Abstract
Background
Our previous study showed that the ribosomal protein L21 (RPL21) may play an important role in the development and survival of pancreatic cancer. In this article, RNA interference (RNAi) experiments were performed with RPL21-specific small interfering RNA (siRNA) to elucidate the mechanism by which RPL21 controls PC PANC-1 and BxPC-3 cell proliferation.
Methods
In the present study, PANC-1, BxPC-3 cells, and BALB/c nude mice were used to investigate antitumor effect and mechanism by which RPL21 controls cell proliferation and apoptosis in vitro and in vivo. The effects of RPL21 knockdown on PANC-1 and BxPC-3 cell proliferation, cell cycle and cell apoptosis in vitro were determined using 3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide assays and flow cytometry assay. The mechanism of RPL21 regulating cell proliferation was investigated using transcriptome sequencing analysis and luciferase reporter assay. The effects of RPL21 knockdown on PANC-1 and BxPC-3 cell proliferation in vivo were determined using BALB/c nude mice tumor model.
Results
In PANC-1 and BxPC-3 cells, the knockdown of RPL21 expression with corresponding siRNA suppressed cell proliferation in vitro and in vivo, inhibited DNA replication, and induced arrests in the G1 phase of the cell cycle. Further results showed that the mini-chromosome maintenance (MCM) protein family (MCM2-7), CCND1 and CCNE1 were down-regulated significantly in PANC-1 and BxPC-3 cells after transfected with RPL21 siRNA, which suggests that the suppression of DNA replication is due to the reduced expression of MCM2-7 family, and the induction of G1 arrest is correlated with the inhibition of CCND1 and CCNE1. Luciferase reporter assay showed that RPL21 controls the DNA replication and G1-S phase progression possibly through the regulation of E2F1 transcription factor in PC cells. Moreover, RPL21 siRNA showed an apoptosis-inducing effect only in BxPC-3 and PANC-1 cells but not in normal HPDE6-C7 cells. The increase of caspase-8 activities and the loss of mitochondrial membrane potential after RPL21 silencing indicates that the RPL21 gene may be involved in caspase-8-related mitochondrial apoptosis.
Conclusion
Our findings suggest that siRNA against the RPL21 gene possesses a potential anti-cancer activity for PC cells by inhibiting their proliferation and DNA replication, as well as inducing cell cycle G1 arrest and cell apoptosis.
Keywords: pancreatic cancer, RPL21, cell proliferation, cell cycle, cell apoptosis
Introduction
Pancreatic cancer (PC) is exceptionally aggressive. Treatment options are limited not just the late diagnosis of the disease (1–3). Similar to other malignancies, its occurrence was a process with multi-factors and multi-steps, including activation of proto-oncogenes, inactivation of tumor-suppressor genes, alterations in cell cycle related proteins and so on. Many cancer-related genes interact with each other and are involved in the progression of PC (4). In the past decade, some proto-oncogenes (EGFR, KRAS, etc.) have been characterized as therapeutic targets for clinic treatment to improve survival rate (5). However, the current strategies haven’t demonstrated significant improvement on the survival rates yet (6). The molecular mechanism of pancreatic cancer remains unclear, and genes associated with PC progression still need to be identified.
Ribosomal proteins (RPs) have been highly conserved throughout evolution, indicating their functional importance to living organisms. In conjunction with ribosomal RNAs (rRNA) and non-ribosomal factors, RPs make up the ribosomal small (40S) and large (60S) subunits that are involved in the cellular process of translation (7). Although, initially considered to be involved only in protein synthesis, but certain RPs can mediate a variety of so called extra-ribosomal functions such as replication, transcription, RNA processing, DNA repair, and even inflammation (8, 9). In recent years, more and more RPs were found dysfunctional in tumors, such as mutation, expression levels alteration and association with differentiation and so on (10, 11). However, the exact roles of RPs in PC development are diverse and still need be further clarified.
In our previous studies, we constructed a long-term suppression model with PANC-1 cells to study the cell response to proto-oncogene KRAS knockdown (12). Several ribosomal protein (RPs) genes (RPL39, RPL21, etc.) were up-regulated in PC cells with long-term suppression of proto-oncogene KRAS. Due to the role of KRAS in the progression of PC, the upregulation of these RPs genes as the compensation of KRAS knockdown suggests that they are possibly critical for PC cells development and survival. RPL39 has been shown to be critical for PC cell proliferation and apoptosis, whereas the apoptosis-related effect occurs only in PC cells but not in normal pancreatic duct epithelial cells (13). These results also indicated a variety of extra-ribosomal functions of RPL39 that are independent of protein biosynthesis. Similarly to RPL39, RPL21 is another up-regulated gene that encodes a ribosomal protein that is a component of the ribosomal 60S subunit and belongs to the ribosomal proteins L21E family. The extra-ribosomal functions of RPL21 involving in PC have not been reported, and the relationship between RPL21 and PC remains unknown.
RNA interference (RNAi) is a powerful tool to silence specific gene functions either by small interfering RNA (siRNA) or by short hairpin RNA (shRNA) (14, 15). Our previous data indicated that the expression of RPL21 might be functionally important in PC (12). Here we tested this hypothesis by selectively reducing the RPL21 expression using siRNA in PC PANC-1 and BxPC-3 cells. Subsequently, global changes in genes modulated in PANC-1 cells have been profiled using transcriptome analysis. The data suggest a possible functional role of RPL21, within a spectrum of altered gene expression, in maintaining the DNA replication and G1-S phase progression of human PC cells. Confirmation of such a scenario would allow selective therapeutic targeting of RPL21 to modulate discrete subsets of cellular proteins that are key promoters of PC cell proliferation.
Materials and Methods
Cell Lines and Culture Condition
Human pancreatic cancer cell lines PANC-1 and BxPC-3 were obtained from the Committee on Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). All cells were cultured in vitro in DMEM (Dulbecco’s modified eagle medium) high glucose medium (Gibco, Novato, CA, United States) supplemented with 10% (v/v) fetal bovine serum (FBS) (Gibco). Cells were incubated at 37°C in a humidified incubator with 5% CO2.
Transfection of siRNA Targeting the RPL21
The gene-specific siRNAs (siL21-1 and siL21-2) against RPL21 (Genbank NM_000982.3) was designed and synthesized by GenePharma Co., Ltd. (Shanghai, China), and the sequences were as follows: siL21-1, 5′- GCACUCUAAGAGCCGAGAUdTdT -3′ (Sense),5′-AUCUCGGCUCUUAGAGUGCdTdT-3′ (Antisense), siL21-2, 5′- GGGAAUGGGUACUGUUCAAdTdT -3′ (Sense), 5′- UUGAACAGUACCCAUUCCCdTdT -3′ (Antisense). The transfection targets of siL21-1 and siL21-2 were BLAST-searched and showed homology and similarity only to RPL21. The Mock-siRNA was also provided as the negative control (NC), which showed no homologous to any known human genes. The sequences of Mock-siRNA were as follows: 5′- UUCUCCG AACGUGUCACGUdTdT -3′ (Sense),5′- ACGUGACACGUUC GGAGAAdTdT -3′ (Antisense). Cells in 6-well plates (1.5 × 105 cells/well) were transfected with siRNAs (40 nM) using lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s guidelines. Usually at 72 h after transfection, gene silencing appears both at mRNA and protein levels, thus the cells were harvested and assayed at 72 h.
Quantitative Real-Time PCR (qPCR)
Total RNAs from cells or xenograft tumor tissues were extracted using TRNzolTM reagent (TIANGEN, Beijing, China) according to the manufacturer’s protocol. The concentration of total RNAs was measured using a UV spectrophotometer. First strand cDNA was synthesized from 2 μg total RNA using the First Strand cDNA Synthesis Kit (Fermentas, Glen Burnie, MD, United States). Quantitative real-time PCR (qPCR) was performed on the Mastercycler ep realplex Real-Time System (Eppendorf, Germany) using the SYBR Green qPCR kit (Fermentas, Glen Burnie, MD, United States). Gene-specific primers designed for detecting RPL21 and GAPDH mRNA were shown in Supplementary Table S1. Melting curves were generated to detect primer-dimer formation and to confirm gene-specific peaks for targets. Expression data were normalized to the amount of GAPDH expressed (16).
Western Blot Analysis
Total protein was extracted and subjected to western blotting analysis as described previously. The following antibodies were used for the western blottings: Primary polyclonal antibodies detecting E2F1, CCND1, CCNE1, MCM2, MCM3, MCM4, MCM5, MCM6, GAPDH, and MCM7 (Sangon Biotechnology, Shanghai, China). Primary monoclonal antibodies detecting RPL21 and β-Actin (Proteintech Group, Rosemont, IL, United States). After incubation with the appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies for 1 h, immunoreactive proteins were visualized with Electrochemiluminescence (ECL) western blot detection reagents (Thermo Fisher Scientific, Waltham, MA, United States).
Cell Proliferation and Colony-Forming Assay
To observe cell proliferation, cells were transfected with Mock-siRNA, siL21-1 and siL21-2 (40 nM). At 24 h after transfection, the cells were trypsinized and seeded into 96-well plates (Corning, NY, United States) at a density of 3000 cells/well in 200 μl media. The plates were incubated in a 37°C humidified incubator. On each day for 5 consecutive days, the number of viable cells was determined by 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay (17).
To detect colony formation, cells were transfected with Mock-siRNA (NC), siL21-1 and siL21-2 (40 nM, 24 h), and then seeded into 35-mm dishes at a density of 1000 cells/dish. The untreated cells served as the control group. Cells were fed with new growth media every 4 days. After 8 days, most of the single cells formed colony with up to more than 50 cells, and the colonies were then dyed with hematoxylin and counted.
EdU Retention Assay
Cells were transfected with siRNAs (Mock-siRNA, siL21-1 and siL21-2) at a concentration of 40 nM for 72 h. The transfected cells were exposed to 50 μM of 5-ethynyl-2′-deoxyuridine (EdU) (RiboBio, Guangzhou, China) for 2 h at 37°C, and the cells were fixed in 4% paraformaldehyde for 30 min at room temperature, followed by the addition of 2 mg/ml glycine to neutralize the reaction. After permeabilization with 0.5% Triton-X, the cells were reacted with Apollo reaction cocktail (RiboBio, Guangzhou, China) for 30 min in the dark at room temperature. Subsequently, the DNA contents of the cells were stained with Hoechst 33342 for 30 min and visualized under a laser scanning confocal microscopy (Leica Microsystems, Buffalo Grove, IL, United States).
Cell Cycle Analysis
Cell Cycle and Apoptosis Analysis Kit (Beyotime, Shanghai, China) was applied for cell cycle analysis. Briefly, the cells were transfected with siRNAs (Mock-siRNA, siL21-1 and siL21-2) at a concentration of 40 nM for 72 h, and then were harvested and fixed with cold 75% ethanol at 4°C overnight. The fixed cells were collected and suspended in phosphate-buffered saline (PBS) buffer containing 10 μg/ml propidium iodide (PI) and 10 μg/ml RNase A, and then incubated for 30 min at room temperature. DNA content was measured by the BD FACSCalibur (BD Biosciences, San Jose, CA, United States), and each histogram was constructed with the data from at 10,000 events. The data were analyzed and expressed as percentages of total gated cells using the Modfit LTTM Software (BD Biosciences, San Jose, CA, United States).
Transcriptome Analysis
Transcriptome sequencing and expression analysis. Human pancreatic cancer PANC-1 cells were transfected with Mock-siRNA (negative control, NC) and siL21-Mix (siL21-1 and siL21-2), respectively, at a concentration of 40 nM for 72 h. Duplicates of total RNA were prepared using TruSeq RNA Sample Prep Kit (Illumina, San Diego, CA, United States). The cDNA and cDNA library were produced, and the HiSeq 2000 Sequencing system (Illumina, San Diego, CA, United States) was used to sequence the cDNA library by Personal bio, Inc (Shanghai, China). Genes significantly regulated by siL21-Mix treatment (>2 or <2-fold change in expression) with known functions in the regulation of G1-S phase of the cell cycle and DNA replication were presented in Supplementary Table S2.
The validation of transcriptome sequencing. Gene expression profiles were validated in RPL21 knockdown cells with qPCR assay to confirm the expression levels of AHR, THBS1, DDIT3 and MKNK2 (up-regulated versus the control), in addition to E2F1, PCNA, CCND1, CCNE1 MCM2, MCM4, MCM5, MCM7, and KIAA0101 (down-regulated versus the control), randomly selected from differential gene after siL21-Mix (siL21-1 and siL21-2) transfection. The gene-specific primers were listed in Supplementary Table S1. All annealing temperatures were 60°C and cycling conditions as described as in section “Quantitative Real-Time PCR (qPCR).”
Luciferase Reporter Assay
The E2F1 expression vector pCMV-E2F1 was constructed using the pcDNA3.1 plasmid (Invitrogen, Carlsbad, CA, United States). The luciferase reporter vectors containing promoter region of genes (E2F1, MCM3, MCM4, CCNE1, CCND1, MCM5, MCM6, MCM3, and MCM7) were constructed using pGL-3 vector (Promega, Madison, WI, United States). The primers of full-length E2F1 gene and promoter region of genes were presented in Supplementary Table S3. For each transfection, the cells were seeded in 6-well plates at 6 × 105 cells/well. Transfection of reporter gene vectors (1 μg/well) was done using lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, United States). The pCMV-E2F1 (1 μg/well) was used as the E2F1 expression vector to co-transfect with the reporter gene vectors, and the DNA amount was kept constant using empty pcDNA3.1 vector. Luciferase activity was measured by the Luciferase Assay System Kit (Promega, Madison, WI, United States) and TECAN Instrument (AG, Switzerland) according to the manufacturer’s protocol. Transfection experiments were done at least in triplicate. For normalization of transfection efficiency, 500 ng of Renilla reniformis luciferase expression plasmid (pRL-TK vector, Promega, Madison, WI, United States) was included in the transfection.
Apoptosis Assay
Flow cytometry (FCM) analysis, the cells were harvested at 72 h after transfection (see transfection section of “Materials and Methods”), washed twice with ice-cold PBS and stained with Annexin V-Fluorescein isothiocyanate (FITC) and propidium iodide (PI) by Apoptosis Detection Kit (Keygen, Nanjing, China) for 10 min in the dark at room temperature. The cells with untreated cells served as the control group, and Mock-siRNA transfection served as the NC group. Then the cells were analyzed with the BD FACSCalibur and FlowJo software at 10,000 events (BD Biosciences, San Jose, CA, United States).
The Caspase-8 activities assay were analyzed using Cell MeterTMCaspase-8 Activity Apoptosis Assay Kit (AAT Bioquest, Sunnyvale, CA, United States) according to the manufacturer’s manual. In brief, PANC-1 and BxPC-3 cells were transfected with siRNAs (Mock-siRNA, siL21-1 and siL21-2) at a concentration of 40 nM for 72 h, then the cells were seeded into black wall, clear bottom 96-well plates and supplemented with 100 μl/well of Caspase-8 assay loading solution (containing caspase-8-specific fluorogenic substrate). Subsequently the plates were incubated with the assay loading solution for 1 h in the dark at room temperature. Then, the fluorescence intensities were measured at corresponding excitation/emission wavelength (Ex/Em = 490/525) with microplate reader (TECAN Instrument, AG, Switzerland).
Mitochondrial membrane potential assay, cell meterTM JC-10 Mitochondrial Membrane Potential Assay Kit was used to measure mitochondrial membrane potential changes according to the manufacturer’s manual. The fluorescence intensities were measured at corresponding excitation/emission wavelength (Ex/Em = 500/525 nm for monomeric form of JC-10, Ex/Em = 540/590 nm for aggregating form of JC-10) with microplate reader (TECAN Instrument, AG, Switzerland). The ratios of fluorescence intensities on Em at 525/590 nm were used for data analysis.
Tumorigenicity in Nude Mice
Eighteen BALB/c nude mice (4–6 weeks old, SPF degree, 20–24 g of body weight) were obtained from Shanghai SLAC Laboratory Animal Co., Ltd (Shanghai, China). A total of 5 × 106 BxPC-3 cells were injected subcutaneously into nude mice in 0.1 ml culture medium. When the tumors reached an average volume of approximately 50 mm3 in about 2 weeks, the mice were randomly divided into three groups (three male and three female mice in each group), namely control, NC and siL21-Mix. All animal experiments were performed by Suzhou Xishan Zhongke Laboratory Animal Co., Ltd. (Suzhou, China) in compliance with the guidelines set by the local government1.
The siRNAs of injection were 2′-O-methyl (2OMe) modified to increase the stability in vivo. RNAi-Mate (GenePharma, Shanghai, China) was used for the in vivo transfection. The siRNA transfection solution containing 10 μg of siRNA was intratumorally injected at multiple sites every 3 days. The mice were closely monitored for 16 days after injection. Tumor growth was monitored by measuring the largest (a) and smallest (b) two perpendicular diameters with a caliper, and calculated the tumor volume: (v) = a × b2 × 0.5236.
The immunohistochemistry (IHC) procedure was performed as described by Li (18). The tumor sections were incubated at 4°C overnight with anti-human PCNA monoclonal antibody (1:100, BlueGene Biotech Co., Ltd., Shanghai, China). After washing with Tris-buffered saline (TBS), those sections were subsequently incubated for 30 min with appropriate HRP-linked secondary antibody. Immunoreactivity was developed with 3,3′-diaminobenzidine tetrahydrochloride (DAB) and CoCl2 enhancer tablets (Sigma-Aldrich, Shanghai, China). Finally, sections were counterstained with hematoxylin.
Statistical Analysis
The statistical analyses were performed with SPSS 12.0 software (SPSS Inc., Chicago, United States). Results were expressed as mean values ± standard deviation (SD). The data were analyzed by Student’s t-test to determine statistical significance. P < 0.05 was considered to be statistically significant.
Results
Knockdown of RPL21 Inhibits PC Cell Proliferation and Colony Formation
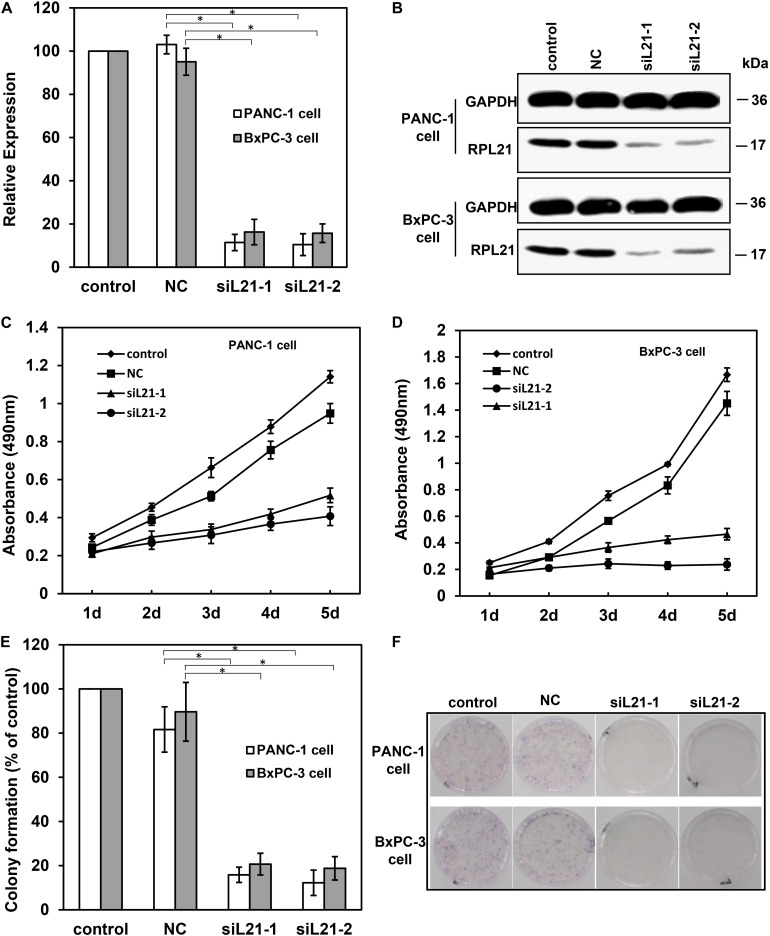
We designed and synthesized two gene-specific siRNAs (siL21-1 and siL21-2) for the post-transcriptional gene silencing against RPL21 gene. As shown in Figure 1A, qPCR results revealed that, at 72 h after transfection of siL21-1 or siL21-2 (40 nM), the expression of RPL21 was significantly decreased at the mRNA levels in PC PANC-1 and BxPC-3 cells as compared to the untreated control group (P < 0.05). No significant difference was observed between negative group (NC, Mock-siRNA transfection) and control groups (Figure 1A). Besides, a same trend was shown in western blot (Figure 1B) compared with qPCR assay. These results indicate that siL21-1 and siL21-2 dramatically reduce the RPL21 expression at mRNA and protein levels in BxPC-3 and PANC-1 cells.
FIGURE 1.
The analysis of cell proliferation on PANC-1 and BxPC-3 cells with siL21-1 and siL21-2 transfection. The quantitative real-time PCR (qPCR) assay (A) and the western blot assay (B) were used to investigate the RNAi effect of siL21-1 and siL21-2 (40 nM, 72 h) in PANC-1 and BxPC-3 cells. GAPDH and β-Actin were used as an internal control for qPCR and western blot analyses, respectively. (C,D) The effect of transfection with siL21-1 and siL21-2 (40 nM) on cell proliferation. The cells were detected by 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay on each day for 5 consecutive days. (E,F) For colony formation, PANC-1 and BxPC-3 cells were trypsinized and seeded into 35-mm dishes (1,000 cells/dish) at 24 h after transfection. After 8 days, cell colonies with more than 50 cells were dyed with hematoxylin and counted. The control, negative control (NC), siL21-1 and siL21-2 represented the untransfected, Mock-siRNA transfected, siL21-1 and siL21-2 transfected, respectively. Data are presented as the mean ± SD (n = 3). * indicates P < 0.05 compared to the control as determined by the Student’s t-test.
To determine the biological effect of silencing RPL21 in PC BxPC-3 and PANC-1 cells, the colony formation and proliferation were measured. MTT assay was used to monitor cell proliferation for 5 consecutive days. As indicated in Figures 1C,D, the MTT analysis showed that transfection of siL21-1 and siL21-2 both inhibited cell proliferation in BxPC-3 and PANC-1 cells) as compared with the control (untransfected) and NC (Mock-siRNA transfected) groups. Consistent with MTT results, the ability of colony formation of PANC-1 and BxPC-3 cells was also decreased after transfection of siL21-1 and siL21-2 (Figure 1F). As shown in Figure 1E, the number of clones in PANC-1 transfected with siL21-1 and siL21-2 were significantly decreased by 84 and 88%, respectively, compared with control groups (untransfected) (P < 0.05), while in BxPC-3 cells, the inhibiting values were 79 and 81% (both P < 0.05). These findings indicate that ribosomal protein gene RPL21 is closely related to proliferation of PC PANC-1 and BxPC-3 cells.
Knockdown of RPL21 Inhibits DNA Replication and Induces G1 Arrest in PC Cells
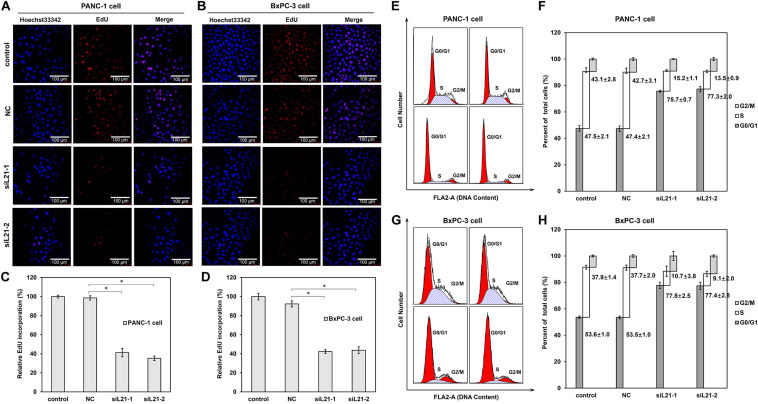
To investigate the mechanisms underlying the altered cell proliferation, EdU incorporation assay was performed to examine the regulatory effect of RPL21 on DNA replication. As shown in Figures 2A,B, both transfections of siL21-1 and siL21-2 drastically inhibited EdU incorporation in PANC-1 and BxPC-3 cells. The statistical analysis results showed that the EdU incorporation after transfection of siL21-1 and siL21-2 were reduced to 41% and 35% in PANC-1 cells, and to 42 and 44% in BxPC-3 cells as compared to untreated control, respectively (Figures 2C,D, all P < 0.05). Since the change of DNA replication tends to affect cell cycle progression (19), we were interested in whether cell cycles would be affected by the inhibition of DNA replication after RPL21 knockdown in PC cells. Further cell cycle analysis revealed that both treatment of siL21-1 and siL21-2 (40 nM, 72 h) resulted in an accumulation in G1 phase and reduction in S phase in PANC-1 and BxPC-3 cells (Figures 2E,G). The fraction of PANC-1 cells in G0/G1 phase was increased from 47.5 to 75.7%, 47.5 to 77.3% after the treatment of siL21-1 and siL21-2, accompanied by a decrease of cells in the S (43.1 to 15.2%, 43.1 to 13.5) phase (Figure 2F) (P < 0.05). In BxPC-3 cells, the fraction of cells in the G0/G1 phase was increased from 37.9 to 77.8%, 37.9 to 77.4% by siL21-1 and siL21-2 treatment (P < 0.05), and simultaneously, we observed a decrease of cells in S (37.9 to 10.7%, 37.9 to 9.1%) phase after the addition of siL21-1 and siL21-2 (Figure 2H) (P < 0.05). Regarding DNA replication and cell cycle, there was no significant difference between NC (Mock-siRNA transfected) and control groups. These changes lead to a logic speculation that the inhibition of proliferation in PC PANC-1 and BxPC-3 cells may be caused by the inhibition of DNA replication and G1 arrest that resulted from silencing of RPL21, which suggest that RPL21 controls proliferation and G1-S phase progression of PC cells.
FIGURE 2.
Effect of RPL21 knockdown on DNA replication and cell cycle progression of PANC-1 and BxPC-3 cells. (A,B) The PANC-1 and BxPC-3 cells after transfection of siRNAs (Mock-siRNA, siL21-1 and siL21-2) (40 nM, 72 h) were incubated with 50 μM of 5-ethynyl-2′-deoxyuridine (EdU). Then the cells were stained with Hoechst 33342 for 30 min and examined by laser scanning confocal microscopy (magnification: ×400). (C,D) Summary graphs of the EdU retention assay for PANC-1 and BxPC-3 cells. (E,G) Human pancreatic cancer cells (PANC-1 and BxPC-3) were seeded in 6-well plates (2 × 105 cells/well) and treated with Mock-siRNA, siL21-1 and siL21-2 (40 nM) for 72 h. Cells were harvested at the indicated time point post transfection and stained with propidium iodide (PI) for DNA cell cycle analysis. (F,H) Percentage of cell cycle distribution. Three independent experiments were of similar results. The untransfected cells serves as a control, and the Mock-siRNA transfected cells serves as a negative control (NC). Each bar represents the mean ± SD of triplicate analysis. * indicates P < 0.05 compared to the control as determined by the Student’s t-test.
RPL21 Controls the Expression of Important G1-S Phase and DNA Replication Regulators in PC Cells
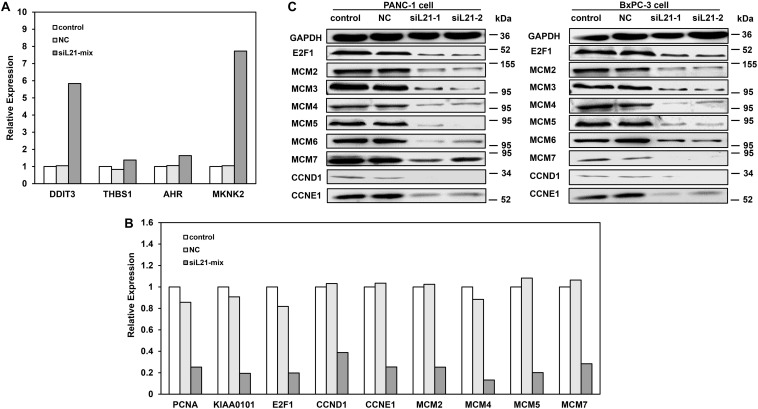
To identify genes contributing to the observed G1 arrest and the inhibition of DNA replication after RPL21 knockdown in PC cells, we did transcriptome sequencing analysis in PANC-1 cells. Comparing with NC groups (PANC-1 cells transfected with Mock-siRNA), 107 genes were found up-regulated and 254 genes were downregulated in siL21-Mix groups (PANC-1 cells transfected with siL21-Mix (siL21-1 and siL21-2) groups) (data was not shown). To verify the transcriptome result, qPCR assay was performed. As shown in Figures 3A,B, for up-regulated genes (AHR, THBS1, DDIT3, and MKNK2) and down-regulated genes (E2F1, PCNA, CCND1, CCNE1 MCM2, MCM4, MCM5, MCM7, and KIAA0101) that we randomly selected, the qPCR results were consistent with the transcriptome sequencing assay. Based on the roles in specific biological functions, the 361 differentially expressed genes were grouped by Gene Ontology (GO) (Supplementary Figure S1). The GO enrichment analysis showed that “cell cycle” in biological process was the most significantly modified, which is consistent with the cell cycle changes that we observed in the previous sections (Figure 2). We then focused and did further analysis on genes that are significantly regulated (<2- or >2-fold change after siL21-Mix transfection) which are known to contribute to G1-S phase progression. The significantly down-regulated genes after siL21-Mix transfection were listed in Supplementary Table S2. Western blot was used to verify the genes (as shown in Supplementary Table S2) that are involved in the regulation of G1-S progression and DNA replication. The results in Figure 3C suggest that the protein expression levels of E2F1, MCM2, MCM3, MCM4, MCM5, MCM6, MCM7, CCND1, and CCNE1 are indeed decreased after the treatment of siL21-1 and siL21-2 in both PANC-1 and BxPC-3 cells. As the replicative helicase, the MCM2-7 family is an evolutionarily conserved group of proteins that are essential for DNA replication (20). Moreover, in mammalian cells, both CCND1 and CCNE1 are positive regulators in G1-S progression (21). This observation suggests that, in PC cells after RPL21 knockdown, the suppression of DNA replication is correlated with the reduced expression of MCM2-7 family, and the G1 arrest is correlated with the reduced expression of CCND1 and CCNE1.
FIGURE 3.
Expression analysis for genes that as the G1-S phase and DNA replication regulators after RPL21 knockdown. (A,B) The validation of transcriptome analysis by quantitative real-time PCR (qPCR). Human pancreatic cancer PANC-1 cells were transfected with siL21-Mix (40 nM, 72 h), the up-regulated genes (AHR, THBS1, DDIT3, and MKNK2) in transcriptome sequencing were confirmed by qPCR. The down-regulated genes (E2F1, PCNA, CCND1, CCNE1 MCM2, MCM4, MCM5, MCM7, and KIAA0101) in transcriptome sequencing were confirmed by qPCR. (C) The PANC-1 and BxPC-3 cells were transfected with Mock-siRNA, siL21-1 and siL21-2 (40 nM, 72 h). The equal amounts (15 μg) of each protein sample were analyzed by western blot with antibodies of E2F1, CCND1, CCNE1, MCM2-7 and GAPDH. The GAPDH served as an internal control. The control, negative control (NC), siL21-1 and siL21-2 represented the untransfected, Mock-siRNA transfected, siL21-1 and siL21-2 transfected, respectively. Three independent experiments were of similar results.
RPL21 Regulates the Cell Cycle and DNA Replication via E2F1 in PC Cells
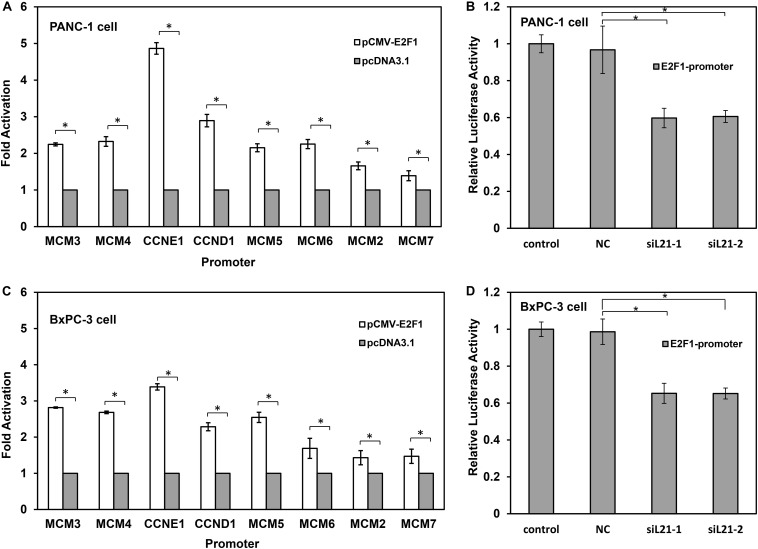
The fact that E2F1 transcription factor is positively correlated with proliferation markers suggests its effector function in G1-S phase progression in PC cells (22). MCM2, MCM3, MCM4, MCM5, MCM6, MCM7, CCND1, and CCNE1 have been shown be regulated by E2F1 (23). The results in Figure 3 showed that, the expression of E2F1 decreased after treatment of siL21-1 and siL21-2 at both mRNA and protein levels in PC cells, which raises a possibility that the G1-S phase progression and DNA replication in PC cells may be regulated by RPL21 via E2F1. To determine the effect of E2F1 on the transcriptional regulation of MCM2, MCM3, MCM4, MCM5, MCM6, MCM7, CCND1, and CCNE1 after RPL21 inhibition, we did luciferase reporter gene assay. As shown in Figures 4A,C, whereas the MCM2-7, CCND1, and CCNE1 promoter reporter vectors were activated by co-transfection of E2F1 expression plasmid (pCMV-E2F1) in PANC-1 and BxPC-3 cells, but were not activated by co-transfection of empty pcDNA3.1 vector. Furthermore, we tested E2F1-dependent transcriptional activity in PANC-1 and BxPC-3 cells using an E2F1 reporter gene constructs (E2F1-promoter). After siL21-1 and siL21-2 treatment, we observed down-regulation of E2F1-dependent transcriptional activity to 59.8 and 60.6% compared with untreated control in PANC-1 cells, and to 65.3 and 65.2% compared with untreated control in BxPC-3 cells (Figures 4B,D, all P < 0.05). These results show that MCM2, MCM3, MCM4, MCM5, MCM6, MCM7, CCND1, and CCNE1 are targets of E2F1 gene in PC cells, and the RPL21 regulates E2F1 binding to the above target genes promoter through the control of E2F1 transcription.
FIGURE 4.
RPL21 regulates the cell cycle and DNA replication via E2F1 in PANC-1 and BxPC-3 cells. (A,C) The luciferase reporter vectors (CCND1, CCNE1 and MCM2-7) were constructed using pGL-3 vectors. 1 μg reporter vector was co-transfected with 1 μg pCMV-E2F1 vector or 1 μg empty pcDNA3.1 vector (control) for each well (6-well plates) containing 6 × 105 PANC-1 and BxPC-3 cells. Luciferase activity was measured 24 h after the transfection. (B,D) The PANC-1 and BxPC-3 cells were transfected with siRNAs with lipofectamine 2000 reagent. The control, NC, siL21-1 and siL21-2 represented the untransfected, Mock-siRNA transfected, siL21-1 and siL21-2 transfected, respectively. The cells after transfection were seeded in 6-well plates at 6 × 105 cells/well, and 2 g E2F1 luciferase reporters (E2F1-promoter) were transfected using lipofectamine 2000 reagent. Luciferase activity was measured 24 h after the transfection. Each bar represents the mean ± SD of triplicate analysis. * indicates P < 0.05 compared to the control as determined by the Student’s t-test.
Silencing of RPL21 Induces Apoptosis of Pancreatic Cancer Cells
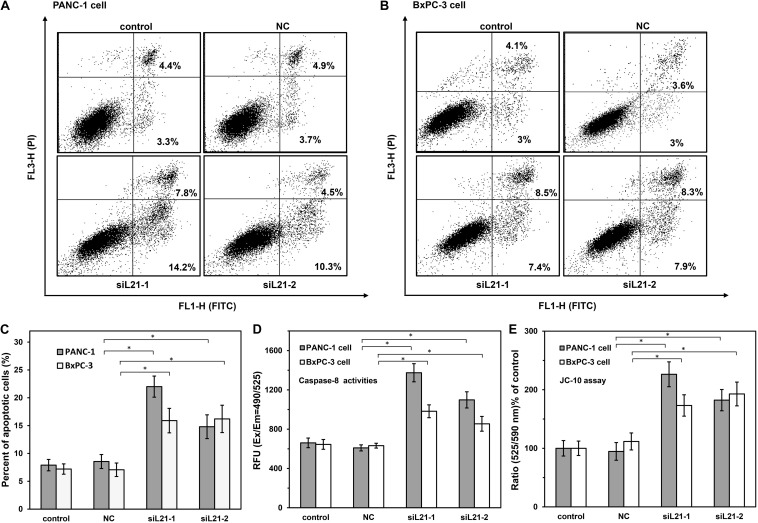
Next, we tested whether cell apoptosis would be induced by RPL21 silencing in PC cells. Apoptosis in BxPC-3 and PANC-1 cells were measured by flow cytometry (FCM) with double-staining of Annexin V-FITC/PI. The rates of early apoptotic cells (lower right area) and late apoptotic cells (upper right area) were shown in Figures 5A,B. And quantitative data from annexin V assay was shown in Figure 5C, siL21-1 and siL21-2 significantly increased the apoptosis rate in BxPC-3 and PANC-1 cells compared to the control group (untransfected) and NC. On the contrary, siL21-1 and siL21-2 did not induce apoptosis in the normal pancreatic duct epithelial HPDE6-C7 cells, compared with control groups (untransfected) and NC (Supplementary Figure S2). These results suggest that silencing RPL21 markedly induces apoptosis only in PC cells but not in pancreatic normal cells, which indicates that RPL21 gene may be a therapeutic target for PC treatment.
FIGURE 5.
The analysis of cell apoptosis on PANC-1 and BxPC-3 cells with siL21-1 and siL21-2 transfection. (A,B) PANC-1 and BxPC-3 cells were transfected with RPL21 siRNA (siL21-1 and siL21-2) and Mock-siRNA (40 nM) for 72 h, respectively, and then analyzed by Annexin V-FITC/PI staining with Flow Cytometry (FCM) analysis. The cells were analyzed with the BD FACSCalibur and FlowJo software at 10,000 events. The lower right area shows early apoptotic cells, and the upper area shows late apoptotic cells. (C) Summary graphs of the flow cytometry (FCM) results. (D) Effects of RPL21 siRNA on Caspase-8 activities. PANC-1 and BxPC-3 cells were transfected with RPL21 siRNA (siL21-1 and siL21-2) and Mock-siRNA (40 nM) for 72 h, respectively, then the cells were seeded into black wall/clear bottom 96-well plates (50,000 cells/well) and were added with caspase-specific fluorogenic substrate. The activities of Caspase-8 were detected at corresponding excitation/emission wavelength (Ex/Em = 490/525 nm) with microplate reader. (E) Effects of RPL21 siRNA on Mitochondrial membrane potential. PANC-1 and BxPC-3 cells were transfected with RPL21 siRNA (siL21-1 and siL21-2) and Mock-siRNA (40 nm, 72 h), respectively, then the cells were harvested and seeded into 96-well plates (60,000 cells/well) followed by the addition of JC-10 dye-loading solution. The ratio of fluorescence intensities on Em at 525/590 was used for Mitochondrial membrane potential analysis. The control, negative control (NC) and siL21-1/siL21-2 represented the untransfected, Mock-siRNA transfected and RPL21 siRNA transfected groups, respectively. Each bar represents the mean ± SD of three separate experiments with triplicate wells per condition, *P < 0.05.
To demonstrate the mechanism of apoptosis induced by knock down of RPL21, we measured the activities of Caspase-3/7, Caspase-8, and Caspase-9 by using of the Caspase-specific fluorogenic substrate. As shown in Figure 5D, compared with control (untreated) and NC (Mock-siRNA transfected) groups, the Caspase-8 (initiators of apoptosis) activities were significantly increased in siL21-1 and siL21-2 transfected groups (both in BxPC-3 and PANC-1 cells, P < 0.05). However, no changes were observed in the Caspase-3/7 (executioners of apoptosis) and Caspase-9 (initiator of apoptosis) activities (Supplementary Figure S3). Next, we tested the changes of mitochondrial membrane potential by JC-10 assay. JC-10 concentrates in the mitochondrial matrix where it forms red fluorescent aggregates (Ex/Em = 540/590 nm) in viable cells. However, in apoptotic and necrotic cells, JC-10 diffuses out of mitochondria. It changes to monomeric form and stains cells in green fluorescence (Ex/Em = 500/525). As shown in Figure 5E, the ratio of fluorescence intensities at Em 525/590 was significantly increased compared to control (untreated) and NC (Mock-siRNA transfected) groups (P < 0.05), indicating that mitochondrial membrane potential was decreased after RPL21 silencing. These results suggested that RPL21 gene may be involved in Caspase-8-related mitochondrial apoptosis in PC cells. Caspase-3/7 did not execute apoptosis in this type of pathway, but the AIF (Apoptosis Inducing Factor) or EndoG (Endonuclease-G) or other factors might be the executioners of apoptosis (24).
Downregulation of RPL21 Inhibits Tumorigenesis of PC Cells in vivo
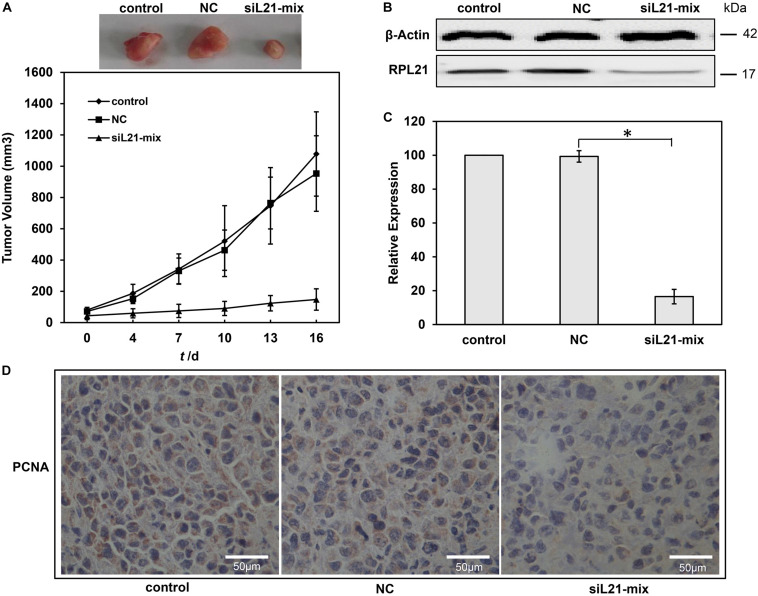
Finally, we determined the effect of knock down of RPL21 on the growth of subcutaneously implanted pancreatic tumors in vivo. Human PC BxPC-3 cells were used to establish tumor xenografts in BALB/c nude mice. The therapeutic effect of siL21-Mix (siL21-1 and siL21-2) was evaluated by intratumorally injection. As shown in Figure 6A, the tumor volumes in the control group were 1078 ± 270 mm3 after 16 days treatment. On the contrary, tumors in the siL21-Mix-treated group grew significantly slower (148 ± 68 mm3 in volume after 16 days). The tumor volume in the NC (Mock-siRNA transfected) group was similar to that of the control group. These results suggest that injection of siL21-Mix significantly inhibited the growth of BxPC-3 tumors in vivo. Then, tumoral RPL21 gene expression from tumor homogenates was evaluated by qPCR and western blot. As shown in Figures 6B,C, RPL21 expression at both mRNA and protein levels in siL21-Mix-treated tumors was significantly decreased compared to control tumors (P < 0.05). Further, we examined the expression of the PCNA that is generally defined as proliferation marker in tumor xenografts. Cell proliferation in tumor section was detected by staining with anti-PCNA monoclonal antibody. As indicated in Figure 6D, there were fewer PCNA positive cells in siL21-Mix-treated tumors, suggesting that the proliferative activities of tumor cells were reduced significantly. These findings demonstrated that the knockdown of RPL21 leads to inhibition of BxPC-3 tumors growth in vivo, which exhibits exceptional applicable potential of RPL21 gene as a therapeutic target for PC.
FIGURE 6.
Effect of RPL21 siRNA on pancreatic cancer growth in vivo. (A) Growth curves of BxPC-3 xenograft tumor in different treated groups. A total of 5 × 106 BxPC-3 cells were injected subcutaneously into BALB/c nude mice. When the tumors reached an average volume of approximately 50 mm3, the mice were randomly divided into three groups (control, NC (negative control), and siL21-Mix). The siRNA transfection solution containing 10 μg of siRNA was injected every 3 days. The mice in control, NC and iL21-Mix groups were injected with transfection solution excluding siRNAs, transfection solution + Mock-siRNA and transfection solution + siL21-1 + siL21-2, respectively. Tumor growth was monitored by measuring the largest (A) and smallest (B) two perpendicular diameters with a caliper, and calculated the tumor volume: (v) = a × b2 × 0.5236. (B)The RPL21 protein levels in different groups with western blot assays. (C) The RPL21 mRNA levels in different xenograft tumor groups with quantitative real-time PCR (qPCR) assays. GAPDH and β-Actin were used as an internal control for qPCR and western blot analyses, respectively. (D) Tumor sections were stained with anti-human PCNA monoclonal antibody to detect proliferating cells. Each bar represents the mean ± SD of three separate experiments with triplicate wells per condition, *P < 0.05.
Discussion
Oncogene KRAS plays an important role in the progression of PC (5, 25). In our previous studies, we found that RPL21 was up-regulated in two stable transfected cell lines (P-M and P-W) with long-term of low KRAS expression (12), which indicated that RPL21 is possibly involved in compensation of KRAS silencing, and also prompted that RPL21 is possibly critical for PC cells survival. In the present article, we confirmed this speculation, and the results showed that the knockdown of RPL21 expression by siRNAs (siL21-1 and siL21-2) significantly inhibited the growth of human PC cells in vitro and in vivo, which suggested that RPL21 is potentially an effective therapeutic target for PC. A pivotal issue about RPL21 as a therapeutic target for PC is the connection between KRAS and the up-regulation of RPL21. Our previous studies showed that the up-regulation of RPL21 gene in PANC-1 cells was because of the long-term suppression rather than the transient suppression of KRAS gene (12), which indicated that RPL21 gene may interact with KRAS gene indirectly. In this study, siL21-1 and siL21-2 showed anti-proliferation and pro-G1 arrest functions both in RAS hyper-activation of cell lines (PC PANC-1 and BxPC-3 cells) and RAS normal-activation of cell lines (normal pancreatic duct epithelial HPDE6-C7 cells). However, siL21-1 and siL21-2 showed pro-apoptosis only in RAS hyper-activation of cell lines (PC PANC-1 and BxPC-3 cells) under the same transfection conditions (Figure 5 and Supplementary Figure S2). These results indicated that the apoptosis-inducing effect by siL21-1 and siL21-2 depends on abnormal regulation of RAS activation/deactivation. Hyper-activation of KRAS gene might be important for RPL21 to regulate cell apoptosis. Our research has established the connection between KRAS and RPL21 genes. Detailed relationship between the ribosomal protein and the oncogene remained unclear. Further dissecting the interaction pathway will reveal the critical signal transduction steps, which would be significantly meaningful for the development of novel treatment for PC in future.
As an integral component of the ribosomal 60S subunit of ribosome, RPL21 primarily plays an important role in protein translation. Intuitively, by knocking down of RPL21, a general decline of ribosome biosynthesis would decrease the protein synthesis, including the cell proliferation-associated proteins and the cell cycle-associated proteins, which may lead to the inhibition of cell proliferation and cell cycle. However, the western blot analysis in this paper (Figure 3C) showed that the level of GAPDH appeared to be unaffected, and further GO enrichment analysis in transcriptome sequencing (Supplementary Figure S1) showed that the genes related to G1-S phase progression were significantly regulated after siL21-Mix (siL21-1 and siL21-2) transfection at mRNA levels. Both evidences suggested RPL21 knockdown did not globally inhibit protein synthesis. Probably alternative genes may be recruited to replace deficient or defective proteins for maintaining structure-function relationships within ribosomes (26). Mammalian cells contain a large number of ribosomes. Transfection of RPL21 siRNA only affects a portion of the ribosome pool. The unaffected ribosomes may still remain normal levels of intracellular proteins for a period of time (27).
For the first time, the present article demonstrated RPL21’s extra-ribosomal functions in proliferation, G1-S phase progression and apoptosis in PC cells. Interestingly, RPL21 gene involved only in proliferation and G1-S progression regulation but not apoptosis (Supplementary Figures S2, S4, S5) in normal pancreatic duct epithelial HPDE6-C7 cells. The differential apoptosis-inducing effect of RPL21 siRNA suggested the possibility of RPL21 gene as a specific therapeutic target for PC treatment. In addition, the gene-specific siRNAs (siL21-1 and siL21-2) were designed to target different regions of the RPL21. Both siL21-1 and siL21-2 showed anti-proliferation, pro-G1 arrest and apoptosis effect in PANC-1 and BxPC-3 cells, therefore the siRNA off-target effect is unlikely in this case. This research emphasizes the important role of RPL21 gene in PC, and highlights the need for further studies in the molecular mechanisms involved in the signaling pathway regulated through RPL21 in PC.
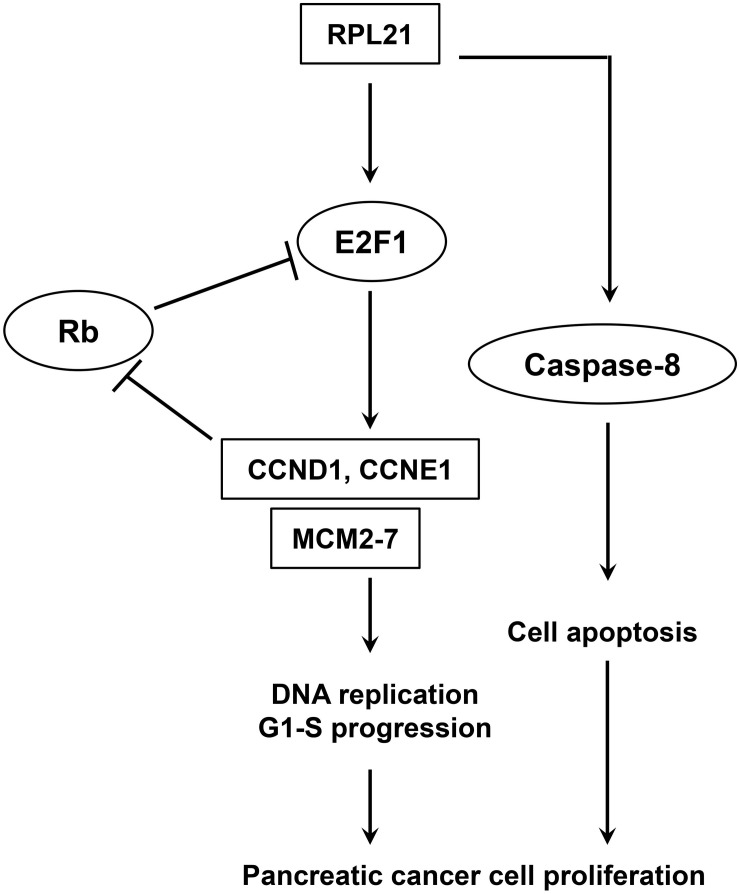
The E2F family of transcription factor consists of eight members that generally associated with transcriptional activation (E2F1, E2F2 and E2F3A) or repression (E2F3B, E2F4, E2F5, E2F6, E2F7, and E2F8) (28). They regulate important cellular responses including cell cycle progression, apoptosis, and DNA damage response (21, 29, 30), and contribute to carcinogenesis of many human tumors (31–34). The accurate transition from G1 to S phase is crucial for the control of eukaryotic cell proliferation, and its misregulation promotes oncogenesis (35, 36). The G1-S phase transition is regulated primarily by D-type cyclins (CCND1, CCND2, or CCND3) in complex with CDK4/CDK6, and E-type cyclins (CCNE1, or CCNE2) in complex with CDK2 (37). During early G1, activator E2F proteins (E2F1, E2F2, or E2F3A) are bound and inhibited by retinoblastoma protein (Rb). When cells are responsive to the mitogenic signals, the CDK4/CCND and CDK2/CCNE complexes Rb, resulting in the activation of E2F proteins and the expression of E2F-responsive genes (38–40). This cluster of genes encodes cell cycle regulators required for G1/S transition (CCNE, CCNA, and CDK1), and components of the DNA replication machinery (CDC6, ORC1, and MCM2-7) (41). The ORC/CDC6/MCM2-7 complex plays a key role for regulated helicase loading in the process of DNA replication. Errors during this process lead to cell cycle arrest at G1-S transition, and further lead to genetic disorders (42). The present paper reported that, in pancreatic cancer (PC) cells, E2F1, CCND1, CCNE1, and all members of the MCM2-7 family were downregulated after inhibition of the RPL21 expression (Figure 3C), leading to the reduction of DNA replication (Figure 2). Further luciferase reporter assay demonstrated that the knockdown of RPL21 decreased the promoter activity of E2F1 (Figures 4B,D). It has been confirmed that MCM2-7, CCND1, and CCNE1 are target genes of E2F1 transcription factor (Figures 4A,C). As shown in Figure 7, after transfection of siL21-1 and siL21-2 in PC cells, the expression of RPL21 is downregulated, resulting in decreased transcription of the transcription factor E2F1. It has been shown previously that the downregulation of CCND1 and CCNE1 also resulted in the inactivation of E2F1 due to the binding of non-phosphorylated Rb and E2F1 (43), which caused the suppression of proliferation and G1-S transition through the regulation of E2F1. Together it is suggested that RPL21 may control DNA replication and G1-S phase progression through E2F1 regulation in PC cells.
FIGURE 7.
Model of E2F1-mediated pancreatic cancer cell proliferation.
The present paper revealed the role of RPL21 on the regulation of E2F1 for the first time. In this work, siL21-1 and siL21-2 reduce the activity of the E2F1 promoter, but the precise molecular mechanism by which RPL21 interacts with E2F1 remains to be unveiled. RPL21 protein may bind to the promoter region of E2F1 gene to regulate its transcription. However, our luciferase reporter assay showed that E2F1 promoter reporter gene construct was not activated by the co-transfection of RPL21 expression vector (pCMV-RPL21) both in PANC-1 and BxPC-3 cells (Supplementary Figure S6). This result suggests that E2F1 promoter region may not contain binding sites of RPL21 protein. The RPL21 protein may bind to other locations of E2F1 gene except the promoter region to regulate the transcription of E2F1. In addition, RPL21 gene may regulate the E2F1 gene transcription indirectly. Some proteins may be used as “messenger” to regulate RPL21-mediated transcription of E2F1. Therefore, how the RPL21 accurately regulate E2F1 transcription will be the focus of our future research. We target to identify protein components in the signal transduction pathways that interact directly with RPL21 that involved in the regulation of E2F1 in our next study.
In this article, we also confirmed that RPL21 gene was involved in Caspase-8-related mitochondrial apoptosis in PC cells. However, the transcriptome sequencing analysis in this study showed no relationship between RPL21 gene and cell apoptosis (Supplementary Figure S1). This is probably because of that, after silencing RPL21 in PC cells, the activities of apoptosis-related protein (such as Caspase-8) changed only at protein levels but not at mRNA levels. It is of interest to understand apoptotic pathways regulated by RPL21 gene. In our research, the Caspase-8 activities were increased, however, the Caspase-3/7 and Caspase-9 activities remained the same after the induction of apoptosis by RPL21 siRNA in PC cells. Under such conditions, Endo G or AIF may be released from the mitochondria as the executioner to induce cell apoptosis (24, 44). Combined with the reducing of mitochondrial membrane potential, in PANC-1 and BxPC-3 cells, RPL21 gene may regulate the apoptosis through the Caspase-8→tBID→EndoG or Caspase-8→tBID→AIF pathway. These studies highlighted the need for further research to a more precisely defined molecular mechanism that involved in apoptotic pathway regulated by RPL21 gene in PC cells.
In summary, the results from this study revealed a new function of the important gene RPL21 in the biological network of PC cells for the first time. The ribosomal protein gene RPL21 controls the PC cell proliferation and G1-S phase progression possibly through the regulation of E2F1 transcription factor. Since little is known about the relationship between RPL21 and human PC cells, our preliminary results provide the first information regarding the possible molecular mechanisms by which RPL21 regulate E2F1 transcription to control PC cell proliferation.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics Statement
This study was carried out in accordance with the principles of the Basel Declaration and recommendations of Guide for the Care and Use of Laboratory Animals and relevant Chinese laws and regulations. The protocol was approved by the Institutional Animal Care and Use Committee (IACUC) of Shanghai Jiao Tong University.
Author Contributions
CL and JZ conceived and designed the experiments. CL, TS, and LH performed the experiments. CL, HL, BZ, and JC collected, analyzed, and interpreted the data. MG, DC, HJ, and YX put forward very valuable comments. CL wrote the manuscript. JZ supervised the study. All authors discussed the results and commented on the manuscript.
Conflict of Interest
CL, LH, and JZ were employed by Jecho Biopharmaceuticals Co., Ltd. HJ, YX, and JZ were employed by Jecho Laboratories, Inc. MG was employed by Shanghai Laiyi Center for Biopharmaceutical R&D. DC was employed by China State Institute of Pharmaceutical Industry. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by grants (No. 2014M551416) from the Chinese Postdoctoral Science Foundation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.01730/full#supplementary-material
References
- 1.Goral V. Pancreatic cancer: pathogenesis and diagnosis. Asian Pac J Cancer Prevent. (2015) 16:5619–24. 10.7314/APJCP.2015.16.14.5619 [DOI] [PubMed] [Google Scholar]
- 2.Zhang L, Sanagapalli S, Stoita A. Challenges in diagnosis of pancreatic cancer. World J Gastroenterol. (2018) 24:2047–60. 10.3748/wjg.v24.i19.2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hosein AN, Beg MS. Pancreatic cancer metabolism: molecular mechanisms and clinical applications. Curr Oncol Rep. (2018) 20:56. 10.1007/s11912-018-0699-5 [DOI] [PubMed] [Google Scholar]
- 4.Pelosi E, Castelli G, Testa U. Pancreatic cancer: molecular characterization, clonal evolution and cancer stem cells. Biomedicines. (2017) 5:65. 10.3390/biomedicines5040065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J, Ji S, Liang C, Qin Y, Jin K, Liang D, et al. Critical role of oncogenic KRAS in pancreatic cancer (Review). Mol Med Rep. (2016) 13:4943–9. 10.3892/mmr.2016.5196 [DOI] [PubMed] [Google Scholar]
- 6.Neoptolemos JP, Kleeff J, Michl P, Costello E, Greenhalf W, Palmer DH. Therapeutic developments in pancreatic cancer: current and future perspectives. Nat Rev Gastroenterol Hepatol. (2018) 15:333–48. 10.1038/s41575-018-0005-x [DOI] [PubMed] [Google Scholar]
- 7.Korobeinikova AV, Garber MB, Gongadze GM. Ribosomal proteins: structure, function, and evolution. Biochem Biokhimiia. (2012) 77:562–74. 10.1134/s0006297912060028 [DOI] [PubMed] [Google Scholar]
- 8.Zimmermann RA. The double life of ribosomal proteins. Cell. (2003) 115:130–2. 10.1016/S0092-8674(03)00804-3 [DOI] [PubMed] [Google Scholar]
- 9.Bhavsar RB, Makley LN, Tsonis PA. The other lives of ribosomal proteins. Hum Genomics. (2010) 4:327–44. 10.1186/1479-7364-4-5-327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. (2009) 34:3–11. 10.1016/j.molcel.2009.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loreni F, Mancino M, Biffo S. Translation factors and ribosomal proteins control tumor onset and progression: how? Oncogene. (2014) 33:2145–56. 10.1038/onc.2013.153 [DOI] [PubMed] [Google Scholar]
- 12.Li C, Ge M, Yin Y, Luo M, Chen D. Silencing expression of ribosomal protein L26 and L29 by RNA interfering inhibits proliferation of human pancreatic cancer PANC-1 cells. Mol Cell Biochem. (2012) 370:127–39. 10.1007/s11010-012-1404-x [DOI] [PubMed] [Google Scholar]
- 13.Li C, Chen D, Luo M, Ge M, Zhu J. Knockdown of ribosomal protein L39 by RNA interference inhibits the growth of human pancreatic cancer cells in vitro and in vivo. Biotechnol J. (2014) 9:652–63. 10.1002/biot.201300321 [DOI] [PubMed] [Google Scholar]
- 14.Guzman-Aranguez A, Loma P, Pintor J. Small-interfering RNAs (siRNAs) as a promising tool for ocular therapy. Br J Pharmacol. (2013) 170:730–47. 10.1111/bph.12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lambeth LS, Smith CA. Short hairpin RNA-mediated gene silencing. Methods Mol Biol. (2013) 942:205–32. 10.1007/978-1-62703-119-6_12 [DOI] [PubMed] [Google Scholar]
- 16.Kenneth JL, Thomas DS. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. (2001) 25:402–8. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 17.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. (1983) 65:55 10.1016/0022-1759(83)90303-4 [DOI] [PubMed] [Google Scholar]
- 18.Li J, Zhang N, Song LB, Liao WT, Jiang LL, Gong LY, et al. Astrocyte elevated gene-1 is a novel prognostic marker for breast cancer progression and overall patient survival. Clin Cancer Res. (2008) 14:3319–26. 10.1158/1078-0432.CCR-07-4054 [DOI] [PubMed] [Google Scholar]
- 19.Diffley JF. Quality control in the initiation of eukaryotic DNA replication. Philos Trans R Soc B Biol Sci. (2011) 366:3545–53. 10.1098/rstb.2011.0073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samson RY, Bell SD. MCM loading–an open-and-shut case? Mol Cell. (2013) 50:457–8. 10.1016/j.molcel.2013.05.008 [DOI] [PubMed] [Google Scholar]
- 21.Bertoli C, Skotheim JM, de Bruin RA. Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol. (2013) 14:518–28. 10.1038/nrm3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamazaki K, Yajima T, Nagao T, Shinkawa H, Kondo F, Hanami K, et al. Expression of transcription factor E2F-1 in pancreatic ductal carcinoma: an immunohistochemical study. Pathol Res Pract. (2003) 199:23–8. 10.1078/0344-0338-00348 [DOI] [PubMed] [Google Scholar]
- 23.Xu X, Bieda M, Jin VX, Rabinovich A, Oberley MJ, Green R, et al. A comprehensive ChIP–chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res. (2007) 17:1550–61. 10.1101/gr.6783507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arnoult D, Gaume B, Karbowski M, Sharpe JC, Cecconi F, Youle RJ. Mitochondrial release of AIF and EndoG requires caspase activation downstream of Bax/Bak-mediated permeabilization. EMBO J. (2003) 22:4385–99. 10.1093/emboj/cdg423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi M, Bien H, Mofunanya A, Powers S. Challenges in Ras therapeutics in pancreatic cancer. Semin Cancer Biol. (2017) 54:101–8. 10.1016/j.semcancer.2017.11.015 [DOI] [PubMed] [Google Scholar]
- 26.Leong WF, Chau JFL, Li B. p53 Deficiency leads to compensatory up-regulation of p16INK4a. Mol Cancer Res. (2009) 7:354–60. 10.1158/1541-7786.MCR-08-0373 [DOI] [PubMed] [Google Scholar]
- 27.Hershey JW. Translational control in mammalian cells. Annu Rev Biochem. (1991) 60:717–55. 10.1146/annurev.bi.60.070191.003441 [DOI] [PubMed] [Google Scholar]
- 28.Crosby ME, Almasan A. Review opposing roles of E2Fs in cell proliferation and death. Cancer Biol Ther. (2004) 3:1208–11. 10.4161/cbt.3.12.1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Engelmann D, Pützer BM. The dark side of E2F1: in transit beyond apoptosis. Cancer Res. (2012) 72:571–5. 10.1158/0008-5472.CAN-11-2575 [DOI] [PubMed] [Google Scholar]
- 30.Wu Z, Zheng S, Yu Q. The E2F family and the role of E2F1 in apoptosis. Int J Biochem Cell Biol. (2009) 41:2389–97. 10.1016/j.biocel.2009.06.004 [DOI] [PubMed] [Google Scholar]
- 31.Hollern DP, Honeysett J, Cardiff RD, Andrechek ER. The E2F transcription factors regulate tumor development and metastasis in a mouse model of metastatic breast cancer. Mol Cell Biol. (2014) 34:3229–43. 10.1128/MCB.00737-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson J, Thijssen B, McDermott U, Garnett M, Wessels LF, Bernards R. Targeting the RB-E2F pathway in breast cancer. Oncogene. (2016) 35:4829–35. 10.1038/onc.2016.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhan L, Zhang Y, Wang W, Song E, Fan Y, Wei B. E2F1: a promising regulator in ovarian carcinoma. Tumour Biol. (2016) 37:2823–31. 10.1007/s13277-015-4770-7 [DOI] [PubMed] [Google Scholar]
- 34.Farra R, Grassi G, Tonon F, Abrami M, Grassi M, Pozzato G, et al. The role of the transcription factor E2F1 in hepatocellular carcinoma. Curr Drug Deliv. (2017) 14:272–81. 10.2174/1567201813666160527141742 [DOI] [PubMed] [Google Scholar]
- 35.Massagué J. G1 cell-cycle control and cancer. Nature. (2004) 432:298–306. 10.1038/nature03094 [DOI] [PubMed] [Google Scholar]
- 36.Ingham M, Schwartz GK. Cell-cycle therapeutics come of age. J Clin Oncol. (2017) 35:2949–59. 10.1200/jco.2016.69.0032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Q, Sakamoto K, Wagner K-U. D-type Cyclins are important downstream effectors of cytokine signaling that regulate the proliferation of normal and neoplastic mammary epithelial cells. Mol Cell Endocrinol. (2014) 382:583–92. 10.1016/j.mce.2013.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Narasimha AM, Kaulich M, Shapiro GS, Choi YJ, Sicinski P, Dowdy SF. Cyclin D activates the Rb tumor suppressor by mono-phosphorylation. eLife (2014) 3:e02872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dick FA, Rubin SM. Molecular mechanisms underlying RB protein function. Nat Rev Mol Cell Biol. (2013) 14:297–306. 10.1038/nrm3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fischer M, Muller GA. Cell cycle transcription control: DREAM/MuvB and RB-E2F complexes. Crit Rev Biochem Mol Biol. (2017) 52:638–62. 10.1080/10409238.2017.1360836 [DOI] [PubMed] [Google Scholar]
- 41.Thwaites MJ, Cecchini MJ, Dick FA. Analyzing RB and E2F during the G1–S transition. Cell Cycle Control. (2014) 1170:449–61. 10.1007/978-1-4939-0888-2_24 [DOI] [PubMed] [Google Scholar]
- 42.Riera A, Fernández-Cid A, Speck C. The ORC/Cdc6/MCM2–7 complex, a new power player for regulated helicase loading. Cell Cycle. (2013) 12:2155. 10.4161/cc.25336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lim S, Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development. (2013) 140:3079–93. 10.1242/dev.091744 [DOI] [PubMed] [Google Scholar]
- 44.Wang X. The expanding role of mitochondria in apoptosis. Genes Dev. (2001) 15:2922–33. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.