Abstract
Ras-related protein Rab-31 (RAB31), a small guanosine 5′-triphosphate-binding protein, is a member of the Rab family and has been demonstrated to serve an oncogenic role in several common types of human cancer. However, the function of RAB31 in osteosarcoma (OS) has not been previously studied. The present study identified that the expression levels of RAB31 were significantly higher in OS tissue samples compared with matched adjacent non-tumor tissue samples, and high RAB31 expression was associated with malignant progression and a poor prognosis for patients with OS. Furthermore, it was identified that the expression levels of RAB31 were increased in OS cell lines compared with normal osteoblast cells. Silencing of RAB31 expression significantly inhibited OS cell proliferation, cell cycle progression, migration and invasion, and significantly increased the rate of cell apoptosis. In addition, the present study used a luciferase reporter assay to demonstrate that RAB31 was a direct target gene of microRNA-26b (miR-26b), which is a known tumor suppressor in OS. The expression levels of RAB31 were negatively associated with miR-26b expression in OS cells. Finally, miR-26b was demonstrated to be significantly decreased in OS tissues compared with adjacent non-tumor tissues, and an inverse correlation was observed between the expression levels of RAB31 and miR-26b in OS tissues. In summary, to the best of our knowledge, the present study is the first to report that RAB31 is a target gene of miR-26b, and silencing of RAB31 may inhibit OS growth and progression.
Keywords: ras-related protein Rab-31, osteosarcoma, microRNA, oncogene
Introduction
In 2015, osteosarcoma (OS) was reported as the most common type of malignancy of the bone, accounting for 5% of all pediatric tumors and 8.9% of cancer-associated mortality in children worldwide (1,2). OS is predominantly caused by metaphysis of the long bones (3). In 2015, the 5-year survival rate of patients with OS was reported to be 60–70% worldwide, due to improvements in chemotherapy and surgical resection (1–4). However, the prognosis of OS patients with distant metastasis remains poor, with a reported 5-year survival rate of only 20% worldwide in 2015 (1–4). Therefore, there is an urgent requirement to investigate the molecular mechanisms underlying OS progression, which may assist with the development of novel treatment strategies for OS.
The small GTPase proteins of the Rab family have been demonstrated to serve key roles in the development and malignant progression of numerous types of human cancer (5,6). Ras-related protein Rab-31 (RAB31), a member of the Rab5 subfamily, is a lipid-anchored protein consisting of 194 amino acids, which localizes to the cytoplasmic side of the cell membrane and serves essential roles in vesicle and granule targeting (7). Previous studies have reported that RAB31 serves an oncogenic role in human cancer types (8–10). For example, Pan et al (8) used glioblastoma and cervical cancer cell lines to investigate the function of RAB31 in cancer progression, which revealed that RAB31 promotes cancer cell proliferation, migration and epithelial-mesenchymal transition, and inhibit cell apoptosis. Knockdown of RAB31 was identified to suppress tumor growth in vivo. Furthermore, Sui et al (9) demonstrated that Rab31 promotes hepatocellular carcinoma progression by inhibiting cell apoptosis induced by the PI3K/AKT/Bcl-2/Bax pathway. Notably, Grismayer et al (10) identified that overexpression of RAB31 in breast cancer cells causes a switch from an invasive to a proliferative phenotype. These findings suggest that the different functions of RAB31 may be tumor-specific. However, to the best of our knowledge, the expression and function of RAB31 in OS has not previously been reported.
MicroRNAs (miRNAs) are a class of small non-coding RNAs that contain 22–25 nucleotides and regulate gene expression by binding to a complementary sequence within the 3′-untranslated region (3′UTR) of their target mRNA, which leads to translation repression or RNA degradation (11,12). By affecting the expression of their target genes, a large number of miRNAs have been demonstrated to serve key roles in various biological processes, including cell survival, differentiation, proliferation, cell cycle, apoptosis, migration and invasion (13–15). Recently, numerous miRNAs have been identified to be dysregulated in OS and to serve a promoting or suppressive role (16–19). For example, miR-195-5p inhibits OS cell proliferation and invasion by suppressing the expression of naked cuticle homolog 1 (19).
Previously, miR-26b has been reported to function as a tumor suppressor (20–23). Li et al (22) reported that miR-26b inhibits the metastasis of colorectal cancer by inhibiting fucosyltransferase 4. miR-26b has also been identified to inhibit proliferation and invasion of neuroglioma cells by downregulating Bcl-2 expression and enhancing caspase-3 activity (23). In OS, miR-26b has been reported to suppress cancer cell proliferation, migration and invasion, and induce OS cell apoptosis by inhibiting glycolysis (20). Furthermore, miR-26b can inhibit OS cell migration and invasion by targeting 6-phosphofructo-2-kinase/fructose-2-6-biphosphatase (PFKFB3), connective tissue growth factor (CTGF) and SMAD family member 1 (Smad1) (21,24). However, to the best of our knowledge, the association between miR-26b and RAB31 has not previously been studied. Therefore, the present study aimed to evaluate the clinical significance of RAB31 expression in OS. In addition, the regulatory role of RAB31 in the malignant phenotype of OS cells was investigated.
Materials and methods
Clinical tissue samples
The present study was approved by the Ethics Committee of The Second Affiliated Hospital of Nanchang University (Nanchang, China) and written informed consent was obtained from all patients. OS tissue samples and matched adjacent non-tumor tissue samples were collected from 54 patients with OS at the Second Affiliated Hospital of Nanchang University between May 2010 and May 2013. The patients included 33 males and 21 females, with a mean age of 18.8 years (range, 10–26 years). The clinical characteristics of the patients are summarized in Table I. No patients received radiotherapy or chemotherapy prior to surgery. All tissues samples were stored at −80°C prior to use.
Table I.
Association between RAB31 expression and clinicopathological characteristics of patients with osteosarcoma.
| Characteristic | Cases (n=54) | Low RAB31 expression (n=33) | High RAB31 expression (n=21) | P-value |
|---|---|---|---|---|
| Age, years | 0.784 | |||
| <20 | 22 | 14 | 8 | |
| ≥20 | 32 | 19 | 13 | |
| Sex | 0.575 | |||
| Male | 33 | 19 | 14 | |
| Female | 21 | 14 | 7 | |
| Tumor size, cm | 0.092 | |||
| <8 | 24 | 18 | 6 | |
| ≥8 | 30 | 15 | 15 | |
| Location | 0.772 | |||
| Femur or tibia | 37 | 22 | 15 | |
| Other | 17 | 11 | 6 | |
| Lung metastasis | 0.001a | |||
| No | 38 | 29 | 9 | |
| Yes | 16 | 4 | 12 | |
| TNM stage | 0.018a | |||
| I/IIA | 19 | 16 | 3 | |
| IIB/III | 35 | 17 | 18 | |
| Serum lactate dehydrogenase | 0.151 | |||
| Normal | 20 | 15 | 5 | |
| Elevated | 34 | 18 | 16 | |
| Serum alkaline phosphatase | 0.168 | |||
| Normal | 22 | 16 | 6 | |
| Elevated | 32 | 17 | 15 |
Statistically significant. RAB31, ras-related protein Rab-31; TNM, tumor-node-metastatsis.
Cell culture
The normal human osteoblastic cell line hFOB 1.19 and the human OS cell lines SAOS2, U2OS, MG63 and HOS were purchased from The Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). hFOB 1.19 cells were cultured in Dulbecco's modified Eagle's medium (DMEM)/F12 (Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2. OS cells were maintained in DMEM with 10% FBS, 100 U/ml penicillin and 100 U/ml streptomycin at 37°C with 5% CO2.
Cell transfection
Cells were cultured at 37°C with 5% CO2 in a 6-well plate at a density of ~1×106 cells/well. When a confluency of 70–80% was reached, the cells were transfected with 100 nM negative control (NC) small interfering (si)RNA (cat. no. AM4611; Thermo Fisher Scientific, Inc.), RAB31 siRNA (cat. no. AM16708; Thermo Fisher Scientific, Inc.), miR-NC (cat. no. 4464058; Thermo Fisher Scientific, Inc.), miR-26b mimic (cat. no. 4464066; Thermo Fisher Scientific, Inc.), NC inhibitor (cat. no. 4464077; Thermo Fisher Scientific, Inc.) or miR-26b inhibitor (cat. no. 4464084; Thermo Fisher Scientific, Inc.) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Following incubation for 48 h at 37°C, subsequent experiments were performed.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from tissue samples and cells (1×106) using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.). RT was performed using miScript Reverse Transcription kit (Qiagen GmbH), according to the manufacturer's protocol. qPCR was conducted using QuantiTect SYBR-Green RT-PCR kit (Qiagen GmbH) with an ABI 7500 fast real-time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling conditions for PCR were as follows: 95°C for 1 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 30 sec. GAPDH or U6 was used as the internal reference for mRNA and miRNA, respectively. Data were quantified using the 2−ΔΔCq method (25). The primer sequences were as follows: RAB31 forward, 5′-GGGGTTGGGAAATCAAGCATC-3′ and reverse, 5′-GCCAATGAATGAAACCGTTCCT-3′; GAPDH forward, 5′-ACAACTTTGGTATCGTGGAAGG-3′ and reverse, 5′-GCCATCACGCCACAGTTTC-3′. Primers for U6 (cat. no. HmiRQP9001) and miR-16 (cat. no. HmiRQP0356) were obtained from Guangzhou FulenGen, Co., Ltd.
Western blot analysis
Tissue samples and cells (1×107) were solubilized with radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology). The protein concentration was measured using a BCA protein assay kit (Beyotime Institute of Biotechnology). Total protein (50 µg) was separated by 10% SDS-PAGE and then transferred to polyvinylidene difluoride membranes (EMD Millipore). The membranes were blocked with 5% non-fat milk in 0.1% TBS/Tween-20 at room temperature for 1 h, followed by incubation with rabbit anti-human RAB31 (dilution, 1:100; cat. no. ab230881; Abcam), or rabbit anti-human GAPDH (dilution, 1:200; cat. no. ab181602; Abcam) primary antibodies at room temperature for 3 h. Subsequently, the membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (dilution, 1:5,000; cat. no. ab6721; Abcam) at room temperature for 1 h. Enhanced chemiluminescent reagent (Pierce; Thermo Fisher Scientific, Inc.) was used to detect the signals on the membranes. ImageJ software (v1.46; National Institutes of Health) was used for densitometry analysis.
Cell proliferation assay
Cell proliferation was investigated using Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.), according to the manufacturer's protocol. A total of 48 h after transfection, cells (3,000 cells/well) were seeded into 96-well plates and cultured for 0, 24, 48 or 72 h. The optical density value at 450 nM was measured using a microplate reader.
Cell apoptosis assay
The transfected cells were harvested and washed twice with ice-cold PBS. Subsequently, 1×106 cells were resuspended with 300 µl binding buffer. Following staining with Annexin V-fluorescein isothiocyanate and propidium iodide (BD Biosciences), according to the manufacturer's protocol, apoptotic cells were analyzed using a flow cytometer and Accuri C6 Software (version 1.0; BD Biosciences).
Wound healing assay
The transfected cells (1×106 cells per well) were cultured in 6-well plates for 24 h. Subsequently, an artificial wound was created using a sterile 10 µl pipette tip in the confluent cell monolayer. Photographs were captured at 0 and 24 h using an inverted light microscope (magnification, ×40).
Cell invasion assay
Matrigel (BD Biosciences) was diluted with serum-free DMEM and added to the upper chamber of a 24-well Transwell™ chamber (Corning Inc.), which was then incubated at 37°C for 1 h. Cells were diluted to a density of 2×105 cells/ml with serum-free DMEM. Subsequently, 100 µl cell suspension was added to the upper chamber and 700 µl DMEM containing 10% FBS was added to the bottom chamber. Following incubation at 37°C for 24 h, the chamber was washed with PBS. The cells were then fixed with 4% paraformaldehyde at room temperature for 30 min, stained with crystal violet at room temperature for 30 min and photographs were obtained using an inverted light microscope (magnification, ×400).
Bioinformatics analysis and luciferase reporter assay
TargetScan software version 7.1 was used for bioinformatics analysis (26). The wild-type (WT) or mutant type (MUT) 3′-UTR of RAB31 was inserted downstream of the luciferase reporter gene in the pMIR-REPORT vector (Thermo Fisher Scientific, Inc.), generating WT-RAB31 and MUT-RAB31. MG63 and U2OS cells were co-transfected with miR-26b mimics/miR-NC, WT-RAB31/MUT-RAB31 and pRL-SV40 (Promega Corporation) expressing Renilla luciferase. Following 48 h incubation at 37°C with 5% CO2, the luciferase activity was measured using the Dual-Luciferase® Reporter Assay System (Promega Corporation).
Statistical analysis
All data are presented as the mean ± standard deviation from a minimum of three independent experiments. Statistical analysis was performed using SPSS 20.0 (IBM Corp.). Student's t-test was used for comparisons between two groups. Multiple comparisons were performed using one-way analysis of variance followed by Tukey's test. The patients with OS were divided into a high RAB31 expression group and low RAB31 expression group using the median expression level of RAB31 (1.91) as the cut-off, and χ2 test was used to perform statistical analysis in Table I. Correlations between the mRNA expression levels of miR-26b and RAB3 were analyzed using Pearson's correlation coefficient. P<0.05 was considered to indicate a statistically significant difference.
Results
Increased expression levels of RAB31 are associated with OS progression
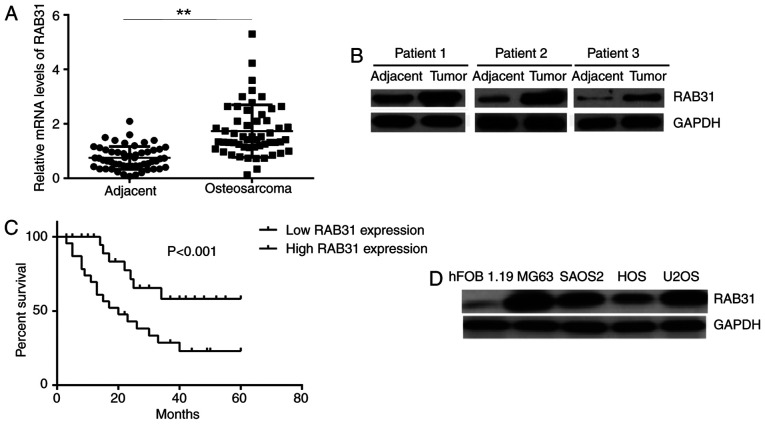
First, the mRNA and protein expression levels of RAB31 were examined in tissue samples from OS patients. RT-qPCR and western blot analysis demonstrated that the expression levels of RAB31 were significantly higher in OS tissues compared with adjacent non-tumor tissues (Fig. 1A and B). The patients with OS were then divided into a high RAB31 expression group and low RAB31 expression group using the median expression level of RAB31 (1.91) as the cut-off. As presented in Table I, high expression of RAB31 was identified to be significantly associated with lung metastasis and advanced clinical stage. Furthermore, the patients with high RAB31 expression exhibited a significantly shorter survival time compared with those with low RAB31 expression (Fig. 1C), which suggested that RAB31 upregulation was associated with a poor prognosis for patients with OS. In addition, the protein expression levels of RAB31 were markedly higher in the OS cell lines SAOS2, U2OS, MG63 and HOS compared with the normal human osteoblastic cell line hFOB 1.19 (Fig. 1D). MG63 and U2OS cells exhibited the highest expression levels of RAB31 protein (Fig. 1D); these cell lines were therefore selected for subsequent experiments.
Figure 1.
RAB31 expression levels in OS tissues and cell lines. (A) mRNA and (B) protein expression levels of RAB31 were higher in OS tissues compared with adjacent non-tumor tissues. **P<0.01. (C) Patients with OS with high RAB31 expression exhibited a significantly shorter survival time compared with patients with low RAB31 expression. (D) Protein expression levels of RAB31 were higher in OS cell lines compared with the osteoblastic cell line hFOB 1.19. RAB31, Ras-related protein Rab-31; OS, osteosarcoma.
Silencing of RAB31 inhibits OS cell proliferation, cell cycle progression, migration and invasion, and induces cell apoptosis
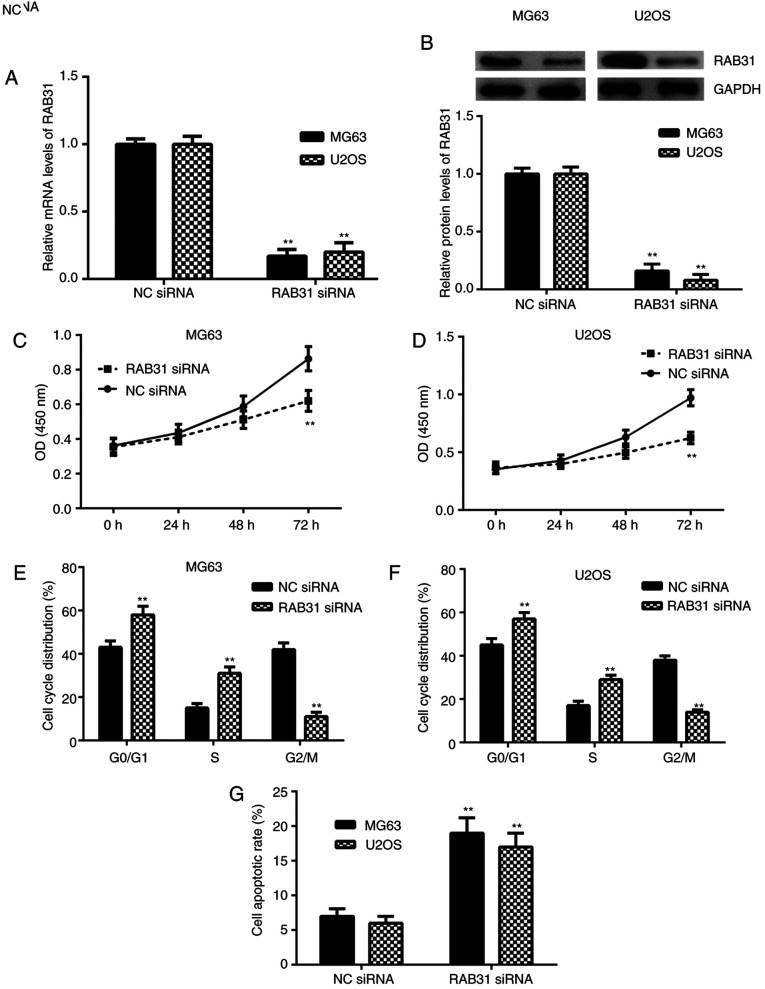
To inhibit the expression of RAB31, MG63 and U2OS cells were transfected with RAB31 siRNA. As presented in Fig. 2A and B, the mRNA and protein expression levels of RAB31 were significantly lower in MG63 and U2OS cells transfected with RAB31 siRNA compared with the NC siRNA group. A CCK-8 assay was then performed to examine cell proliferation. As presented in Fig. 2C and D, knockdown of RAB31 significantly inhibited OS cell proliferation. The cell cycle distribution and cell apoptosis were then examined by flow cytometry. The data from this experiment indicated that silencing of RAB31 was significantly associated with cell cycle arrest at the G1 stage and significantly promoted OS cell apoptosis (Fig. 2E-H). The aforementioned results suggested that RAB31 may serve a promoting role in OS growth.
Figure 2.
Effects of RAB31 silencing in OS cells. (A) mRNA and (B) protein expression levels of RAB31 were significantly lower in MG63 and U2OS cells transfected with RAB31 siRNA compared with cells transfected with NC siRNA. (C) Knockdown of RAB31 significantly inhibited proliferation of MG63 and (D) U20S cells. (E) Knockdown of RAB31 caused cell cycle arrest at the G1 stage in MG63 and (F) U2OS cells. (G) Knockdown of RAB31 induced MG63 and U2OS cell apoptosis. Data are presented as the mean ± standard deviation. **P<0.01 vs. NC siRNA for the same cell line. RAB31, Ras-related protein Rab-31; siRNA, small interfering RNA; NC, negative control; OD, optical density.
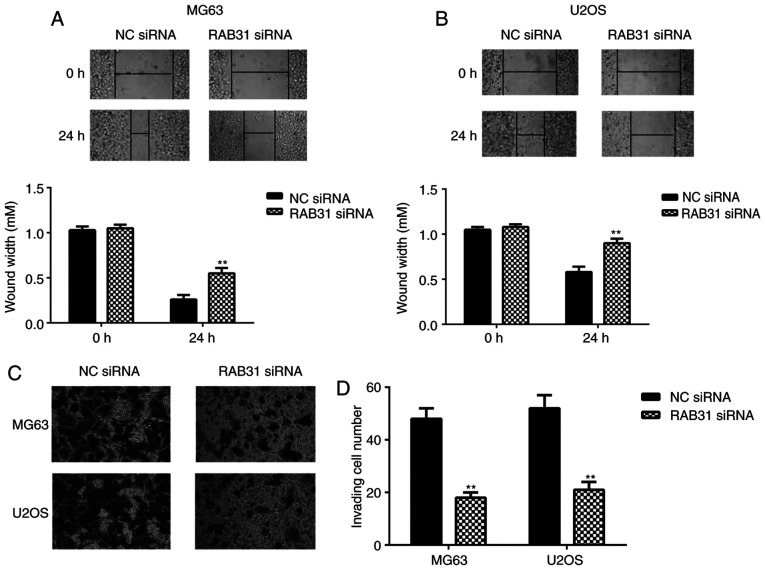
To study the potential role of RAB31 in OS metastasis, a wound healing assay and Transwell assay were performed to examine cell migration and invasion, respectively. As presented in Fig. 3, silencing of RAB31 expression significantly inhibited the migration and invasion of MG63 and U2OS cells compared with the NC siRNA group. In summary, RAB31 silencing inhibited OS cell proliferation, cell cycle progression, migration and invasion, and induced cell apoptosis.
Figure 3.
Effects of RAB31 silencing in OS migration and invasion. (A) Silencing of RAB31 expression significantly inhibited the migration of MG63 and (B) U2OS cells compared with the NC siRNA groups. (C) Representative images and (D) quantification showing that silencing of RAB31 significantly inhibited the invasion of MG63 and U2OS cells compared with the NC siRNA groups. Data are presented as the mean ± standard deviation. **P<0.01 vs. NC siRNA. RAB31, Ras-related protein Rab-31; siRNA, small interfering RNA; NC, negative control. Magnification for cell migration ×40 and for cell invasion ×400.
RAB31 is a target gene of miR-26b in OS cells
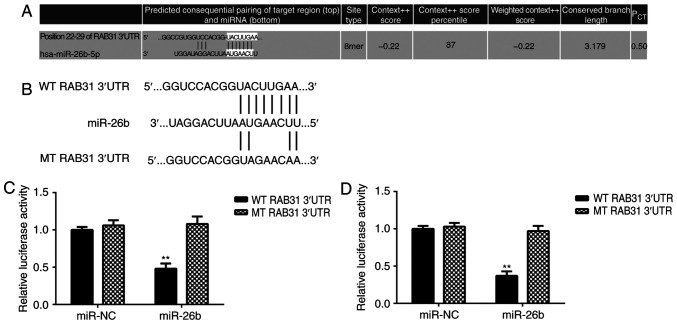
TargetScan software data predicted that RAB31 is a putative target gene of miR-26b (Fig. 4A). To confirm the association between RAB31 and miR-26b, wild-type (WT) and mutant (MT) RAB31 luciferase reporter plasmids were generated (Fig. 3B) and a luciferase reporter gene assay was performed. The data demonstrated that transfection with miR-26b mimic significantly inhibited the luciferase activity in MG63 and U2OS cells transfected with WT RAB31 3′UTR reporter plasmid; however, no effect was observed on the luciferase activity in cells transfected with the MT RAB31 3′UTR reporter plasmid (Fig. 4C and D). This indicates that RAB31 is a target gene of miR-26b in OS cells.
Figure 4.
RAB31 is a target gene of miR-26b in OS cells. (A) TargetScan analysis results predicted that RAB31 may be a putative target gene of miR-26b. (B) WT and MT RAB31 luciferase reporter plasmids were generated. (C) Transfection with miR-26b mimic significantly inhibited the luciferase activity in MG63 and (D) U2OS cells expressing the WT RAB31 3′UTR reporter plasmid; however, no effect was observed in cells expressing the MT reporter plasmid. Data are presented as the mean ± standard deviation. **P<0.01 vs. miR-NC. RAB31, Ras-related protein Rab-31; miR, microRNA; NC, negative control; WT, wild-type; MT, mutant; 3′UTR, 3′-untranslated region.
RAB31 is negatively regulated by miR-26b in OS cells
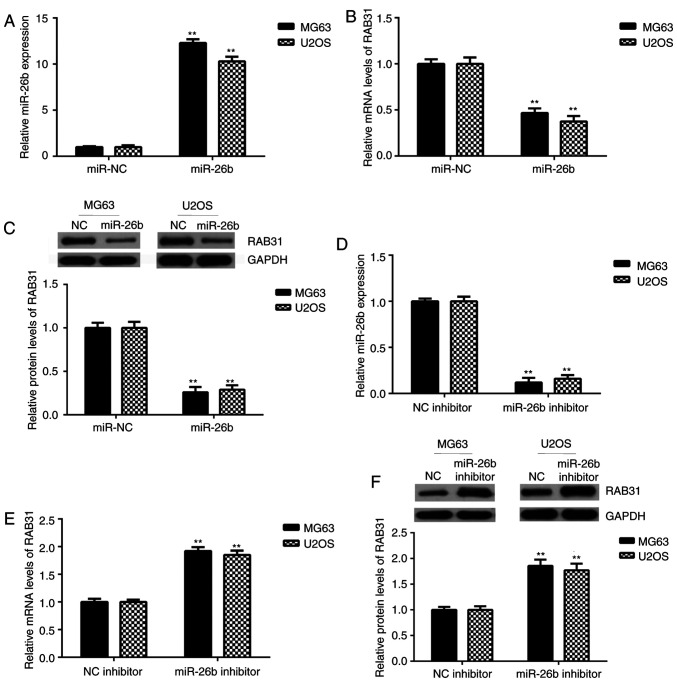
The effects of miR-26b on RAB31 expression were then investigated in OS cells. MG63 and U2OS cells were transfected with miR-26b mimic or miR-NC. Following transfection, miR-26b expression was significantly upregulated in the miR-26b mimic group compared with the miR-NC group (Fig. 5A). Further experiments revealed that the mRNA and protein expression levels of RAB31 were significantly decreased following miR-26b overexpression (Fig. 5B and C). To further confirm these findings, MG63 and U2OS cells were transfected with miR-26b inhibitor or NC inhibitor. RT-qPCR analysis revealed that miR-26b was significantly downregulated in the miR-26b inhibitor-transfected group compared with the NC inhibitor-transfected group (Fig. 5D). As presented in Fig. 5E and F, the mRNA and protein expression levels of RAB31 were significantly increased in the miR-26b inhibitor-transfected group compared with the NC inhibitor-transfected group. These data indicated that RAB31 was negatively regulated by miR-26b in OS cells.
Figure 5.
RAB31 is negatively regulated by miR-26b in OS cells. (A) miR-26b was significantly upregulated in MG63 and U2OS cells following transfection with miR-26b mimic compared with the miR-NC group. **P<0.01 vs. miR-NC. (B) mRNA and (C) protein expression levels of RAB31 were significantly decreased following miR-26b overexpression. **P<0.01 vs. miR-NC. (D) miR-26b was significantly downregulated in MG63 and U2OS cells following transfection with miR-26b inhibitor when compared with the NC inhibitor group. **P<0.01 vs. NC inhibitor. (E) mRNA and (F) protein expression levels of RAB31 were significantly decreased following miR-26b inhibition. **P<0.01 vs. NC inhibitor. Data are presented as the mean ± standard deviation. miR, microRNA; NC, negative control; RAB31, Ras-related protein Rab-31.
miR-26b expression is inversely correlated with RAB31 expression in OS tissues
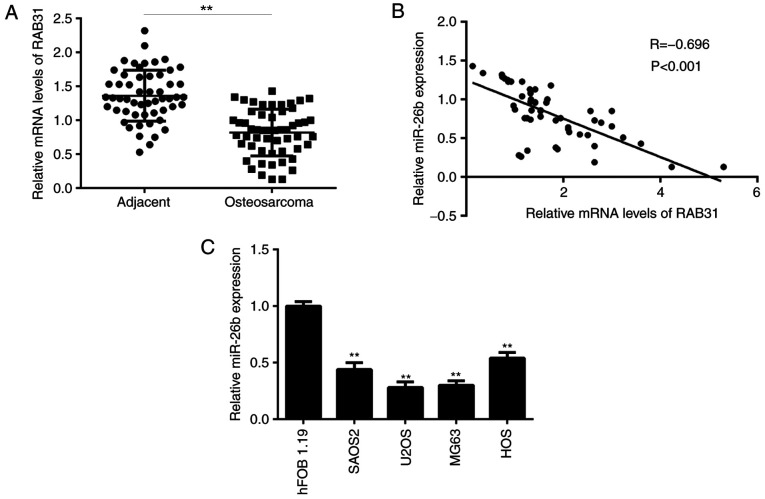
RT-qPCR analysis identified that miR-26b levels were significantly lower in OS tissues compared with matched adjacent non-tumor tissues (Fig. 6A). Additionally, an inverse correlation was identified between the miR-26b and RAB31 expression levels in OS tissues (Fig. 6B), which suggested that the increased expression levels of RAB31 in OS tissues may be due to downregulation of miR-26b. Furthermore, miR-26b levels were demonstrated to be decreased in OS cell lines compared with hFOB 1.19 cells (Fig. 6C).
Figure 6.
miR-26b expression is inversely correlated with RAB31 expression in OS tissues. (A) miR-26b levels are significantly lower in osteosarcoma tissues compared with adjacent non-tumor tissues. **P<0.01 vs. adjacent. (B) A significant inverse correlation was identified between miR-26b and RAB31 expression levels in osteosarcoma tissues. (C) miR-26b levels were significantly lower in osteosarcoma cell lines compared with the osteoblastic cell line hFOB 1.19. **P<0.01 vs. hFOB 1.19. Data are presented as the mean ± standard deviation. miR, microRNA; RAB31, Ras-related protein Rab-31.
Discussion
To the best of our knowledge, the function of RAB31 in OS has previously been unclear. The present study revealed that RAB31 was upregulated in OS tissues and cell lines, and that high RAB31 expression levels were associated with malignant progression and a poor prognosis for patients with OS. Silencing of RAB31 expression significantly inhibited OS cell proliferation, cell cycle progression, migration and invasion, and induced cell apoptosis. RAB31 was then identified as a direct target gene of miR-26b. Additionally, the expression of RAB31 was negatively regulated by miR-26b in OS cells. Furthermore, miR-26b was downregulated in OS tissues and reversely correlated to the expression levels of RAB31.
Initially, RAB31 was reported to be upregulated in breast cancer and high expression of RAB31 is significantly associated with worse outcome for patients (27). Furthermore, the mucin 1 C-terminal subunit (MUC1-C) oncoprotein and RAB31 have been demonstrated to function in an autoinductive loop that contributes to overexpression of MUC1-C in breast cancer cells (28). Grismayer et al (10) reported that RAB31 overexpression increases breast cancer cell proliferation, reduces adhesion and invasion in vitro, and results in decreased lung metastasis formation in vivo. In addition, RAB31 has been revealed to promote hepatocellular carcinoma progression by inhibiting tumor cell apoptosis induced by the PI3K/AKT/Bcl-2/Bax pathway (9). However, no previous study has focused on the function of RAB31 in OS.
The present study first examined the expression levels of RAB31 in OS tissues and cell lines. The data demonstrated that RAB31 was significantly upregulated in OS tissues compared with adjacent non-tumor tissues. Furthermore, it was identified that high expression of RAB31 was significantly associated with lung metastasis and advanced clinical stage in patient with OS, which suggested that an upregulation of RAB31 may contribute to OS progression. Additionally, patients with OS exhibiting high RAB31 expression had shorter survival times compared with patients exhibiting low RAB31 expression, which indicates that RAB31 may be used as a novel predicator for the prognosis of patients with OS. Consistent with the clinical data, RAB31 was also significantly upregulated in OS cell lines compared with the normal human osteoblastic hFOB 1.19 cell line. To further clarify the function of RAB31 in OS, two common OS cell lines MG63 and U2OS were used to perform in vitro experiments. The results demonstrated that silencing of RAB31 expression significantly reduced the proliferation, cell cycle progression, migration and invasion of OS cells, and induced cell apoptosis. Therefore, the current results suggested that RAB31 may have a promoting role in regulating the malignant phenotype of OS cells. These findings indicate that targeting RAB31 may be a promising strategy for the treatment of OS.
The mechanism underlying RAB31 expression in OS was further investigated in the current study. As a large number of genes are regulated by miRNAs (11,12), bioinformatics analysis was used to predict the putative miRNAs that may directly target RAB31. Among the predicted miRNAs, miR-26b was selected as it has been reported to function as a tumor suppressor in OS (24). Duan et al (24) identified that miR-26b inhibits OS metastasis by targeting CTGF and Smad1. Zheng et al (21) reported that miR-26b represses OS cell migration and invasion by inhibiting the expression of PFKFB3. In addition, miR-26b has been demonstrated to inhibit OS cell proliferation, migration and invasion, and induce cell apoptosis by inhibiting PFKFB3-driven glycolysis (20). To further clarify the association between miR-26b and RAB31, the present study performed a luciferase reporter gene assay, which confirmed that RAB31 was a target gene of miR-26b. In addition, it was identified that the expression of RAB31 was negatively regulated by miR-26b in OS cells and that miR-26b levels were inversely correlated with the expression levels of RAB31 in OS tissues. These findings suggested that downregulation of miR-26b may contribute to the upregulation of RAB31 in OS. A limitation of the current study is that the effects of miR-26b on proliferation, apoptosis, migration and invasion were not investigated; this should be performed in future studies.
In conclusion, to the best of our knowledge, the present study was the first to demonstrate that RAB31 was upregulated in OS, was significantly associated with malignant progression and a poor prognosis for patients with OS, and that RAB31 silencing inhibited the malignant phenotype of OS cells. Furthermore, a novel association between RAB31 and miR-26b, a tumor suppressor in OS, was identified. Therefore, the present results suggested that RAB31 may be a promising therapeutic target for OS.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
QW designed the study and wrote the manuscript. QF collected sample tissues and performed statistical analysis. YX and XL performed the experiments. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of The Second Affiliated Hospital of Nanchang University (Nanchang, China). Written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Saraf AJ, Fenger JM, Roberts RD. Osteosarcoma: Accelerating Progress Makes for a Hopeful Future. Front Oncol. 2018;8:4. doi: 10.3389/fonc.2018.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harrison DJ, Geller DS, Gill JD, Lewis VO, Gorlick R. Current and future therapeutic approaches for osteosarcoma. Expert Rev Anticancer Ther. 2018;18:39–50. doi: 10.1080/14737140.2018.1413939. [DOI] [PubMed] [Google Scholar]
- 5.Shaughnessy R, Echard A. Rab35 GTPase and cancer: Linking membrane trafficking to tumorigenesis. Traffic. 2018;19:247–252. doi: 10.1111/tra.12546. [DOI] [PubMed] [Google Scholar]
- 6.Wang S, Hu C, Wu F, He S. Rab25 GTPase: Functional roles in cancer. Oncotarget. 2017;8:64591–64599. doi: 10.18632/oncotarget.19571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bao X, Faris AE, Jang EK, Haslam RJ. Molecular cloning, bacterial expression and properties of Rab31 and Rab32. Eur J Biochem. 2002;269:259–271. doi: 10.1046/j.0014-2956.2001.02645.x. [DOI] [PubMed] [Google Scholar]
- 8.Pan Y, Zhang Y, Chen L, Liu Y, Feng Y, Yan J. The Critical role of Rab31 in cell proliferation and apoptosis in cancer progression. Mol Neurobiol. 2016;53:4431–4437. doi: 10.1007/s12035-015-9378-9. [DOI] [PubMed] [Google Scholar]
- 9.Sui Y, Zheng X, Zhao D. Rab31 promoted hepatocellular carcinoma (HCC) progression via inhibition of cell apoptosis induced by PI3K/AKT/Bcl-2/BAX pathway. Tumour Biol. 2015;36:8661–8670. doi: 10.1007/s13277-015-3626-5. [DOI] [PubMed] [Google Scholar]
- 10.Grismayer B, Sölch S, Seubert B, Kirchner T, Schäfer S, Baretton G, Schmitt M, Luther T, Krüger A, Kotzsch M, et al. Rab31 expression levels modulate tumor-relevant characteristics of breast cancer cells. Mol Cancer. 2012;11:62. doi: 10.1186/1476-4598-11-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moss EG. MicroRNAs: Hidden in the genome. Curr Biol. 2002;12:R138–R140. doi: 10.1016/S0960-9822(02)00708-X. [DOI] [PubMed] [Google Scholar]
- 12.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L, Xu J, Yang G, Li H, Guo X. miR-202 inhibits cell proliferation, migration, and invasion by targeting EGFR in human bladder cancer. Oncol Res. 2018;26:949–957. doi: 10.3727/096504018X15149787144385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang C, Wu K, Wang S, Wei G. Long non-coding RNA XIST promotes osteosarcoma progression by targeting YAP via miR-195-5p. J Cell Biochem. 2018;119:5646–5656. doi: 10.1002/jcb.26743. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Y, Yang C, Wang K, Liu X, Liu Q. MicroRNA-33b inhibits the proliferation and migration of osteosarcoma cells via targeting hypoxia-inducible factor-1α. Oncol Res. 2017;25:397–405. doi: 10.3727/096504016X14743337535446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang R, Xia T. Long non-coding RNA XIST regulates PDCD4 expression by interacting with miR-21-5p and inhibits osteosarcoma cell growth and metastasis. Int J Oncol. 2017;51:1460–1470. doi: 10.3892/ijo.2017.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qu Q, Chu X, Wang P. MicroRNA-195-5p suppresses osteosarcoma cell proliferation and invasion by suppressing naked cuticle homolog 1. Cell Biol Int. 2017;41:287–295. doi: 10.1002/cbin.10723. [DOI] [PubMed] [Google Scholar]
- 20.Du JY, Wang LF, Wang Q, Yu LD. miR-26b inhibits proliferation, migration, invasion and apoptosis induction via the downregulation of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3 driven glycolysis in osteosarcoma cells. Oncol Rep. 2015;33:1890–1898. doi: 10.3892/or.2015.3797. [DOI] [PubMed] [Google Scholar]
- 21.Zheng WD, Zhou FL, Lin N. MicroRNA-26b inhibits osteosarcoma cell migration and invasion by down-regulating PFKFB3 expression. Genet Mol Res. 2015;14:16872–16879. doi: 10.4238/2015.December.14.14. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, Sun Z, Liu B, Shan Y, Zhao L, Jia L. Tumor-suppressive miR-26a and miR-26b inhibit cell aggressiveness by regulating FUT4 in colorectal cancer. Cell Death Dis. 2017;8:e2892. doi: 10.1038/cddis.2017.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li YP, Dai WM, Huang Q, Jie YQ, Yu GF, Fan XF, Wu A, Mao DD. Effects of microRNA-26b on proliferation and invasion of glioma cells and related mechanisms. Mol Med Rep. 2017;16:4165–4170. doi: 10.3892/mmr.2017.7121. [DOI] [PubMed] [Google Scholar]
- 24.Duan G, Ren C, Zhang Y, Feng S. MicroRNA-26b inhibits metastasis of osteosarcoma via targeting CTGF and Smad1. Tumour Biol. 2015;36:6201–6209. doi: 10.1007/s13277-015-3305-6. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:4. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kotzsch M, Sieuwerts AM, Grosser M, Meye A, Fuessel S, Meijer-van Gelder ME, Smid M, Schmitt M, Baretton G, Luther T, et al. Urokinase receptor splice variant uPAR-del4/5-associated gene expression in breast cancer: Identification of rab31 as an independent prognostic factor. Breast Cancer Res Treat. 2008;111:229–240. doi: 10.1007/s10549-007-9782-6. [DOI] [PubMed] [Google Scholar]
- 28.Jin C, Rajabi H, Pitroda S, Li A, Kharbanda A, Weichselbaum R, Kufe D. Cooperative interaction between the MUC1-C oncoprotein and the Rab31 GTPase in estrogen receptor-positive breast cancer cells. PLoS One. 2012;7:e39432. doi: 10.1371/journal.pone.0039432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.