Abstract
The discovery and development of new and potentially nonaddictive pain therapeutics requires rapid, yet clinically relevant assays of nociception in preclinical models. A reliable and scalable automated scoring system for nocifensive behavior of mice in the formalin assay would dramatically lower the time and labor costs associated with experiments and reduce experimental variability. Here, we present a method that exploits machine learning techniques for video recordings that consists of three components: key point detection, per frame feature extraction using these key points, and classification of behavior using the GentleBoost algorithm. This approach to automation is flexible as different model classifiers or key points can be used with only small losses in accuracy. The adopted system identified the behavior of licking/biting of the hind paw with an accuracy that was comparable to a human observer (98% agreement) over 111 different short videos (total 284 min) at a resolution of 1 s. To test the system over longer experimental conditions, the responses of two inbred strains, C57BL/6NJ and C57BL/6J, were recorded over 90 min post formalin challenge. The automated system easily scored over 80 h of video and revealed strain differences in both response timing and amplitude. This machine learning scoring system provides the required accuracy, consistency, and ease of use that could make the formalin assay a feasible choice for large-scale genetic studies.
Keywords: Formalin nociception assay, neural network, licking behavior, automated behavior recognition, machine vision, computer vision
Introduction
The phenomenon of pain is a complex combination of physical information, emotional context, and personal subjective experience.1 It is not possible to directly measure pain in animals, as we do not have access to their subjective experiences, consequently many methods have been developed that quantify “nocifensive” behaviors, which are defined as behavioral responses to painful stimuli. Most nociception assays depend on a quick motor withdrawal reflex in response to a brief mechanical or thermal stimulation, and this simple movement is relatively easy to define and recognize,2–5 but such assays lack similarity to clinical pain. In mice, these assays are genetically poorly correlated with more clinically relevant chronic pain assays6,7 and are more closely associated with startle and reactivity traits.8 In contrast, the formalin test, originally developed for use with rats by Dubuisson and Dennis,9 was designed to monitor complex actions over an extended period, in response to chemically induced, localized inflammation. The irritant formalin is usually injected in one hind paw and then the animal is observed for nocifensive behaviors such as licking, biting, lifting, flicking, or clutching the paw.10–12 Formalin typically produces a biphasic response, with a short intense acute reaction (Phase I; from 0–10 min postinjection), a brief interphase of low response and then a sustained (Phase II) response, starting at about 10–15 min postinjection, increasing to a peak and then gradually subsiding, with an elevated response often still maintained at 60 min or more postinjection.13–15 This assay is a commonly used form of nonstimulus evoked spontaneous nocifensive behavior and the sustained nature of the behaviors are particularly pertinent to biological understanding of chronic pain.
The formalin assay, although well-accepted, relies on individual observers which makes it labor intensive, time consuming, and subjective as the different nocifensive behaviors observed are not always uniformly defined and recorded.4,15 Rating scales are subject to interobserver variability and some behaviors, such as favoring or lifting, are reportedly hard to score reliably in mice.16,17 Consequently, mouse behaviors scored are often restricted to licking/biting behaviors because they are easy to recognize and record.12,14–16 To reduce scoring bias, the formalin assay is usually videoed and then scoring is subsequently completed by one or more observers, typically an hour of video of a single mouse will take between 1.5 and 2 h to fully score. Time sampling methods that score prescribed portions of data have been developed10,15 to reduce the required manual effort. Sampling produces similar results to the full scoring methods10 but still requires considerable time investment and observer training.
Automated scoring can overcome some of these obstacles, and several studies have shown that nocifensive behaviors can be effectively scored. Methods to score the formalin assay for rats have included the use of force detectors,18 electromagnetic field detectors,19 and video20–22 with some success. Typically, these early studies of automation validated performance by demonstrating that their model could distinguish behaviors induced at different levels through the use of analgesics or formalin doses. Manual scoring comparisons were used to give general patterns of responses, but details of accuracy were not provided in terms of precision/recall or sensitivity/specificity measures which are the accepted standards for modeling papers today. Although we cannot fully assess the accuracy of these early methods, they did establish that in principle, these behaviors can be detected and scored automatically.
A video-based automated system is readily adoptable because the experiments are already typically recorded on video, the equipment is inexpensive and accessible, and it causes no additional stress by restricting animal movement. Jourdan et al.21 used video recordings of rats to measure change in pixel color to assess gross and fine motor actions. They were able to use these movements to distinguish between periods of locomotion and periods of smaller movements, which included grooming, licking, and biting. Mice present additional challenges to automated video scoring systems as they are considerably smaller than rats and move very quickly. However, recent advances in machine learning have led to the development of systems that can assess tiny differences in mouse facial expressions,23 grooming specific behaviors in mice,24 and with very high-speed video, accurately assess the rapid withdrawal reflex action used in many nociception assays.25 The behaviors of licking and biting induced by formalin are well-defined, easy to label, and are therefore well-suited for machine learning classification of mouse nociception.
In the first study to automate mouse nocifensive licking behavior, Hayashi et al.26 used two marked color points, on the abdomen and snout, to measure the changing distances between the points during an intracolonic-induced pain assay. They showed that a simple distance measure, estimated by tracking two points, could provide sufficient information to infer abdomen licking. All behaviors are essentially instantiated as a series of movements, and these can be represented as body parts changing in position over time. The licking behavior of the mouse in the formalin assay can take several distinct postural configurations, as the mouse may bend down toward the paw, hold the paw up, or rapidly move the paw, and it is likely that more than two points would be needed to accurately capture all behaviors. Machine learning techniques, using convolutional neural networks, can identify and track multiple specific body parts on animals thereby eliminating the need to add physical markers, fur bleaching, or dyes.27–30 The ability of these networks to accurately label numerous body parts allows for the calculation of many relative positions and the more complex representation of body needed for the formalin assay.
The genetic tractability of the mouse makes it an essential component in studies of pain and analgesia, and therefore, the development of automated nociception scoring in mice is critical for large-scale studies. Recent innovations in machine learning allow for accurate classification of specific mouse behaviors over the full length of any recorded video.31 Advantages of such a system providing scalability, clearly include the savings of time, labor, and information with no restricting sampling methods required. Equally important however, refining the method of scoring the formalin assay would result in greater reliability and reproducibility by improving the consistency of the measurements. The use of machine learning gives new opportunities to address the ethical requirements of replacement, reduction, and refinement. Here, we present an automated scoring system, based on supervised machine learning methods using recorded formalin assays performed on laboratory mice. The system was validated with extensive comparison to manual scoring. To assess the applicability to the widely used C57BL/6J-derived strains and to the International Mouse Phenotyping Consortium’s extensive collection of C57BL/6N-derived deletion mutants, a comparison of both strains was performed.
Methods
Animals
Mice were single sex, group-housed (3–5) with ad lib water and food under a 12-h light–dark schedule, and experiments were conducted in the light phase. Video data from 166 mice were used in training, testing, and validation of the model (The Jackson Laboratory: C57BL/6NJ=JR005304: male n = 53, female = 37, C57BL/6J=JR000664: male = 46, female = 30). Mice (age 11–17 weeks) were tested in 25 sessions between August 2018 and March 2019 and at the conclusion of each experimental session all mice were euthanized. All procedures and protocols were approved by The Jackson Laboratory Animal Care and Use Committee and were conducted in compliance with the National Institutes of Health Guideline for Care and Use of Laboratory Animals.
Video data collection
Video data of mouse behavior in response to a hind paw formalin injection were collected and used in training, testing, and validation of the automatic scoring system. A clear acrylic enclosure (22 cm L × 21.6 cm W × 12.7 cm H; IITC Life Science, catalog number 433) containing four testing arenas separated by opaque black walls (see Figures 1 and 2) was placed on a clear glass surface. A video camera (black and white, Bosch, Dinion) was placed directly below (16 cm) the glass floor of the enclosure to provide the best view of the paws and recording, under the control of Noldus software (Noldus media recorder v4), began with the empty enclosure. Four enclosures, each with one dedicated camera, were set up such that a total of 16 mice could be run simultaneously. The lighting varied between the four enclosures but was optimized to reduce glare and reflections with the addition of a white polycarbonate cover for the top of each enclosure (23.5 cm L × 12.1 cm W × 1 cm H; manufactured in-house). Video was recorded (30 frames per second: 704 × 480 pixels) for 90 min after the last mouse entered the arena. The choice to extend the video beyond 60 min was to ensure that any strain differences in the timing of peak behavior would be captured.
Figure 1.
Model components. Labels (a) and associated images (b) were used as input to train the DeepLabCut tracker. (a) The video frames were manually annotated for training. Each mouse was labeled (x,y pixel coordinates) with 12 points (mouth, nose, right front paw, left front paw, three points on each hind paw—outer, inner, and base; abdomen; and tailbase) and the inner enclosure walls were marked (at the ends and cross point). (b) Input to the model was consecutive video frames. Each enclosure had four arenas; in this image, the mouse in arena 4 is still in recovery from the anesthetic, and the other arenas show active mice. (c) The output of the keypoint tracker is 12 points per mouse, shown here with “skeleton” connections and “body/head” circles to orient the reader. (d) Relative location measures (66 paired distances and 15 angles; only one of each shown in the figure) were calculated for the body parts for every video frame. The putative frame of interest is highlighted and the measures for the preceding and following frames are shown (24 consecutive frames). The statistical inputs were calculated over windows of three different sizes (6, 11, and 21 frames), and these per frame features were used as the input to the classifier model.
Figure 2.
Tracking error. The tracking errors for a single test frame of the body-part tracking (average error of these four mice for body parts listed above are: 2.65, 1.84, 2.89, 1.01, 2.77, 1.67, 2.89, 3.19, 2.50, 2.36, 2.03, 3.88; the average grid error was 1.26 pixels).
Formalin was administered while the mice were under anesthesia to maximize the consistency of both the injection site and the volume delivered and to reduce stress for the mice. The right hind paw of the mouse was injected (intra-plantar) with 30 µl of 2.5% Formalin solution in saline (Formaldehyde solution (Sigma-Aldrich; Product number: 15512); Sterile saline solution (Henry Schein; Product number: 002477)) under gas anesthesia (4% isoflurane; Henry Schein Isothesia; Product number: 1169567762). The mouse was then transferred into the first testing arena, and the procedure was repeated with the next three mice for this enclosure. Typically, mice regained consciousness from the anesthesia within 1 min of being placed in the testing arena and were fully ambulatory within 3 min.
Building the automated scoring system
The model has distinct tasks to solve and was accordingly divided into three separate modules. First, the model was trained to use key point detection to track the body parts of mice in the arena for every video frame using “DeepLabCut”29 (https://github.com/AlexEMG/DeepLabCut) (Figure 1(a) to (c)). Next, features were extracted on a per frame basis, utilizing the concepts established for the JAABA model of analyzing statistical metrics over different time windows to detect behaviors32 (Figure 1(d)). Finally, the model took these frame-based features for a single mouse as input and classified each video frame as showing the behavior of licking/biting or not.
Module 1 key point detection
Training data
To create a training set for key point detection, frames were pseudo-randomly selected from eight videos of mice covering the four different enclosures and ensuring representation of early (up to 30 min), middle (30–60) and late (60–90) portions of the recordings. Labels were manually applied to the desired points on 370 frames using imageJ (https://imagej.nih.gov/ij/). Each mouse was labeled with 12 points shown in Figure 1(a) (mouth, nose, right front paw, left front paw, 3 points on each hind paw—outer, inner, and base; mid-abdomen; and tail base) and the inner walls of each arena were labeled with 5 points (Figure 1(a)). Thus, each frame was labeled with 53 points. The point tracker was trained to find all 53 points per frame, and therefore, it was not necessary to crop or manipulate the video frame to locate a single arena. The location of the grid walls was included for training purposes to verify that all 12 mouse points were located within a single arena. Any point missing or obscured was labeled as location x = 0, y = 0, and all labeled frames were visually rechecked for accuracy. Examples of empty arenas were included in training. To increase the number of frames for training, the 370 frames were reflected and rotated so that every mouse appeared in each of the four locations for a total of 1480 labeled frames. To increase the variability in lighting conditions used for training, approximately 11% of the 1480 frames were augmented with the addition of Gaussian noise (40 frames) or alterations of contrast (39 frames), brightness (39 frames), or gamma filtering (40 frames) using imageJ (see Appendix 1). The augmented frames were pseudo-randomly chosen and distributed evenly across the original 370 and each of the reflection and rotation conditions. After these adjustments, the set of labeled frames was divided randomly into a training set (85%) and a test set for validation (15%).
Pose estimation using DeepLabCut
DeepLabCut is based on the pose detection architecture of “Deepercut”33 and takes advantage of a pretrained Residual Network (ResNet50) for body-part detection. Residual network architecture uses convolution layers to learn specific visual features and the skipping function minimizes information loss, thereby enhancing extraction of global rules.34 DeepLabCut was chosen as the key point detector primarily for the ease of implementation, using Python and Tensorflow, but it could be replaced with another tracker if desired.
Tensorflow (https://www.tensorflow.org/versions; 1.2.1 Cuda 8; CudNN 6) was used to train the ResNet50 architecture on a GPU (Tesla P100). The model was trained for 750,000 iterations attaining training accuracy of 1.9 pixels and test error of approximately 4.4 pixels error averaged over all test frames and points. An error rate of less than five pixels is very similar to the results obtained by Mathis for mouse tracking.29 Figure 2 shows an example of a single test frame (average error of 2.4 pixels). In arena 4, the right front paw is missed by 4.3 pixels, which is the approximate size of the average error over all test frames. The stability of performance was verified by repeating the training with a different training and testing set (train error 1.9 and test error 4.3).
The trained model was locked and subsequently used to track the experimental videos. The videos were approximately 100–120 min long (ranging from 1.6 to 2.2 GB), each frame was 337,920 pixels (704 × 480), and the speed to label 53 points varied between 36 and 37 frames per second (on Tesla GPU). Tracking of the four mice in a video was effectively slightly faster than the video recording speed of 30 frames a second.
Module 2: Per frame feature extraction
The (x, y) pixel coordinates for each specified location, as well as a likelihood estimate that is based on agreement of score maps indicating the probability that this body part is at this pixel, were used for behavior classification. The likelihood feature was particularly useful for determining the presence of a mouse. When the arena was empty, all 12 points were located with very low probabilities (e.g., >0.0001), and as soon as the mouse was place in the arena, all points dramatically jumped up in likelihood estimations. The threshold of an average probability of 0.8 across the 12 points was used to indicate that a mouse was present.
Since the number of mice per enclosure varied (1–4), each mouse was classified independently. The 12 key points of interest (Figure 1(c)) were used to generate pairwise Euclidean distances between body parts (66 pairs) and the angles between selected trios of body parts (15 angles; see Appendix 2). These parameters represent relative body-part location information for a single frame. Change in the relative positions of body parts over time constitutes action and behaviors can be regarded as a series of actions. It is therefore essential to examine the consecutive video frames to see the changes across each time-based vector parameter. Figure 1(d) shows an example of one paired distance (from key points LeftFront to RightHindout) and one angle (between points RightHindout, abdomen, LeftHindOut) over 24 consecutive frames with the frame of interest marked in the center.
Kabra et al.32 instantiated a GentleBoost classifier (Matlab: Mathworks) that used moving windows to calculate statistical metrics of parameters on a per frame rate, based on an ellipse fit to the animal, over multiple frames to classify a range behaviors. Their JAABA classifier tested different window sizes depending on the behavior and organism. We used this as a guide and examined our videos and chose windows sizes of 6, 11, and 21 frames (200,367,700 ms) for licking behavior. Each window was moved along the parameter vector, and the statistics of the parameter were calculated within that time frame. For window sizes 11 and 21, the frame of interest was in the middle, and for window size 6, it was preceded by two frames (see Figure 1(d)). A total of 1047 different metrics were calculated for each frame of video and served as the input data to the classifier. Statistical metrics were mean, standard deviation, median, and median absolute deviation for each distance pair. A second measure of distance was included that reported no value (NaN) if the mouth or nose fell below 0.1 likelihood, and the mean was calculated for this and for the angles. The 12 likelihood estimations for the frame of interest were also included as input without windowing.
Module 3: Behavior classification
Training data
To create a training set for behavior classification, utilizing the extracted frame-based features, an experienced human observer annotated videos labeling the onset and offset of licking behavior. Training data were taken from 40 different videos, to cover all enclosures, arenas and sizes, or sex of mice. A total of 9300 frames were used for training with no more than 10 s (300 frames) per video. The video was initially manually annotated (Noldus) to determine the behavior of licking with a resolution of a second. This was subsequently reanalyzed frame by frame to obtain exact onset and offset video frames of licking behavior (Matlab). No distinction was made between licking and biting behaviors, any contact between the mouth and the right hind paw was scored as licking. To obtain a well-balanced training set, frames were selected using stratified random sampling from clear licking (22%) and nonlicking video segments (78%). The bias toward no licking behavior was intentional because this behavior does not occur equally in an experimentally recorded video. The selected training frames were not consecutive. The classifier evaluates each frame based only on the metrics provided for that frame, and it is the window-based statistics that give the context of what happened before and after the frame. Several different classifiers were trained and tested (Matlab: Mathworks) to determine the most effective model (see Table 1).
Table 1.
Results of classifier models on the validation data set.
| Classifier | Precision (%) | Recall (%) | False positive (%) | Total accuracy (%) |
|---|---|---|---|---|
| 12 point GentleBoost | 96 | 95 | 1 | 97.9 |
| 12 point GentleBoost PCA 99% | 94 | 90 | 2 | 96.7 |
| 12 point GentleBoost PCA 95% | 93 | 89 | 2 | 96.2 |
| 8 point GentleBoost | 95 | 94 | 1 | 97.6 |
| 8 point GentleBoost PCA 99% | 92 | 89 | 2 | 96.4 |
| 8 point GentleBoost PCA 95% | 94 | 87 | 2 | 95.6 |
| 5 point GentleBoost | 93 | 93 | 2 | 96.9 |
| 5 point GentleBoost PCA 99% | 92 | 88 | 2 | 95.7 |
| 5 point GentleBoost PCA 95% | 87 | 85 | 3 | 94.1 |
| 12 point Fine KNN | 95 | 94 | 1 | 97.6 |
| 12 point Ensemble KNN | 95 | 94 | 1 | 97.6 |
| 12 point Cubic SVM | 93 | 92 | 2 | 96.8 |
[Precision = true positive/(true positive + false positive): (or what proportion of frames identified as licking are truly licking)] [Recall = true positive/(true positive + false negative): (or what proportion of true licking frames were found)] [False positive = what proportion of “no licking” frames incorrectly identified as licking). PCA: principal component analysis; SVM: support vector machine; KNN: k nearest neighbor.
For validation purposes, 111 short videos, with a wide range of timing relative to injection, were annotated for behaviors. The most common nocifensive behavior observed was licking/biting, with paw flicking/shaking observed at a relatively low level. The total annotated time was 17,029 s, of which 1673 s were labeled as licking (in 318 bouts of varying lengths), and flicking was noted for approximately 8 s (in 21 events). Although flicking represented about 6% of events recorded, each instance was of short duration and thus total flicking time comprised less than 0.5% of nocifensive behavior (8/1681) and less than 0.05% (8/17,029) of annotated video. The classifier was therefore trained and tested only on licking/biting behavior, as this was the most common and reliable behavior to score.
GentleBoost classifier models
GentleBoost (gentle adaptive boost35) is an ensemble supervised learner based on minimizing exponential loss using decision trees and was utilized by Kabra et al. to classify a wide range of behaviors.32 The GentleBoost algorithm is well-suited for a dichotomous categorical response and is easy to implement. The classifier used 30 weighted learners, each of which fitted a regression model to the predictors and labels using a maximum of 20 splits and a learning rate of 0.1. Five-fold cross-validation was used to limit overlearning and to provide estimates of training. The large number of inputs obviously have significant redundancy, and therefore, the GentleBoost model was also trained with the implementation of principal component analysis (PCA), accounting for either 99% (65 inputs) or 95% (11 inputs) of variation. Table 1 shows the final results of all the tested classifiers for precision (proportion of frames correctly classified as licking), recall (proportion of licking frames correctly identified), false positives (proportion of incorrectly labeled frames as licking), and total overall accuracy. These metrics are helpful to distinguish the performance of models in accurately labeling the behavior when the licking is present and also when it is absent. High values in precision–recall dimensions indicate that the model is able to correctly find licking without missing occurrences of behavior, regardless of how rare; this view is particularly useful when the two behaviors are unequally distributed. Low false-positive rates indicate that when the licking is not present, then the model does not report the behavior; this shows that the model does not label everything as licking in order to prevent missing the behavior. The GentleBoost model performs very well on all metrics. PCA reduction of parameters resulted in diminished performance for precision and recall, with a slight increase in false-positive rates (Table 1).
To determine if 12 points were necessary for optimal performance, the GentleBoost model was retrained with inputs calculated from 8 points (removed both front paws and the inner point on both hind paws) or 5 points (removed the mouth, both front paws and the inner and outer points from both hind paws). Reducing the number of points resulted in a slight of loss of performance, but the 8-point model was very similar to the full 12-point model. PCA for the eight-point and five-point models resulted in clear loss of precision and recall as well as a small increase in false-positive rates.
The full GentleBoost classifier using all 12 mouse body parts and all statistical parameters (1047 inputs) had the best performance (Table 1). However, labeling all 12 points for training the tracking module is a time-consuming endeavor and could be reduced to fewer points if the slight loss of performance was an acceptable trade-off. The loss of performance with PCA is not worth the efficiency benefits as the cost of evaluating the full 12-point classifier is very low (prediction speed approximately 10,000 observations per second). Using Matlab on a CPU (laptop) to open excel file data, calculate inputs, classify behavior, calculate bins, and save results to three different formats (HDF5 file, excel spreadsheet, and backup Matlab output structure file) took approximately 20–25 s per mouse. A smaller parameter list is more efficient, but even a small loss in accuracy in detecting licking does not seem warranted given the low cost of keeping all parameters.
Other classifier models
Two other classifiers using all the 1047 input parameters performed almost as well as the full GentleBoost model (Table 1); a k-nearest neighbor (KNN) classifier (neighbors = 1, Euclidian distance, equal distance weight, ties broken to smallest; prediction speed 110 observations/s) and an ensemble subspace KNN classifier (30 learners, subspace dimension = 624, prediction speed 8.7 observations/s) but were less efficient in implementation of prediction. A support vector machine with a cubic kernel was more efficient than the KNN models (1600 observations/s) but slightly less accurate.
Model parameters
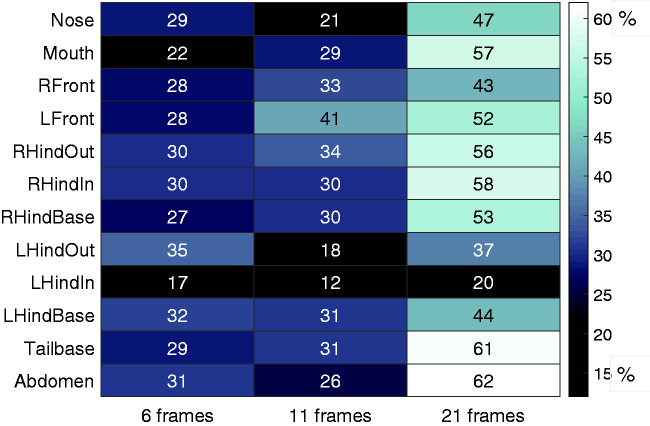
The 12-point GentleBoost model has 1047 inputs, but only 385 actually contributed information to the classifier and as is typical for this ensemble classifier, each useful input contributed a small amount of information, and there were no dominant cues. Each of the 12 body parts and three time windows are included multiple times in the 1047 inputs, and the heatmap of Figure 3 shows the percentage of useful representation as a proportion of all possible opportunities for that variable. The time window with greatest information was the 21 frames (700 ms) with approximately 46% of all used cues in this window. Licking duration generally extends well beyond a second, and the 700 ms window appears to be sufficient for capturing the ongoing behavior, and the shorter windows may be more useful for transitions between behaviors.
Figure 3.
Input value to classifier model. Heatmap of body parts and window sizes highlighting actual contribution to the decision of the classifier as a percentage of their possible contribution. The figure shows the relative importance of each window size and each body part to the model.
Examination of the relative information content of body-part points can be used to determine the most valuable points to keep for a model of this type (see Figure 3). Not surprisingly, all points on the right hind paw, mouth, and nose appear to provide useful information about contact between right hind paw and mouth. Points on the tail and abdomen seem to provide pertinent information about body shape and position. Although the base point on the left hind paw is useful as a relative comparison (average 36% use), it may not be necessary to include all three points for the left hind paw, as the outer (average 30%) and inner (average 16%) points were the least used. Inclusion of the front paws, however, appears to be warranted, as the mouse often uses the front paws to hold the hind paw while licking.
Results
Classifier validation: 111 short videos
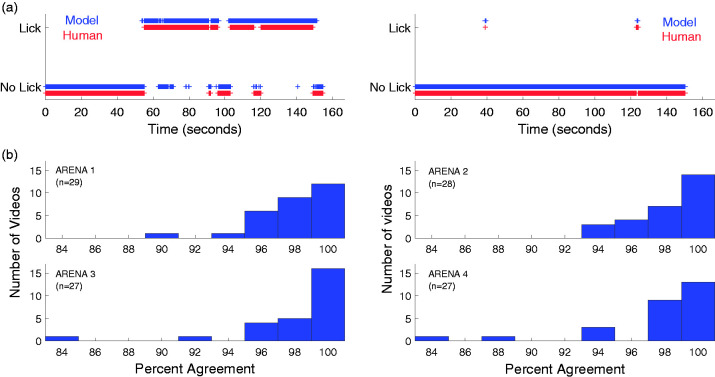
The GentleBoost classifier performance was tested on 111 new short video clips (from 111 different mice: with 71 completely novel videos and new clips from 40 training videos) for a combined total of about 284 min of testing. Each video was manually annotated for licking behavior, for a single mouse in an arena, with temporal resolution of a second (Noldus). Mice were from all enclosures, and an approximately equal number of arenas were annotated (see Figure 4). The temporal resolution of human classification was poorer than the model, and therefore, a match of licking behavior was recorded if the model was within ±15 frames of the human (i.e., 1 sec). Classifications were compared for each second of annotated videos for a total of 17,029 tests (283 min and 49 sec) and resulted in approximately 98% accuracy. Figure 4(a) shows the direct comparisons of the human and model licking classification for two videos (mouse 54 and mouse 56: all the 111 mice are shown in Supplemental Figures 1 to 4). Kabra et al.32 also found that a similar number of comparisons for the classification of two mouse behaviors, “following” (20,015 frames) and “walking” (12,223 frames) was sufficient to validate performance, while using only four mice for testing and training.
Figure 4.
Video validation: classifier performance on novel test videos. (a) Classification of licking and no licking by human (red) and model (blue) for each second of two short example videos (approximately 2.5 min each). The percent agreement between human and model was 93.7% for the left panel (mouse 56) and 99.6% for the right panel (mouse 54). All 111 mice are shown in Supplemental Figures 1 to 4 grouped by arena. (b) The agreement (percentage) between model and human for each of 111 videos displayed as histograms by arena. The average agreement over all arenas was 98%.
To summarize the performance for each video, the percentage of video seconds in agreement between the model and human observer was calculated and displayed (Figure 4(b)) as histograms of the number of videos at a given performance level. Forty-three video clips had no licking behavior, and the average agreement over these videos was 98.8% which indicates a low false-positive level. Figure 4(b) shows that matching on two videos was less accurate with performance in the range of 84% agreement. Close examination of these videos revealed ambiguous behavior, and it was difficult for a human observer to ascertain if the mouse was licking or not. For example, on one video, the mouth was apparently in contact with the right hind paw, but it was obscured by the tail, so licking could be marked only by inference and not by observation, and the model does not score missing information. The other video showed a lot of grooming of the leg and paw area, and it was difficult to score purely paw licking. These behaviors were not typical but are hard to classify and different human observers do not agree well under these circumstances.
Previous models of automated scoring have demonstrated their utility by showing that their models can reliably detect differences in the amount of licking in response to formalin dose changes19 or analgesia20 for groups of animals. Without additional pharmacological manipulation, our validation videos already show a wide range of response levels (confirmed by human annotation) and can therefore be clustered to determine how well the model can distinguish between groups of animals with known differences. Typically, behavior in a formalin test is not reported at the precise resolution of a second and is instead more broadly reported as the sum of behaviors over a time bin of one to several minutes.12,15 To demonstrate the ability of the model to detect different levels of response, each of the videos was truncated at 2 min, and the sum of licking in seconds was calculated for the human observer and the model. The level of summed licking across the 111 mice ranged from no licking, as might be seen in an experiment using saline control or analgesics, to a moderate-high level of approximately 70% licking. The 111 videos are shown ranked from lowest to highest level of summed licking, as determined by the human observer (Figure 5(a)). Similar levels of behavior, based on the human observations, were then clustered together to create groups of videos in categories of low to high behavior. Bootstrap sampling using an experimentally realistic sample size (n = 6) was used to determine if these categories could be reliably differentiated by the observer and model (10,000 bootstrapped samples for t-test comparisons). The example shown in Figure 5(a) used categories, based on summed licking performance, grouped with approximately 20 s mean difference (human observer determined mean and standard deviation of seconds of summed licking for each category I–IV: 1.6 (2.6), 20.7 (4.0), 35.6 (5.7), 63.3 (12.9); model means and standard deviations: 2.5 (3.4), 22.9 (10.3), 38.2 (13.7), 62.2 (11.8)). This 20 s category spacing was easily distinguished by both observer and model scorings (>9800/10,000 significant tests) for every neighboring group comparison. The windows were then shifted to produce smaller group differences to determine the smallest difference that could be reliably detected for this data set. The human scored data could reliably distinguish between groups with an average of 11 s of separation (80% significant tests) and the model required about 16 s of separation (80% significant tests). The videos were ranked by human observer, and therefore, the created categories for the human observer were less variable and should be more readily differentiated statistically than the model categories. The lowest response level category had the lowest variability for both the model and human observer scores, and the highest response level had the greatest variability. However, the most interesting comparisons are between adjacent clusters of moderate response levels with similar levels of variability and these consistently show that effect sizes of about 15–16 s for the model categories are necessary for differentiation. This bootstrapping test shows that the model is able to distinguish biologically realistic differences6,14 of approximately 13% in the level of licking/biting between groups of videos which would be sufficient to detect known strain differences14 or analgesic effects.15
Figure 5.
Human observer validation. (a) Sum of licking in seconds was calculated for 2 min for each of 111 videos for both Observer 1 (red square) and the model (blue circle). The videos are displayed ranked from low to high levels of licking ordered by the human observations. The boxes I–IV represent an example of categorization of behavior response level imposed on the data with approximately a 20-s separation: (I) very low level (human mean 1.6 s: model mean 2.5 s), (II) low-mid level (human mean 20.7: model mean 22.9), (III) moderate level (human mean 35.6: model mean 38.2), and (IV) high level (human mean 63.3: model mean 62.2). Observer 2 annotated 14 of these videos and the summed results for each (green open square) are superimposed on the plot of Observer 1 rankings (located at rank: 65, 73, 82, 83, 87, 88, 94, 95, 100, 102, 106, 107, 108, 110). Mice ranked at 95 and 110 have identical scores, and the markers are therefore indistinguishable, scores for mice at 73, 82, and 106 differ by only 1 s for one of the marks. (b) to (d) Comparison of manual scoring by two human operators with the classifier model for three mice. The data are shown as summed licking in 5 min bins for 60 min postinjection (bins range from 5–10 to 55–60).
Interobserver validation
Fourteen of the 111 validation videos were annotated by an additional observer (see Figure 5(a); Observer 2). These 14 videos were intentionally selected to include examples of good agreement between Observer 1 and model (no difference) and poor agreement (e.g., 21 s discrepancy) and ranged from low (3 s of licking) to high (76 s) response level. To show the full range, it is necessary to include videos with poor agreement even though they are not typical of the validation data. The average difference between the model and Observer 1, for the 111 video validation set, is 1 s, but the average absolute discrepancy is 4.5 s. The discrepancy between the two scores (Observer 1 vs. model) as described here emphasizes the size of the difference by using absolute discrepancy (abs (Observer 1 − model)) because the size of the difference is retained, and the direction of the difference is removed. The total absolute discrepancy over all 14 videos (1680 s) was 41 s between the two human observers (2.3%), 63 s between model and Observer 1 (3.75%), and 53 s between model and Observer 2 (3.1%). Ten of the 14 videos had very good agreement between both human observers and the model with no more than 4 s absolute difference in 2 min for any video and an average of 1% disagreement or less across all 10 videos. Several points in Figure 5(a) show identical scoring for model and observers; of the remaining videos with greater discrepancy in scoring, two videos showed improved agreement using scoring between the model and Observer 2, and two showed improvement using both human observers (see Figure 5(a)). The correlations between the recorded scores of licking in seconds for the 14 videos were consistently high (Observer 1: Observer 2 Pearson r = 0.98; Observer 1: Model Pearson r = 0.95; Observer 2: Model Pearson r = 0.97).
To further test experimenter interobserver reliability, videos (60 min) of three mice were annotated by two observers, visualized using Noldus Media Recorder 4 software. The observations were summed in 5 min intervals, and the correlation between observers was generally good (Pearson r = 1.0, 0.82, and 0.97). Both human observers agree about what constitutes licking but can disagree on exactly when to start and stop recording the behavior, sometimes licking bouts were scored as continuous by one observer and a series of short bouts by the other. For observations of mouse 9, this resulted in several substantially different measures, for example, two 5-min bins were recorded as 67 and 60 s by Observer 1 and 197 and 151 s by Observer 2 (see Figure 5(c)). Both human observers were compared to the model, and again agreement was quite high (Observer 1 Pearson r = 0.98, 0.75, 0.95 and Observer 2 Pearson r = 0.98, 0.96, 0.99). For mouse 9, the model appeared to be in better agreement with Observer 2, with the aforementioned 5-min bins recorded as 213 and 141 s of licking (see Figure 5(c)). Observer 2 was the more experienced of the two people and was training Observer 1 in behavioral annotation; these three mice were human scored months before the model was developed. After completing training, Observer 1 ultimately conducted all the manual annotation of the videos used for model training, testing, and validation.
Strain comparison validation
Bryant et al. used manual scoring methods for the formalin test to compare the licking response of C57BL/6NCrl compared to C57BL/6J mice in Phase I and Phase II.36 They found that male C57BL/6NCrl showed a reduced licking response in the Phase II of nociception response (measured as 20–45 min), but there was no significant difference for females. To validate the utility of the automatic video classification under experimental conditions, the formalin test was conducted comparing similar mouse strains (The Jackson Laboratory: C57BL/6NJ male n = 45, female= 30: C57BL/6J male = 46, female =30). As the mice in our study were anesthetized for the injection, the first 5 min of the nociception response, known as Phase I, was atypical and not included in the analysis. All of the data were run through the system, but the model determined the start frame for each mouse and then skipped 9000 frames (5 min) before binning the data into seventeen 5-min bins (5–10: 85–90) of cumulative licking in seconds.
Figure 6(a) shows the summed licking behavior in the time bin equivalent to that used in the Bryant study (20–45 min postinjection) and as predicted male C57BL/6NJ showed reduced licking compared to C57BL/6J male mice. The female mice, however, showed the reverse pattern with C57BL/6NJ licking more (Sex by Strain interaction F(1,147) = 9.99 p = 0.0019; Holms–Sidak multiple comparisons for male and female, p = 0.042). The time course of the responses over the full 90 min reveals differences more clearly between the sexes and strains (Figure 6(b) and (c)). The curves differ in both timing and amplitude of licking and the choice of how the data are binned for analysis will determine if differences between sexes or strains are detected. Figure 6(d) and (e) compare bootstrapped statistical analysis (alpha level 0.05) for two different bin choices with increasing sample size. The 20- to 45-min bin, selected by Bryant and replicated here, shows that as sample size increases, the probability of finding a significant difference between the strains also increases, for both males and females (Figure 6(d)). For females, this bin sizing appears to maximize the onset timing difference of the response between strains. The larger bin size of 10–60 minimizes the timing difference for females, and bootstrapped comparisons show that an increase of sample size does not alter showing statistical significance (Figure 6(e)). The female probability remains near 5% which would be the level due to chance (alpha of 0.05); the females have the same amount of licking. However, the male probability of detecting a difference increases with sample size, the males appear to differ in the amount of licking, in both the amplitude and duration of peak behavior.
Figure 6.
Comparison of male and female mice of the two strains: C57BL/6J and C57BL/6NJ. (a) Licking summed in a single bin over the 20–45 min postinjection period; showing mean and SEM separated by sex (significant 2-way ANOVA sex × strain interaction 0.05; *significant post hoc comparison between strains for each sex). (b) and (c) Mean and SEM of licking summed in 5 min bins and displayed for 90 min postinjection (bins range from 5–10 min to 85–90 min) for females (b) and males (c). (d) and (e) Percentage from bootstrapping of significant t-tests (alpha = 0.05) showing the difference between the strains by sample size for metrics binned over the periods of 20–45 min (d) and of 10–60 min (e).
Sample sizes of 12 or greater yield reasonable probabilities of showing differences but for lower, more typical experimental sample sizes (6–8), the odds are above chance but not high. The licking behavior in response to formalin appears quite variable; the automated classification accuracy of six extremely high or low licking mice were manually verified, and it was determined that the recorded variability is behavioral, some mice lick more than others. This nociception assay will reveal strong effects with small numbers of animals, but minor differences are likely to be obscured with variability.
Discussion
The results of automated classification of licking behavior in the formalin assay using machine learning are comparable to that of human observers. The automated system accuracy, over many small videos, had approximately 98% agreement with a human observer on a second by second basis, and it was also highly correlated with bin scoring over both long and short videos with two human observers. The efficiency in using automated scoring is considerable, the human observer takes about 9 h to score four mice in an enclosure compared to less than 2 h for the automated system. Additionally, the automated system can be run in parallel, assessing multiple videos at the same time with the same consistency for 24 h a day, seven days a week. Automation allows the formalin assay to be scaled for high-throughput nociception phenotyping to assess different genetic strains. Accurate and reliable automatic classification of scoring for licking/biting behavior becomes easy to implement with this system.
We used a generalizable framework for analysis of nocifensive behavior, consisting of three steps: key point detection, per frame feature extraction using these key points, and classification of behavior using the GentleBoost algorithm. Different components could be substituted in the model, another key point tracker could be used, such as LEAP30 or DeepPoseKit,28 the window sizes could be changed to remove the smaller window sizes that did not provide much information and the choice of classifier is quite open. The GentleBoost classifier performed best and was highly efficient but other classifiers could be substituted with very little loss of accuracy. This automated system is a generalized solution with considerable flexibility in application.
The localization of individual body parts results in a remarkable reduction of data from 337,920 pixels per video frame to 159 numbers of interest (x, y point coordinates and likelihoods of 53 points), making the task of tracking body parts over long videos manageable. The position of body parts relative to each other, measured in distances and angles, is a simple and low cost way to generate pose features for each frame. The average errors of localization of points of interest in test frames were below five pixels which appear to be sufficiently accurate for the task. The scoring is conducted over many thousands of frames (153,000 frames for 85 min), and even if a few frames contain larger errors, it will have negligible impact on the total scores. The use of temporal windows to assess change across time also reduces the impact of a single frame and helps to smooth disparities between consecutive frames.
The automated scoring system is limited to detecting a single class of behavior, but it does so as well as the trained human observers. Many studies have reported that licking/biting is the most important nocifensive behavior for the formalin test, and therefore, this was the focus of the study.14,16 Neither the classifier nor human observers distinguished between licking and biting of the paw which would require better spatial and temporal resolution in the video recording. These behaviors may represent different biological contexts, biting can be a response to both itch and painful stimulation,37,38 and the ability to differentiate biting from licking could be informative. However, our current system was designed for high-throughput scoring of formalin testing and in order to test four mice per enclosure, the video resolution was necessarily limited. Behaviors such as elevation and flinching were not reliably scored by the two human observers who showed substantial disagreement and could not reconcile their differences. Previous studies suggest that the inclusion of these additional measures could improve reliability in formalin scoring, but we found that these behaviors were not reliably scored. The supervised machine learning model requires consistent information for training, and this was not available for these other behaviors. In our data set, flicking the paw represented a small proportion of nocifensive behavior, but it could still provide interesting contextual information, and we anticipate exploring this rapid movement behavior in a different model system in the future.
It is clear that there can be considerable intermouse response variability, the variation is very small for the early portion of Phase II, but once the animals respond more vigorously, then the individual variability increases. There are many factors that have been implicated in the variability of this test including strain, sex, environment, dose, injection quantity, anesthesia, site of injection, time of day, experimenter effects, and observer bias.15,39–41 The automated scoring system removed inconsistent observer biases and fatigue, but substantial variability remains in the data. The formalin test is a widely accepted assay, but if sample sizes of mice are small, then it may be difficult to reliably detect small differences.
Many studies use a single summed bin to examine the Phase II period, and although this strategy is likely to be the best choice for revealing a general difference in summed lick duration, it risks losing information about phase differences in the timing of behavior. The choice of bin duration and starting time to analyze Phase II vary widely across studies, for example, 10–30,42 10–60,14 10–90,43 10–45,44 15–45,45 20–45,36 or 20–60.46 Bootstrapping with different bins indicates that bin choice could contribute to inconsistent results of studies if there is a temporal difference between the strains, sexes, or drug treatments of interest. The mice in this study were anesthetized which may also influence the timing of the behavior as the early part of the response is clearly reduced.47 Both the automated system and Bryant36 showed that C57BL/6N males lick less regardless of the bin choice, but bin size for females heavily influenced the outcome. Experiments using either C57BL/6N or C57BL/6NJ as control mice need to consider the sexes separately, as they show clear differences in timing over Phase II. Machine learning-enabled scoring allows experimenters to easily extend the length of the formalin experiment without incurring lengthy video annotation costs. Differences in timing should be evident over the longer duration (60 or 90 min) allowing for the possibility of anesthesia effects and for informed choices about bin size.
Clearly defined behaviors are well-suited for machine learning classification and the consistency that automated scoring can bring to formalin studies is highly desirable. The assay is well-established but is conducted, scored, and analyzed with many differences in protocol making comparisons between studies difficult. Automated scoring is an important refinement in examining nocifensive behaviors that improves reliability and speed with which the assay can be performed. The scoring system utilizes video along with established machine learning techniques and can be made readily accessible to nociception researchers for large-scale experimentation including mouse genetic studies, preclinical compound evaluation, and other applications.
Supplemental Material
Supplemental material, sj-pdf-1-mpx-10.1177_1744806920958596 for Machine learning-based automated phenotyping of inflammatory nocifensive behavior in mice by Janine M Wotton, Emma Peterson, Laura Anderson, Stephen A Murray, Robert E Braun, Elissa J Chesler, Jacqueline K White and Vivek Kumar in Molecular Pain
Supplemental material, sj-pdf-2-mpx-10.1177_1744806920958596 for Machine learning-based automated phenotyping of inflammatory nocifensive behavior in mice by Janine M Wotton, Emma Peterson, Laura Anderson, Stephen A Murray, Robert E Braun, Elissa J Chesler, Jacqueline K White and Vivek Kumar in Molecular Pain
Supplemental material, sj-pdf-3-mpx-10.1177_1744806920958596 for Machine learning-based automated phenotyping of inflammatory nocifensive behavior in mice by Janine M Wotton, Emma Peterson, Laura Anderson, Stephen A Murray, Robert E Braun, Elissa J Chesler, Jacqueline K White and Vivek Kumar in Molecular Pain
Supplemental material, sj-pdf-4-mpx-10.1177_1744806920958596 for Machine learning-based automated phenotyping of inflammatory nocifensive behavior in mice by Janine M Wotton, Emma Peterson, Laura Anderson, Stephen A Murray, Robert E Braun, Elissa J Chesler, Jacqueline K White and Vivek Kumar in Molecular Pain
Acknowledgments
The authors wish to acknowledge Justin Gardin, Brian Geuther, and Keith Sheppard for assistance with technical aspects of image handling and for invaluable advice about machine learning techniques and Jennifer Wright for aid with graphics. The authors also wish to thank Erin Young and Kyle Baumbauer for their assistance in planning and setting up the formalin challenge protocol.
Appendix 1. Adjustments made to approximately 11% of images using ImageJ.
| Gaussian noise |
Brightness |
Contrast |
Gamma filter |
||||
|---|---|---|---|---|---|---|---|
| parameter value |
Number of frames |
Parameter value |
Number of frames |
Parameter value |
Number of frames |
Parameter value |
Number of frames |
| 10% add | 20 | 220 | 8 | 220 | 8 | 0.3 | 12 |
| 15% add | 20 | 235 | 12 | 235 | 12 | 0.5 | 12 |
| 285 | 12 | 285 | 12 | 1.5 | 12 | ||
| 300 | 7 | 301 | 7 | 2 | 4 | ||
Appendix 2. Angles calculated between three body-part points with the angle subtended around the midpoint.
| Point1 | Midpoint | Point2 |
|---|---|---|
| Nose | RHout | LHout |
| Nose | RHbase | LHbase |
| Nose | RHin | LHin |
| Mouth | RHout | LHout |
| Mouth | RHbase | LHbase |
| Mouth | RHin | LHin |
| Tailbase | RHout | LHout |
| Tailbase | RHbase | LHbase |
| Tailbase | RHin | LHin |
| Abdomen | RHout | LHout |
| Abdomen | RHbase | LHbase |
| Abdomen | RHin | LHin |
| LF | RHout | RF |
| LF | RHbase | RF |
| LF | RHin | RF |
Code and Data Availability
Links to the training data sets, pretrained models, and associated codes are available at our website (https://www.kumarlab.org/data/).
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Research reported in this publication was supported by the Office of The Director, National Institutes of Health under Award Number UM1OD023222. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Additional funding was provided by The Jackson Laboratory, Director’s Innovation Fund (to VK) and R21DA048634 from National Institute of Drug Abuse (VK).
ORCID iDs
Janine M Wotton https://orcid.org/0000-0002-1899-9452
Vivek Kumar https://orcid.org/0000-0001-6643-7465
Supplemental Material
Supplemental material for this article is available online.
References
- 1.Dubin AE, Patapoutian A. Nociceptors: the sensors of the pain pathway. J Clin Invest 2010; 120: 3760–3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrot M. Tests and models of nociception and pain in rodents. Neuroscience 2012; 211: 39–50. [DOI] [PubMed] [Google Scholar]
- 3.Graham DM. Methods for measuring pain in laboratory animals. Lab Anim (NY) 2016; 45: 99–101. [DOI] [PubMed] [Google Scholar]
- 4.Jourdan D, Ardid D, Eschalier A. Automated behavioural analysis in animal pain studies. Pharmacol Res 2001; 43: 103–110. [DOI] [PubMed] [Google Scholar]
- 5.Sewell RDE. Neuropathic pain models and outcome measures: a dual translational challenge. Ann Transl Med 2018; 6: S42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mogil JS, Wilson SG, Bon K, Lee SE, Chung K, Raber P, Pieper JO, Hain HS, Belknap JK, Hubert L, Elmer GI, Chung JM, Devor M. Heritability of nociception I: responses of 11 inbred mouse strains on 12 measures of nociception. Pain 1999; 80: 67–82. [DOI] [PubMed] [Google Scholar]
- 7.Lariviere WR, Wilson SG, Laughlin TM, Kokayeff A, West EE, Adhikari SM, Wan Y, Mogil JS. Heritability of nociception. III genetic relationships among commonly used assays of nociception and hypersensitivity. Pain 2002; 97: 75–86. [DOI] [PubMed] [Google Scholar]
- 8.Philip VM, Duvvuru S, Gomero B, Ansah TA, Blaha CD, Cook MN, Hamre KM, Lariviere WR, Matthews DB, Mittleman G, Goldowitz D, Chesler EJ. High-throughput behavioral phenotyping in the expanded panel of BXD recombinant inbred strains. Genes Brain Behav 2010; 9: 129–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubuisson D, Dennis SG. The formalin test: a quantitative study of the analgesic effects of morphine, meperidine, and brain stem stimulation in rats and cats. Pain 1977; 4: 161–174. [DOI] [PubMed] [Google Scholar]
- 10.Abbot FV, Orcvirk R, Najafee R, Franklin KBJ. Improving the efficiency of the formalin test. Pain 1999; 83: 561–569. [DOI] [PubMed] [Google Scholar]
- 11.Abbot FV, Saddi G-M, Franklin KBJ. The formalin test: scoring properties of first and second phases of pain response in rats. Pain 1995; 60: 91–102. [DOI] [PubMed] [Google Scholar]
- 12.Sufka KJ, Watson GS, Nothdurft RE, Mogil JS. Scoring the mouse formalin test: validation study. Eur J Pain 1998; 2: 351–358. [DOI] [PubMed] [Google Scholar]
- 13.McNamara CR, Mandel-Brehm J, Bautista DM, Siemens J, Deranian KL, Zhao M, Hayward NJ, Chong JA, Julius D, Moran MM, Fanger CM. Trpa1 mediates formalin-induced pain. Proc Natl Acad Sci USA 2007; 104: 13525–13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mogil JS, Lichtensteiger CA, Wilson SG. The effect of genotype on sensitivity to inflammatory nociception characterization of resistant (a/J) and sensitive (C57BL/6J) inbred mouse strains. Pain 1998; 76: 115–125. [DOI] [PubMed] [Google Scholar]
- 15.Saddi G-M, Abbot FV. The formalin test in the mouse: a parametric analysis of scoring properties. Pain 2000; 89: 53–63. [DOI] [PubMed] [Google Scholar]
- 16.Hunskaar S, Fasmer OB, Hole K. Formalin test in mice, a useful technique for evaluating mild anelgesics. J Neuro Methods 1985; 14: 69–76. [DOI] [PubMed] [Google Scholar]
- 17.Murray CW, Porreca F, Cowan A. Methodolical refinements to the mouse paw formalin test: an animal model of tonic pain. J Pharmacol Methods 1988; 20: 175–186. [DOI] [PubMed] [Google Scholar]
- 18.Jett MF, Michelson S. The formalin test in rats: validation of an automated system. Pain 1996; 64: 19–25. [DOI] [PubMed] [Google Scholar]
- 19.Xie Y-F, Wang J, Huo F-Q, Jia H, Tang J-S. Validation of a simple automated movement detection system for formalin test in rats. Acta Pharmacol Sin 2005; 26: 39–45. [DOI] [PubMed] [Google Scholar]
- 20.Gregoire S, Etienne M, Gaulmin M, Caussade F, Neuzeret D, Ardid D. New method to discriminate sedative and analgesic effects of drugs in the automated formalin test in rats. Neurosci Res 2012; 72: 194–198. [DOI] [PubMed] [Google Scholar]
- 21.Jourdan D, Ardid D, Bardin L, Bardin M, Neuzeret D, Lanphouthacoul L, Eschalier A. A new automated method of pain scoring in the formalin test in rats. Pain 1997; 71: 265–270. [DOI] [PubMed] [Google Scholar]
- 22.Sakiyama Y, Sujaku T, Furuta A. A novel automated method for measuring the effect of analgesics on formalin-evoked licking behavior in rats. J Neurosci Methods 2008; 167: 167–175. [DOI] [PubMed] [Google Scholar]
- 23.Tuttle AH, Molinaro MJ, Jethwa JF, Sotocinal SG, Prieto JC, Styner MA, Mogil JS, Zylka MJ. A deep neural network to assess spontaneous pain from mouse facial expressions. Mol Pain 2018; 14: 1744806918763658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.VK personal communication.
- 25.Abdus-Saboor I, Fried NT, Lay M, Burdge J, Swanson K, Fischer R, Jones J, Dong P, Cai W, Guo X, Tao YX, Bethea J, Ma M, Dong X, Ding L, Luo W. Development of a mouse pain scale using Sub-second behavioral mapping and statistical modeling. Cell Rep 2019; 28: 1623–1634.e1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayashi E, Kobayashi T, Shiroshita Y, Kuratani K, Kinoshita M, Hara H. An automated evaluation system for analyzing antinociceptive effects on intracolonic capsaicin-induced visceral pain-related licking behavior in mice. J Pharmacol Toxicol Methods 2011; 64: 119–123. [DOI] [PubMed] [Google Scholar]
- 27.Dell AI, Bender JA, Branson K, Couzin ID, de Polavieja GG, Noldus LP, Perez-Escudero A, Perona P, Straw AD, Wikelski M, Brose U. Automated image-based tracking and its application in ecology. Trends Ecol Evol (Amst) 2014; 29: 417–428. [DOI] [PubMed] [Google Scholar]
- 28.Graving JM, Chae D, Naik H, Li L, Koger B, Costelloe BR, Couzin ID. DeepPoseKit, a software toolkit for fast and robust animal pose estimation using deep learning. Elife 2019; 8: e47994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathis A, Mamidanna P, Cury KM, Abe T, Murthy VN, Mathis MW, Bethge M. DeepLabCut: markerless pose estimation of user-defined body parts with deep learning. Nat Neurosci 2018; 21: 1281–1289. [DOI] [PubMed] [Google Scholar]
- 30.Pereira TD, Aldarondo DE, Willmore L, Kislin M, Wang SS, Murthy M, Shaevitz JW. Fast animal pose estimation using deep neural networks. Nat Methods 2019; 16: 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geuther BQ, Deats SP, Fox KJ, Murray SA, Braun RE, White JK, Chesler EJ, Lutz CM, Kumar V. Robust mouse tracking in complex environments using neural networks. Commun Biol 2019; 2: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kabra M, Robie AA, Rivera-Alba M, Branson S, Branson K. JAABA: interactive machine learning for automatic annotation of animal behavior. Nat Methods 2013; 10: 64–67. [DOI] [PubMed] [Google Scholar]
- 33.Insafutdinov E, Pishchulin L, Andres B, Andriluka M, Schiele B. DeeperCut: a deeper, stronger, and faster multi-person pose estimation model In: Liebe B, Matas J, Sebe N, Welling M (eds) Computer vision ECCV. New York: Springer, 2016. [Google Scholar]
- 34.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: IEEE conference on computer vision and pattern recognition (CVPR) Piscataway: IEEE, 2016, pp. 770–778.
- 35.Friedman J, Hastie T, Tibshirani R. Additive logistic regression: a statistical view of boosting. Ann Statist 2000; 28: 337–407. [Google Scholar]
- 36.Bryant CD, Bagdas D, Goldberg LR, Khalefa T, Reed ER, Kirkpatrick SL, Kelliher JC, Chen MM, Johnson WE, Mulligan MK, Imad Damaj M. C57BL/6 substrain differences in inflammatory and neuropathic nociception and genetic mapping of a major quantitative trait locus underlying acute thermal nociception. Mol Pain 2019; 15: 174480691882504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.LaMotte RH, Shimada SG, Sikand P. Mouse models of acute, chemical itch and pain in humans. Exp Dermatol 2011; 20: 778–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.LaMotte RH, Dong X, Ringkamp M. Sensory neurons and circuits mediating itch. Nat Rev Neurosci 2014; 15: 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Capone F, Aloisi AM. Refinement of pain evaluation techniques. The formalin test. Ann Ist Super Sanita 2004; 40: 223–229. [PubMed] [Google Scholar]
- 40.Mogil JS, Ritchie J, Sotocinal SG, Smith SB, Croteau S, Levitin DJ, Naumova AK. Screening for pain phenotypes: analysis of three congenic mouse strains on a battery of nine nociceptive assays. Pain 2006; 126: 24–34. [DOI] [PubMed] [Google Scholar]
- 41.Mogil JS. Laboratory environmental factors and pain behavior: the relevance of unknown unknowns to reproducibility and translation. Lab Anim (NY) 2017; 46: 136–141. [DOI] [PubMed] [Google Scholar]
- 42.Pinho-Ribeiro FA, Zarpelon AC, Fattori V, Manchope MF, Mizokami SS, Casagrande R, Verri WA., Jr. Naringenin reduces inflammatory pain in mice. Neuropharmacology 2016; 105: 508–519. [DOI] [PubMed] [Google Scholar]
- 43.Wilson SG, Chesler EJ, Hain H, Rankin AJ, Schwarz JZ, Call SB, Murray MR. Indentification of quantitative trait loci for chemical/inflammatory nociception in mice. Pain 2002; 96: 385–391. [DOI] [PubMed] [Google Scholar]
- 44.Takasaki I, Nakamura K, Shimodaira A, Watanabe A, Du Nguyen H, Okada T, Toyooka N, Miyata A, Kurihara T. The novel small-molecule antagonist of PAC1 receptor attenuates formalin-induced inflammatory pain behaviors in mice. J Pharmacol Sci 2019; 139: 129–132. [DOI] [PubMed] [Google Scholar]
- 45.Taves S, Berta T, Liu DL, Gan S, Chen G, Kim YH, Van de Ven T, Laufer S, Ji RR. Spinal inhibition of p38 MAP kinase reduces inflammatory and neuropathic pain in male but not female mice: sex-dependent microglial signaling in the spinal cord. Brain Behav Immun 2016; 55: 70–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu F, Ma J, Liu P, Chu Z, Lei G, Jia XD, Wang JB, Dang YH. Hint1 gene deficiency enhances the supraspinal nociceptive sensitivity in mice. Brain Behav 2016; 6: e00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lopes DM, Cater HL, Thakur M, Wells S, McMahon SB. A refinement to the formalin test in mice. F1000Res 2019; 8: 891. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-mpx-10.1177_1744806920958596 for Machine learning-based automated phenotyping of inflammatory nocifensive behavior in mice by Janine M Wotton, Emma Peterson, Laura Anderson, Stephen A Murray, Robert E Braun, Elissa J Chesler, Jacqueline K White and Vivek Kumar in Molecular Pain
Supplemental material, sj-pdf-2-mpx-10.1177_1744806920958596 for Machine learning-based automated phenotyping of inflammatory nocifensive behavior in mice by Janine M Wotton, Emma Peterson, Laura Anderson, Stephen A Murray, Robert E Braun, Elissa J Chesler, Jacqueline K White and Vivek Kumar in Molecular Pain
Supplemental material, sj-pdf-3-mpx-10.1177_1744806920958596 for Machine learning-based automated phenotyping of inflammatory nocifensive behavior in mice by Janine M Wotton, Emma Peterson, Laura Anderson, Stephen A Murray, Robert E Braun, Elissa J Chesler, Jacqueline K White and Vivek Kumar in Molecular Pain
Supplemental material, sj-pdf-4-mpx-10.1177_1744806920958596 for Machine learning-based automated phenotyping of inflammatory nocifensive behavior in mice by Janine M Wotton, Emma Peterson, Laura Anderson, Stephen A Murray, Robert E Braun, Elissa J Chesler, Jacqueline K White and Vivek Kumar in Molecular Pain
Data Availability Statement
Links to the training data sets, pretrained models, and associated codes are available at our website (https://www.kumarlab.org/data/).