Abstract
The initial colonization of the neonatal intestinal tract is influenced by delivery mode, feeding, the maternal microbiota, and a host of environmental factors. After birth, the composition of the infant’s microbiota undergoes a series of significant changes particularly in the first weeks and months of life ultimately developing into a more stable and diverse adult-like population in childhood. Intestinal dysbiosis is an alteration in the intestinal microbiota associated with disease and appears to be common in neonates. The consequences of intestinal dysbiosis are uncertain, but strong circumstantial evidence and limited confirmations of causality suggest that dysbiosis early in life can influence the health of the infant acutely, as well as contribute to disease susceptibility later in life.
Subject terms: Intestinal diseases, Inflammatory diseases
Introduction
The intestinal microbiota consists of trillions of living microbes that co-exist with the host. Bacteria represent the most abundant colonizers, but archaea, fungi, and viruses are also abundant in healthy individuals. The complexity of interactions between the triad of microbiota, environmental factors, and the host is underscored by evidence that this massive microbial community (1) competes for and generates nutrients, (2) influences and is shaped by the host innate and adaptive systems, and (3) protects against and triggers acute and chronic disease. Mounting evidence further suggests that the intestinal microbiota influences mucus production, maturation and continuity of the epithelial monolayer separating the intestinal lumen from the submucosa, essential qualities of inflammatory responses, and physiological homeostasis [1]. Interactions between gut microbes and the developing immune system appear to be particularly important early in life [2].
The paradigm that intestinal microbes easily fit into circumscribed categories such as symbionts or mutualists (both the host and the microbe benefit from this co-existence), commensals (the microbe benefits and the host is unharmed), and pathogens (the microbe benefits and the host is harmed) is being replaced because of an understanding that cooperative factors that allow nonpathogenic microbes to colonize and benefit the host share essential qualities similar to virulence factors that can drive pathogenicity [3, 4]. Thus, many intestinal microbes are more precisely characterized as pathobionts, capable of causing harm in some situations while being tolerated (or even beneficial) in others.
Intestinal dysbiosis describes alterations in the intestinal microbiota associated with disease. In some cases, the compositional shift and associated phenotype can be ascribed to changes in a small collection of microbes. Disease causality for such changes can be tested in model systems by demonstrating transmissibility of host disease phenotype with microbiota transfer [5, 6]. In other cases, an association is more nuanced. Measures of dysbiosis often cannot confirm a causal role of the specific microbiome pattern in a specific disease [7–9]. Dysbiosis may be the result of disease, or merely coincident, rather than causative. In addition, in newborns the converse maybe true. Alterations that are not immediately associated with pathology may potentially have far reaching consequences later in childhood via microbiome–immune interactions. Herein, we review the importance of the intestinal microbiota in neonatal development and the consequences of alterations in this complex microbial community in newborns.
Dysbiosis and development
Injury to an organ during its morphogenesis can have consequences for both organ recovery and development, as well as on function of downstream systems reliant on that organ. Formative years for the microbiome include the first 3 years of life with age-dependent shifts in dominant taxa driven by environment, diet, and medication [10–12]. The intestinal immune system develops in parallel with the child’s microbiome and is affected by both structural components of microbes and products of microbial metabolism [12–14]. The importance of this early period is demonstrated in animal models where microbiome disruption during the early life window causes specific immune/health alterations that are not elicited when the disruption is triggered at a mature age [15, 16].
Animal models have been helpful in analyzing the impact of the intestinal microbiota on development, particularly experiments in animals raised in germ-free conditions. The impact of the microbiota begins in utero as evidenced by the observation that offspring of germ-free dams are at high risk for metabolic syndrome even if raised in conventional conditions [17]. Mice raised in germ-free conditions have poor growth, decreased weight and mucosal surface area of the small intestine, decreased intestinal motility, increased mucus production, increased size of the cecum and mild diarrhea [18]. Germ-free mice also demonstrate altered neurodevelopment with deficiencies in stress responsivity, anxiety-like behaviors, sociability, and cognition [19]. The elegance of this approach is fully evident with colonization of germ-free animals with single microbes or specific microbial communities (gnotobiotic or “known microbiota”) and analysis of the resulting phenotype. Comparisons of germ-free, gnotobiotic, and wild-type animals demonstrate the importance of the intestinal microbiota in intestinal development [20] and an essential causal role of intestinal dysbiosis in severe acute malnutrition [21], obesity [22], nonalcoholic liver disease [23], Alzheimer’s disease [24], and colitis-associated colorectal cancer [25], to name a few.
Prevalence of neonatal intestinal dysbiosis
To address the question of how commonly intestinal dysbiosis occurs in early life presumes an understanding of the “normal” microbiota of the infant which unfortunately is lacking. The following assumptions seem reasonable: (1) microbial communities are dynamic and shaped by host factors, environmental factors, and available nutrients, (2) the composition of the intestinal microbiota of our ancestors’ neonates was shaped by the maternal vaginal, fecal, oral, and milk microbiota, by nutrients available in human milk, and by horizontal transmission within families and small communities that shared food, eating utensils and toileting facilities, and (3) the introduction of modern hygiene practices, antibiotics, antiseptics, and other medications, modern diets such as the so-called “Western” high fat/high sugar diet, and infant formula has fundamentally changed the intestinal microbiota of humans from that of our ancestors.
The earliest attempts to describe the fecal microbiota of the breast-fed infant were published by Tissier [26] and Logan [27]. Both described a predominance of Gram-positive, anaerobic, Y-shaped, nonmotile bacteria in healthy infants. These bacteria were characterized as a single species, Lactobacillus bifidus, until the 1960s when the multiple species were grouped in a single genus Bifidobacterium (family Bifidobacteriaceae, order Bifidobacteriales, phylum Actinobacteria). Bifidobacterium species selectively ferment carbohydrates; some produce the glycosidases necessary to digest plant oligosaccharides (e.g., Bifidobacterium adolescentis, B. longum subsp longum) and others the glycosidases necessary to digest milk oligosaccharides (e.g., B. longum subsp infantis, B. bifidum and some strains of B. breve). These differences are encoded in the bacterial genome and have been confirmed by biochemical and molecular characterization of bacterial glycosidases and transport proteins [28–30]. Microbes capable of consuming milk components have a competitive advantage in the infant gut. Given the abundance of oligosaccharides in human milk, the relatively few species of bacteria capable of consuming these glycans, such as those mentioned from the Bifidobacterium genus, can multiply and their abundance “blooms.” The observation that mothers produce large quantities of human milk oligosaccharides with vast structural diversity that do not serve as a direct nutrition source for the infant, given their lack of the necessary glycosidases, suggests either co-evolution of human milk and a limited number of bacterial species millions of years ago, or a very clever design, or perhaps some of both. The end result is that the mother is shaping the microbiota of her infant through selective nutrients and antimicrobials in her milk (Fig. 1).
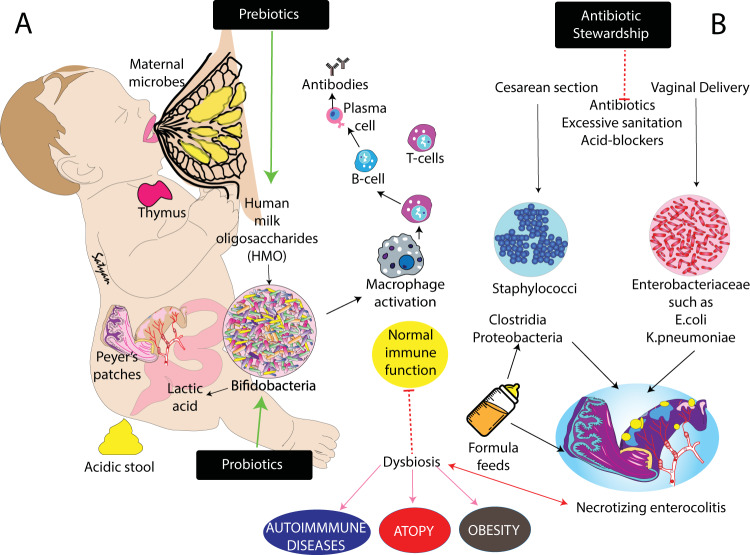
Fig. 1. Neonatal intestinal dysbiosis and disease risk.
a Normal colonization of the infant gut is shaped by environmental factors, maternal microbes and breast milk (human milk oligosaccharides). Colonizing microbes influence development of the intestinal innate and adaptive immune systems. b Several factors result in dysbiosis increasing risk for neonatal conditions such as necrotizing enterocolitis and long-term health issues such as autoimmune diseases, atopy and obesity. Copyright Satyan Lakshminrusimha.
However, this ancient mechanism of maternal influence on infant microbiota via breastmilk, may be weakened by modern practices. A recent review of 14 studies published between 1926 and 2017, which included more than 300 healthy breastfed infants, documented over that time period an increase in fecal pH from 5.0 to 6.5 [31]. This change in pH is consistent with a decrease in Bifidobacteriaceae (which produce lactic acid and lower fecal pH) and increases in Clostridiaceae and Enterobacteriaceae (both associated with increased fecal pH).
Such changes could have immediate health consequences (Figs. 1, 2). For example, the mentioned Enterobacteriaceae (phylum Proteobacteria) are Gram-negative facultative anaerobes that include two species well known to neonatologists: Escherichia coli and Klebsiella pneumoniae. Virulence factors include flagella that provide motility, fimbriae that allow attachment to the host cell surface, and toxins that interfere with homeostasis. Many Enterobacteriaceae trigger a host immune response through recognition of microbial patterns such as lipopolysaccharide in the cell wall or flagella. E. coli and K. pneumoniae, in particular, can cause devastating overt disease in neonates. An abundance of Enterobacteriaceae has been described as one hallmark of dysbiosis [32].
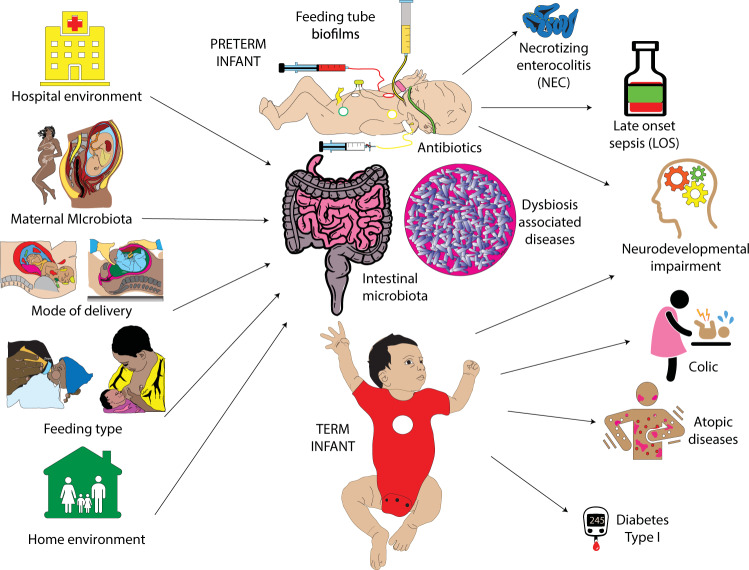
Fig. 2. Contributors and consequences of intestinal dybiosis.
Factors influencing the intestinal microbiota in preterm and term infants are presented at the left and top of the figure with diseases associated with intestinal dybiosis to the right.
Studies of the fecal microbiota of the healthy term infant have shown recognizable patterns although with a high degree of individual variation. The broad patterns now emerging suggest a first wave of bacterial colonists influenced by mode of delivery (Enterobacteriaceae in babies born vaginally and Staphylococcaceae in babies born by cesarean section) and a second wave influenced by feeding type (Bifidobacteriaceae in breast-fed infants and a more diverse population in formula-fed infants including Staphylococcaceae, Clostridiaceae, Enterococcaceae, Bifidobacteriaceae, and Bacteroidaceae) [33]. In the very preterm infant, the pattern is more complex (again with a high degree of individual variation): initial colonization with Staphylococcaceae followed by Enterobacteriaceae and then Clostrideaceae with Bifidobacteriaceae and Lactobacillaceae characteristically delayed or absent [34, 35]. In both term and preterm infants, cessation of breastfeeding and introduction of complementary foods begin a steady increase in microbial diversity resulting in a more complex microbiota, ultimately resulting in stable communities characteristic of adults.
In broad terms, the presence of low numbers of Bifidobacteriaceae and high numbers of Enterobacteriaceae and Clostridiaceae can serve as a reasonable starting place to define dysbiosis in the first months of life. This pattern is common in several settings, including very preterm infants, term infants requiring prolonged hospitalization or antibiotics, and in well-appearing infants in some geographic areas with increased risk for dysbiosis-associated diseases [36]. Microbiota composition “immaturity” is a term coined for a delay of the patterned progression of the microbiota. It is characteristic of severe acute malnutrition in children [37] and may be another useful definition of dysbiosis in the infant [38–40].
Causes of neonatal intestinal dysbiosis
As noted above, in term infants, both mode of delivery and feeding type (human milk vs. formula vs. a combination) significantly influence the composition of the infant microbiota [41, 42]. Differences in the infant microbiota with home delivery vs. hospital delivery have been reported [43]. The mother’s diet and the microbes in her milk (generally similar to the microbiota of maternal skin and feces) also influence the infant fecal microbiota though generally to a modest extent [44–46]. Many other factors can influence the microbiota and some can cause dysbiosis. Exposure to intrapartum antibiotics for prevention of Group B streptococcal sepsis and/or due to cesarean section is extremely common and impacts the neonatal intestinal microbiota [47, 48]. In addition to antibiotics [49], other medications commonly administered to mothers and/or infants that alter the intestinal microbiota include acid blocking agents [50, 51], selective serotonin reuptake inhibitors, metformin and laxatives [52, 53]. Recent demonstrations of altered infant microbiota with exposure to environmental toxins [54, 55], maternal smoking [56], and proximity to furry animals [42, 57] underscore the many factors affecting this community.
Very preterm infants are a unique population in that they are “new” from an evolutionary standpoint, have prolonged hospital stays (with multiple environmental exposures), are frequently exposed to antibiotics, and have immature innate and adaptive immune systems. In very preterm infants, the primary determinants of the composition of the intestinal microbiota are the post-menstrual age and age in weeks [34, 35], however, multiple other factors are also influential including antenatal corticosteroids, mode of delivery, antibiotic exposure, feeding type, feeding tube dwell time and biofilms, gender, and stress [58–61]. Table 1 presents several examples of alterations in the infant fecal microbiota related to perinatal exposures [43, 47–49, 56, 57, 60, 62–67].
Table 1.
Alterations in the infant fecal microbiota related to perinatal exposures.
| Intervention | Change in the neonatal intestinal microbiota and metabolites |
|---|---|
| Intrapartum antibiotics [47, 48, 62] | ↓ Bacteroides and Bacteroidetes |
| ↓ Parabacteroides | |
| ↓ Bifidobacterium and Actinobacteria | |
| ↑ Proteobacteria | |
| ↑ Veillonella, Enterococcus and Firmicutes | |
| ↑ Clostridia | |
| ↓ Alpha diversity | |
| ↓ Fecal acetate | |
| ↑ Antibiotic resistance genes | |
| Postnatal antibiotics (term infant) [49] | ↓ Bacteroidetes |
| Postnatal antibiotics (preterm) [63, 64] | ↓ Alpha diversity |
| ↑ Antibiotic resistance genes | |
| Cesarean delivery [65] | ↓ Actinobacteria |
| ↓ Bacteroidetes | |
| ↑ Firmicutes | |
| Home birth [43] | ↑ Bifidobacterium |
| ↑ Bacteroides | |
| ↑ Streptococcus | |
| ↑ Lactobacillus | |
| ↓ Clostridium | |
| ↓ Enterobacteriaceae | |
| Very preterm birth [66] | ↑ Proteobacteria |
| ↓ Firmicutes | |
| ↓ Bifidobacterium | |
| ↓ Short chain fatty acids | |
| Mother’s own milk [60] | ↑ Bifidobacteriaceae |
| ↓ Staphylococcaceae | |
| ↓ Clostridiaceae | |
| ↓ Pasteurellaceae | |
| Maternal smoking [56] | ↑ Ruminococcus |
| ↑ Akkermansia | |
| ↑ Bacteroides | |
| ↑ Staphylococcus | |
| Furry pet in the home [57] | ↓ Streptococcaceae |
| ↑ Oscillospira and Ruminococcus | |
| Disinfectant [67] | ↑ Lachnospiraceae |
| ↓ Haemophilus | |
| Eco-friendly disinfectant [67] | ↓ Enterobacteriaceae |
A dysbiosis index
A “dysbiosis index” has been described for new onset pediatric Crohn’s disease (CD) based on the ratio of bacteria increased in patients with CD (Enterobacteriaceae, Pasteurellacaea, Veillonellaceae, Fusobacteriaceae, Neisseriaceae, and Gemellaceae) to the bacteria decreased in patients with CD (Erysipelotrichales, Bacteroidales, Clostridiales, and Bifidobacteriaceae) [7, 68]. This index has diagnostic validity and correlated with symptom severity but did not predict response to treatment. Gastrointestinal dysbiosis indices have subsequently been described with varying diagnostic and prognostic value for stroke [69] and Helicobacter pylori infection [70]. A commercially available GA-map™ Dysbiosis Test (Genetic Analysis, Oslo, Norway) determines the degree of deviation of the microbiota composition of a patient sample compared with a healthy control adult population and generates a dysbiosis index score that correlates with fecal calprotectin (a marker of intestinal inflammation) [8]. An elevated dysbiosis index with this product has been demonstrated in adults with ankylosing spondylitis [8], irritable bowel syndrome, inflammatory bowel disease [71], and Sjogren’s syndrome [72]. A dysbiosis index has not yet been described for diseases affecting the term or preterm infant. Given the discussion above of patterned progression of the microbiota, a dysbiosis index for neonates would need to incorporate gestational age and age at the time of stool collection.
Consequences of neonatal intestinal dysbiosis
The list of disease processes associated with intestinal dysbiosis is long and diverse (Table 2 provides several examples). For the majority, a causal link between intestinal dysbiosis and the dysbiosis-associated disease has not been established, however, for some causality and underlying mechanisms have been demonstrated. Examples of the latter include type 1 diabetes [73, 74] and atopic disease/asthma [75, 76]. Studies of associations between antibiotic exposure in utero or in the perinatal period and the diseases of childhood obesity and the triad of atopic dermatitis, asthma and allergic rhinitis have been mixed [77–80]. Among diseases of infants, compelling data for an association with intestinal dysbiosis come from studies of NEC, late onset sepsis (LOS) and infant colic (Fig. 2).
Table 2.
Disease processes associated with intestinal dysbiosis.
| Acute/subacute | Chronic |
| Necrotizing enterocolitis in preterm infants | Type 1 diabetes |
| Late onset neonatal sepsis | Type 2 diabetes |
| Antibiotic associated diarrhea | Chronic kidney disease |
| Clostridium difficile colitis | Inflammatory bowel disease |
| Infant colic | Dyslipidemia |
| Severe acute malnutrition | Coronary artery disease |
| Breast cancer | |
| Colon cancer | |
| Atopic dermatitis | |
| Non-alcoholic fatty liver | |
| Parkinson’s disease | |
| Alzheimer’s disease | |
| Obesity | |
| Depression | |
| Schizophrenia |
For NEC, careful studies of the fecal microbiota prior to disease onset demonstrated a pattern at the phylum level of increased Proteobacteria and decreased Firmicutes and Bacteroidetes [81]. Delving deeper, increased Enterobacteriaceae at the family level [82], and Klebsiella at the genus level were reported as well as increased fimbriae-expressing bacteria [82]. Several studies have demonstrated that empiric treatment of the preterm infant with antibiotics for more than 4 days increased risk of later development of NEC [83, 84]. Administration of acid-blocking agents to preterm infants is also associated with both fecal dysbiosis [50] and increased risk of NEC [85]. It is likely that dysbiosis in hospitalized infants is influenced by the microbes on NICU surfaces [86, 87] and may explain some of the wide variation in NEC incidence between hospitals. A recent careful analysis demonstrated a NICU “room specific microbiota” that is shaped by the infant gut microbes [88].
For LOS, the case for intestinal dysbiosis in disease pathogenesis is less clear. Most LOS in the NICU has been attributed to infections involving skin microbes and central catheters. However, recent investigation of LOS identified identical organisms in the feces and blood cultures of infants with LOS, which suggests that translocation of gut microbes could be a common cause of LOS [89]. In rodent models, an abundance of specific intestinal microbes is protective against induced sepsis, while their absence is associated with increased disease severity [90]. In preterm infants, an abundance of Bifidobacterium species appears to be protective, or a marker of protection, against LOS [89, 91].
Lastly, infant colic is common and associated with increased risk of long term learning and behavioral challenges [92]. Several studies have demonstrated an association between intestinal dysbiosis and infant colic, including association of specific taxa with severity [93].
Mechanisms
A detailed discussion of mechanisms by which intestinal dysbiosis triggers local and systemic disease is beyond the scope of this review and has been reviewed by others [94–102]. Rather, we will touch briefly on well-established mechanisms and some promising novel proposed mechanisms and then consider in more detail the role of short chain fatty acids produced by probiotic and commensal bacteria. Key well-established mechanistic demonstrations, include (1) alteration of intestinal permeability by gut microbes [103], (2) alteration of local and systemic inflammation by gut microbes (e.g., through stimulation of Toll-like receptors and related cytokines and through alteration of T cell and neutrophil responses) [13, 104], and (3) alteration of intestinal maturation by gut microbes and their metabolites [105, 106]. More recent discoveries that shed light on mechanisms by which gut microbes impact host disease susceptibility, include (1) the alteration by gut microbes of passage of antigens from the intestinal lumen to the lamina propria through goblet cell-associated passages [107], (2) the induction by early life gut microbes of erythroid differentiation regulator-1 (Erdr1) a regulator of cellular differentiation and survival that is not detected in mice raised in germ-free conditions or in germ-free mice colonized with fecal microbiota from control mice at weaning, suggesting that there is a window of development early in life in which the microbiota (directly or via metabolites) can induce Erdr1 to alter intestinal development and capacity for regenerative repair [108], and (3) the stimulation by gut microbes of production by the host enterochromaffin cells of serotonin, the primary neurotransmitter of the enteric nervous system [109].
Short chain fatty acids (SCFAs, most commonly acetate, propionate, and butyrate) are the products of fermentation of dietary fiber by several gut bacteria (Table 3). SCFAs are an important energy source for colonocytes [32] and modulate insulin sensitivity, glucose and lipid homeostasis, and systemic inflammation [110, 111]. A recent large scale analysis of genotype, fecal microbiota, and fecal SCFA analysis in adults demonstrated causal associations between increased fecal butyrate and improved insulin response after oral glucose tolerance testing and between decreased fecal propionate and increased risk of type 2 diabetes [112]. SCFAs impact glucose metabolism through induction of intestinal gluconeogenesis [113], however, the role of SCFAs in energy metabolism appears to be even broader with reported effects of SCFAs on appetite regulation and the gut–brain axis [114]. In mice, SCFAs produced by maternal gut microbes influence differentiation of the neural, intestinal, and pancreatic cells of the embryo through activation of embryonic G protein-coupled receptors [17]. In rabbits, the cecal microbiota is altered early in the transition from suckling to weaning resulting in increased production of butyrate which impacts intestinal permeability [105].
Table 3.
Bacteria that produce short chain fatty acids and lactate.
| Genus (phylum) | SFCA |
|---|---|
| Bifidobacterium (Actinobacteria) | Acetate, lactate |
| Akkermansia (Verrucomicrobia) | Acetate, propionate |
| Faecalibacterium (Firmicutes) | Butyrate |
| Clostridium (Firmicutes) | Butyrate |
| Eubacterium (Firmicutes) | Butyrate |
| Roseburia (Firmicutes) | Butyrate |
| Bacteroides (Bacteroidetes) | Acetate, propionate |
| Lactobacillus (Firmicutes) | Lactate |
Patients with CD and ulcerative colitis have low numbers of SCFA-producing bacteria and low levels of fecal SCFAs. Mechanisms by which SCFAs protect the host against inflammatory bowel disease include support of enterocyte proliferation, induction of tight junction proteins to increase barrier function, induction of antimicrobial peptides, and a range of anti-inflammatory effects [115].
SCFAs and lactic acid are produced in abundance when human milk is combined with a microbe that can consume human milk oligosaccharides, such as B. longum ssp infantis which results in decreased fecal pH and suppression of facultative anaerobes such as Enterobacteriaceae [116]. The immature intestine may respond differently to SCFAs than the mature intestine. While SCFAs have demonstrated anti-inflammatory effects on human fetal organoids and fetal mouse intestine [117], butyrate appears to trigger NEC-like lesions in germ-free lactose-fed quails [118] and in a piglet NEC model, formula containing fermented E. coli had high levels of acetate and propionate and was associated with NEC-like lesions [119]. Tryptophan in human milk is metabolized by gut microbes, including B. longum ssp infantis, into indole-3-lactic acid, which binds to the aryl-hydrocarbon receptor in immature intestinal tissue and inhibits the pro-inflammatory cytokine IL8 [120].
Attempts to attenuate neonatal intestinal dysbiosis
Antibiotic stewardship
In the United States, 30–35% of term infants are exposed to antibiotics in the immediate perinatal period, either for Group B Streptococcus prophylaxis or concerns for maternal intra-amniotic infection (Fig. 1) [121]. This approach has decreased infant mortality, however, the long-term consequences of the resultant intestinal dysbiosis are uncertain. Unfortunately, a much-anticipated maternal vaccination against Group B Streptococcus and key strains of E. coli [122] does not appear imminent but would potentially lead to decreased antibiotic exposure. Minimizing unnecessary antibiotic exposure in the first months of life may have value in decreasing risk for dysbiosis and its associated disease processes as well as decreasing costs and emergence of antibiotic resistant bacteria [123]. A review of 127 California NICUs showed a wide range of antibiotic usage (from 2.4 to 97.1% of patient-days) with a median of 24% of patient days. NICUs with high antibiotic usage had similar rates of documented infection to NICUs with low antibiotic usage [124] suggesting an opportunity to decrease antibiotic administration. A follow-up study by the same group showed a 22% decline in NICU antibiotic use from 2013 to 2016 (42,960 fewer antibiotic days) with a greater decline in NICUs with an established antibiotic stewardship program [125]. This and other studies suggest that further benefits may be gained by antibiotic stewardship [123].
Prebiotics
Prebiotic glycans are dietary supplements that provide a nutrient substrate to stimulate growth of commensal bacteria. Human milk oligosaccharides, discussed above, are highly selective in that only a limited number of bacteria can utilize them as a source of nutrition (predominantly Bifidobacterium and Bacteroides species). In contrast, many other commercial prebiotics are consumed by a variety of gut microbes and thus may stimulate the growth of both commensals and pathogens. Human milk oligosaccharides are highly tolerant of heat and therefore resistant to pasteurization (and abundant in donor human milk). While the benefits of human milk are related to multiple bioactive components, it is likely that the prebiotic effects of the complex carbohydrates are at least partly responsible for observed decreases in NEC with human milk feeding. A recent meta-analysis of 18 randomized placebo controlled trials showed a decrease in the incidence of sepsis and death and length of hospital stay with administration of prebiotic supplements, but no difference in the incidence of NEC [126].
Probiotics
Probiotics are dietary supplements that contain live microbes. The evidence for benefit from probiotic administration is perhaps most compelling for reduction of NEC and death in preterm infants. A large number of randomized placebo-controlled clinical trials of probiotics in preterm infants have been performed. Several meta-analyses of these trials have demonstrated a significant decrease in NEC, death and/or length of hospital stay in preterm infants with probiotic administration [127, 128]. In addition, meta-analyses of observational cohort studies of probiotic administration to preterm infants have shown similar reductions in NEC and death to those seen in the clinical trials [129, 130]. A recent meta-analysis of randomized controlled trials of probiotic administration to mothers during pregnancy demonstrated a reduction in atopic eczema, preterm delivery, NEC, and death in the offspring [131]. A meta-analysis of randomized controlled trials in term infants with colic demonstrated a significant decrease in crying time with probiotic administration. The probability of a >50% reduction in crying time was close to 2-fold higher [132]. The European Society for Paediatric Gastroenterology and Hepatology recently released a conditional recommendation for probiotic use in preterm infants for the prevention of NEC [133] based on a strain-specific network analysis of clinical trials [134]. The recent recommendations of the American Gastroenterological Association summarized the limited evidence for benefits of probiotic administration in adults and the compelling evidence for benefits in prevention of NEC in preterm infants [135]. Risks associated with probiotics include contamination of the probiotic product [136] and sepsis from the probiotic microbes [137]. It appears that harm from probiotic administration to infants is an uncommon complication given that NEC, sepsis and death are either equal or lower in infants receiving probiotics compared to those receiving placebo or no probiotic in most studies. Those advocating against routine administration of probiotics to preterm infants note that regulatory oversight of probiotic manufacture is inadequate and that questions remain regarding the preferred probiotic strain or combination of strains and the optimum dose [138].
Conclusion
In the last two decades there has been a virtual explosion of knowledge gained on the role played by the colonizing microbiota in mediating host physiology and maintaining homeostasis. Unfavorable changes in the composition of the microbiota, dysbiosis, can in some cases lead to overt disease, but often has more subtle and insidious consequences. In well-appearing term infants in developed countries, intestinal dysbiosis is common and is likely due to diversion from exclusive breast-feeding, cesarean sections, antibiotics and other medications, a “Western” diet and modern sanitation practices. In preterm infants cared for in modern NICUs, intestinal dysbiosis appears to be almost universal. The consequences of intestinal dysbiosis in infants are uncertain, but strong circumstantial evidence and limited confirmations of causality suggest that many acute and chronic diseases that are increasing over time are related to intestinal dysbiosis. Interventions to limit or reverse maternal and early infant intestinal dysbiosis including antibiotic stewardship, increased breastfeeding support, vaccine development to reduce use of prophylactic antibiotics and informed use of probiotic microbes and prebiotic glycans offer promise in reversing these trends.
Acknowledgements
This work is supported by the National Institutes of Health HD096241 (M.A.U.), HD088753 (S.M.), HD072929 (S.L.), AI32738 and AI125926 (C.L.B.).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dominguez-Bello MG, Godoy-Vitorino F, Knight R, Blaser MJ. Role of the microbiome in human development. Gut. 2019;68:1108–14.. doi: 10.1136/gutjnl-2018-317503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science. 2016;352:539–44.. doi: 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayres JS. Cooperative microbial tolerance behaviors in host-microbiota mutualism. Cell. 2016;165:1323–31.. doi: 10.1016/j.cell.2016.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanchez KK, Chen GY, Schieber AMP, Redford SE, Shokhirev MN, Leblanc M, et al. Cooperative metabolic adaptations in the host can favor asymptomatic infection and select for attenuated virulence in an enteric pathogen. Cell. 2018;175:146–58.e15. doi: 10.1016/j.cell.2018.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sorbara MT, Pamer EG. Interbacterial mechanisms of colonization resistance and the strategies pathogens use to overcome them. Mucosal Immunol. 2019;12:1–9. doi: 10.1038/s41385-018-0053-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koh A, Bäckhed F. From association to causality: the role of the gut microbiota and its functional products on host metabolism. Mol Cell. 2020;78:584–96.. doi: 10.1016/j.molcel.2020.03.005. [DOI] [PubMed] [Google Scholar]
- 7.Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell. Host Microbe. 2014;15:382–92.. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klingberg E, Magnusson MK, Strid H, Deminger A, Stahl A, Sundin J, et al. A distinct gut microbiota composition in patients with ankylosing spondylitis is associated with increased levels of fecal calprotectin. Arthritis Res Ther. 2019;21:248. doi: 10.1186/s13075-019-2018-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, et al. Population-level analysis of gut microbiome variation. Science. 2016;352:560–4. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 10.Yatsunenko TRF, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bittinger K, Zhao C, Li Y, Ford E, Friedman ES, Ni J, et al. Bacterial colonization reprograms the neonatal gut metabolome. Nat Microbiol. 2020;5:838–47.. doi: 10.1038/s41564-020-0694-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vangay P, Ward T, Gerber JS, Knights D. Antibiotics, pediatric dysbiosis, and disease. Cell. Host Microbe. 2015;17:553–64. doi: 10.1016/j.chom.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deshmukh HS, Liu Y, Menkiti OR, Mei J, Dai N, O’Leary CE, et al. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat Med. 2014;20:524–30.. doi: 10.1038/nm.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rooks MG, Garrett WS. Gut. microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16:341–52.. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–93.. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–21.. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimura I, Miyamoto J, Ohue-Kitano R, Watanabe K, Yamada T, Onuki M, et al. Maternal gut microbiota in pregnancy influences offspring metabolic phenotype in mice. Science. 2020;367:eaaw8429. doi: 10.1126/science.aaw8429. [DOI] [PubMed] [Google Scholar]
- 18.Gordon HA, Pesti L. The gnotobiotic animal as a tool in the study of host microbial relationships. Bacteriol. Rev. 1971;35:390–429. doi: 10.1128/br.35.4.390-429.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luczynski P, McVey Neufeld KA, Oriach CS, Clarke G, Dinan TG, Cryan JF. Growing up in a bubble: using germ-free animals to assess the influence of the gut microbiota on brain and behavior. Int J Neuropsychopharmacol. 2016;19:pyw020. doi: 10.1093/ijnp/pyw020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilks M. Bacteria and early human development. Early Hum Dev. 2007;83:165–70.. doi: 10.1016/j.earlhumdev.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 21.Gehrig JL, Venkatesh S, Chang HW, Hibberd MC, Kung VL, Cheng J, et al. Effects of microbiota-directed foods in gnotobiotic animals and undernourished children. Science. 2019;365:eaau4732. doi: 10.1126/science.aau4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31.. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 23.Safari Z, Gerard P. The links between the gut microbiome and non-alcoholic fatty liver disease (NAFLD) Cell. Mol life Sci. 2019;76:1541–58.. doi: 10.1007/s00018-019-03011-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang C, Li G, Huang P, Liu Z, Zhao B. The gut microbiota and Alzheimer’s disease. J Alzheimers Dis. 2017;58:1–15. doi: 10.3233/JAD-161141. [DOI] [PubMed] [Google Scholar]
- 25.Lee YP, Chiu CC, Lin TJ, Hung SW, Huang WC, Chiu CF, et al. The germ-free mice monocolonization with Bacteroides fragilis improves azoxymethane/dextran sulfate sodium induced colitis-associated colorectal cancer. Immunopharmacol Immunotoxicol. 2019;41:207–13.. doi: 10.1080/08923973.2019.1569047. [DOI] [PubMed] [Google Scholar]
- 26.Tissier H. Recherches sur la flore intestinale des nourrissons. C R Mem Soc Biol. 1899;51:943. [Google Scholar]
- 27.Logan WR. The intestinal flora of infants and young children. J Pathol. 1913;18:527–51.. [Google Scholar]
- 28.Garrido D, Barile D, Mills DA. A molecular basis for bifidobacterial enrichment in the infant gastrointestinal tract. Adv Nutr. 2012;3:415S–21S. doi: 10.3945/an.111.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruiz-Moyano S, Totten SM, Garrido DA, Smilowitz JT, German JB, Lebrilla CB, et al. Variation in consumption of human milk oligosaccharides by infant gut-associated strains of Bifidobacterium breve. Appl Environ Microbiol. 2013;79:6040–9. doi: 10.1128/AEM.01843-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zivkovic AM, German JB, Lebrilla CB, Mills DA. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4653–8. doi: 10.1073/pnas.1000083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henrick BM, Hutton AA, Palumbo MC, Casaburi G, Mitchell RD, Underwood MA, et al. Elevated fecal pH indicates a profound change in the breastfed infant gut microbiome due to reduction of bifidobacterium over the past century. mSphere. 2018;3:e00041–18. doi: 10.1128/mSphere.00041-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Litvak Y, Byndloss MX, Tsolis RM, Baumler AJ. Dysbiotic Proteobacteria expansion: a microbial signature of epithelial dysfunction. Curr Opin Microbiol. 2017;39:1–6. doi: 10.1016/j.mib.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 33.Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, et al. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev. 2017;81:e00036–17. doi: 10.1128/MMBR.00036-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.La Rosa PS, Warner BB, Zhou Y, Weinstock GM, Sodergren E, Hall-Moore CM, et al. Patterned progression of bacterial populations in the premature infant gut. Proc Natl Acad Sci USA. 2014;111:12522–7. doi: 10.1073/pnas.1409497111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korpela K, Blakstad EW, Moltu SJ, Strommen K, Nakstad B, Ronnestad AE, et al. Intestinal microbiota development and gestational age in preterm neonates. Sci Rep. 2018;8:2453. doi: 10.1038/s41598-018-20827-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vatanen T, Plichta DR, Somani J, Munch PC, Arthur TD, Hall AB, et al. Genomic variation and strain-specific functional adaptation in the human gut microbiome during early life. Nat Microbiol. 2019;4:470–9. doi: 10.1038/s41564-018-0321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raman AS, Gehrig JL, Venkatesh S, Chang HW, Hibberd MC, Subramanian S, et al. A sparse covarying unit that describes healthy and impaired human gut microbiota development. Science. 2019;365:eaau4735. doi: 10.1126/science.aau4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Younge NE, Newgard CB, Cotten CM, Goldberg RN, Muehlbauer MJ, Bain JR, et al. Disrupted maturation of the microbiota and metabolome among extremely preterm infants with postnatal growth failure. Sci Rep. 2019;9:8167. doi: 10.1038/s41598-019-44547-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J, Li Y, Feng Y, Pan L, Xie Z, Yan Z, et al. Patterned progression of gut microbiota associated with necrotizing enterocolitis and late onset sepsis in preterm infants: a prospective study in a Chinese neonatal intensive care unit. PeerJ. 2019;7:e7310. doi: 10.7717/peerj.7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galazzo G, van Best N, Bervoets L, Dapaah IO, Savelkoul PH, Hornef MW, et al. Development of the microbiota and associations with birth mode, diet, and atopic disorders in a longitudinal analysis of stool samples, collected from infancy through early childhood. Gastroenterology. 2020;158:1584–96.. doi: 10.1053/j.gastro.2020.01.024. [DOI] [PubMed] [Google Scholar]
- 41.Madan JC, Hoen AG, Lundgren SN, Farzan SF, Cottingham KL, Morrison HG, et al. Association of cesarean delivery and formula supplementation with the intestinal microbiome of 6-week-old infants. JAMA Pediatr. 2016;170:212–9. doi: 10.1001/jamapediatrics.2015.3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stewart CJ, Ajami NJ, O’Brien JL, Hutchinson DS, Smith DP, Wong MC, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. 2018;562:583–8. doi: 10.1038/s41586-018-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Combellick JL, Shin H, Shin D, Cai Y, Hagan H, Lacher C, et al. Differences in the fecal microbiota of neonates born at home or in the hospital. Sci Rep. 2018;8:15660. doi: 10.1038/s41598-018-33995-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Savage JH, Lee-Sarwar KA, Sordillo JE, Lange NE, Zhou Y, O’Connor GT, et al. Diet during pregnancy and infancy and the infant intestinal microbiome. J Pediatr. 2018;203:47–54.e4. doi: 10.1016/j.jpeds.2018.07.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lundgren SN, Madan JC, Emond JA, Morrison HG, Christensen BC, Karagas MR, et al. Maternal diet during pregnancy is related with the infant stool microbiome in a delivery mode-dependent manner. Microbiome. 2018;6:109. doi: 10.1186/s40168-018-0490-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams JE, Carrothers JM, Lackey KA, Beatty NF, Brooker SL, Peterson HK, et al. Strong multivariate relations exist among milk, oral, and fecal microbiomes in mother-infant dyads during the first six months postpartum. J Nutr. 2019;149:902–14.. doi: 10.1093/jn/nxy299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Azad MB, Konya T, Persaud RR, Guttman DS, Chari RS, Field CJ, et al. Impact of maternal intrapartum antibiotics, method of birth and breastfeeding on gut microbiota during the first year of life: a prospective cohort study. Int J Obstet Gynaecol. 2016;123:983–93.. doi: 10.1111/1471-0528.13601. [DOI] [PubMed] [Google Scholar]
- 48.Coker MO, Hoen AG, Dade E, Lundgren S, Li Z, Wong AD, et al. Specific class of intrapartum antibiotics relates to maturation of the infant gut microbiota: a prospective cohort study. Int J Obstet Gynaecol. 2020;127:217–27.. doi: 10.1111/1471-0528.15799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eck A, Rutten N, Singendonk MMJ, Rijkers GT, Savelkoul PHM, Meijssen CB, et al. Neonatal microbiota development and the effect of early life antibiotics are determined by two distinct settler types. PloS ONE. 2020;15:e0228133. doi: 10.1371/journal.pone.0228133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gupta RW, Tran L, Norori J, Ferris MJ, Eren AM, Taylor CM, et al. Histamine-2 receptor blockers alter the fecal microbiota in premature infants. J Pediatr Gastroenterol Nutr. 2013;56:397–400. doi: 10.1097/MPG.0b013e318282a8c2. [DOI] [PubMed] [Google Scholar]
- 51.Levy EI, Hoang DM, Vandenplas Y. The effects of proton pump inhibitors on the microbiome in young children. Acta Paediatr. 2020;109:1531–8. doi: 10.1111/apa.15213. [DOI] [PubMed] [Google Scholar]
- 52.Vich Vila A, Collij V, Sanna S, Sinha T, Imhann F, Bourgonje AR, et al. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat Commun. 2020;11:362. doi: 10.1038/s41467-019-14177-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jackson MA, Verdi S, Maxan ME, Shin CM, Zierer J, Bowyer RCE, et al. Gut microbiota associations with common diseases and prescription medications in a population-based cohort. Nat Commun. 2018;9:2655. doi: 10.1038/s41467-018-05184-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iszatt N, Janssen S, Lenters V, Dahl C, Stigum H, Knight R, et al. Environmental toxicants in breast milk of Norwegian mothers and gut bacteria composition and metabolites in their infants at 1 month. Microbiome. 2019;7:34. doi: 10.1186/s40168-019-0645-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laue HE, Moroishi Y, Jackson BP, Palys TJ, Madan JC, Karagas MR. Nutrient-toxic element mixtures and the early postnatal gut microbiome in a United States longitudinal birth cohort. Environ Int. 2020;138:105613. doi: 10.1016/j.envint.2020.105613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McLean C, Jun S, Kozyrskyj A. Impact of maternal smoking on the infant gut microbiota and its association with child overweight: a scoping review. World J Pediatr. 2019;15:341–9. doi: 10.1007/s12519-019-00278-8. [DOI] [PubMed] [Google Scholar]
- 57.Tun HM, Konya T, Takaro TK, Brook JR, Chari R, Field CJ, et al. Exposure to household furry pets influences the gut microbiota of infant at 3-4 months following various birth scenarios. Microbiome. 2017;5:40. doi: 10.1186/s40168-017-0254-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.D’Agata AL, Wu J, Welandawe MKV, Dutra SVO, Kane B, Groer MW. Effects of early life NICU stress on the developing gut microbiome. Dev Psychobiol. 2019;61:650–60.. doi: 10.1002/dev.21826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ho TTB, Groer MW, Kane B, Yee AL, Torres BA, Gilbert JA, et al. Dichotomous development of the gut microbiome in preterm infants. Microbiome. 2018;6:157. doi: 10.1186/s40168-018-0547-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parra-Llorca A, Gormaz M, Alcantara C, Cernada M, Nunez-Ramiro A, Vento M, et al. Preterm gut microbiome depending on feeding type: significance of donor human milk. Front Microbiol. 2018;9:1376. doi: 10.3389/fmicb.2018.01376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taft DH, Salinero LK, Vongbhavit K, Kalanetra KM, Masarweh C, Yu A, et al. Bacterial colonization and antimicrobial resistance genes in neonatal enteral feeding tubes. FEMS Microbiol Ecol. 2019;95:fiz039. doi: 10.1093/femsec/fiz039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nogacka A, Salazar N, Suárez M, Milani C, Arboleya S, Solís G, et al. Impact of intrapartum antimicrobial prophylaxis upon the intestinal microbiota and the prevalence of antibiotic resistance genes in vaginally delivered full-term neonates. Microbiome. 2017;5:93. doi: 10.1186/s40168-017-0313-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gasparrini AJ, Crofts TS, Gibson MK, Tarr PI, Warner BB, Dantas G. Antibiotic perturbation of the preterm infant gut microbiome and resistome. Gut Microbes. 2016;7:443–9. doi: 10.1080/19490976.2016.1218584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gibson MK, Wang B, Ahmadi S, Burnham CA, Tarr PI, Warner BB, et al. Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome. Nat Microbiol. 2016;1:16024. doi: 10.1038/nmicrobiol.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rutayisire E, Huang K, Liu Y, Tao F. The mode of delivery affects the diversity and colonization pattern of the gut microbiota during the first year of infants’ life: a systematic review. BMC Gastroenterol. 2016;16:86. doi: 10.1186/s12876-016-0498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dahl C, Stigum H, Valeur J, Iszatt N, Lenters V, Peddada S, et al. Preterm infants have distinct microbiomes not explained by mode of delivery, breastfeeding duration or antibiotic exposure. Int J Epidemiol. 2018;47:1658–69.. doi: 10.1093/ije/dyy064. [DOI] [PubMed] [Google Scholar]
- 67.Tun MH, Tun HM, Mahoney JJ, Konya TB, Guttman DS, Becker AB, et al. Postnatal exposure to household disinfectants, infant gut microbiota and subsequent risk of overweight in children. Can Med Assoc J. 2018;190:E1097–e107. doi: 10.1503/cmaj.170809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shaw KA, Bertha M, Hofmekler T, Chopra P, Vatanen T, Srivatsa A, et al. Dysbiosis, inflammation, and response to treatment: a longitudinal study of pediatric subjects with newly diagnosed inflammatory bowel disease. Genome Med. 2016;8:75. doi: 10.1186/s13073-016-0331-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xia GH, You C, Gao XX, Zeng XL, Zhu JJ, Xu KY, et al. Stroke dysbiosis index (SDI) in gut microbiome are associated with brain injury and prognosis of stroke. Front Neurol. 2019;10:397. doi: 10.3389/fneur.2019.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guo Y, Zhang Y, Gerhard M, Gao JJ, Mejias-Luque R, Zhang L, et al. Effect of Helicobacter pylori on gastrointestinal microbiota: a population-based study in Linqu, a high-risk area of gastric cancer. Gut. 2019. 10.1136/gutjnl-2019-319696. [DOI] [PMC free article] [PubMed]
- 71.Casen C, Vebo HC, Sekelja M, Hegge FT, Karlsson MK, Ciemniejewska E, et al. Deviations in human gut microbiota: a novel diagnostic test for determining dysbiosis in patients with IBS or IBD. Alimentary Pharmacol Therap. 2015;42:71–83. doi: 10.1111/apt.13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mandl T, Marsal J, Olsson P, Ohlsson B, Andreasson K. Severe intestinal dysbiosis is prevalent in primary Sjogren’s syndrome and is associated with systemic disease activity. Arthritis Res Ther. 2017;19:237. doi: 10.1186/s13075-017-1446-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vatanen T, Kostic AD, d’Hennezel E, Siljander H, Franzosa EA, Yassour M, et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. 2016;165:842–53.. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vatanen T, Franzosa EA, Schwager R, Tripathi S, Arthur TD, Vehik K, et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature. 2018;562:589–94.. doi: 10.1038/s41586-018-0620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chua HH, Chou HC, Tung YL, Chiang BL, Liao CC, Liu HH, et al. Intestinal dysbiosis featuring abundance of ruminococcus gnavus associates with allergic diseases in infants. Gastroenterology. 2018;154:154–67.. doi: 10.1053/j.gastro.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 76.Lee MJ, Kang MJ, Lee SY, Lee E, Kim K, Won S, et al. Perturbations of gut microbiome genes in infants with atopic dermatitis according to feeding type. J Allergy Clin Immunol. 2018;141:1310–9. doi: 10.1016/j.jaci.2017.11.045. [DOI] [PubMed] [Google Scholar]
- 77.Lamont RF, Moller Luef B, Stener Jorgensen J. Childhood inflammatory and metabolic disease following exposure to antibiotics in pregnancy, antenatally, intrapartum and neonatally. F1000Res. 2020;9:144. doi: 10.12688/f1000research.19954.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dowhower Karpa K, Paul IM, Leckie JA, Shung S, Carkaci-Salli N, Vrana KE, et al. A retrospective chart review to identify perinatal factors associated with food allergies. Nutr J. 2012;11:87. doi: 10.1186/1475-2891-11-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Metz TD, McKinney J, Allshouse AA, Knierim SD, Carey JC, Heyborne KD, et al. Exposure to group B Streptococcal antibiotic prophylaxis and early childhood body mass index in a vaginal birth cohort. J Matern Fetal Neonatal Med. 2020;33:3318–3323. doi: 10.1080/14767058.2019.1571575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wohl DL, Curry WJ, Mauger D, Miller J, Tyrie K. Intrapartum antibiotics and childhood atopic dermatitis. J Am Board Fam Med. 2015;28:82–9. doi: 10.3122/jabfm.2015.01.140017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pammi M, Cope J, Tarr PI, Warner BB, Morrow AL, Mai V, et al. Intestinal dysbiosis in preterm infants preceding necrotizing enterocolitis: a systematic review and meta-analysis. Microbiome. 2017;5:31. doi: 10.1186/s40168-017-0248-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Olm MR, Bhattacharya N, Crits-Christoph A, Firek BA, Baker R, Song YS, et al. Necrotizing enterocolitis is preceded by increased gut bacterial replication, Klebsiella, and fimbriae-encoding bacteria. Sci Adv. 2019;5:eaax5727. doi: 10.1126/sciadv.aax5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Esmaeilizand R, Shah PS, Seshia M, Yee W, Yoon EW, Dow K. Antibiotic exposure and development of necrotizing enterocolitis in very preterm neonates. Paediatr Child Health. 2018;23:e56–e61. doi: 10.1093/pch/pxx169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Esaiassen E, Fjalstad JW, Juvet LK, van den Anker JN, Klingenberg C. Antibiotic exposure in neonates and early adverse outcomes: a systematic review and meta-analysis. J Antimicrob. Chemother. 2017;72:1858–70.. doi: 10.1093/jac/dkx088. [DOI] [PubMed] [Google Scholar]
- 85.Guillet R, Stoll BJ, Cotten CM, Gantz M, McDonald S, Poole WK, et al. Association of H2-blocker therapy and higher incidence of necrotizing enterocolitis in very low birth weight infants. Pediatrics. 2006;117:e137–42. doi: 10.1542/peds.2005-1543. [DOI] [PubMed] [Google Scholar]
- 86.Hewitt KM, Mannino FL, Gonzalez A, Chase JH, Caporaso JG, Knight R, et al. Bacterial diversity in two Neonatal Intensive Care Units (NICUs) PloS ONE. 2013;8:e54703. doi: 10.1371/journal.pone.0054703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bokulich NA, Mills DA, Underwood MA. Surface microbes in the neonatal intensive care unit: changes with routine cleaning and over time. J Clin Microbiol. 2013;51:2617–24.. doi: 10.1128/JCM.00898-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brooks B, Olm MR, Firek BA, Baker R, Geller-McGrath D, Reimer SR, et al. The developing premature infant gut microbiome is a major factor shaping the microbiome of neonatal intensive care unit rooms. Microbiome. 2018;6:112. doi: 10.1186/s40168-018-0493-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stewart CJ, Embleton ND, Marrs ECL, Smith DP, Fofanova T, Nelson A, et al. Longitudinal development of the gut microbiome and metabolome in preterm neonates with late onset sepsis and healthy controls. Microbiome. 2017;5:75. doi: 10.1186/s40168-017-0295-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Singer JR, Blosser EG, Zindl CL, Silberger DJ, Conlan S, Laufer VA, et al. Preventing dysbiosis of the neonatal mouse intestinal microbiome protects against late-onset sepsis. Nat Med. 2019;25:1772–82.. doi: 10.1038/s41591-019-0640-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Masi AC, Stewart CJ. The role of the preterm intestinal microbiome in sepsis and necrotising enterocolitis. Early Hum Dev. 2019;138:104854. doi: 10.1016/j.earlhumdev.2019.104854. [DOI] [PubMed] [Google Scholar]
- 92.Hemmi MH, Wolke D, Schneider S. Associations between problems with crying, sleeping and/or feeding in infancy and long-term behavioural outcomes in childhood: a meta-analysis. Arch Dis Child. 2011;96:622–9. doi: 10.1136/adc.2010.191312. [DOI] [PubMed] [Google Scholar]
- 93.Loughman A, Quinn T, Nation ML, Reichelt A, Moore RJ, Van TTH, et al. Infant microbiota in colic: predictive associations with problem crying and subsequent child behavior. J Dev Orig Health Dis. 2020:1–11. 10.1017/S2040174420000227. [DOI] [PubMed]
- 94.Huttenhower C, Kostic AD, Xavier RJ. Inflammatory bowel disease as a model for translating the microbiome. Immunity. 2014;40:843–54.. doi: 10.1016/j.immuni.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mayer EA, Tillisch K, Gupta A. Gut/brain axis and the microbiota. J Clin Investig. 2015;125:926–38.. doi: 10.1172/JCI76304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 97.Schroeder BO, Bäckhed F. Signals from the gut microbiota to distant organs in physiology and disease. Nat Med. 2016;22:1079–89.. doi: 10.1038/nm.4185. [DOI] [PubMed] [Google Scholar]
- 98.Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. Dysbiosis and the immune system. Nat Rev Immunol. 2017;17:219–32.. doi: 10.1038/nri.2017.7. [DOI] [PubMed] [Google Scholar]
- 99.Maruvada P, Leone V, Kaplan LM, Chang EB. The human microbiome and obesity: moving beyond associations. Cell. Host Microbe. 2017;22:589–99.. doi: 10.1016/j.chom.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 100.Bach JF. The hygiene hypothesis in autoimmunity: the role of pathogens and commensals. Nat Rev Immunol. 2018;18:105–20.. doi: 10.1038/nri.2017.111. [DOI] [PubMed] [Google Scholar]
- 101.Brown JM, Hazen SL. Microbial modulation of cardiovascular disease. Nat Rev. 2018;16:171–81.. doi: 10.1038/nrmicro.2017.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tilg H, Zmora N, Adolph TE, Elinav E. The intestinal microbiota fuelling metabolic inflammation. Nat Rev Immunol. 2020;20:40–54. doi: 10.1038/s41577-019-0198-4. [DOI] [PubMed] [Google Scholar]
- 103.Iacob S, Iacob DG. Infectious threats, the intestinal barrier, and its trojan horse: dysbiosis. Front Microbiol. 2019;10:1676. doi: 10.3389/fmicb.2019.01676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fouhse JM, Yang K, More-Bayona J, Gao Y, Goruk S, Plastow G, et al. Neonatal exposure to amoxicillin alters long-term immune response despite transient effects on gut-microbiota in piglets. Front Immunol. 2019;10:2059. doi: 10.3389/fimmu.2019.02059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Beaumont M, Paës C, Mussard E, Knudsen C, Cauquil L, Aymard P, et al. Gut microbiota derived metabolites contribute to intestinal barrier maturation at the suckling-to-weaning transition. Gut Microbes. 2020;11:1268–86. doi: 10.1080/19490976.2020.1747335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Suh SH, Choe K, Hong SP, Jeong SH, Mäkinen T, Kim KS, et al. Gut microbiota regulates lacteal integrity by inducing VEGF-C in intestinal villus macrophages. EMBO Rep. 2019;20:e46927. doi: 10.15252/embr.201846927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kulkarni DH, McDonald KG, Knoop KA, Gustafsson JK, Kozlowski KM, Hunstad DA, et al. Goblet cell associated antigen passages are inhibited during Salmonella typhimurium infection to prevent pathogen dissemination and limit responses to dietary antigens. Mucosal Immunol. 2018;11:1103–13.. doi: 10.1038/s41385-018-0007-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Abo H, Chassaing B, Harusato A, Quiros M, Brazil JC, Ngo VL, et al. Erythroid differentiation regulator-1 induced by microbiota in early life drives intestinal stem cell proliferation and regeneration. Nat Commun. 2020;11:513. doi: 10.1038/s41467-019-14258-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fung TC, Vuong HE, Luna CDG, Pronovost GN, Aleksandrova AA, Riley NG, et al. Intestinal serotonin and fluoxetine exposure modulate bacterial colonization in the gut. Nat Microbiol. 2019;4:2064–73.. doi: 10.1038/s41564-019-0540-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Canfora EE, Meex RCR, Venema K, Blaak EE. Gut microbial metabolites in obesity, NAFLD and T2DM. Nature reviews. Endocrinology. 2019;15:261–73. doi: 10.1038/s41574-019-0156-z. [DOI] [PubMed] [Google Scholar]
- 111.Lau WL, Vaziri ND. Gut microbial short-chain fatty acids and the risk of diabetes. Nat Rev Nephrol. 2019;15:389–90.. doi: 10.1038/s41581-019-0142-7. [DOI] [PubMed] [Google Scholar]
- 112.Sanna S, van Zuydam NR, Mahajan A, Kurilshikov A, Vich Vila A, Vosa U, et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet. 2019;51:600–5. doi: 10.1038/s41588-019-0350-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156:84–96. doi: 10.1016/j.cell.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 114.Dalile B, Van Oudenhove L, Vervliet B, Verbeke K. The role of short-chain fatty acids in microbiota-gut-brain communication. Nat Rev Gastroenterol Hepatol. 2019;16:461–78. doi: 10.1038/s41575-019-0157-3. [DOI] [PubMed] [Google Scholar]
- 115.Parada Venegas D, De la Fuente MK, Landskron G, Gonzalez MJ, Quera R, Dijkstra G, et al. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol. 2019;10:277. doi: 10.3389/fimmu.2019.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Frese SA, Hutton AA, Contreras LN, Shaw CA, Palumbo MC, Casaburi G, et al. Persistence of supplemented Bifidobacterium longum subsp. infantis EVC001 in breastfed infants. mSphere. 2017;2:e00501–17. doi: 10.1128/mSphere.00501-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zheng N, Gao Y, Zhu W, Meng D, Walker WA. Short chain fatty acids produced by colonizing intestinal commensal bacterial interaction with expressed breast milk are anti-inflammatory in human immature enterocytes. PloS ONE. 2020;15:e0229283. doi: 10.1371/journal.pone.0229283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Waligora-Dupriet AJ, Dugay A, Auzeil N, Nicolis I, Rabot S, Huerre MR, et al. Short-chain fatty acids and polyamines in the pathogenesis of necrotizing enterocolitis: kinetics aspects in gnotobiotic quails. Anaerobe. 2009;15:138–44.. doi: 10.1016/j.anaerobe.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 119.Roy SK, Meng Q, Sadowitz BD, Kollisch-Singule M, Yepuri N, Satalin J, et al. Enteral administration of bacteria fermented formula in newborn piglets: a high fidelity model for necrotizing enterocolitis (NEC) PloS ONE. 2018;13:e0201172. doi: 10.1371/journal.pone.0201172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Meng D, Sommella E, Salviati E, Campiglia P, Ganguli K, Djebali K, et al. Indole-3-lactic acid, a metabolite of tryptophan, secreted by Bifidobacterium longum subspecies infantis is anti-inflammatory in the immature intestine. Pediatr Res. 2020. 10.1038/s41390-019-0740-x. [DOI] [PMC free article] [PubMed]
- 121.Mukhopadhyay S, Wade KC, Puopolo KM. Drugs for the prevention and treatment of sepsis in the newborn. Clin Perinatol. 2019;46:327–47.. doi: 10.1016/j.clp.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Caballero-Flores G, Sakamoto K, Zeng MY, Wang Y, Hakim J, Matus-Acuña V, et al. Maternal immunization confers protection to the offspring against an attaching and effacing pathogen through delivery of IgG in breast milk. Cell. Host Microbe. 2019;25:313–23.e4. doi: 10.1016/j.chom.2018.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mukhopadhyay S, Sengupta S, Puopolo KM. Challenges and opportunities for antibiotic stewardship among preterm infants. Arch Dis Child. 2019;104:F327–f32.. doi: 10.1136/archdischild-2018-315412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schulman J, Dimand RJ, Lee HC, Duenas GV, Bennett MV, Gould JB. Neonatal intensive care unit antibiotic use. Pediatrics. 2015;135:826–33.. doi: 10.1542/peds.2014-3409. [DOI] [PubMed] [Google Scholar]
- 125.Schulman J, Profit J, Lee HC, Duenas G, Bennett MV, Parucha J, et al. Variations in neonatal antibiotic use. Pediatrics. 2018;142:e20180115. doi: 10.1542/peds.2018-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chi C, Buys N, Li C, Sun J, Yin C. Effects of prebiotics on sepsis, necrotizing enterocolitis, mortality, feeding intolerance, time to full enteral feeding, length of hospital stay, and stool frequency in preterm infants: a meta-analysis. Eur J Clin Nutr. 2019;73:657–70.. doi: 10.1038/s41430-018-0377-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sun J, Marwah G, Westgarth M, Buys N, Ellwood D, Gray PH. Effects of probiotics on necrotizing enterocolitis, sepsis, intraventricular hemorrhage, mortality, length of hospital stay, and weight gain in very preterm infants: a meta-analysis. Adv Nutr. 2017;8:749–63.. doi: 10.3945/an.116.014605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Deshpande G, Jape G, Rao S, Patole S. Benefits of probiotics in preterm neonates in low-income and medium-income countries: a systematic review of randomised controlled trials. BMJ Open. 2017;7:e017638. doi: 10.1136/bmjopen-2017-017638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dermyshi E, Wang Y, Yan C, Hong W, Qiu G, Gong X, et al. The “Golden Age” of probiotics: a systematic review and meta-analysis of randomized and observational studies in preterm infants. Neonatology. 2017;112:9–23. doi: 10.1159/000454668. [DOI] [PubMed] [Google Scholar]
- 130.Olsen R, Greisen G, Schroder M, Brok J. Prophylactic probiotics for preterm infants: a systematic review and meta-analysis of observational studies. Neonatology. 2016;109:105–12.. doi: 10.1159/000441274. [DOI] [PubMed] [Google Scholar]
- 131.Kuang L, Jiang Y. Effect of probiotic supplementation in pregnant women: a meta-analysis of randomised controlled trials. Br J Nutr. 2020;123:870–80.. doi: 10.1017/S0007114519003374. [DOI] [PubMed] [Google Scholar]
- 132.Skonieczna-Zydecka K, Janda K, Kaczmarczyk M, Marlicz W, Loniewski I, Loniewska B. The effect of probiotics on symptoms, gut microbiota and inflammatory markers in infantile colic: a systematic review, meta-analysis and meta-regression of randomized controlled trials. J Clin Med. 2020;9:999. doi: 10.3390/jcm9040999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.van den Akker CHP, van Goudoever JB, Shamir R, Domellof M, Embleton ND, Hojsak I, et al. Probiotics and preterm infants: a position paper by the european society for paediatric gastroenterology hepatology and nutrition committee on nutrition and the european society for paediatric gastroenterology hepatology and nutrition working group for probiotics and prebiotics. J Pediatr Gastroenterol Nutr. 2020;70:664–80.. doi: 10.1097/MPG.0000000000002655. [DOI] [PubMed] [Google Scholar]
- 134.van den Akker CHP, van Goudoever JB, Szajewska H, Embleton ND, Hojsak I, Reid D, et al. Probiotics for preterm infants: a strain-specific systematic review and network meta-analysis. J Pediatr Gastroenterol Nutr. 2018;67:103–22.. doi: 10.1097/MPG.0000000000001897. [DOI] [PubMed] [Google Scholar]
- 135.Su GL, Ko CW, Bercik P, Falck-Ytter Y, Sultan S, Weizman AV, et al. AGA clinical practice guidelines on the role of probiotics in the management of gastrointestinal disorders. Gastroenterology. 2020. 10.1053/j.gastro.2020.05.059. [DOI] [PubMed]
- 136.Lewis ZT, Shani G, Masarweh CF, Popovic M, Frese SA, Sela DA, et al. Validating bifidobacterial species and subspecies identity in commercial probiotic products. Pediatr Res. 2016;79:445–52.. doi: 10.1038/pr.2015.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chiang MC, Chen CL, Feng Y, Chen CC, Lien R, Chiu CH. Lactobacillus rhamnosus sepsis associated with probiotic therapy in an extremely preterm infant: pathogenesis and a review for clinicians. J Microbiol Immunol Infect. 2020. 10.1016/j.jmii.2020.03.029 [DOI] [PubMed]
- 138.Pell LG, Loutet MG, Roth DE, Sherman PM. Arguments against routine administration of probiotics for NEC prevention. Curr Opin Pediatrics. 2019;31:195–201. doi: 10.1097/MOP.0000000000000730. [DOI] [PubMed] [Google Scholar]