Abstract
DNA methylation plays an important role in a wide range of developmental and physiological processes in plants. It is primarily catalyzed and regulated by cytosine-5 DNA methyltransferases (C5-MTases) and a group of DNA glycosylases that act as demethylases. To date, no genome-scale analysis of the two kiwifruit (Actinidia chinensis) families has been undertaken. In our study, nine C5-MTases and seven DNA demethylase genes were identified in the kiwifruit genome. Through selective evolution analysis, we found that there were gene duplications in C5-MTases and demethylases, which may have arisen during three genome doubling events followed by selection during evolution of kiwifruit. Expression analysis of DNA methylases (C5-MTases) and demethylases identified changes in transcripts of DNA methylation and demethylation genes during both vegetative and reproductive development. Moreover, we found that some members of the two methylase/demethylase families may also be involved in fruit ripening and the regulation of softening. Our results help to better understand the complex roles of methylation/demethylation in plants and provide a foundation for analyzing the role of DNA methylation modification in kiwifruit growth, development and ripening.
Keywords: DNA methylases, C5-MTases, DNA demethylases, Actinidia chinensis, kiwifruit
Introduction
The accurate qualitative, quantitative and temporal regulation of gene expression accomplished by cis-regulatory elements and trans-acting factors is indispensable for normal development in plants and animals. In addition to the conventional genetic basis of inheritance and gene expression, epigenetic processes cause heritable changes in gene function that occur by covalent modification of DNA without alteration in the base sequence (Holliday, 2006). One major epigenetic modification is DNA methylation, which occurs by the addition of methyl groups to the C-5 site of cytosine, the N-6 site of adenine, and the N-7 site of guanine (Jeltsch, 2002) in DNA molecules. These epigenetic regulatory mechanisms are widely found in plants and animals (Cao et al., 2014; Wang et al., 2016) and play an important role in the regulation of gene expression. Previous studies have shown that DNA methylation of cytosine at the c-5 site is important in various biological processes, including genome stability, gene imprinting, growth and development, stress response, and biosynthetic regulation of secondary metabolites (Hsieh and Fischer, 2005; FitzGerald et al., 2008; Yelina et al., 2015). In plants, DNA methylation occurs primarily in three distinct sequence contexts: symmetric CG, CHG, and asymmetric CHH sites (where H =A, T or C) (Law and Jacobsen, 2010).
Dynamic mechanisms of DNA methylation in plants include: de novo methylation, DNA methylation maintenance, and DNA demethylation. Cytosines in all sequence contexts can be de novo methylated through the well-known RNA-directed DNA methylation pathway (RdDM), in which 24-nt siRNAs guide the DNA methyltransferase domains rearranged methyltransferase 2 (DRM2) to methylate target loci (Matzke and Mosher, 2014). In Arabidopsis, pathways have been identified that indicate DNA methyltransferase 1 (MET1), as well as chromomethylase 3 (CMT3), can maintain CG and CHG methylation states, whereas chromomethylase 2 (CMT2) can maintain CHH methylation (Law and Jacobsen, 2010). The dynamic regulation of DNA methylation levels also requires the participation of DNA demethylases, which can excise 5-meC from sequence contexts (Zhu, 2009). Four DNA demethylases have been reported in Arabidopsis, namely Repressor of Silencing 1 (ROS1), DEMETER (DME), DEMETER-LIKE (DML2) and DML3 (Gong et al., 2002; Gehring et al., 2006; Ortega-Galisteo et al., 2008).
Kiwifruit (Actinidia chinensis) is an important economic crop and a member of the Actinidiaceae family that consists of three genera and approximately 360 species (Huang et al., 2013). In other plants, DNA methylation has been shown to be involved in many aspects of plant growth and development, such as resistance response, leaf polarity, flowering time, fruit ripening and resistance to disease (Hsieh and Fischer, 2005; FitzGerald et al., 2008; Yelina et al., 2015). The importance of methylation has been demonstrated, for example, by the finding that DNA methylase inhibitors induce early ripening of tomato fruit (Zhong et al., 2013). In contrast, absence of demethylase gene function delays the ripening process (Lang et al., 2017). Despite the fact that C5-MTases and DNA demethylases play a fundamental role in the determination of DNA methylation pattern in the epigenome, little is known about the control of DNA methylation status in many plants, including kiwifruit. To investigate the putative role of DNA methylation in kiwifruit developmental process, we first identified C5-MTases and DNA demethylases family genes in the genome.
In our study, the protein sequences of C5-MTases and DNA demethylases across the whole genome of kiwifruit (Pilkington et al., 2018) were determined and the phylogenetic relationship among C5-MTases and demethylases in kiwifruit were analyzed. We also measured the expression of C5-MTase and demethylase encoding genes in various tissues/developmental stages in kiwifruit. In addition, to further understand the functions of selected members, three-dimensional (3D) structural modeling was performed. Our results provide a foundation for analyzing the role of DNA methylation modification in kiwifruit.
Materials and Methods
Data Collection and Identification of C5-MTases and DNA Demethylases in Kiwifruit
Protein sequences of C5-MTases and DNA demethylases in Arabidopsis (Gong et al., 2002; Xiao et al., 2006; Henderson et al., 2010; Liu et al., 2015) were downloaded from the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/). Gene IDs for all protein sequences are shown in Supplementary Data 1. Using Blastp (2.9.0) (E < 0.001) (Camacho et al., 2009), putative protein sequences from the whole genome of kiwifruit (Pilkington et al., 2018) with homology to methylase and demethylase families in Arabidopsis were identified. We then used Hmmsearch (3.2.1) (Wheeler and Eddy, 2013) to scan all previously identified sequences (Gong et al., 2002; Xiao et al., 2006; Henderson et al., 2010; Liu et al., 2015). Finally, we identified nine C5-MTases and seven DNA demethylases. Furthermore, names to C5-MTases and DNA demethylases in kiwifruit were assigned based on names of best hit proteins by NCBI-blastp (Non-redundant protein sequences database) (Letunic et al., 2015). The isoelectric point (pI) and molecular weight (Mw) of all C5-MTases and DNA demethylases proteins were analyzed by the online program Expasy (http://web.expasy.org/compute_pi/) (Gasteiger et al., 2005).
Analysis of Conserved Motif, Domain, Gene Structure, and 3D Model
Full length amino acid sequences of C5-MTases and DNA demethylases in kiwifruit were analyzed using the MEME (5.05) program (http://meme.nbcr.net/meme/) to study their conserved motifs (parameter settings: pattern 10; minimum pattern width 6; maximum pattern width 50). Both Pfam (32.0) (http://pfam.xfam.org/) and SMART (http://smart.embl-heidelberg.de/) were used for protein domain identification in kiwifruit, Arabidopsis, tomato, and rice. Tbtools was used to draw the gene structure of the C5-MTases and DNA demethylases from the kiwifruit genome (Chen et al., 2018). SWISS-MODEL (http://www.swissmodel.expasy.org/interactive) was used for building a AcCMT3, AcCMT4, AcCMT5, and AtCMT1 homologous protein model (at least 186 models for each protein were generated using the “building model” engine, and the best model was selected based on the best global model quality estimation) (Guex et al., 2009; Benkert et al., 2011; Bienert et al., 2016; Bertoni et al., 2017; Waterhouse et al., 2018).
Multiple Sequence Alignment and Phylogenetic Tree Construction
Multiple sequence alignments of the C5-MTase and DNA demethylase amino acid sequences were performed with MEGA X (10.0.5) (Kumar et al., 2018). Neighbor-Joining (NJ) and maximum likelihood (ML) trees were constructed using MEGA X (10.0.5) with aligned protein sequences (Poisson correction and bootstrap = 1000 replicates) (Yang, 2007). Visualization of the phylogenetic tree was accomplished by JalView (Waterhouse et al., 2009; Troshin et al., 2011) and Evolview v2 (https://www.evolgenius.info/) (Zhang et al., 2012; He et al., 2016).
Analysis of Selection Pressure
Blastp (E<1e-5) and MCScanX (Wang et al., 2012) were used for colinear analysis. Visualization of the data was accomplished by Circos plot (Connors et al., 2009). We used DnaSP (v6.12.03) (Rozas et al., 2017) to calculate the Ka/Ks values of the corresponding C5-MTases and DNA demethylases in kiwifruit.
Analysis of the Promoter Cis-Regulating Elements
The 2000 bp of kiwifruit genomic DNA sequence upstream of the transcriptional start sites of C5-MTases and DNA demethylases were extracted from the kiwifruit genome. The online tool PlantCare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002) was used to analyze the promoter sequences.
Analysis of Gene Expression
A plant RNA extraction kit (V1.5) (DNase I) (Chengdu Biofit Biotechnologies CO., LTD) was used to extract the total RNA of roots, young stems, stems, young leaves, leaves, flowers, fruits of 40 days after anthesis, fruits of 140 days after anthesis, fruits of 4 days after harvest, and fruits 12 days after harvest of kiwifruit. The roots, young stems and young leaves were obtained from tissue-cultured seedlings. cDNA was obtained by reverse transcription according to the PrimeScript™RT reagent Kit with gDNA Eraser (Perfect Real Time) (Takara biomedical technology (Beijing) co., LTD., Beijing, China). Real-time quantitative (RT) PCR was performed as described by Pirrello et al., 2006 (Pirrello et al., 2006). AcActin1 (EF063572) genes (Wang et al., 2019) were used as a standardized internal control and relative mRNA levels of genes were calculated as 2-ddct values. Primers for amplification were designed using the software PerlPrimer v1.1.21 (Marshall, 2004) (Supplementary Data 2). Three independent biological replicates were used for this experiment.
Results
Identification and Analysis of C5-MTase and DNA Demethylase Genes in Kiwifruit
In our study, C5-MTases and DNA demethylases were identified from the kiwifruit genome using the sequences of C5-MTases and DNA demethylases of Arabidopsis as BLAST queries against the kiwifruit genome. We then used HMMER to verify whether the identified C5- MTases contained a typical DNA methylase domain (PF00145). By using this method, nine C5-MTase and seven DNA demethylase genes were identified in kiwifruit (Table 1). The number of genes is similar to that in Arabidopsis and tomato species (Gong et al., 2002; Xiao et al., 2006; Hsieh et al., 2009; Henderson et al., 2010; Cao et al., 2014). The polypeptide lengths of identified C5-MTase genes (CMT1, CMT2, CMT3, CMT4, CMT5, DRM2X1, DRM2X2, DRM2X3, and MET) in kiwifruit ranges from 558 to 1055 amino acids and the predicted molecular weights are from 63.11 to 119.70 (kDa). The seven demethylase genes (DME1, DME2, DME3, DME4, DML, ROS1X1, and ROS1X2) code polypeptides composed of 1202 to 1980 amino acids. The molecular weights of these demethylases ranges from 135.51 to 220.26 (kDa).
Table 1.
Basic Information of C5-MTase and DNA Demethylase Genes in kiwifruit.
| Protein name | Gene ID | Chromosome_location | Protein | ORF length(bp) | ||||
|---|---|---|---|---|---|---|---|---|
| PI | Mw(kDa) | Length(aa) | ||||||
| Cytosine-5 DNA methyltransferases | ||||||||
| MET | Acc06669 | Chr6 | 11943186 | 11957240 | 5.69 | 101.17 | 902 | 2706 |
| CMT1 | Acc10156 | Chr9 | 4688362 | 4699999 | 5.46 | 93.73 | 830 | 2490 |
| CMT2 | Acc22670 | Chr20 | 4250119 | 4266395 | 6.44 | 119.70 | 1055 | 3165 |
| CMT3 | Acc29050 | Chr25 | 16658886 | 16677120 | 6.45 | 105.67 | 923 | 2769 |
| CMT4 | Acc30443 | Chr26 | 19829011 | 19840660 | 5.15 | 92.97 | 821 | 2463 |
| CMT5 | Acc32510 | Chr28 | 15169385 | 15181698 | 5.06 | 105.30 | 928 | 2784 |
| DRM2X1 | Acc02430 | Chr2 | 13147187 | 13161342 | 5.13 | 66.53 | 591 | 1581 |
| DRM2X2 | Acc05665 | Chr5 | 11554590 | 11564191 | 5.87 | 63.11 | 558 | 1674 |
| DRM2X3 | Acc30617 | Chr27 | 1537771 | 1548409 | 5.33 | 65.84 | 585 | 1554 |
| Demethylases | ||||||||
| DME1 | Acc04130 | Chr3 | 18269904 | 18284006 | 8.23 | 220.26 | 1980 | 5940 |
| DME2 | Acc08438 | Chr8 | 1986494 | 1997536 | 6.33 | 189.93 | 1706 | 5118 |
| DME3 | Acc27569 | Chr24 | 9270345 | 9285217 | 8.3 | 216.44 | 1953 | 5859 |
| DME4 | Acc16912 | Chr22 | 13533612 | 13546816 | 6,15 | 208.53 | 1868 | 5604 |
| DML | Acc00310 | Chr1 | 3905070 | 3916337 | 9.24 | 158.76 | 1405 | 4215 |
| ROS1X1 | Acc03168 | Chr3 | 6425862 | 6443713 | 9.31 | 138.30 | 1223 | 3669 |
| ROS1X2 | Acc08944 | Chr8 | 11005987 | 11014368 | 9.26 | 135.51 | 1202 | 3606 |
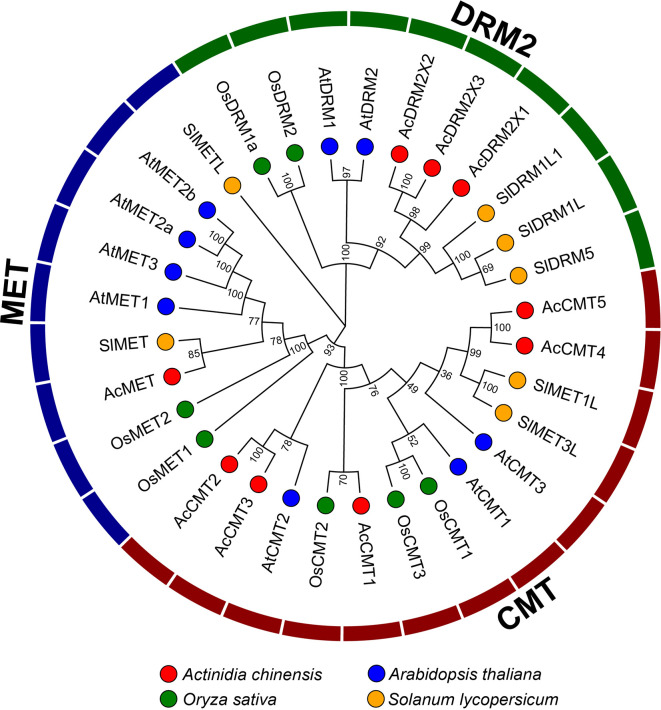
To investigate the phylogenetic relationship of C5-MTase and DNA demethylase genes in kiwifruit, we constructed a phylogenetic tree using the neighbor-joining (NJ) method based on multiple sequence alignments (Figure 1). The results showed that the C5-MTase families of the four plants are divided into three distinct branches, namely CMT, MET, and DRM2. The evolutionary branch of CMT contains five kiwifruit proteins (CMT1, CMT2, CMT3, CMT4, and CMT5), while the evolutionary branch of MET contains a single protein in kiwifruit (MET), and the evolutionary branch DRM contains three proteins in kiwifruit (DRM2X1, DRM2X2, and DRM2X3).
Figure 1.
Neighbor-joining phylogeny of C5-MTases.
Interestingly, C5-MTases in kiwifruit were found to be closer to that in tomato. Moreover, it was also found that proteins in dicotyledonous plants (kiwifruit, Arabidopsis, and tomato) and monocotyledon (rice) are significantly different. These results suggested that the duplication of C5-MTases might be different between dicotyledonous and monocotyledonous plants.
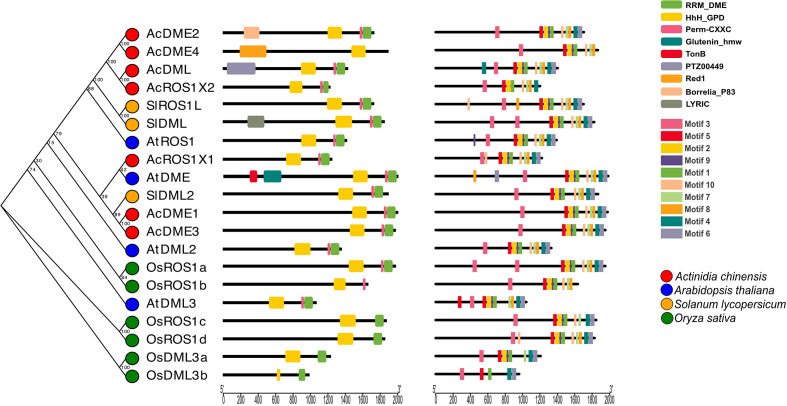
The characteristic conserved key domain of demethylase proteins is HhH_GPD and most also possess RRM_DME and Perm-CXXC domains (Figure 2). For further analysis of the evolutionary relationships among plant demethylase families, we constructed a rootless evolutionary tree using 20 full-length protein sequences from four plant species (kiwifruit, Arabidopsis, tomato, and rice) and analyzed their motifs. We found that DNA demethylases share similar conserved domains (Figure 2), indicating that the demethylase gene family has been relatively conserved in different plant species. However, the monocotyledons and dicotyledons are clearly separated in the evolutionary tree, indicating that the evolution of DNA demethylase might have occurred separately in monocotyledons and dicotyledons. This is consistent with previously constructed phylogenetic trees using the DNA glycosylase domain of DMLs from flowering plants (Zemach et al., 2010). These results also showed the conservation and diversity of DNA demethylase between monocotyledons and dicotyledons.
Figure 2.
DNA demethylase gene structure, phylogenetic relationships and conserved motifs in four species.
Motif and Domain Analysis of C5-MTases and DNA Demethylases in Kiwifruit
We found that members of the CMT protein group not only contain a DNA methylase domain, but also a chromosome domain located in the DNA methylase domain and a bromine-adjacent homologous domain (BAH). The MET group members include a DNA-methylase domain and two bromine-adjacent homologous domains (BAH) (Xiao et al., 2006; Henderson et al., 2010; Song et al., 2013; Cao et al., 2014; Wang et al., 2016). The seven demethylase genes all contain a HhH_GPD domain, a RRM_DME domain, and a Perm-CXXC domain, except DME4 which only has a HhH_GPD domain (Supplementary Data 3) (Gong et al., 2002; Liu et al., 2015; Wang et al., 2016).
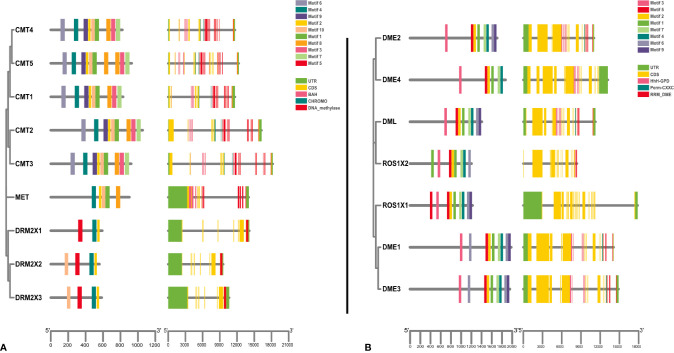
To further explore the conservation and diversity of C5-MTases and DNA demethylases in kiwifruit, we used MEME to analyze their protein motifs (E ≤ 0.01) (Supplementary Data 4). All C5-MTase genes were found to contain motif 2 and motif 4. Specifically, all members of the MET group contained motif 1, motif 2, motif 4, motif 8, and motif 10. All members of the CMT group contained motif 1, motif 2, motif 3, motif 4, motif 6, motif 7, Motif 8, motif 9, and motif 10. All members of the DRM group contained motif 2, motif 4, and motif 5 (Figure 3). As shown in Figure 3A, the conservation of C5-MTases amino acid sequences in kiwifruit was higher than that the DNA sequence. All DNA demethylase genes contain motif 1, motif 2, motif 3, motif 4, motif 5, and motif 8, which demonstrates a high degree of sequence conservation (Figure 3B).
Figure 3.
Gene structure of kiwifruit C5-MTase and demethylase genes and phylogenetic relationships between conserved motifs of each group of kiwifruit proteins. (A) C5-MTase; (B) Demethylase.
Homology Analysis of C5-MTases and DNA Demethylases in Kiwifruit
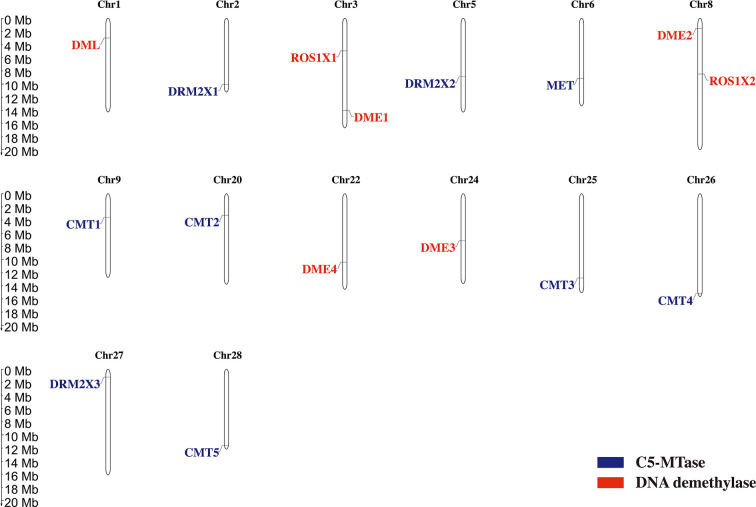
The distribution of genes across chromosomes can influence their downstream functions. The location of C5-MTases and DNA demethylases in kiwifruit chromosomes were determined according to the genome and gene annotation (Figure 4). Nine C5-MTase genes in kiwifruit are dispersed across chromosome 2 (DRM2X1), chromosome 5 (DRM2X2), chromosome 6 (MET), chromosome 9 (CMT1), chromosome 20 (CMT2), chromosome 25 (CMT3), chromosome 26 (CMT4), chromosome 27 (DRM2X3), and chromosome 28 (CMT5). Seven kiwifruit DNA demethylase genes are dispersed across chromosome 1 (DML), chromosome 3 (ROS1X1 and DME1), chromosome 8 (DME2 and ROS1X2), chromosome 22 (DME4), and chromosome 24 (DME3).
Figure 4.
Chromosome localization of C5-MTase and demethylase genes in kiwifruit.
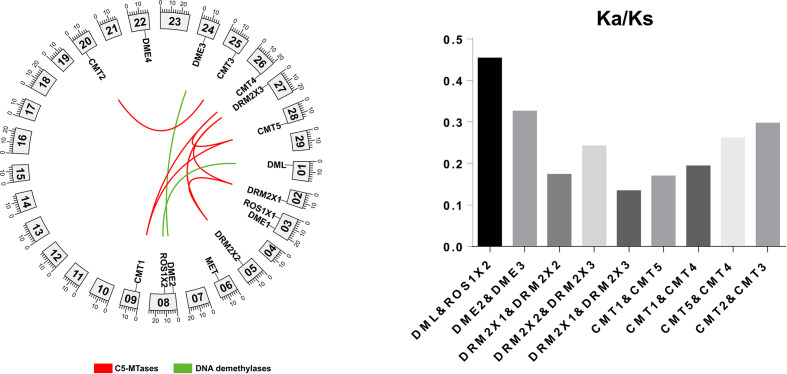
The potential mechanisms and evolutionary relationships between C5-MTases and DNA demethylases in the kiwifruit genome were studied using Blastp. The results were analyzed by colinearity analysis using MCScanX as shown in Figure 5. Five groups of genes were found to possess colinearity including DRM2X1, DRM2X2, and DRM2X3; CMT1, CMT4, and CMT5; CMT2 and CMT3; DME2 and DME3; DML and ROS1X2. The level of colinearity in C5-MTases (33.3%) was close to that of DNA demethylases (28.6%). The emergence of these homologous gene pairs may be associated with three gene doubling events that have occurred in kiwifruit (Huang et al., 2013).
Figure 5.
Homology analysis of C5-MTases and DNA demethylases.
To analyze the constraints that have governed C5-MTases and DNA demethylases evolution in kiwifruit, we investigated the Ka/Ks ratios for five duplicated pairs of genes. The results showed that all Ka/Ks ratios are lower than 1.0 (Figure 5), which suggested that these homologous genes have undergone purifying selection (Zhang et al., 2006).
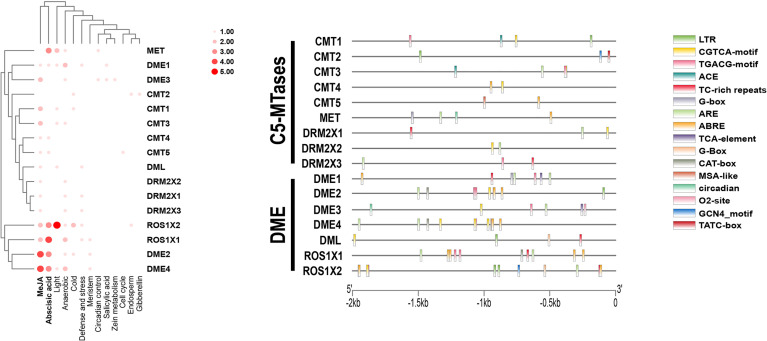
Cis-Acting Elements Analysis of C5-MTases and DNA Demethylases Promoters in Kiwifruit
The 2000 bp of kiwifruit genomic DNA sequence upstream of the transcriptional start sites of C5-MTases and DNA demethylases in kiwifruit were analyzed and Cis-acting elements of all promoters are shown in Supplementary Data 5. All promoter sequences contained a CAAT-box, a conventional cis-element. More specifically, most C5-MTases and DNA demethylases promoters contained the jasmonic acid-inducible CGTCA-motif, TGACG-motif, and abscisic acid-induced ABRE, and anaerobic inducing elements (ARE) (Figure 6). This suggests that C5-MTases and DNA demethylases may be responsive to both biotic and abiotic stressors, which is consistent with previous studies in peanuts (Wang et al., 2016).
Figure 6.
Cis-acting elements in the promoters of C5-MTases and demethylases.
Expression of C5-MTases and DNA Demethylases in Kiwifruit
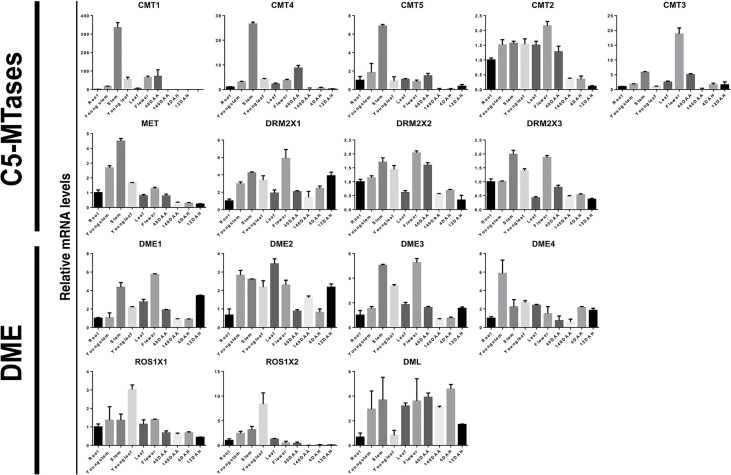
C5-MTases are responsible for the establishment and maintenance of DNA methylation (Niederhuth and Schmitz, 2013). DNA demethylases, on the other hand, plays a major role in the process of DNA demethylation (Zhu, 2009). For further exploration of their putative role in plant growth and development, using qRT-PCR we analyzed the expression patterns of C5-MTases and DNA demethylases in different tissues and ripening stages including roots, young stems, stems, young leaves, leaves, flowers, fruit at 40 days after anthesis, fruits at 140 days after anthesis, fruits at 4 days after harvest, fruits at 12 days after harvest (Figure 7).
Figure 7.
Relative expression levels of kiwifruit C5-MTase and demethylase genes in different tissues. 40DAA: Fruit 40 days after anthesis; 140DAA: Fruit 140 days after anthesis; 4DAH: Fruit 4 days after harvest; 12DAH: Fruit 12 days after harvest. Values represent the average of three biological replicates, and vertical lines represent the SD of the average.
It is noteworthy that MET, CMT1, CMT4, and CMT5 have similar expression patterns. These genes are homologous and are all highly expressed in the stems of kiwifruit plants. However, their expression levels were relatively low during early developmental of the stem, suggesting that a large amount of DNA methylation only occurs for a certain period during plant stem development. The expression of CMT2 and CMT3 was more prominent in flowers and DRM2X1, DRM2X2, as well as DRM2X3 were more prominently expressed in stems and flowers than in early developmental stages. Interestingly, the expression of DRM2 were more prominent in fruit than that of MET and CMT. In particular, although DRM2X1, DRM2X2, and DRM2X3 are homologous genes, the expression pattern of DRM2X1 in fruits was different to that of DRM2X2 and DRM2X3. Relatively high levels of DNA demethylases were found in young stems, stems, young leaves, and flowers, but transcripts of DNA demethylases were rather low in roots.
Discussion
In plants, DNA methylation has been figured out to be involved in different processes of plant growth and development (Hsieh and Fischer, 2005; FitzGerald et al., 2008; Yelina et al., 2015). Despite the fact that C5-MTases and DNA demethylases play a fundamental role in the determination of DNA methylation pattern in the epigenome, little is known about the control of DNA methylation status in kiwifruit. In our study, nine C5-MTases and seven DNA demethylases were identified in kiwifruit.
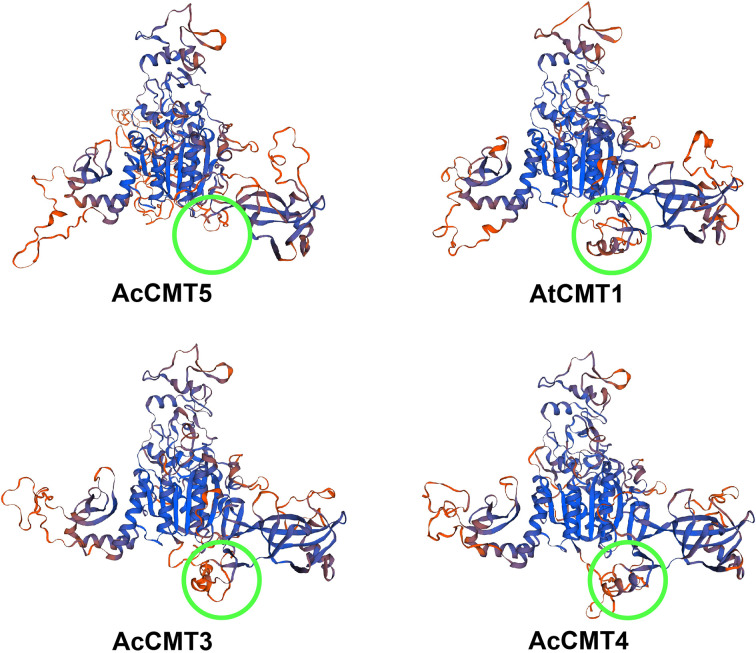
Phylogenetic analysis of C5-MTases from four plant species showed that the duplication of C5-MTases might be different between dicotyledonous and monocotyledonous plants (Figure 1). As shown in Figure 2, the C5-MTases of kiwifruit could be clearly divided into three groups: MET, CMT, and DRM2. Compared with other CMT members, CMT5 lacked part of the BAH domain which contained aromatic cages comprised of three conserved amino acids (Y, W, Y) capturing methylated-lysine.
To further investigate the difference in structures between CMT5 and other CMTs in kiwifruit, we built 3D models for AcCMT3, AcCMT4, AcCMT5, and AtCMT1 (Figure 8). From these models, we found that the BAH domain of AcCMT5 was incomplete and part of the domain was missing when compared with the BAH structural domain of AcCMT3 and AcCMT4. This suggests that the ability of AcCMT5 to bind methylated lysine might be different from the other CMTs.
Figure 8.
3D structure of AtMET1, AcMET3, AcMET4, and AcMET5.
Analysis of cis-acting elements has shown that C5-MTase and DNA demethylase genes are strongly associated with biological and abiotic stressors. This is consistent with changes in the expression of C5-MTase and DNA demethylase genes after kiwifruit is harvested, suggesting that postharvest storage might be affected by DNA methylation. Based on chromosomal localization and collinear analysis, we found that these gene pairs (DRM2X1, DRM2X2, DRM2X3; CMT4, CMT4, CMT5; CMT2, CMT3; DME2, DME3; DML and ROS1X2) in C5-MTases and DNA demethylases in kiwifruit are homologous. Their positions on chromosomes are similar, which is likely related to the three chromosome multiplication events of kiwifruit over the course of its evolutionary history. Through selection pressure analysis, we found these enzymatic proteins are strongly regulated under purifying selection and play an important conserved role in these organisms. Gene expression analysis of C5-MTases and DNA demethylases across different tissues and developmental stages showed that C5-MTases were more prominent in stems and flowers, while DNA demethylases had higher expression in young stems, stems, young leaves, and flowers. This suggests that DNA methylation dynamics are important in both vegetative and reproductive development.
Conclusion
In our study, nine C5-MTases and seven DNA demethylases were identified in the kiwifruit genome. Through a comprehensive analysis of its gene structure, chromosome localization, and protein structure, we found similar results to C5-MTases and DNA demethylases previously recognized in Arabidopsis. Additionally, we found that expression level of DNA methylases were more prominent in stems and flowers, and DNA demethylases were expressed in the young stems, stems, young leaves, and flowers of kiwifruit. We also found that kiwifruit maintained a certain level of expression of C5-MTases and DNA demethylases after harvest, suggesting that DNA methylation dynamics may affect fruit ripening and postharvest. These results help to understand the complexity of these two gene families and lay a foundation for analyzing the role of DNA methylation modification in fruit ripening and fruit shelf life of kiwifruit.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.
Author Contributions
ML and YZ designed the experiments. YZ, XH, and WX interpreted the results. YZ, XH, HZ, and HD wrote the paper. WX and SW participated in the data mining. XH, HW, and DS helped in kiwifruit materials collection and qRT-PCR analysis. ZZ, BY, JW, and DG helped improve the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Key R&D Program of China (2016YFD0400100), the Science and technology innovation talent project of Sichuan province (2018RZ0144) to ML and National Natural Science Foundation of China (No. 30670204) to JW. This project is also supported by the College student innovation and entrepreneurship training program of Sichuan University (201910610874).
Conflict of Interest
Authors ZZ and BY were employed by company Sichuan Dexin Guoyuan Biological Technology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial of financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.514993/full#supplementary-material
References
- Benkert P., Biasini M., Schwede T. (2011). Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27, 343–350. 10.1093/bioinformatics/btq662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoni M., Kiefer F., Biasini M., Bordoli L., Schwede T. (2017). Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci. Rep. 7, 1–15. 10.1038/s41598-017-09654-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienert S., Waterhouse A., de Beer, Tjaart A. P., Tauriello G., Studer G., Bordoli L., et al. (2016). The SWISS-MODEL Repository—new features and functionality. Nucleic Acids Res. 45 (D1), D313–D319. 10.1093/nar/gkw1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., et al. (2009). BLAST+: architecture and applications. BMC Bioinf. 10, 421. 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D., Ju Z., Gao C., Mei X., Fu D., Zhu H., et al. (2014). Genome-wide identification of cytosine-5 DNA methyltransferases and demethylases in Solanum lycopersicum. Gene 550, 230–237. 10.1016/j.gene.2014.08.034 [DOI] [PubMed] [Google Scholar]
- Chen C., Chen H., He Y., Xia R. (2018). TBtools, a Toolkit for Biologists integrating various biological data handling tools with a user-friendly interface. bioRxiv 289660. 10.1101/289660 [DOI]
- Connors J., Krzywinski M., Schein J., Gascoyne R., Horsman D., Jones S. J., et al. (2009). Circos : An information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. 10.1101/gr.092759.109.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzGerald J., Luo M., Chaudhury A., Berger F. (2008). DNA Methylation Causes Predominant Maternal Controls of Plant Embryo Growth. PloS One 3 (5), e2298. 10.1371/journal.pone.0002298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E., Hoogland C., Gattiker A., Duvaud S. E., Wilkins M. R., Appel R. D., et al. (2005). "Protein Identification and Analysis Tools on the ExPASy Server," in in The Proteomics Protocols Handbook. Ed. Walker J. M. (Totowa, NJ: Humana Press; ), 571–607. 10.1385/1-59259-890-0:571 [DOI] [Google Scholar]
- Gehring M., Huh J., Hsieh T.-F., Penterman J., Choi Y., Harada J., et al. (2006). DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124, 495–506. 10.1016/j.cell.2005.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z., Morales-Ruiz T., Ariza R. R., Roldán-Arjona T., David L., Zhu J. K. (2002). ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111, 803–814. 10.1016/S0092-8674(02)01133-9 [DOI] [PubMed] [Google Scholar]
- Guex N., Peitsch M. C., Schwede T. (2009). Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 30 (S1), S162–S173. 10.1002/elps.200900140 [DOI] [PubMed] [Google Scholar]
- He Z., Zhang H., Gao S., Lercher M., Chen W.-H., Hu S. (2016). Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 44:gkw370. 10.1093/nar/gkw370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I. R., Deleris A., Wong W., Zhong X., Chin H. G., Horwitz G. A., et al. (2010). The De novo cytosine methyltransferase DRM2 requires intact UBA domains and a catalytically mutated paralog DRM3 during RNA-directed DNA methylation in Arabidopsis thaliana. PloS Genet. 6, 1–11. 10.1371/journal.pgen.1001182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R. (2006). Epigenetics: A Historical Overview. Epigenet. Off. J. DNA Methyl. Soc. 1, 76–80. 10.4161/epi.1.2.2762 [DOI] [PubMed] [Google Scholar]
- Hsieh T.-F., Fischer R. L. (2005). Biology of Chromatin Dynamics. Annu. Rev. Plant Biol. 56, 327–351. 10.1146/annurev.arplant.56.032604.144118 [DOI] [PubMed] [Google Scholar]
- Hsieh T. F., Ibarra C. A., Silva P., Zemach A., Eshed-Williams L., Fischer R. L., et al. (2009). Genome-wide demethylation of Arabidopsis endosperm. Science 324, 1451–1454 . 10.1126/science.1172417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Ding J., Deng D., Tang W., Sun H., Liu D., et al. (2013). Draft genome of the kiwifruit Actinidia chinensis. Nat. Commun. 4, 2640. 10.1038/ncomms3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeltsch A. (2002). Beyond Watson and Crick: DNA Methylation and Molecular Enzymology of DNA Methyltransferases. ChemBioChem 3 (4), 274–293. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang Z., Wang Y., Tang K., Tang D., Datsenka T., Cheng J., et al. (2017). Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. PNAS 114 (22). 10.1073/pnas.1705233114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J. A., Jacobsen S. E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220. 10.1038/nrg2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M., Déhais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. 10.1093/nar/30.1.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Doerks T., Bork P. (2015). SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 43, D257–D260. 10.1093/nar/gku949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., How-Kit A., Stammitti L., Teyssier E., Rolin D., Mortain-Bertrand A., et al. (2015). A DEMETER-like DNA demethylase governs tomato fruit ripening. Proc. Natl. Acad. Sci. U. States America 112, 10804–10809. 10.1073/pnas.1503362112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall O. (2004). PerlPrimer: Cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinf. (Oxford Engl.) 20, 2471–2472. 10.1093/bioinformatics/bth254 [DOI] [PubMed] [Google Scholar]
- Matzke M. A., Mosher R. A. (2014). RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15 (6), 394–408. 10.1038/nrg3683 [DOI] [PubMed] [Google Scholar]
- Niederhuth C., Schmitz R. (2013). Covering Your Bases: Inheritance of DNA Methylation in Plant Genomes. Mol. Plant 7 (3), 472–480. 10.1093/mp/sst165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Galisteo A., Morales-Ruiz T., Ariza R., Roldan Arjona T. (2008). Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol. Biol. 67, 671–681. 10.1007/s11103-008-9346-0 [DOI] [PubMed] [Google Scholar]
- Pilkington S. M., Crowhurst R., Hilario E., Nardozza S., Fraser L., Peng Y., et al. (2018). A manually annotated Actinidia chinensis var. chinensis (kiwifruit) genome highlights the challenges associated with draft genomes and gene prediction in plants. BMC Genomics 19, 1–19. 10.1186/s12864-018-4656-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrello J., Jaimes-Miranda F., Sanchez-Ballesta M., Tournier B., Khalil Q., Regad F., et al. (2006). Sl-ERF2, a Tomato Ethylene Response Factor Involved in Ethylene Response and Seed Germination. Plant Cell Physiol. 47, 1195–1205. 10.1093/pcp/pcj084 [DOI] [PubMed] [Google Scholar]
- Rozas J., Ferrer-Mata A., Sanchez-DelBarrio J. C., Guirao-Rico S., Librado P., Ramos-Onsins S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. 10.1093/molbev/msx248 [DOI] [PubMed] [Google Scholar]
- Song Q. X., Xiang L., Li Q. T., Chen H., Hu X. Y., Ma B., et al. (2013). Genome-Wide analysis of DNA methylation in soybean. Mol. Plant 6, 1961–1974. 10.1093/mp/sst123 [DOI] [PubMed] [Google Scholar]
- Troshin P., Procter J., Barton G. (2011). Java bioinformatics analysis web services for multiple sequence alignment—JABAWS:MSA. Bioinf. (Oxford Engl.) 27, 2001–2002. 10.1093/bioinformatics/btr304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Tang H., Debarry J. D., Tan X., Li J., Wang X., et al. (2012). MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, 1–14. 10.1093/nar/gkr1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Gao C., Bian X., Zhao S., Zhao C., Xia H., et al. (2016). Genome-wide identification and comparative analysis of cytosine-5 DNA methyltransferase and demethylase families in wild and cultivated peanut. Front. Plant Sci. 7, 7. 10.3389/fpls.2016.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Tang W., Hu Y., Zhang Y., Sun J., Guo X., et al. (2019). A MYB/bHLH complex regulates tissue-specific anthocyanin biosynthesis in the inner pericarp of red-centered kiwifruit Actinidia chinensis cv. Hongyang. Plant J. 99 (2), 359–378. 10.1111/tpj.14330 [DOI] [PubMed] [Google Scholar]
- Waterhouse A., Procter J., Martin D., Clamp M., Barton G. (2009). Jalview version 2: A Multiple Sequence Alignment and Analysis Workbench. Bioinf. (Oxford Engl.) 25, 1189–1191. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., et al. (2018). SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. 10.1093/nar/gky427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler T. J., Eddy S. R. (2013). Nhmmer: DNA homology search with profile HMMs. Bioinformatics 29, 2487–2489. 10.1093/bioinformatics/btt403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W., Custard R. D., Brown R. C., Lemmon B. E., Harada J. J., Goldberg R. B., et al. (2006). DNA methylation is critical for Arabidopsis embroyogenesis and seed viability. Plant Cell 18, 805–814. 10.1105/tpc.105.038836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (2007). PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Yelina N., Lambing C., Hardcastle T., Zhao X., Henderson I. (2015). DNA methylation epigenetically silences crossover hot spots and controls chromosomal domains of meiotic recombination in Arabidopsis. Genes Dev. 29, 2183–2202. 10.1101/gad.270876.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A., Kim M. Y., Silva P., Rodrigues J. A., Dotson B., Brooks M. D., et al. (2010). Local DNA hypomethylation activates genes in rice endosperm. Proc. Natl. Acad. Sci. U S A 107 (43), 18729–18734 . 10.1073/pnas.1009695107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Li J., Zhao X.-Q., Wang J., Wong G., Yu J. (2006). KaKs_Calculator: Calculating Ka and Ks Through Model Selection and Model Averaging. Genomics Proteomics Bioinf. 4, 259–263. 10.1016/S1672-0229(07)60007-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Gao S., Lercher M., Hu S., Chen W.-H. (2012). EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 40, W569–W572. 10.1093/nar/gks576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Fei Z., Chen Y.- R., Zheng Y., Huang M., Vrebalov J., et al. (2013). Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 31. 10.1038/nbt.2462 [DOI] [PubMed] [Google Scholar]
- Zhu J.-K. (2009). Active DNA methylation mediated by DNA glycosylases. Annu. Rev. Genet. 43, 143–166. 10.1146/annurev-genet-102108-134205 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.