to the accuracy obtained from the five-fold cross-validation test. To demonstrate the effect of sequence length on predictive performance, distinct models based on different lengths ranging from 61 to 301 bp were tested as well. Finally, an easy-to-use webserver for ncPro-ML was developed, which is freely available at http://www.bio-bigdata.cn/ncPro-ML/. The flowchart on building ncPro-ML was shown in Fig. 1.
Fig. 1.
The flowchart for building ncPro-ML.
2. Materials and methods
2.1. Benchmark dataset
In this work, the promoter sequences of ncRNA from Homo sapiens and Mus musculus genome were obtained from the publicly available Eukaryotic Promoter Database (EPDnew) [17]. Compared with other TSS annotation databases, i.e. refTSS [18] and DBTSS/DBKERO [19], the EPD contains non-redundant collection promoters with stronger support from experimental data. To avoid the inclusion of noisy sequences, sequences which contains uncertain bases were removed. Considering that non-promoters do not have TSS, thus they were extracted from the downstream region of the promoter sequences. Thus, the dataset can be formulated as following,
| (1) |
where is the positive dataset including promoter sequences. All these sequences are long from (ξ-20) bp upstream to 20 bp downstream of the TSS (TSS is regarded at the 0th site). is the negative dataset including non-promoter sequences. They are also long, but start from 1000 bp downstream of the TSS. To demonstrate the effect of sequence length on predictive performance, a series of datasets based on different sequence lengths ranging from 61 to 221 bp with a step 20 bp, and 261 bp and 301 bp were built, which were formulated as following,
| (2) |
The detail information about these datasets were given in Table 1. For the promoter and non-promoter sequences, 1170 and 1539 sequences of each length for human and mouse are used to train the model, and the rest are used as independent testing datasets to validate the performance of the model.
Table 1.
Detail information on the datasets used in this study.
| Organism | Dataset Name | Promoter | number | Non-promoter | number |
|---|---|---|---|---|---|
| human | 61 bp | P40-20 | 2339 | N1000-1060 | 2339 |
| 81 bp | P60-20 | N1000-1080 | |||
| 101 bp | P80-20 | N1000-1100 | |||
| 121 bp | P100-20 | N1000-1120 | |||
| 141 bp | P120-20 | N1000-1140 | |||
| 161 bp | P140-20 | N1000-1160 | |||
| 181 bp | P160-20 | N1000-1180 | |||
| 201 bp | P180-20 | N1000-1200 | |||
| 221 bp | P200-20 | N1000-1220 | |||
| 261 bp | P240-20 | N1000-1260 | |||
| 301 bp | P280-20 | N1000-1300 | |||
| mouse | 61 bp | P40-20 | 3077 | N1000-1060 | 3076 |
| 81 bp | P60-20 | N1000-1080 | |||
| 101 bp | P80-20 | N1000-1100 | |||
| 121 bp | P100-20 | N1000-1120 | |||
| 141 bp | P120-20 | N1000-1140 | |||
| 161 bp | P140-20 | N1000-1160 | |||
| 181 bp | P160-20 | N1000-1180 | |||
| 201 bp | P180-20 | N1000-1200 | |||
| 221 bp | P200-20 | N1000-1220 | |||
| 261 bp | P240-20 | N1000-1260 | |||
| 301 bp | P280-20 | N1000-1300 |
2.2. Feature representation algorithms
A given DNA sequence with L bp is defined as,
| (3) |
where Ri∈{A,C,G,T} indicates the nucleotide at i-th position in the sequence. In this study, we utilized eight sequence-based feature representation algorithms to encode the sequences in the dataset.
2.2.1. Binary and k-mer frequency (BKF)
For the nucleotide binary profile, the nucleotides A, C, G and T are encoded by using the vectors (1, 0, 0, 0), (0, 1, 0, 0), (0, 0, 1, 0) and (0, 0, 0, 1), respectively. Accordingly, a sequence can be represented by a 4L-dimensional feature vector. k-mer frequency is another way of representing DNA sequences, which refers to the frequency of all the possible k-tuple nucleotides in a given sequence. In this study, k was set to 2, 3 and 4. Thus, we could obtain three vectors with the dimension of 16, 64, 256, respectively. By combining the nucleotide binary profile and k-tuple nucleotide frequency, a sequence will be encoded by a (4L + 16 + 64 + 256) dimension vector.
2.2.2. Dinucleotide binary profile and frequency (DBPF)
Dinucleotide binary profile (DBP) and dinucleotide frequency were also widely used for sequence representation. For DBP, each dinucleotide type is encoded as a 4-dimensional vector containing 0 and 1. For instance, AA, AC, AG, were represented by (0, 0, 0, 0), (0, 0, 1, 0) and (0, 1, 0, 0), and so forth. The dinucleotide frequency is defined as following,
| (4) |
where |Xi| is the length of sub-sequence (R1R2…Ri) in the sequence D, and is the occurrence frequency of the dinucleotide in the -length sub-sequence. Therefore, for a given sequence, the dimension of the vector based on DBPF is 4×(L-1) + L-1).
2.2.3. Dinucleotide physical-chemical properties (DPCP)
Physicochemical properties are also important information for genomic functional elements identifications and were incorporated into promoter prediction[20], [21]. Inspired by those works, 15 different physicochemical properties, namely PC1, F-roll; PC2, F-tilt; PC3, F-twist; PC4, F-slide; PC5, F-shift; PC6, F-rise; PC7, roll; PC8, tilt; PC9, twist; PC10, slide; PC11, shift; PC12, rise; PC13, energy; PC14, enthalpy; and PC15, entropy, were employed to encode sequences in the dataset. The values of the 15 properties for each dinucleotide were provided in Supplementary Table S1. Since the values of different properties vary greatly, their original values were normalized to the range of [0, 1] by using the max–min normalization method. The DPCP is formulated as following,
| (5) |
where i is one of the 16 dinucleotides, f(i) is the frequency of the i-th dinucleotide in a sequence and the X represents one of the 15 physicochemical properties. Based on DPCP, a given sequence can be encoded as a 240 (16 × 15)-dimensional vector.
2.2.4. Trinucleotide physical-chemical properties (TPCP)
Similar to DPCP, the following 11 physical-chemical properties: PC1, bendability (DNase); PC2, bendability (consensus); PC3, trinucleotide GC content; PC4, nucleosome positioning; PC5, consensus (roll); PC6, consensus (rigid); PC7, DNase I (rigid); PC8, molecular weight (daltons); PC9, nucleosome (rigid); PC10, nucleosome; and PC11, DNase I were used to define TPCP. The values of these 11 physicochemical properties for each trinucleotide are listed in Supplementary Table S2. These values were normalized as described above before performing the following calculation. The TPCP is formulated as
| (6) |
where X is one of the 11 physicochemical properties, i is one of the trinucleotides and f(i) is the frequency of the i-th trinucleotide in a sequence. Then, a sequence can be encoded as a 704 (64 × 11) -dimensional vector.
2.2.5. Electron-ion interaction pseudopotentials of trinucleotide (triEIIP)
The EIIP was an effective feature encoding method which has been widely used bioinformatics [22], [23], [24]. The EIIP values of the four nucleotides are given in Supplementary Table S3. The composition of each sequence can be represented as a 64-dimensional feature vector E as follows,
| (7) |
where EIIPxyz = EIIPx + EIIPy + EIIPy + EIIPz, is the EIIP value of the nucleotide , and , is the frequency of trinucleotide xyz in the sequence.
2.2.6. Ring-function-hydrogen-chemical properties (RFHCP)
The deoxyribonucleic acid is composed of four nucleic acids that have different chemical properties in terms of ring structures, strength of hydrogen bonds and chemical functionality [25]. Considering the number of rings, A and G are grouped together because they both contains two rings and the others are in one group which only have one ring. In terms of hydrogen bond, C and G can be distributed in the same group since they form strong hydrogen bonds, whereas A and T form weak hydrogen bonds and thus belong to the other group. In the aspect of the chemical functionality, A and C can be classified into the amino group, while G and T can be classified into the keto group. Accordingly, three coordinates (x, y, z) were used to represent the chemical properties of the four nucleotides. The x , y and z stand for the ring structure, the hydrogen bond and the chemical functionality, respectively. Each nucleotide i in the sequence can be encoded by , where
| (8) |
Moreover, the density di of a nucleotide at position i was defined as following [26],
| (9) |
where |Ni| is the length of subsequence (R1R2…Ri) in the sequence D. By integrating the two schemes, a sequence can be encoded by a 4 × L- dimensional vector.
2.2.7. Pseudo dinucleotide composition (PseDNC)
PseDNC can reflect both short-range and long-range sequence-order information by calculating the dinucleotide nucleotide composition and the correlation of physics-chemical properties from a consider sequence [27]. In this study, we used six types of local structural parameters (Slide, Shift, Rise, Twist, Tilt and Roll) to characterize the spatial arrangements of any two successive base pairs. For a given sequence, it can be denoted as a dimension vector formulated as following,
| (10) |
where
| (11) |
where is the normalized frequency of the u-th k-tuple nucleotide composition, is the weight factor range from 0.1 to 1 with a step 0.1 and is the number of the total counted ranks or tiers of the correlations along a DNA sequence. In this study, we set a search strategy for ranges from 1 to 10. The j-th tire structural correlation factor that reflects the local structure correlation between all the j-th most contiguous dinucleotide along a DNA sequence and can be given by
| (12) |
Θ(RiRi+1;Ri+jRi+j+1) is the correlation function and can be defined by
| (13) |
where the Pv(riRi+1) is the value of the v-th DNA local structural property for the dinucleotide RiRi+1 at position i in the sequence.
2.2.8. Multivariate mutual information (MMI)
The feature encoding method of multivariate mutual information (MMI) was proposed by Pan et al. and has been widely used in the field of bioinformatics [23], [28]. In order to use MMI on a DNA sequence, we first define 2-tuple nucleotide composition set T2 and 3-tuple nucleotide composition set T3 as follows.
| (14) |
According to the formula described by Pan et al.[28], the MMI can be defined as follows:
| (15) |
where f(Ri) is the frequency of Ri in the sequence, the f(RiRj) is the frequency of categories RiRj appearing in the T2 feature on a sequence and f(RiRjRk) frequency of categories RiRjRk appearing in the T3 feature on a sequence. Accordingly, a sequence is represented by 10 + 20 = 30 features generated according to Eq. (14).
2.3. Feature selection
Feature selection is a key step to find the most useful features to improve the classification accuracy and reduce the number of features. For eliminating redundant and irrelevant features, we first applied the F-score method to calculate the importance of features and yielded a feature ranking list regarding their classification importance. And then, we used the sequential forward search (SFS) strategy to find the optimal feature representations [29], [30]. For the strategy of SFS, features from the ranked feature list was added one by one from higher to lower rank to select the sub-features. Then, the SVM based models were trained and tested based on the sub-features by using a 5-fold cross-validation. Finally, the sub-features with the best performance was recognized as the optimal feature set.
2.4. Building promoter recognition models based on SVM
SVM is a powerful supervised-learning algorithm based on the statistical learning theory and has widely applied to handle many biological problems, such as recognizing special peptides [31], [32], [33] and protein [34], disease diagnosis [35]. In this study, the LIBSVM package [36] was used to train the SVM and built a model that could discriminate between ncRNA promoter and non-promoter sequence, and the most commonly used Radial Basis Function (RBF) was selected as its kernel function. To achieve the optimal performance, we optimized the SVM using a grid search approach to filter the regularization parameter C and kernel parameter γ. The search ranges for both of the parameters are given as following,
| (16) |
2.5. Performance measures
The performance of the proposed method was evaluated by using four commonly used metrics, namely sensitivity (Sn), specificity (Sp), accuracy (Acc) and the Mathew’s correlation coefficient (MCC). They are calculated as follows:
| (17) |
In equation (17), TP, TN, FP and FN represent the numbers of true positives, true negatives, false positives and false negatives, respectively.
Besides, the receiver operating characteristic curve (ROC) was also employed to evaluate the overall performance of the proposed model. ROC is an objective metric that designed to simultaneously display the true positive rate against the false positive rate at every possible classification threshold and has been widely used in diverse fields. The value of the area under ROC curve (AUC), which ranges from 0.5 to 1, can reduce the ROC performance to a single scalar value representing expected performance. The higher the value of the AUC the better performance is implied.
3. Results and discussion
3.1. Feature optimization
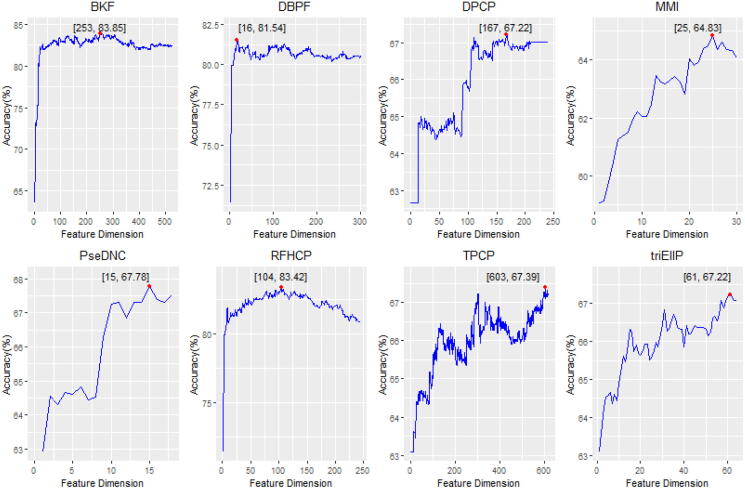
In this study, we generated eleven feature representations by using eight kinds of feature encoding schemes that represents sequence information in different sides. For some feature encoding schemes, the longer the sequences, the greater the dimension of the feature vector. Take BKF as an example, the feature dimension was 4 × L + 16 + 64 + 256, a total 1540-D features will be generated when the sequence with 301 nucleotides. Such a problem may lead to an increase in classifiers training time and a reduction of their predictive performance. To address these issues, we conducted a 5-fold cross-validation test for each feature representation scheme based on optimal features obtained by using the feature selection strategy. To intuitively analyze the results, in Fig. 2, we plotted the variation of Acc versus the increment of feature dimension for identifying human ncRNA promoters based on dataset S61. The red point in the figure is the highest Acc for each feature representations. It was found that the maximum Acc of 83.85% was achieved when 253 BKF derived optimal sub-features were used. This result demonstrates that the feature dimension is greatly reduced and the accuracy of the model is significantly improved by using the feature optimization strategy. The results of feature selection process for human and mouse based on different datasets were shown in the Supplementary Figure S1 to Figure S21.
Fig. 2.
The variation of Acc versus the increment of feature dimension for identifying human ncRNA promoters based on the 61 bp dataset.
3.2. Construct ncPro-ML by integrating multiple models
Multiple model integration method is an important pattern classification technique to obtain better performance and can avoid the potential deviation generated by a single classifier [37]. Therefore, we combined these eight models according to the weighted sum of their prediction scores, where the weights were normalized by the Acc of a single model divided by the sum of the Accs of the eight models. For example, based on the human dataset , the weight of 0.14 was obtained for the model based on feature BKF. Similarly, the weight of 0.1369, 0.1178, 0.1121, 0.1181, 0.1387, 0.1183 and 0.1182 were obtained for the models based on feature DBPF, DPCP, MMI, PseDNC, RFHCP, TPCP and triEIIP, respectively. For the final model, the prediction score was the sum of the prediction score of the eight models based on their weights. Finally, eight integrated models were constructed based on different lengths datasets for human and mouse. The weights of the eight models in different lengths datasets for human and mouse were listed in Supplementary Table S4 and Table S5.
3.3. Effect of sequences length on model performance
We have built eleven datasets including different sequence length ranging from 61 to 301 nucleotides for human and mouse. The best accuracy produced by the feature selection process for each feature representation method of different datasets were shown in Fig. 3. The number of features for BKF, RFHCP and DBPF were larger than others, and increased with the length of the training sequences. As it can be seen in the Fig. 3, those models built based on BKF, RFHCP and DBPF obtained better predictive performance than the models based on other kinds of features.
Fig. 3.
The accuracy of models based on different features and datasets in human and mouse.
Although the variation of the performance based on different datasets are not significant for the eight kinds of features in both human and mouse, the best predictive accuracy was obtained by using BKF based on dataset for human and based on dataset for mouse. Especially, the TPCP has a very high predictive accuracy for human on dataset . Taken together, the performance of eight models remained relatively stable for all datasets with different lengths.
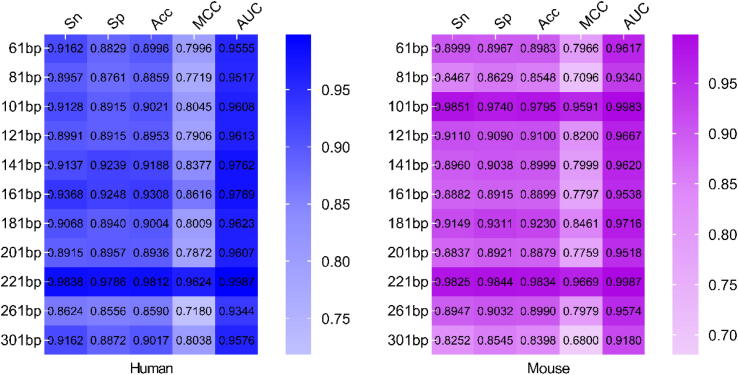
According to the weights of each model based on different datasets, we constructed eight integrated predictors by adding weights to a model for human and mouse, respectively. Due to the consistency among the accuracy of different datasets for each model, we evaluated the performance of the eight predictors by using self-test to determine the best sequence length for human and mouse. Where the self-test refers to using the training datasets to validate the constructed model. The results obtained from the experiments to verify the impact of the sequence length variation on the predictors performance are shown in Fig. 4. For human and mouse, the eight predictors trained based on different datasets all yielded a better predictive performance.
Fig. 4.
The performance of predictors based on different datasets. The performance was measured in term of Sn, Sp, Acc, MCC and AUC.
From these results, we chose the predictor training based on the as the final predictor for human and mouse. The two predictors obtained the highest Acc of 98.12% and 98.34%, and were used to build ncPro-ML for identifying ncRNA promoters in human and mouse, respectively. Moreover, we compared the performance of SVM with that of different machine learning based methods, namely Naive Bayes, Random Forest, Random Tree, Logistic, k-nearest neighbor (KNN) and SVM based on the datasets S221. The results from five-fold cross validation test demonstrated that the SVM based method yielded the best performance in term of Acc (Figure S22).
3.4. Performance assessment of ncPro-ML based on the independent datasets
To assess the generalization ability and robustness of the predictor, ncPro-ML was validated on the independent datasets. The performances of the predictor based on the independent datasets for identifying promoters in human and mouse were shown in Fig. 5A and B. The predictor achieved the accuracy of 81.65% with the sensitivity of 81.27%, specificity of 82.04% and MCC of 0.633 for human, and accuracy 83.09% with the sensitivity of 81.66%, specificity of 84.52% and MCC of 0.6621 for mouse. The corresponding AUC is 0.8930 and 0.9036 for human and mouse, respectively. These results indicate that the proposed method is reliable for identifying ncRNA promoters in human and mouse.
Fig. 5.
Performance of ncPro-ML based on independent datasets (A and B) and cross-species datasets (C and D). In A and B, the Mouse and Human represent the model human and mosue in ncPro-ML, respectively. In C and D, the ncPro-ML(Human) denote using human model in ncPro-ML to perform the mouse independent testing datasets, and vice versa.
To demonstrate generalization ability of ncPro-ML, the cross-species validation was also performed. Accordingly, the model trained in one species (human or mouse) was tested on the independent datasets of the other species. The predictive results were shown in Fig. 5 C and D.
The human and mouse specific model achieved the Acc of 82.08% and 77.16% to identify promoters in mouse and human independent datasets, respectively. The corresponding AUCs were 0.8885 and 0.8531. The excellent performance of ncPro-ML indicates that the proposed predictor can server as a powerful tool for the discovery of new ncRNA promoters.
4. Conclusion
Accurate identification of promoters is essential for understanding the mechanism of the gene regulation process and is also a fundamental step for functional annotation of a new genome. Therefore, numerous computational approaches have been proposed by using different machine learning methods. However, to the best of our knowledge, there is no predictor specifically for identifying the ncRNA promoters. To address this challenge, we proposed the first machine-learning based method ncPro-ML to identify ncRNA promoters in human and mouse. In order to make ncPro-ML yield excellent performance, for both human and mouse, eleven datasets composed of sequences with different lengths were constructed to evaluate the sequence length required for training a predictor with the best performance. The performance of ncPro-ML on independent datasets indicate that ncPro-ML is good enough for identify ncRNA promoters in human and mouse. In addition, results from cross-species evaluation demonstrate that ncPro-ML have the ability to identify ncRNA promoters in other species as well. For the convenience of scientific community, a user-friendly web server for ncPro-ML was provided at http://www.bio-bigdata.cn/ncPro-ML/. We hope it could become a useful tool for identifying ncRNA promoters.
Funding
This work was supported by the National Nature Scientific Foundation of China (No. 31771471), the Natural Science Foundation for Distinguished Young Scholar of Hebei Province (No. C2017209244), and the Xinglin Scholar Research Premotion Project of Chengdu University of TCM (NO. ZRQN2019015).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.09.001.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Matsui M., Corey D.R. Non-coding RNAs as drug targets. Nat Rev Drug Discov. 2017;16:167–179. doi: 10.1038/nrd.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang W., Zhang H., Yang H. Computational resources associating diseases with genotypes, phenotypes and exposures. Brief Bioinform. 2019;20:2098–2115. doi: 10.1093/bib/bby071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kimura T. Metal-mediated epigenetic regulation of gene expression. Yakugaku Zasshi. 2017;137:273–279. doi: 10.1248/yakushi.16-00230-4. [DOI] [PubMed] [Google Scholar]
- 4.Engreitz J.M., Haines J.E., Perez E.M., Munson G., Chen J., Kane M., McDonel P.E., Guttman M., Lander E.S. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539:452–455. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ponting C.P., Oliver P.L., Reik W. Evolution and Functions of Long Noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 9.Wang K., Chang H. Molecular Mechanisms of Long Noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong C.-M., Tsang F.-C., Ng I.-L. Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nat Rev Gastroenterol Hepatol. 2018;15:137–151. doi: 10.1038/nrgastro.2017.169. [DOI] [PubMed] [Google Scholar]
- 11.Matsumine H., Yamamura Y., Hattori N., Kobayashi T., Kitada T., Yoritaka A., Mizuno Y. A Microdeletion of D6S305 in a Family of Autosomal Recessive Juvenile Parkinsonism (PARK2) Genomics. 1998;49:143–146. doi: 10.1006/geno.1997.5196. [DOI] [PubMed] [Google Scholar]
- 12.Kim J.-W., Zeller K.I., Wang Y., Jegga A.G., Aronow B.J., O'Donnell K.A., Dang C.V. Evaluation of Myc E-Box Phylogenetic Footprints in Glycolytic Genes by Chromatin Immunoprecipitation Assays. MCB. 2004;24:5923–5936. doi: 10.1128/MCB.24.13.5923-5936.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahl J.A., Collas P. A rapid micro chromatin immunoprecipitation assay (microChIP) Nat Protoc. 2008;3:1032–1045. doi: 10.1038/nprot.2008.68. [DOI] [PubMed] [Google Scholar]
- 14.Oubounyt M., Louadi Z., Tayara H. DeePromoter: Robust Promoter Predictor Using Deep Learning. Front Genet. 2019;10:286. doi: 10.3389/fgene.2019.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang S., Cheng X., Li Y., Wu M., Zhao Y. Image-based promoter prediction: a promoter prediction method based on evolutionarily generated patterns. Sci Rep. 2018;8 doi: 10.1038/s41598-018-36308-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin H., Deng E.Z., Ding H. iPro54-PseKNC: a sequence-based predictor for identifying sigma-54 promoters in prokaryote with pseudo k-tuple nucleotide composition. Nucleic Acids Res. 2014;42:12961–12972. doi: 10.1093/nar/gku1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meylan P., Dreos R., Ambrosini G. EPD in 2020: enhanced data visualization and extension to ncRNA promoters. Nucleic Acids Res. 2020;48:D65–D69. doi: 10.1093/nar/gkz1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abugessaisa I., Noguchi S., Hasegawa A., Kondo A., Kawaji H., Carninci P., Kasukawa T. refTSS: A Reference Data Set for Human and Mouse Transcription Start Sites. J Mol Biol. 2019;431:2407–2422. doi: 10.1016/j.jmb.2019.04.045. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki A., Kawano S., Mitsuyama T. DBTSS/DBKERO for integrated analysis of transcriptional regulation. Nucleic Acids Res. 2018;46:D229–D238. doi: 10.1093/nar/gkx1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brick K., Watanabe J., Pizzi E. Core promoters are predicted by their distinct physicochemical properties in the genome of Plasmodium falciparum. Genome Biol. 2008;9:R178. doi: 10.1186/gb-2008-9-12-r178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abeel T., Saeys Y., Bonnet E., Rouze P., Van de Peer Y. Generic eukaryotic core promoter prediction using structural features of DNA. Genome Res. 2008;18(2):310–323. doi: 10.1101/gr.6991408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nair A.S., Sreenadhan S.P. A coding measure scheme employing electron-ion interaction pseudopotential (EIIP) Bioinformation. 2006;1:197–202. [PMC free article] [PubMed] [Google Scholar]
- 23.Wei L., Su R., Luan S. Iterative feature representations improve N4-methylcytosine site prediction. Bioinformatics. 2019;35:4930–4937. doi: 10.1093/bioinformatics/btz408. [DOI] [PubMed] [Google Scholar]
- 24.He W., Jia C., Zou Q. 4mCPred: machine learning methods for DNA N4-methylcytosine sites prediction. Bioinformatics. 2019;35:593–601. doi: 10.1093/bioinformatics/bty668. [DOI] [PubMed] [Google Scholar]
- 25.Chen W., Yang H., Feng P. iDNA4mC: identifying DNA N4-methylcytosine sites based on nucleotide chemical properties. Bioinformatics. 2017;33:3518–3523. doi: 10.1093/bioinformatics/btx479. [DOI] [PubMed] [Google Scholar]
- 26.Chen W., Feng P., Song X., Lv H., Lin H. iRNA-m7G: Identifying N7-methylguanosine Sites by Fusing Multiple Features. Mol Ther Nucleic Acids. 2019;18:269–274. doi: 10.1016/j.omtn.2019.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen W., Lin H., Chou K.-C. Pseudo nucleotide composition or PseKNC: an effective formulation for analyzing genomic sequences. Mol BioSyst. 2015;11:2620–2634. doi: 10.1039/c5mb00155b. [DOI] [PubMed] [Google Scholar]
- 28.Pan G., Jiang L., Tang J. A Novel Computational Method for Detecting DNA Methylation Sites with DNA Sequence Information and Physicochemical Properties. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ru B., 'T Hoen P.A.C., Nie F. PhD7FASTER: predicting clones propagating faster from the Ph.D.-7 phage display peptide library. J Bioinform Comput Biol. 2014;12 doi: 10.1142/S021972001450005X. [DOI] [PubMed] [Google Scholar]
- 30.Liu K., Chen W. iMRM:a platform for simultaneously identifying multiple kinds of RNA modifications. Bioinformatics. 2020;36:3336–3342. doi: 10.1093/bioinformatics/btaa155. [DOI] [PubMed] [Google Scholar]
- 31.Tang Q., Nie F., Kang J., Ding H., Zhou P., Huang J. NIEluter: Predicting peptides eluted from HLA class I molecules. J Immunol Methods. 2015;422:22–27. doi: 10.1016/j.jim.2015.03.021. [DOI] [PubMed] [Google Scholar]
- 32.He B., Kang J., Ru B., Ding H., Zhou P., Huang J. SABinder: A Web Service for Predicting Streptavidin-Binding Peptides. Biomed Res Int. 2016;2016:1–8. doi: 10.1155/2016/9175143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li N., Kang J., Jiang L., He B., Lin H., Huang J. PSBinder: A Web Service for Predicting Polystyrene Surface-Binding Peptides. Biomed Res Int. 2017;2017:1–5. doi: 10.1155/2017/5761517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang J., Fang Y., Yao P., Li N., Tang Q., Huang J. NeuroPP: A Tool for the Prediction of Neuropeptide Precursors Based on Optimal Sequence Composition. Interdiscip Sci Comput Life Sci. 2019;11:108–114. doi: 10.1007/s12539-018-0287-2. [DOI] [PubMed] [Google Scholar]
- 35.Kang J., Yu S., Lu S., Xu G., Zhu J., Yan N.a., Luo D., Xu K., Zhang Z., Huang J. Use of a 6-miRNA panel to distinguish lymphoma from reactive lymphoid hyperplasia. Sig Transduct Target Ther. 2020;5 doi: 10.1038/s41392-019-0097-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang C.-C., Lin C.-J. LIBSVM: A library for support vector machines. ACM Trans Intell Syst Technol. 2011;2(3):1–27. [Google Scholar]
- 37.Tang Q., Kang J., Yuan J. DNA4mC-LIP: a linear integration method to identify N4-methylcytosine site in multiple species. Bioinformatics. 2020;36:3327–3335. doi: 10.1093/bioinformatics/btaa143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.