Graphical abstract
Keywords: Genome-wide, Noninvasive prenatal diagnosis, NIPD, NIPT, Monogenic disorders
Abstract
Noninvasive prenatal diagnosis (NIPD) is a risk-free alternative to invasive methods for prenatal diagnosis, e.g. amniocentesis. NIPD is based on the presence of fetal DNA within the mother’s plasma cell-free DNA (cfDNA). Though currently available for various monogenic diseases through detection of point mutations, NIPD is limited to detecting one mutation or up to several genes simultaneously. Noninvasive prenatal whole exome/genome sequencing (WES/WGS) has demonstrated genome-wide detection of fetal point mutations in a few studies. However, Genome-wide NIPD of monogenic disorders currently has several challenges and limitations, mainly due to the small amounts of cfDNA and fetal-derived fragments, and the deep coverage required. Several approaches have been suggested for addressing these hurdles, based on various technologies and algorithms. The first relevant software tool, Hoobari, recently became available. Here we review the approaches proposed and the paths required to make genome-wide monogenic NIPD widely available in the clinic.
1. Introduction
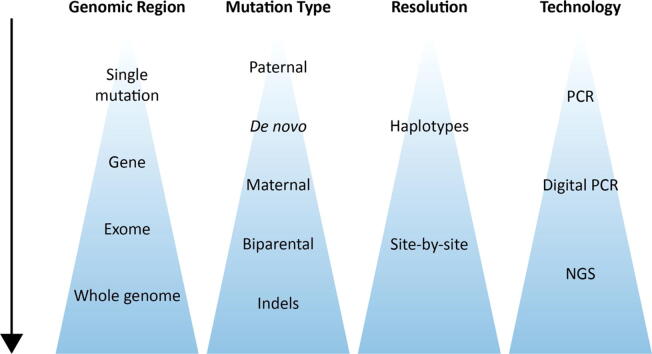
Prenatal diagnosis is a broad field that integrates several medical areas, from obstetrics and gynecology, to genetics and pediatrics. Statistics, computer science and other exact sciences enable the ongoing development of technologies and algorithms for risk-free diagnosis, through noninvasiveness. Noninvasive diagnosis is important in any medical field, as a fundamental principle in medicine is primum non nocere, i.e. first do no harm. Noninvasive prenatal diagnosis (NIPD) refers to genetic diagnosis of the fetus through circulating cell-free DNA (cfDNA) that is extracted from maternal plasma. Some of this cfDNA originates from placental cells, thus enabling to infer the fetal inheritance. NIPD is rapidly becoming an alternative to invasive techniques for prenatal diagnosis, e.g. chorionic villus sampling (CVS) and amniocentesis, which carry a risk of miscarriage [1], [2]. The first clinical uses of NIPD were for chromosomal-level phenomena, e.g. detection of aneuploidies [3], [4], and fetal sex determination [5]. The most notable example is NIPD for Down syndrome, a test that has shown high sensitivity and specificity, not only for the population at risk, but also in the general population [6]. These tests are based on comparing the amount of cfDNA that originates from each chromosome. Deviations from a reference amount, which can be calculated by various methods, indicate the number of copies of each chromosome. To distinguish a true deviation from noise, the fractional concentration of fetal DNA within the cfDNA, i.e. the fetal fraction, is also calculated. Similar quantification methods have enabled the NIPD of sub-chromosomal deletions and duplications, which is also clinically available nowadays [7], [8], [9]. Higher-resolution utilizations of cfDNA, i.e. at the level of single genes and mutations, have also become available (Fig. 1). These include Rhesus-D (RhD) blood typing [10], point mutations of paternal origin [11], [12], [13] and de novo mutations, all of which can be deduced based on the absence or presence of foreign alleles in the maternal plasma.
Fig. 1.
Trends in NIPD of monogenic diseases.
The inheritance of point mutations that derive from the mother is harder to infer, as the cfDNA in maternal plasma is a mixture of both fetal and maternal DNA fragments. Moreover, since the fetus shares half its genome with the mother, completely separating the fetal and maternal DNA is impossible. Specifically, in positions where the mother is heterozygous, both of the possibly-inherited alleles are present in the maternal plasma, and thus the fetal DNA is not distinguishable. Therefore, methods based solely on detecting the presence of an allele in the blood are not useful, and quantification is required. However, this poses several challenges. First, the amounts of cfDNA in the plasma are low, which makes its quantification difficult. Second, a strong skew towards maternal-derived fragments in the cfDNA make the specific exploration of the fetal DNA more challenging. Both these parameters are also lower in earlier stages of pregnancy, which are more clinically relevant. For example, the average fetal fraction is 10% at the end of the first trimester [14]. Solving these challenges requires either an ultra-accurate counting technology or DNA amplification. The first method for inferring the inheritance of maternal mutations was based on digital PCR (dPCR), an accurate quantification technology that enables detecting minute allelic imbalances [15]. In this method, termed relative mutation dosage (RMD) analysis, a genomic locus in which the mother is heterozygous, is examined within the cfDNA. Skewing the ratio between the two alleles towards one of them indicates homozygosity of the fetus to this allele. The expected allelic imbalance in case of homozygosity is proportional to the fetal fraction. The RMD approach was subsequently validated over various mutations and diseases [16], [17], [18].
The emergence of next generation sequencing (NGS) was an important step in the study of monogenic disorders. NGS-based methods, such as whole-exome sequencing (WES) and whole-genome sequencing (WGS), have yielded the discovery of countless variants, many of which are pathogenic and responsible for monogenic diseases [19], [20], [21], [22], [23]. Invasive prenatal WES/WGS, performed using amniocentesis, enables the diagnosis of monogenic diseases already in the earliest stages of life, thus affording early treatment in some cases, and pregnancy termination if required, in others [24], [25]. Interest in achieving noninvasive prenatal WES/WGS has been growing steadily, but the aforementioned methods to detect point mutations are based on technologies that are not feasible over large scales. This is especially true regarding maternal inheritance, and also de novo mutations. The RMD approach, for example, was based on dPCR. Accordingly, detecting maternal mutations was not suitable for genome-wide inference, since it required designing specific primer sets for each mutation. Clearly, genome-wide detection of de novo mutations is one of the most powerful tools enabled through WES/WGS analysis, but it requires accurate and deep sequencing. Compared to tests for specific mutations and small regions, searching the entire genome for rare mutations is substantially more difficult due to the absence of prior knowledge. Separating a true de novo mutation from sequencing noise becomes challenging in such setting.
The progress achieved in NIPD and the increasing availability of NGS eventually resulted in the first successful attempts for noninvasive prenatal WGS [26], [27], [28]; this enabled genome-wide NIPD of monogenic diseases. However, in the decade that has passed since the first achievements of genome-wide monogenic NIPD, this method has not become clinically available. Accurate genome-wide noninvasive genotyping has been shown to depend on the fetal fraction and the sequencing depth; these can both become limitations even with NGS-based approaches. This is mostly due to the low availability and high costs of the required technologies. Improving the computational and algorithmic aspects of these approaches can help overcome such problems.
While most of the literature describes the clinical implementation of NIPD, few studies suggest novel algorithms. In this review, we focus on the methods suggested for genome-wide NIPD of monogenic diseases, and explain the various approaches that have been suggested over the last decade. We then discuss the future of this field, and the research and clinical gaps to be filled to achieve clinical availability.
2. Methods for genome-wide NIPD of monogenic disorders
2.1. The first cfDNA-based reconstruction of the fetal genome
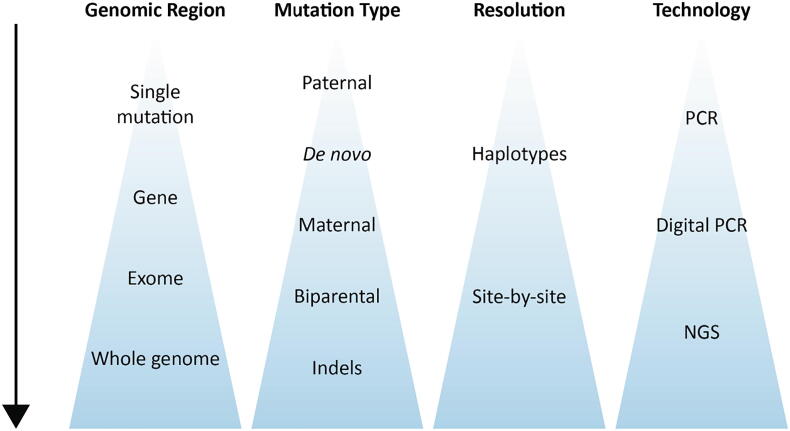
Following the success in NIPD of single point mutations, several approaches for genome-wide NIPD of monogenic diseases were demonstrated over the last decade (Fig. 2; Table 1). Lo et al. [26] were the first to reveal the entire fetal genome using cfDNA, and to define the limitations of this task. This laid the infrastructure for the field. In their study, the parents and fetus (i.e. the CVS sample) were genotyped using a single-nucleotide polymorphism (SNP) array. The cfDNA underwent WGS to a depth of 65-fold. SNPs were utilized based on the parental genotype. Positions at which both parents are homozygous enable estimating the sequencing error rate. Positions at which the parents are both homozygous but for different alleles, together with those in which only the father is heterozygous, enable calculating the fetal fraction. At positions where only the father is heterozygous, the fetal genotype is deduced based on the presence or absence of the paternal-specific allele in the cfDNA. Deducing maternal inheritance is more challenging, as both maternal alleles are always present in cfDNA. Thus, inference is performed by measuring the slight allelic imbalance that occurs in the cfDNA if the fetus is homozygous. This can be performed using dPCR to analyze a given position, similar to the RMD approach. However, such method does not fit an NGS-based genome-wide analysis, as the coverage over a given SNP is too low. Thus, Lo et al. suggested measuring the number of reads covering a haplotype by a method termed relative haplotype dosage (RHDO) analysis (Fig. 3). To increase the resolution to the degree possible, a sequential probability ratio test (SPRT) test was used. This method enables hypothesis testing as data accumulates along a specific region, and the inherited haplotype is determined when enough data exists to reach a pre-defined statistical threshold. SPRT was also used in the RMD approach, in which data accumulated through dPCR runs.
Fig. 2.
A timeline of milestone methods for NIPD of monogenic diseases.
Table 1.
Approaches for noninvasive prenatal sequencing of the fetal genome.
| Lo et al, 2010 [26] | Kitzman et al, 2012 [28] | Fan et al, 2012 [27] | Chan et al, 2016 [34] | Rabinowitz et al, 2019 [37] | |
|---|---|---|---|---|---|
| Sequencing method | WGS | WGS | WGS, WES | WGS | WGS, WES |
| Maternal allele inference | Haplotype-based SPRT (i.e. RHDO) | HMM + Viterbi | Haplotype-counting + Poisson-based test | Site-by-site SPRT (i.e. GRAD, Genome-wide RMD) | Naïve-Bayes + random forest |
| Maternal inference resolution | Haplotypes | Haplotypes | WGS: Haplotypes, WES: Site-by-site | Site-by-site | Site-by-site |
| Maternal haplotyping approach | Using the fetus, or a family member | Clone-pool dilution sequencing | Direct deterministic phasing | – | – |
| Paternal allele inference | Site-by-site | Site-by-site log-odds ratio | Haplotype imputation or site-by-site | Site-by-site | Site-by-site |
| Both parents heterozygous | – | Partially, using haplotypes | – | – | Site-by-site |
| Indels | – | – | – | – | V |
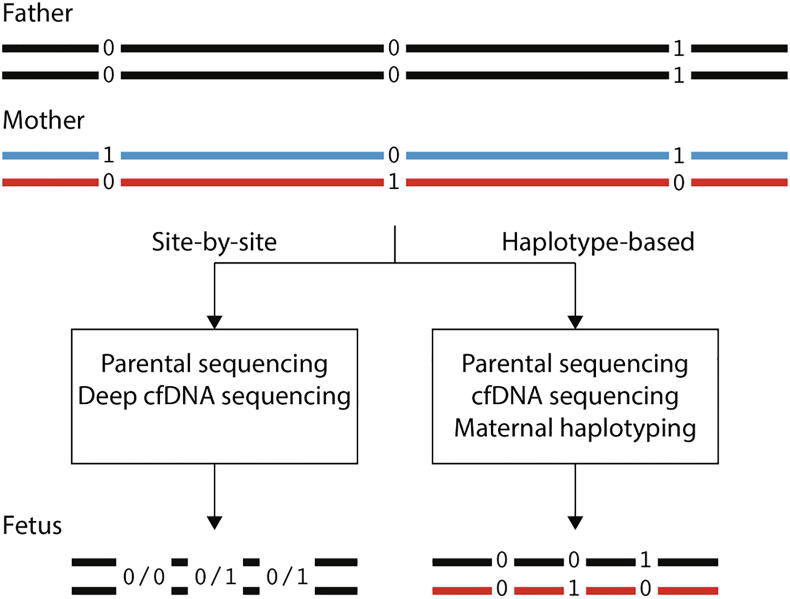
Fig. 3.
Two main approaches for genome-wide noninvasive fetal genotyping. The parental diploid genomes are represented as parallel lines, with the parental genotype in maternal-only heterozygous loci (the father is homozygous). Zero and one represent the reference and alternate alleles, respectively. 0/0 and 0/1 represent homozygous and heterozygous genotypes, respectively. Red and blue colors represent the two maternal haplotypes. In the haplotype-based approach, the maternal genome is phased through various methods, and this enables inference of the fetal haplotypes. For biparental loci, i.e. sites in which both parents are heterozygous, haplotyping of both parents is required (not shown here). In the site-by-site approach, deeper sequencing replaces the requirement for haplotype information, thus the fetus is genotyped, but its haplotypes are not deduced. This method is applicable for biparental loci. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The RHDO entails several limitations. To carry out RHDO analysis, the maternal haplotype information is first deduced using the maternal genotypes and the fetal genotypes from the CVS sample. In clinical settings, fetal information cannot be acquired, and information from another family member can be used instead. Thus, obtaining fetal haplotype information through an invasive procedure is a main disadvantage of this method [27], [28]. Moreover, the invasively obtained fetal genotypes are used for inferring the parental haplotypes, which are later used for predicting the fetal genotypes; this creates circular inference. Another limitation is the use of SNP arrays, which results in a relatively small number of SNPs tested. Using WES or WGS would reveal millions of SNPs and would also enable inferring the inheritance of small insertions-deletions (indels). Moreover, de novo mutations, which cause a substantial fraction of autosomal dominant disorders, were not addressed in this method. In addition, due to lack of paternal haplotype information, analysis was not carried out over biparental loci, i.e. loci in which both parents are heterozygous. These positions are relevant for autosomal recessive conditions in the setting of consanguinity or of a strong founder effect. In general, the requirement to assemble parental haplotype information while dealing with possible recombination events complicates RHDO analysis. Essentially, Lo et al.’s approach is not entirely noninvasive, and does not assess certain parts of the fetal genome.
2.2. Improvements in haplotype-based noninvasive fetal genotyping
During the several years after the publication of Lo et al.’s study, efforts were invested to improve the haplotype-based approach. These attempts focused on direct haplotyping of the mother using various technologies, thus avoiding a need to study another family member. In addition, the focus on various algorithms aimed to cope with the problem of recombination events.
In 2012, two separate teams, Kitzman et al. [28] and Fan et al. [27] published novel approaches for genome-wide NIPD of monogenic diseases. Similar to RHDO, Kitzman et al. [28] presented a method based on haplotypes, which were analyzed using a Hidden Markov model (HMM). Moreover, a sample from another family member or from the fetus is not required to infer the maternal haplotypes. Instead, they used clone-pool dilution sequencing. This method includes fragmentation of the maternal genome, cloning onto fosmids (small DNA molecules that are used as vectors) and culturing within Escherichia coli [29]. They also attempted to determine fetal inheritance at the allele rather than haplotype level. At positions where only the father is heterozygous, they used a log-odds test, which yielded 96.8% accuracy. However, at positions where only the mother is heterozygous (the father is homozygous), the site-by-site approach resulted in only 64.4% accuracy. Positions where both parents are heterozygous were partially analyzed, as no haplotype information of the father was available. They used only the maternal haplotype information to demonstrate that these positions could potentially be assessed. In addition, Kitzman et al. demonstrated a means for discovering de novo mutations and defined the challenges involved. A given human genome contains only ~60 de novo mutations, and the number of sequencing and mapping errors in a cfDNA sample is considerable. Thus, reaching a satisfactory level of sensitivity would result in very low specificity. To improve the specificity, a series of filtering criteria were applied, including criteria related to the predicted deleteriousness of the assessed SNPs.
Fan et al. [27] also used a method that is based on haplotype-counting. To obtain the maternal haplotypes, they used direct deterministic phasing. This method involves microfluidic separation and amplification of individual metaphase chromosomes, obtained from 3 to 4 cells by culturing maternal blood, and a subsequent genome-wide SNP array. Paternal haplotypes were reconstructed using paternal-specific alleles in the plasma, followed by imputation at linked positions, using haplotypes of normal population documented by the 1000 Genome Project. This research group was the first to effectively apply these principles to infer the maternal inheritance of single alleles, rather than haplotypes, on a genome-wide scale. This also enables following the inheritance of de novo mutations. To reach the depth required for such analysis, they used WES, thus sequencing only 2% of the genome. The acquired median depth was ~200× for the first and second trimesters cases, and 631x for the third trimester. They then calculated the minor allele fraction, defined as the second largest nucleotide fraction divided by the sum of the two largest nucleotide fractions, at every position of interest in the cfDNA. The fetal and maternal genotypes were inferred based on the measured minor allele fraction and the fetal fraction, and regardless of paternal genotype information. For instance, for a fetal fraction of , a minor allele fraction of would suggest that the fetus is homozygous and the mother is heterozygous. This method was quite accurate for loci where only the father is heterozygous, but notably less accurate at positions where the mother is heterozygous. In the latter, accuracy depended on high fetal fraction and sequencing depth. Nevertheless, even in a lenient scenario of a third trimester case sequenced to 631x median coverage with a fetal fraction of 26%, separation between fetal heterozygosity and homozygosity at such positions was limited. Various handcrafted filters were applied to improve sensitivity and specificity, including a stringent threshold for the depth of coverage and filtering out misaligned regions. It was suggested that a deeper coverage can reduce the bias that is caused by the numerous PCR amplification cycles required for WES.
In another attempt to noninvasively sequence the fetal genome, Chen et al. presented a method that combines principles from the aforementioned haplotype-based studies, and discussed the preferred haplotyping method [30]. Other novel direct haplotyping methods were also suggested, and these continued to improve. One of these is a microfluidics-based linked-read sequencing technology that enables genome-wide phasing, and which was followed by RHDO [31].
The reliance on haplotype information is the main limitation shared among all the aforementioned methods. These methods either require haplotype information from another family member to reconstruct maternal haplotypes, or they rely on complex experimental technologies that are not widely available, that require significant expertise and that are time consuming. In addition, none of these methods could phase the entire parental genomes, due to various reasons, such as a low density of informative genomic markers [31], [32]. The use of haplotypes also requires dealing with recombination events and phasing errors. When these occur near a mutation, they may result in incorrect fetal genotype classification [31], [32]. Recombination assessment comprises a prominent portion of the computational efforts in the aforementioned methods. Targeted phasing of the region-of-interest may be performed, yet this would not enable genome-wide NIPD [33]. Haplotype-based methods are also not adequate for genome-wide detection of de novo mutations, which require ultra-deep sequencing of the maternal plasma [28]. Finally, haplotype-level inference of the fetal genome has low resolution compared to the single nucleotide-level, as the mean haplotype block length used in the aforementioned studies ranged from 300 kb to over 1 Mb [34]. Although haplotyping-based methods have many limitations, site-by-site methods may be infeasible in early stages of pregnancy due to their dependence on the fetal fraction and sequencing depth, especially for maternal-specific SNPs [30]. Even the deep coverage achieved through WES resulted in low performance in early stages of pregnancy, partly due to the substantial DNA amplification needed.
2.3. Genome-wide site-by-site genotyping
The original rationale for the haplotype-based methods was the deep coverage required for inference of the maternal inheritance. As NGS became more affordable, attempts were made to reach deeper coverage using reliable sequencing protocols, e.g. PCR-free WGS of cfDNA. Later, these attempts were combined with novel algorithms and machine learning methods, and enabled application to other types of loci.
To address the limitations and challenges of haplotype-based analysis, Chan et al. demonstrated that site-by-site inference of maternal-specific alleles is possible over the entire genome, rather than the exome, with high accuracy [34]. To this end, they used ultra-deep WGS instead of the previously used WES. This also enabled relying on PCR-free protocols, thus eliminating sequencing errors. To assess the inheritance of maternal-specific alleles, they used their previously described RMD method [15]. This time, they applied it over the entire genome using NGS technology and termed it genome-wide relative allelic dosage (GRAD) analysis. In addition, they presented improvements to genome-wide de novo mutation analysis, using deeper coverage, careful mapping to the reference genome and other filtering criteria. Their study outlined the two main frontiers of genome-wide NIPD of monogenic diseases, namely, complete site-by-site inference of maternal-specific alleles and de novo mutation detection.
Despite the advances of GRAD analysis, several limitations arise. First, since the samples were taken from the second and third trimester, the high cfDNA levels enabled reaching deep coverage using WGS. The high fetal fraction values in such advanced stages of pregnancy also enable more accurate results, but the clinical relevance is low compared to the first trimester. Second, although paternal DNA was used to deduce the inheritance of paternal-specific alleles, it cannot be used in GRAD. Paternal DNA can potentially improve the results, since paternal homozygosity and heterozygosity have different effects on the prior knowledge of the fetal genotype. Third, the effect of the paternal genotype was not demonstrated, as no results were presented for loci where both parents were heterozygous. Fourth, the sequential test has no advantage in the setting of an NGS-based RMD method, since the information is not cumulative. Finally, accuracy was calculated from only 6.5 × 105 sites where the mother was heterozygous and the father was homozygous. However, ~3 million heterozygous SNPs are expected to be found in the genome of an individual, with ~1.3 million being maternal-only heterozygous SNPs [28], [35].
Another haplotype-free algorithm, termed pseudo tetraploid genotyping, was later presented by Yin et al. [36]. This algorithm used population-wide allelic frequency information from the 1000 Genome Project to compute the prior probabilities of each possible fetal genotype. The method included seven possible combinations of maternal and fetal genotypes, without sequencing the father. The most probable genotype is inferred using an expectation maximization algorithm, which is based on the assumption that the coverage of the alternate allele at a given site follows a negative binomial distribution. The method was evaluated using WES of one family, and a causative mutation was correctly predicted. The accuracy at positions where the mother was heterozygous was in the range of 54.77–62.23%. At positions where the mother was homozygous and the fetus heterozygous, i.e. where the fetus inherited the foreign allele from the father, the accuracy was 64.24–81.54%. Similar to the WES attempt performed by Fan et al, this low accuracy could be a result of the amplification process.
We recently presented a novel framework for genome-wide NIPD of monogenic diseases that does not rely on parental haplotype information [37]. Our approach consists of a Bayesian algorithm that utilizes characteristics that facilitate distinguishing fetal from maternal fragments, e.g. the fragment length. We applied our method to first trimester cases, since these are the most clinically relevant. We sequenced two families using WES, similar to Fan et al.; and one using WGS, similar to Chan et al. We managed to achieve the most accurate results that have been documented, and over the largest number of loci. Similar to other approaches, accuracy was calculated as the number of correctly predicted genotypes within the total number of positions where at least one parent is heterozygous. This improvement was prominent especially over maternal-heterozygous loci. We also showed the feasibility of NIPD over indels and biparental loci. Indels are the second most common type of variants and can be deleterious, especially when they affect the reading frame [38], [39]. Our larger goal, aside from the new algorithm and its results, is to introduce the widely practiced concepts of NGS-based variation analysis to the NIPD field. We suggest that this is a unique case of variant calling. To this end, we implemented our algorithm as Hoobari, the first software tool for noninvasive prenatal variant calling. Hoobari’s output is a variant call format (VCF) file that is compatible with existing tools for downstream analyses. This change of paradigm inspired us to introduce other common practices of variant calling to NIPD, such as a variant recalibration step. This process is typically achieved using a machine learning algorithm that leverages previously analyzed and verified data. We showed that even with only two previously analyzed families, Hoobari’s results can be improved by this technique, especially for indels and biparental loci.
Our study and method have some limitations. We did not analyze de novo mutations, multi-allelic loci and X-linked inheritance, yet the algorithm can be generalized to include them. The fetal inheritance of indels and biparental loci had lower accuracy. This could result from the higher alignment error rate, and the larger number of possible alleles in the parents. The additional machine learning algorithm requires larger datasets to be more generalizable, and careful selection of features is required to facilitate the training. WES cases resulted in lower accuracy, which can be explained by the amplification steps required for preparation of the WES library and for the low-input protocols [27]. When fragment lengths are considered components of the algorithm, the effect of amplification over the length distributions can be crucial. WES was previously shown to be less accurate than WGS, even for exome variants [40]. The implementation of more accurate WES methods or other wide panels might improve accuracy of the method without a reliance on deep WGS.
To conclude the current situation, several approaches exist for genome-wide NIPD of monogenic diseases. These methods aim to reconstruct the fetal genome using cfDNA in maternal plasma. While the detection of paternal-specific alleles is straightforward, positions where the mother is heterozygous remain challenging. Early solutions to this problem relied on haplotyping of the parents, while recent solutions are based on ultra-deep WES/WGS, which also enable genome-wide detection of de novo mutations.
3. The future of genome-wide NIPD of monogenic disorders
Most recent attempts to perform genome-wide NIPD of monogenic diseases do not require phasing of the parents. However, as genome-wide direct phasing methods become affordable and less technically-demanding, the haplotype-based approach seems closer to becoming clinically available [41], [42]. Haplotype-based methods enable sequencing the cfDNA to a relatively shallow depth, which also results in lower costs. Although some regions of the genome are not covered, the haplotype-based method is currently more accurate than other methods. Another advantage is the ability to resolve fetal compound heterozygosity. As explained above, among the disadvantages of haplotype-based methods is the requirement of haplotype information of another family member or the reliance on time-consuming technologies that require expertise. Moreover, these methods have low resolution, and typically miss certain regions; recombination events may result in incorrect genotyping; and de novo mutations cannot be detected. Site-by-site approaches successfully address these limitations.
Population-based imputation and phasing can potentially assist in haplotyping of the parents. However, as described by Fan et al, imputation accuracy is dependent on the density of markers [27]. Regions where paternal-only heterozygous loci were not found or were lacking, or where the paternal alleles were associated with more than one haplotype observed in the population, resulted in haplotypes that could not be confidently imputed. As stated by Fan et al, such loci can be completely determined by deeper sequencing and application of a site-by-site approach. Another limitation of population-based phasing is the requirement for high quality datasets for different populations. Even with reference datasets that are ever increasing in size, different populations and the variance within them, result in lacking or misleading haplotype information. Eventually, when parental information is available, it results in higher accuracy, without relying on population-based imputation. This is true for both haplotype-based methods and for using population-based allelic frequencies in site-by-site methods.
Once available, site-by-site methods have greater potential for wide use, for several reasons. First, the costs of WGS and WES have been constantly declining, thus genome-wide NIPD using a site-by-site approach is becoming more affordable. Second, NGS platforms have been available for several years, enabling the performance of sequencing in numerous facilities worldwide. This facilitates implementing a site-by-site approach that relies on currently available infrastructure, rather than introducing new and unfamiliar technologies. Most importantly, the site-by-site approach is potentially more accurate and enables detecting more types of mutations. The major algorithmic improvements in site-by-site methods enable further lowering the required depth of coverage without lowering the accuracy, thus reducing costs even more [43].
Additional improvements in site-by-site methods depend on several factors. The most important of them is the need for available data. The standardization and wide use of WES/WGS analysis is due to publicly available verified data sources, such as the 1000 Genomes Project [44]. Pipelines for variant detection were benchmarked using datasets with high confidence variants [45]. Both these data sources were published as part of the Genome in a Bottle Consortium (GIAB), which aims to develop technical infrastructure (reference standards, reference methods, and reference data) to enable translation of WGS to clinical practice. The state of affairs of noninvasive prenatal variant detection is different, for several reasons. First, all the aforementioned attempts for genome-wide NIPD of monogenic diseases rely on small cohorts and case reports. Each study used its own cohort of families, with particular gestational age, fetal fraction, sequencing protocols, settings and platforms. Second, methods were rarely compared by testing that used the same samples. Each method was evaluated using particular metrics, and over a certain set of variants. Third, these methods were presented as part of proof-of-concept studies, which do not include statistical risk analysis or conformance with a standardized protocol. For these reasons, developing reference standards for genome-wide monogenic NIPD is currently impossible. Creating such standards will not only accelerate the introduction of this field to the clinic, but also promote further research. For example, since novel methods for noninvasive prenatal genotyping rely on machine learning algorithms, a large number of families that are analyzed for training will enable creating robust and generalizable models. Therefore, for further development of genome-wide monogenic NIPD, such datasets will have to be created and become available to researchers. This can be achieved through collaborations between laboratories, consortiums (new or existing, e.g. GIAB), or the involvement of non-profit, possibly governmental organizations. The largest cohort to date is currently being created as part of an ongoing clinical trial, but even this dataset will consist of only 20 families [42]. The goal should be a much larger number of families, and we believe that this will enable complete resolution of noninvasive prenatal genotyping.
Aside from monogenic diseases that result from SNPs and indels, cfDNA have several other clinical applications in the context of prenatal diagnostics. These applications include other types of genetic abnormalities, such as aneuploidies, sex determination, RhD blood typing, and large and small size sub-chromosomal deletions and duplications. Additional subjects that require a solution are multiallelic SNPs (rather than the common biallelic ones), mutations in sex chromosomes, repeat expansions and structural rearrangements. The same test can also assess several maternal conditions, such as preeclampsia [46], [47], [48] and maternal malignancy [49], and possibly pregnancy-induced hypertension and gestational diabetes mellitus [50]. Ideally, one test could cover all these cfDNA utilizations. A recently presented example of this unified approach [51] demonstrated a single test for aneuploidy, copy number variation and single‐gene disorder screening. A handcrafted NGS panel was used, which consists of regions of common mutation hotspots for several monogenic disorders. Theoretically, all the aforementioned methods can be similarly applied over an NGS panel. However, such panels do not cover the whole genome, and thus, often fail to detect mutations [52]. An intermediate option would be to use WES or wide panels that cover all genes that are known to cause Mendelian diseases. However, as explained, the amplification required in such approaches causes high error rate and creates bias. Moreover, since these problems arise during amplification and library preparation, deeper sequencing coverage does not always result in improved accuracy [53]. Sequencing noise can be reduced by introducing special techniques during the library preparation and sequencing steps. Such techniques were recently presented, and some are already commercially used. In one study, a panel of 30 genes was used to detect paternal and de novo mutations [54]. To reduce sequence noise, a unique molecular indexing (UMI, or molecular barcoding) technique was used. In another study, synthesized DNA molecules (Quantitative Counting Templates, QCTs) were spiked into the cfDNA sample prior to amplification, to enable accurate counting in downstream analysis [53]. Although these techniques were tested over small NGS panels, they can be used over larger parts of the genome, and with other algorithms as well.
As the technology for genome-wide monogenic NIPD rapidly becomes available, clinical partners will be needed. The precedent of NIPD for other genetic conditions, e.g. aneuploidies and single point mutations, implies how genome-wide monogenic NIPD will develop. Presumably, in the near future, any consideration of prenatal WES/WGS should address the noninvasive alternative, while recognizing its advantages and limitations [55]. For example, while CVS and amniocentesis tests are only possible in specific time windows during pregnancy, NIPD may be performed at almost any stage of pregnancy. NIPD is also relevant to cases of low adherence to invasive methods, as it contributes high-confidence information before the decision to perform amniocentesis. Although NIPD is currently less accurate than invasive techniques, the fetal inheritance revealed by NIPD is more informative than traditional markers, as shown with Down Syndrome. In general, a non-risk test should first be suggested to patients, regardless of adherence. This, however, requires exploring the exact advantages and accuracy, which will need to be ascertained through clinical trials, and eventually using large cohorts.
4. Conclusion
The last decade has witnessed rapid advancements in genome-wide NIPD of monogenic diseases. Experience with other NIPD applications suggests that such approach will likely become more commonly used in the coming years, for translational research, and as a clinical tool for diagnosis. While prenatal WES of amniotic fluid or a CVS sample is presently becoming more accepted, the reliance on invasive methods remains a hindrance. Performing prenatal WES noninvasively is likely to yield greater demand and to accelerate its acceptance.
Genome-wide NIPD of monogenic diseases requires advanced NGS methodologies. As the sequencing costs continue to decrease, and algorithms improve, various approaches are becoming available for this task. Classical methods are based on attaining the haplotype information of the parents using various techniques and following the inheritance of haplotypes by the fetus. Subsequently developed methods are based on a site-by-site approach, in which the fetal inheritance is deduced at the level of a single nucleotide. The possibility has recently been shown of improving such approaches by implementation of principles from standard variant calling pipelines, and by using unique characteristics of the fetal-derived cfDNA (i.e. fragmentomics) in a probabilistic manner. This concept enables rapid development of noninvasive fetal genotyping to achieve complete genome-wide NIPD of monogenic diseases.
We expect that in the next few years, both haplotype-based and site-by-site NIPD approaches will continue to improve and eventually reach the clinic. To further advance current algorithms, especially the recent machine learning-based ones, larger amounts of data will be required. These will be based on the convergence of computational research and medical evaluations in an increasing effort by both researchers and clinicians.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. The Shomron Laboratory is supported by the Israel Science Foundation (ISF; 1852/16); Israeli Ministry of Defense, Office of Assistant Minister of Defense for Chemical, Biological, Radiological and Nuclear (CBRN) Defense; Foundation Fighting Blindness; The Edmond J. Safra Center for Bioinformatics at Tel Aviv University; Zimin Institute for Engineering Solutions Advancing Better Lives; Eric and Wendy Schmidt Breakthrough Innovative Research Award; Tel Aviv University Richard Eimert Research Fund on Solid Tumors; Djerassi-Elias Institute of Oncology; Canada-Montreal Friends of Tel Aviv University; Harold H. Marcus; Amy Friedkin; Natalio Garber; Kirschman Dvora Eleonora Fund for Parkinson's Disease; Joint funding between Tel Aviv University and Yonsei University; Israeli Ministry of Science and Technology, Israeli–Russia joint funding; Aufzien Family Center for the Prevention and Treatment of Parkinson’s Disease; and a generous donation from the Adelis Foundation.
References
- 1.Agarwal K., Alfirevic Z. Pregnancy loss after chorionic villus sampling and genetic amniocentesis in twin pregnancies: a systematic review. Ultrasound Obstet Gynecol. 2012;40(2):128–134. doi: 10.1002/uog.10152. [DOI] [PubMed] [Google Scholar]
- 2.Akolekar R., Beta J., Picciarelli G., Ogilvie C., D’Antonio F. Procedure-related risk of miscarriage following amniocentesis and chorionic villus sampling: a systematic review and meta-analysis. Ultrasound Obstet Gynecol. 2015;45(1):16–26. doi: 10.1002/uog.14636. [DOI] [PubMed] [Google Scholar]
- 3.Fan H.C., Blumenfeld Y.J., Chitkara U., Hudgins L., Quake S.R. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci USA. 2008;105(42):16266–16271. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lo Y.M.D., Lun F.M.F., Chan K.C.A., Tsui N.B.Y., Chong K.C., Lau T.K. Digital PCR for the molecular detection of fetal chromosomal aneuploidy. Proc Natl Acad Sci USA. 2007;104(32):13116–13121. doi: 10.1073/pnas.0705765104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hill M., Finning K., Martin P., Hogg J., Meaney C., Norbury G. Non-invasive prenatal determination of fetal sex: translating research into clinical practice. Clin Genet. 2011;80(1):68–75. doi: 10.1111/j.1399-0004.2010.01533.x. [DOI] [PubMed] [Google Scholar]
- 6.van der Meij K.R.M., Sistermans E.A., Macville M.V.E., Stevens S.J.C., Bax C.J., Bekker M.N. TRIDENT-2: national implementation of genome-wide non-invasive prenatal testing as a first-tier screening test in the Netherlands. Am J Human Genet. 2019;105(6):1091–1101. doi: 10.1016/j.ajhg.2019.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Srinivasan A., Bianchi D.W., Huang H., Sehnert A.J., Rava R.P. Noninvasive detection of fetal subchromosome abnormalities via deep sequencing of maternal plasma. Am J Hum Genet. 2013;92(2):167–176. doi: 10.1016/j.ajhg.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu T., Yeniterzi S., Rajkovic A., Hogge W.A., Dunkel M., Shaw P. High resolution non-invasive detection of a fetal microdeletion using the GCREM algorithm. Prenat Diagn. 2014;34(5):469–477. doi: 10.1002/pd.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neofytou M.C., Tsangaras K., Kypri E., Loizides C., Ioannides M., Achilleos A. Targeted capture enrichment assay for non-invasive prenatal testing of large and small size sub-chromosomal deletions and duplications. PLoS ONE. 2017;12(2) doi: 10.1371/journal.pone.0171319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finning K.M., Martin P.G., Soothill P.W., Avent N.D. Prediction of fetal D status from maternal plasma: introduction of a new noninvasive fetal RHD genotyping service. Transfusion. 2002;42(8):1079–1085. doi: 10.1046/j.1537-2995.2002.00165.x. [DOI] [PubMed] [Google Scholar]
- 11.Amicucci P., Gennarelli M., Novelli G., Dallapiccola B. Prenatal diagnosis of myotonic dystrophy using fetal DNA obtained from maternal plasma. Clin Chem. 2000;46(2):301–302. [PubMed] [Google Scholar]
- 12.Saito H., Sekizawa A., Morimoto T., Suzuki M., Yanaihara T. Prenatal DNA diagnosis of a single-gene disorder from maternal plasma. Lancet. 2000;356(9236):1170. doi: 10.1016/S0140-6736(00)02767-7. [DOI] [PubMed] [Google Scholar]
- 13.Dennis Lo Y.M., Chiu R.W.K. Prenatal diagnosis: progress through plasma nucleic acids. Nat Rev Genet. 2007;8(1):71–77. doi: 10.1038/nrg1982. [DOI] [PubMed] [Google Scholar]
- 14.Ashoor G., Syngelaki A., Poon L.C.Y., Rezende J.C., Nicolaides K.H. Fetal fraction in maternal plasma cell-free DNA at 11–13 weeks’ gestation: relation to maternal and fetal characteristics. Ultrasound Obstet Gynecol. 2013;41(1):26–32. doi: 10.1002/uog.12331. [DOI] [PubMed] [Google Scholar]
- 15.Lun F.M.F., Tsui N.B.Y., Chan K.C.A., Leung T.Y., Lau T.K., Charoenkwan P. Noninvasive prenatal diagnosis of monogenic diseases by digital size selection and relative mutation dosage on DNA in maternal plasma. Proc Natl Acad Sci USA. 2008;105(50):19920–19925. doi: 10.1073/pnas.0810373105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perlado S., Bustamante-Aragonés A., Donas M., Lorda-Sánchez I., Plaza J., de Alba M.R. Fetal genotyping in maternal blood by digital PCR: towards NIPD of monogenic disorders independently of parental origin. PLoS ONE. 2016;11(4) doi: 10.1371/journal.pone.0153258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsui N.B.Y., Kadir R.A., Chan K.C.A., Chi C., Mellars G., Tuddenham E.G. Noninvasive prenatal diagnosis of hemophilia by microfluidics digital PCR analysis of maternal plasma DNA. Blood. 2011;117(13):3684–3691. doi: 10.1182/blood-2010-10-310789. [DOI] [PubMed] [Google Scholar]
- 18.Barrett A.N., McDonnell T.C.R., Chan K.C.A., Chitty L.S. Digital PCR analysis of maternal plasma for noninvasive detection of sickle cell anemia. Clin Chem. 2012;58(6):1026–1032. doi: 10.1373/clinchem.2011.178939. [DOI] [PubMed] [Google Scholar]
- 19.Vodo D., Sarig O., Jeddah D., Malchin N., Eskin-Schwarz M., Mohamad J. Punctate palmoplantar keratoderma: an unusual mutation causing an unusual phenotype. Br J Dermatol. 2018;178(6):1455–1457. doi: 10.1111/bjd.16502. [DOI] [PubMed] [Google Scholar]
- 20.Mohamad J., Sarig O., Godsel L.M., Peled A., Malchin N., Bochner R. Filaggrin 2 deficiency results in abnormal cell-cell adhesion in the cornified cell layers and causes peeling skin syndrome type A. J Invest Dermatol. 2018;138(8):1736–1743. doi: 10.1016/j.jid.2018.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malki L., Sarig O., Romano M.-T., Méchin M.-C., Peled A., Pavlovsky M. Variant PADI3 in central centrifugal cicatricial alopecia. N Engl J Med. 2019;380(9):833–841. doi: 10.1056/NEJMoa1816614. [DOI] [PubMed] [Google Scholar]
- 22.Tatour Y., Tamaiev J., Shamaly S., Colombo R., Bril E., Rabinowitz T. A novel intronic mutation of PDE6B is a major cause of autosomal recessive retinitis pigmentosa among Caucasus Jews. Mol Vis. 2019;25:155–164. [PMC free article] [PubMed] [Google Scholar]
- 23.Mohamad J, Sarig O, Malki L, Rabinowitz T, Assaf S, Malovitski K, et al. Loss-of-function variants in SERPINA12 underlie autosomal recessive palmoplantar keratoderma. Journal of Investigative Dermatology [Internet]. 2020 [cited 2020 Apr 7]; Available from: http://www.sciencedirect.com/science/article/pii/S0022202X20312549. [DOI] [PubMed]
- 24.Lord J., McMullan D.J., Eberhardt R.Y., Rinck G., Hamilton S.J., Quinlan-Jones E. Prenatal exome sequencing analysis in fetal structural anomalies detected by ultrasonography (PAGE): a cohort study. Lancet. 2019;393(10173):747–757. doi: 10.1016/S0140-6736(18)31940-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petrovski S., Aggarwal V., Giordano J.L., Stosic M., Wou K., Bier L. Whole-exome sequencing in the evaluation of fetal structural anomalies: a prospective cohort study. Lancet. 2019;393(10173):758–767. doi: 10.1016/S0140-6736(18)32042-7. [DOI] [PubMed] [Google Scholar]
- 26.Lo YMD, Chan KCA, Sun H, Chen EZ, Jiang P, Lun FMF, et al. Maternal Plasma DNA Sequencing Reveals the Genome-Wide Genetic and Mutational Profile of the Fetus. Science Translational Medicine. 2010;2(61):61ra91–61ra91. [DOI] [PubMed]
- 27.Fan H.C., Gu W., Wang J., Blumenfeld Y.J., El-Sayed Y.Y., Quake S.R. Non-invasive prenatal measurement of the fetal genome. Nature. 2012;487(7407):320–324. doi: 10.1038/nature11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitzman JO, Snyder MW, Ventura M, Lewis AP, Qiu R, Simmons LE, et al. Noninvasive whole-genome sequencing of a human fetus. Sci Transl Med 2012;4(137):137ra76. [DOI] [PMC free article] [PubMed]
- 29.Chan L.L., Jiang P. Bioinformatics analysis of circulating cell-free DNA sequencing data. Clin Biochem. 2015;48(15):962–975. doi: 10.1016/j.clinbiochem.2015.04.022. [DOI] [PubMed] [Google Scholar]
- 30.Chen S., Ge H., Wang X., Pan X., Yao X., Li X. Haplotype-assisted accurate non-invasive fetal whole genome recovery through maternal plasma sequencing. Genome Med. 2013 Feb 27;5(2):18. doi: 10.1186/gm422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hui W.W.I., Jiang P., Tong Y.K., Lee W.-S., Cheng Y.K.Y., New M.I. Universal haplotype-based noninvasive prenatal testing for single gene diseases. Clin Chem. 2017;63(2):513–524. doi: 10.1373/clinchem.2016.268375. [DOI] [PubMed] [Google Scholar]
- 32.Wei X., Lv W., Tan H., Liang D., Wu L. Development and validation of a haplotype-free technique for non-invasive prenatal diagnosis of spinal muscular atrophy. J Clin Lab Anal. 2020;34(2) doi: 10.1002/jcla.23046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vermeulen C., Geeven G., de Wit E., Verstegen M.J.A.M., Jansen R.P.M., van Kranenburg M. Sensitive monogenic noninvasive prenatal diagnosis by targeted haplotyping. Am J Hum Genet. 2017 Sep 7;101(3):326–339. doi: 10.1016/j.ajhg.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan K.C.A., Jiang P., Sun K., Cheng Y.K.Y., Tong Y.K., Cheng S.H. Second generation noninvasive fetal genome analysis reveals de novo mutations, single-base parental inheritance, and preferred DNA ends. Proc Natl Acad Sci USA. 2016;31:201615800. doi: 10.1073/pnas.1615800113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics. 2014;30(20):2843–2851. doi: 10.1093/bioinformatics/btu356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yin X., Du Y., Zhang H., Wang Z., Wang J., Fu X. Identification of a de novo fetal variant in osteogenesis imperfecta by targeted sequencing-based noninvasive prenatal testing. J Hum Genet. 2018;63(11):1129–1137. doi: 10.1038/s10038-018-0489-9. [DOI] [PubMed] [Google Scholar]
- 37.Rabinowitz T., Polsky A., Golan D., Danilevsky A., Shapira G., Raff C. Bayesian-based noninvasive prenatal diagnosis of single-gene disorders. Genome Res. 2019 doi: 10.1101/gr.235796.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mullaney J.M., Mills R.E., Pittard W.S., Devine S.E. Small insertions and deletions (INDELs) in human genomes. Hum Mol Genet. 2010;19(R2):R131–R136. doi: 10.1093/hmg/ddq400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neuman J.A., Isakov O., Shomron N. Analysis of insertion-deletion from deep-sequencing data: software evaluation for optimal detection. Brief Bioinf. 2013;14(1):46–55. doi: 10.1093/bib/bbs013. [DOI] [PubMed] [Google Scholar]
- 40.Belkadi A., Bolze A., Itan Y., Cobat A., Vincent Q.B., Antipenko A. Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants. Proc Natl Acad Sci USA. 2015;112(17):5473–5478. doi: 10.1073/pnas.1418631112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Che H, Villela D, Dimitriadou E, Melotte C, Brison N, Neofytou M, et al. Noninvasive prenatal diagnosis by genome-wide haplotyping of cell-free plasma DNA. Genet Med [Internet]. 2020 Feb 6 [cited 2020 Apr 2]; Available from: http://www.nature.com/articles/s41436-019-0748-y. [DOI] [PubMed]
- 42.Non-invasive Prenatal Diagnosis of Monogenic Disorders by Linked-reads Technology – Full Text View – ClinicalTrials.gov [Internet]. [cited 2020 Mar 30]. Available from: https://clinicaltrials.gov/ct2/show/NCT03622892.
- 43.Hardy T. The role of prenatal diagnosis following preimplantation genetic testing for single-gene conditions: a historical overview of evolving technologies and clinical practice. Prenatal Diagnosis [Internet]. [cited 2020 Mar 22];n/a(n/a). Available from: https://obgyn.onlinelibrary.wiley.com/doi/abs/10.1002/pd.5662. [DOI] [PubMed]
- 44.1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. [DOI] [PMC free article] [PubMed]
- 45.Hwang S., Kim E., Lee I., Marcotte E.M. Systematic comparison of variant calling pipelines using gold standard personal exome variants. Sci Rep. 2015;7(5):17875. doi: 10.1038/srep17875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hahn S., Rusterholz C., Hösli I., Lapaire O. Cell-free nucleic acids as potential markers for preeclampsia. Placenta. 2011;32(Suppl):S17–S20. doi: 10.1016/j.placenta.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 47.Zhong X.Y., Laivuori H., Livingston J.C., Ylikorkala O., Sibai B.M., Holzgreve W. Elevation of both maternal and fetal extracellular circulating deoxyribonucleic acid concentrations in the plasma of pregnant women with preeclampsia. Am J Obstet Gynecol. 2001;184(3):414–419. doi: 10.1067/mob.2001.109594. [DOI] [PubMed] [Google Scholar]
- 48.Lazar L., Rigó J., Nagy B., Balogh K., Makó V., Cervenak L. Relationship of circulating cell-free DNA levels to cell-free fetal DNA levels, clinical characteristics and laboratory parameters in preeclampsia. BMC Med Genet. 2009;10(1):120. doi: 10.1186/1471-2350-10-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carlson L.M., Hardisty E., Coombs C.C., Vora N.L. Maternal malignancy evaluation after discordant cell-free DNA results. Obstet Gynecol. 2018;131(3):464–468. doi: 10.1097/AOG.0000000000002474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thurik F.F., Ruiter M.L., Javadi A., Kwee A., Woortmeijer H., Page-Christiaens G.C.M.L. Absolute first trimester cell-free DNA levels and their associations with adverse pregnancy outcomes. Prenat Diagn. 2016;36(12):1104–1111. doi: 10.1002/pd.4940. [DOI] [PubMed] [Google Scholar]
- 51.Luo Y., Jia B., Yan K., Liu S., Song X., Chen M. Pilot study of a novel multi-functional noninvasive prenatal test on fetus aneuploidy, copy number variation, and single-gene disorder screening. Mol Genet Genomic Med. 2019;7(4) doi: 10.1002/mgg3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scotchman E., Chandler N.J., Mellis R., Chitty L.S. Noninvasive prenatal diagnosis of single-gene diseases: the next frontier. Clin Chem. 2020;66(1):53–60. doi: 10.1373/clinchem.2019.304238. [DOI] [PubMed] [Google Scholar]
- 53.Tsao D.S., Silas S., Landry B.P., Itzep N.P., Nguyen A.B., Greenberg S. A novel high-throughput molecular counting method with single base-pair resolution enables accurate single-gene NIPT. Sci Rep. 2019;9(1):1–14. doi: 10.1038/s41598-019-50378-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang J., Li J., Saucier J.B., Feng Y., Jiang Y., Sinson J. Non-invasive prenatal sequencing for multiple Mendelian monogenic disorders using circulating cell-free fetal DNA. Nat Med. 2019;25(3):439. doi: 10.1038/s41591-018-0334-x. [DOI] [PubMed] [Google Scholar]
- 55.Best S., Wou K., Vora N., Van der Veyver I.B., Wapner R., Chitty L.S. Promises, pitfalls and practicalities of prenatal whole exome sequencing. Prenat Diagn. 2018;38(1):10–19. doi: 10.1002/pd.5102. [DOI] [PMC free article] [PubMed] [Google Scholar]