Abstract
BACKGROUND
Guanine nucleotide-binding protein, alpha stimulating (GNAS) mutations are characteristic of intraductal papillary mucinous neoplasms (IPMNs). Pancreatic ductal adenocarcinomas (PDACs) harboring GNAS mutations originate in IPMNs. GNAS is a complex imprinted locus that produces five transcripts regulated by differential methylated regions, NESP55, GNASAS, GNASXL, GNAS1A, and GNAS.
AIM
To evaluate if methylation changes in the differential methylated regions of GNAS locus contributed to malignant progression of pancreatic cysts.
METHODS
GNAS locus methylation was analyzed in archival pancreatic cyst fluid (PCF) obtained by endoscopic ultrasound with fine-needle aspiration by methylation specific–multiplex ligation dependent probe amplification. Results were normalized and analyzed using Coffalyser.Net software.
RESULTS
Fifty-two PCF samples obtained by endoscopic ultrasound with fine-needle aspiration and previously characterized for KRAS and GNAS mutations were studied. The final diagnoses were surgical (11) and clinicopathological (41), including 30 benign cysts, 14 pre-malignant cyst, and eight malignant cysts. Methylation changes at NESP55, GNASAS, GNAS1A, and especially GNASXL were more frequent in malignant cysts, and NESP55 and GNASAS were useful for diagnosis. A combined variable defined as “GNAS locus methylation changes” was significantly associated with malignancy (6/8 malignant cysts and only 2/20 benign cysts) and improved classification. Hypermethylation in both maternally (NESP55) and paternally (GNASXL) derived promoters was found in 3/3 PDACs.
CONCLUSION
This is the first study to identify methylation changes in the GNAS locus, improving the diagnosis of malignant pancreatic cysts and suggesting a role in progression to PDAC.
Keywords: Intraductal papillary mucinous neoplasms, Pancreas cyst, Methylation, Biomarker, GNAS locus, Pancreatic neoplasm
Core Tip: Pancreatic cystic lesions are a clinical dilemma due to risk of malignancy. Somatic mutations of guanine nucleotide-binding protein, alpha stimulating (GNAS) are characteristic of intraductal papillary mucinous neoplasms. We found methylation changes in differential methylated regions at the GNAS locus in pancreatic cyst fluid predominantly of malignant cysts. Methylation changes in GNAS locus may improve the diagnosis of malignant cysts and shed light on the development of novel therapeutic approaches for pancreatic cancer.
INTRODUCTION
Pancreatic cystic lesions (PCLs) constitute a clinical dilemma due to indeterminate risk of malignancy, including benign cysts (BCs), pre-malignant cysts (PMCs), and malignant cysts (MCs)[1]. Intraductal papillary mucinous neoplasms (IPMNs) and mucinous cystic neoplasms (MCNs) are cystic precursors of pancreatic ductal adenocarcinoma (PDAC), allowing early diagnosis[2].
Somatic mutations in guanine nucleotide-binding protein, alpha stimulating (GNAS) are characteristic of IPMNs[3,4], but their role in carcinogenesis is unclear, with early occurrence precluding prediction of dysplasia[5,6]. However, if detected in PDACs, somatic mutations in GNAS are specific for an IPMN origin[3].
GNAS is a complex imprinted locus in the long arm of chromosome 20 (20q13.32)[7], which encodes the α-subunit of the stimulatory heterotrimeric G protein (Gsα), a ubiquitous signaling protein translated from GNAS exons 1-13. This locus encodes four monoallelic (NESP55, AS, XL, 1A) and one biallelic (Gsα) transcript, due to differentially methylated regions (DMRs) in paternal and maternal alleles, denominated imprinting[8,9]. Paternal methylation of NESP55 and maternal methylation of AS, XL, and 1A lead, respectively, to maternal and paternal allele expressions, with Gsα biallelically expressed in most tissues, due to absent methylation[10].
Epigenetic alterations in the GNAS locus have not been previously evaluated in PCLs. Methylation of DMRs may occur at the somatic level and modulate Gsα expression[10,11], leading us to hypothesize that methylation changes in DMRs at the GNAS locus could contribute to tumor progression of PCLs. To test our hypothesis, we performed a longitudinal cohort pilot study of PCLs and analyzed GNAS locus methylation in pancreatic cyst fluid (PCF) samples.
MATERIALS AND METHODS
Case selection
All patients gave informed consent, and the study was approved by the Ethics Committee and Institutional Scientific Board (UIC/1143).
For this study we performed molecular analysis in samples of 52 patients with more than 1 mL of PCF stored in the biorepository of our hospital, with sample processing and storage described in a previous publication[12]. Clinical data, including demographics, cyst characteristics, and treatment decision, have been prospectively registered.
After undergoing endoscopic ultrasound with fine needle aspiration, patients were evaluated in clinics, and referred for surgery (surgical cohort, surgical pathology diagnosis) or imaging surveillance, palliation, or endoscopic drainage (clinical cohort, clinico-cytological diagnosis) when surgery was not clinically indicated and a surgical pathology specimen was not available for diagnosis. The diagnostic criteria for the clinical cohort were determined a priori by one of the investigators (SF) after reviewing imaging features, PCF levels of CEA, and cytology analysis of PCLs, all with a prolonged imaging and clinical follow-up (of at least 24 mo). To evaluate GNAS locus methylation distribution and the performance of methylation analysis for cyst diagnosis, PCLs were further classified into one of three groups: Group 1) Benign cysts (BCs), including neoplastic benign and inflammatory cysts (serous cystadenomas (SCAs), pseudocysts, and lymphangiomas); Group 2) Mucinous pre-malignant cysts (PMCs), including IPMNs and MCNs with low grade atypia (LG); Group 3) High-risk/malignant cysts (MCs), including cystic PDACs, IPMNs with adenocarcinoma (ADC) or high grade atypia (HG), MCN-HG, and neuroendocrine cystic tumors (NETs).
Patients and specimens
The samples studied were predominantly from female patients (35/52, 67%) with a mean age of 59 ± 15 years (29-91); 22 PCLs were in the head, 20 in the body, nine in the tail, and one case of multiple pancreatic locations. The mean cyst size was 3.9 ± 2.3 cm (1-10), CEA level in PCF was > 192 ng/mL in 17/52 (33%), and malignant/atypical cytology was present in 11/52 (21%) PCF samples, as shown in Table 1.
Table 1.
Demographics and clinical characteristics of the study population
| Characteristics | Value |
| Female gender, n (%) (n = 52) | 35 (67.3) |
| Mean age at EUS-FNA, y, mean ± SD (interval) | 59.1 ± 14.8 (29-91) |
| Cyst location, n (%) (n = 52) | |
| Head | 22 (42.3) |
| Body | 20 (38.5) |
| Tail | 9 (17.3) |
| Multiple cyst locations | 1 (1.9) |
| Cyst size, cm, mean ± SD (interval) | 3.9 ± 2.3 (1-10) |
| Cyst size > 3 cm, n (%) | 29 (55.8) |
| Cyst with nodule/mass, n (%) | 18 (34.6) |
| EUS imaging, n (%) (n = 52)1 | |
| No high risk features | 13 (25) |
| 1 high risk feature | 29 (55.8) |
| ≥ 2 risk features | 10 (19.2) |
| PCF CEA, n (%) (n = 52) | |
| CEA < 192 ng/mL | 31 (59.6) |
| CEA ≥ 192 ng/mL | 17 (32.7) |
| No result available | 4 (7.7) |
| PCF cytology, n (%) (n = 52) | |
| Non-diagnostic | 27 (51.9) |
| Negative for malignancy | 14 (26.9) |
| Suspicious/malignant | 10 (19.2) |
| NET | 1 (2) |
| Treatment decision, n (%) (n = 52) | |
| Follow up | 34 (65.4) |
| Surgery | 11 (21.2) |
| Endoscopic drainage | 1 (1.9) |
| Palliation (symptomatic or chemotherapy) | 6 (11.5) |
High-risk features: cyst size ≥ 3 and solid component or thick wall or dilated Wirsung (> 10 mm). CEA: Carcinoembryonic antigen; EUS-FNA: Endoscopic ultrasound with fine needle aspiration; NET: Neuroendocrine tumor; PCF: Pancreatic cyst fluid; SD: standard deviation.
These 52 PCF samples obtained by endoscopic ultrasound with fine-needle aspiration (EUS-FNA) have been previously characterized for KRAS and GNAS mutations[12], which were present in nine and two samples, respectively.
The final diagnoses, 11 surgical and 41 clinicopathological, encompassed 30 BCs (SCAs, pseudocysts, and lymphangiomas), 14 PMCs (IPMNs and MCNs), and eight MCs (one cystic PDAC, one IPMN-ADC, one NET, and five mucinous-malignant).
Methylation analysis and categorization
For this study, DNA was extracted from 0.250 mL of archival PCF. Methylation analysis of the GNAS locus was performed by methylation specific–multiplex ligation dependent probe amplification (MS-MLPA) (SALSA MS-MLPA ME031-B1, MRC-Holland®, Amsterdam, The Netherlands), according with the manufacturer’s instructions. MS-MLPA fragments were analyzed on the Applied Biosystems® 3130 Genetic Analyzer (ThermoFisher Scientific, Waltham, MA, United States) using the GeneMapper® software. Results were normalized and analyzed using Coffalyser.Net software (MRC-Holland®).
We studied methylation in four DMRs, NESP55, GNASAS, GNASXL, and GNAS1A, and in the biallelic expressed Gsα, including two exonic regions. DMRs were classified as hypermethylated or hypomethylated, according to the percentage of methylation obtained using the Coffalyser.net software recommended by the manufacturer, if methylation percentage was, respectively, above or below the reference values plus or minus twice the standard deviation (SD). The normal methylation of NESP55 is approximately 50%, as only the paternal allele is methylated, similarly to the percentage of methylation in GNASXL, GNASAS, and GNAS1A, as only the maternal alleles are expected to be methylated. The methylation of Gsα exon 1 is usually absent, as neither maternal nor paternal allele is methylated. Methylation of Gsα exonic regions (exons 9 and 13) is usually near 100%, as both maternal and paternal alleles are methylated. The MS-MLPA kit comprised three methylation sensitive probes for NESP55, three for GNASAS, five for GNASXL, two for GNAS1A, and four for Gsα methylation evaluation.
Statistical analysis
The methylation levels obtained for each of the individual DMRs and for each individual MS-MLPA probe were calculated and converted into a categorical variable defined as: (1) Hypomethylated if methylation level obtained was below the cut-off level minus twice the SD; (2) Hypermethylated if the methylation level obtained was above the cut-off level plus twice the SD; and (3) Normally methylated if neither criteria (1) or (2) were met. A combined variable, including hypermethylation at upstream DMRs or intragenic hypomethylation of GNAS locus, defining “GNAS locus methylation changes” pattern was created. For (epi)genotype-phenotype associations, Fisher’s exact test and chi-square test were performed as well as Kendall’s rank correlation adjusted for age and gender, using partial correlation. Methylation analysis in mucinous and malignant cysts was also represented by boxplot, and Mann-Whitney was used to assess the difference of median methylation values. The diagnostic accuracy of PCF biomarkers was assessed by receiver operating characteristics curve analysis. Statistics were performed using SPSS Statistical software, version 23 (Armonk, NY, United States), with a P value < 0.05 considered as statistically significant.
RESULTS
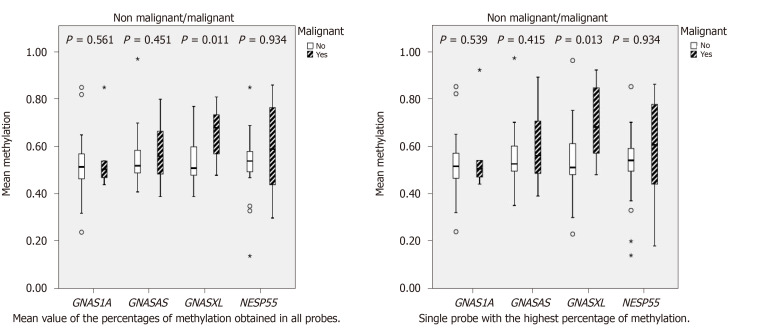
GNAS locus methylation was informative in 38/52 (73%) PCF samples, with the remaining (14/52) non-informative due to inadequate quality/quantity of DNA and rarely, to copy-number variation (probe ratios below 0.7 or above 1.3, regarded as indicative of heterozygous deletion or duplication, respectively, according with the manufacturer (Coffalyser.Net software, MRC-Holland®). Methylation changes at NESP55, GNASAS, GNAS1A, and especially GNASXL were more frequent in MCs (Table 2), presenting wider methylation levels of these DMRs compared to non-malignant cysts, which showed methylation levels around 50% in imprinted alleles (Figure 1).
Table 2.
Frequency of GNAS locus methylation changes in malignant, mucinous, and benign cysts, n (%)
| Informative cyst fluid methylation analysis, 38 samples | Malignant, n = 8 | Mucinous pre-malignant, n = 10 | Benign, n = 20 | P value |
| NESP55 hypermethylation | 3 (37.5) | 0 (0.0 ) | 1 (5.0) | 0.053 |
| GNASAS hypermethylation | 3 (37.5) | 1 (10.0) | 3 (15.0) | 0.065 |
| GNASXL hypermethylation | 4 (50) | 0 (0.0) | 2 (10.0) | 0.004 |
| GNAS1A hypermethylation | 1 (12.5) | 0 (0.0) | 0 (0) | 0.0355 |
| GNAS locus methylation changes | 6 (75.0) | 0 (0.0) | 2 (6.7) | 0.000 |
GNAS locus methylation changes: DMR hypermethylation or GNAS intragenic hypomethylation. GNAS: Guanine nucleotide-binding protein, alpha stimulating.
Figure 1.
Methylation analysis of non-malignant and malignant cysts. GNAS: Guanine nucleotide-binding protein, alpha stimulating.
Based on the influence of methylation changes at DMRs in the modulation of GNAS transcription[10,11] and on the suggested role for hypomethylated exons in transcription regulation and its overlap with predicted enhancers[13], we defined a combined variable documenting “GNAS locus methylation changes”: (1) Presence of hypermethylation in at least two DMRs or in one DMR for all MLPA probes; or (2) Presence of intragenic hypomethylation of GNAS in at least two exonic regions. Notably, “GNAS locus methylation changes” was significantly associated with malignancy (6/8 MCs and only 2/20 BCs) (Table 2), and it is of note that one of these two BCs was later diagnosed as pancreatic cancer.
We further analyzed the correlation between methylation changes and malignancy, while controlling for gender and age. We found a strong significant positive rank correlation between malignancy and GNAS methylation changes (r = 0.837, P < 0.001) and a moderate rank correlation with GNASAS hypermethylation (r = 0.431, P = 0.015), GNASXL hypermethylation (r = 0.434, P = 0.011), and NESP55 hypermethylation (r = 0.539, P = 0.003), which was sustained after controlling for gender and age using partial correlation analysis (Table 3).
Table 3.
Correlation between methylation status and malignancy with partial correlation controlling for patients’ gender and age
|
Rank correlation |
Kendall |
Partial |
||
|
Possible confounders |
Gender |
Age |
||
| Malignant cysts | Correlation | P value | Correlation | Correlation |
| NESP55 hypermethylation | 0.539 | 0.003 | 0.545 | 0.519 |
| GNASAS hypermethylation | 0.431 | 0.015 | 0.459 | 0.584 |
| GNASXL hypermethylation | 0.434 | 0.011 | 0.461 | 0.356 |
| GNAS1A hypermethylation | 0.160 | 0.361 | 0.191 | 0.147 |
| GNAS locus methylation changes | 0.837 | <0.001 | 0.870 | 0.825 |
GNAS: Guanine nucleotide-binding protein, alpha stimulating.
Moreover, the “GNAS locus methylation changes” variable improved MCs classification in samples with clinicopathological diagnosis (possible diagnostic uncertainty) as well as surgical diagnosis (definitive diagnosis but limited number of cases), further supporting our results.
Interestingly, simultaneous hypermethylation in NESP55 and GNASXL DMRs was detected exclusively in 3/3 PDACs. Hypomethylation in two exonic GNAS regions (exons 9 and 13) was detected in the only NET in this series.
Additionally, “GNAS locus methylation changes” was associated with symptoms, KRAS/GNAS mutations, and malignant/atypical cytology but not with patient gender, age, or CEA level in PCF (Table 4), with the area under the curve analysis revealing better performance than cytology for diagnosis of MCs (Table 5).
Table 4.
Frequencies of distinct clinical features and pancreatic cystic fluid analysis in the two groups, with or without GNAS locus methylation changes
| Cyst fluid samples | GNAS locus methylation changes | No GNAS locus methylation changes | P value |
| Female | 63% | 75% | 0.486 |
| Age > 65 yr | 50% | 40% | 0.216 |
| Symptoms | 63% | 17% | 0.008 |
| CEA > 192 ng/mL | 63% | 25% | 0.133 |
| KRAS/GNAS mutation | 63% | 11% | 0.008 |
| Cytology, malignant/atypical | 63% | 7% | 0.003 |
GNAS locus methylation changes: DMR hypermethylation or GNAS intragenic hypomethylation. CEA: Carcinoembryonic antigen; GNAS: Guanine nucleotide-binding protein, alpha stimulating.
Table 5.
Area under the curve for diagnosis of mucinous and malignant cysts
| Variables |
Mucinous cysts |
Malignant cysts |
||||||
| AUC | P value |
Confidence interval |
AUC | P value |
Confidence interval |
|||
| Lower limit | Upper limit | Lower limit | Upper limit | |||||
| CEA in mg/dL | 0.889 | 0.002 | 0.720 | 1.000 | 0.812 | 0.038 | 0.579 | 1.000 |
| Cytology | 0.598 | 0.443 | 0.349 | 0.847 | 0.771 | 0.072 | 0.571 | 0.970 |
| Mutation (KRAS/GNAS) | 0.833 | 0.009 | 0.634 | 1.000 | 0.841 | 0.023 | 0.615 | 1.000 |
| Met_NESP55 | 0.620 | 0.35 | 0.370 | 0.869 | 0.759 | 0.085 | 0.481 | 1.000 |
| Met_AS | 0.590 | 0.483 | 0.339 | 0.841 | 0.741 | 0.108 | 0.461 | 1.000 |
| Met_XL | 0.474 | 0.841 | 0.228 | 0.721 | 0.629 | 0.389 | 0.357 | 0.902 |
| Met_1A | 0.513 | 0.92 | 0.262 | 0.764 | 0.565 | 0.667 | 0.261 | 0.868 |
| GNAS locus methylation changes | 0.645 | 0.256 | 0.400 | 0.891 | 0.971 | 0.002 | 0.901 | 1.000 |
AUC: Area under the curve; CEA: Carcinoembryonic antigen; GNAS: Guanine nucleotide-binding protein, alpha stimulating; Met: Methylation changes.
DISCUSSION
Aberrant DNA methylation in PCF of IPMNs progressing to high-grade dysplasia and carcinoma has been described[14], but GNAS locus methylation was not studied therein. We report for the first time methylation changes in the GNAS locus, namely hypermethylation of GNASXL, NESP55, GNASAS, and GNAS1A in PCLs. Notably, hypermethylation of GNASXL, and especially the combined variable “GNAS locus methylation changes”, was associated with malignancy, suggesting the potential to be used for diagnosis of MCs and for monitoring cancer progression, if confirmed in larger series. Indeed, hypermethylation of GNASXL has been associated to GNAS locus gain of function[10], and although its possible association with malignant progression remains poorly understood, GNAS oncogenic potential appears to be unquestionable[3-5,10,15]. Moreover, somatic DNA methylation has been shown to drive transcription within the imprinted GNAS cluster[11], further supporting our results. NESP55 also appears to regulate imprinting at the GNAS complex locus, and its hypermethylation in the maternal allele may lead, similarly to maternal deletion, as previously described, to subsequent modulation of GNAS[10].
Herein, hypermethylation in both maternally (NESP55) and paternally (GNASXL) derived promoters, and therefore overall increase of methylation in these two DMRs, was detected exclusively in PDAC, further suggesting a role of GNAS in malignant progression of PCL. Interestingly, the detection of exonic GNAS hypomethylation in the pancreatic NET is in agreement with the recent findings showing that pancreatic NETs are genetically and phenotypically related to pancreatic ductal adenocarcinoma, having a closer relationship to ductal adenocarcinomas than to neuroendocrine tumors G3[16]. In agreement with the role of GNAS in the progression to PDAC is also the recent finding that overexpression of mutant GNAS, resulting in constitutive activation of Gsα, in a mouse model of KrasG12D-driven pancreatic cancer, led to the formation of moderately differentiated PDAC that were locally invasive and increased mitogen-activated protein kinase activation[17].
Although copy-number alterations, which could in part explain some of the methylation changes found, were detected in only one case, we cannot exclude the presence of uniparental disomy (UPD) associated copy-neutral loss of heterozygosity (LOH), as previously described by Bastepe et al[18] to explain GNAS methylation changes. An analysis of LOH in the GNAS locus would be needed to evaluate uniparental disomy (UPD) associated copy-neutral LOH (which can often be segmental) and investigate if some of these methylation alterations may indeed reflect epigenetic alterations or could instead be explained (at least in part) by acquired UPD. Nevertheless, independent of their cause (epigenetic or acquired UPD), the resulting methylation alterations detected in the GNAS locus DMRs appear to be related to malignant progression and may improve MCs diagnosis. Our study may contribute to the current epigenetic landscape of PCs, similar to recent studies documenting a role for methylation markers in discriminating pancreatic neoplasia[19,20], possibly offering an opportunity for early diagnosis for pancreatic cancer.
Ultimately, the significant association of GNAS locus methylation changes to malignant behavior suggests a role for modulation of GNAS expression in the malignant progression of PCs, which may be relevant for the development of novel therapeutic approaches for pancreatic cancer. Due to small sample size and poor DNA yield, the final analysis was based on eight samples with HGD/cancer. Although the small sample size and lack of validation in an independent sample are significant limits regarding the present study, our pilot data may be the basis for exploring GNAS methylation in larger, well-characterized sets of samples that may represent future validation studies. Finally, as gene methylation may affect gene expression, additional evaluation of GNAS transcripts in PCF may elucidate their function in PCLs.
ARTICLE HIGHLIGHTS
Research background
Pancreatic cystic lesions (PCLs) constitute a clinical dilemma due to indeterminate risk of malignancy. Intraductal papillary mucinous neoplasms (IPMNs) and mucinous cystic neoplasms are cystic precursors of pancreatic ductal adenocarcinoma (PDAC), possibly allowing early diagnosis. Somatic mutations in GNAS are characteristic of IPMNs, but their role in carcinogenesis is unclear. GNAS is a complex imprinted locus that encodes the α-subunit of the stimulatory heterotrimeric G protein (Gsα), an ubiquitous signaling protein. This locus encodes four monoallelic (NESP55, AS, XL, 1A) and one biallelic (Gsα) transcript(s), due to differentially methylated regions (DMRs) in paternal and maternal alleles, denominated imprinting. Paternal methylation of NESP55 and maternal methylation of AS, XL, and 1A lead, respectively, to maternal and paternal allele expressions, with Gsα biallelically expressed in most tissues, due to absent methylation.
Research motivation
GNAS somatic mutations are characteristic of IPMNs, although epigenetic alterations in the GNAS locus have not been previously evaluated in PCLs. Methylation of DMRs at the GNAS locus may occur at the somatic level and modulate Gsα expression.
Research objectives
In this study, we evaluate if methylation changes in DMRs at the GNAS locus could contribute to tumor progression of PCLs.
Research methods
We performed a longitudinal cohort study of PCLs with GNAS locus methylation analysis performed in PCF samples obtained by endoscopic ultrasound with fine needle aspiration.
Research results
Fifty-two PCF samples obtained by endoscopic ultrasound with fine needle aspiration and previously characterized for KRAS and GNAS mutations were studied. The final diagnoses were surgical (11) and clinicopathological (41), including 30 benign cysts, 14 pre-malignant cyst, and eight malignant cysts. Methylation changes at NESP55, GNASAS, GNAS1A, and especially GNASXL were more frequent in malignant cysts and were useful for their diagnosis. A combined variable defined as “GNAS locus methylation changes” was significantly associated with malignancy (6/8 malignant cysts and only 2/20 benign cysts) and improved classification. Hypermethylation in both maternally (NESP55) and paternally (GNASXL) derived promoters was found in 3/3 PDACs.
Research conclusions
This is the first study to identify methylation changes in the GNAS locus that improved the diagnosis of malignant PCs and suggest a role in progression to PDAC.
Research perspectives
Although the small sample size and lack of validation in an independent sample are significant limits regarding the present study, our pilot data may be the basis for exploring GNAS methylation in larger, well-characterized sets of samples. As methylation status may impact gene expression, additional evaluation of GNAS transcripts in PCF may elucidate their function in pancreatic cystic neoplasms.
Footnotes
Institutional review board statement: The study was approved by the Ethics Committee and Institutional Scientific Board (UIC/1143).
Informed consent statement: All patients gave informed consent.
Conflict-of-interest statement: The authors declare no potential financial interests.
STROBE statement: Guidelines of STROBE statement have been adopted.
Manuscript source: Unsolicited manuscript
Corresponding Author's Membership in Professional Societies: American Society for Gastrointestinal Endoscopy, No. 112613; European Society of Gastrointestinal Endoscopy, No.11655.
Peer-review started: April 15, 2020
First decision: July 5, 2020
Article in press: August 1, 2020
Specialty type: Oncology
Country/Territory of origin: Portugal
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Cheng J, Hori T S-Editor: Gong ZM L-Editor: Filipodia P-Editor: Li JH
Contributor Information
Sandra Faias, Department of Gastroenterology, Instituto Português de Oncologia de Lisboa Francisco Gentil, EPE, Lisboa 1099-023, Portugal; Faculty of Health Sciences, University of Beira Interior, Covilhã 6200-506, Portugal. sandrarfaias@hotmail.com.
Marlene Duarte, Unidade de Investigação em Patobiologia Molecular (UIPM), Instituto Português de Oncologia de Lisboa Francisco Gentil, EPE, Lisboa 1099-023, Portugal.
Luísa Pereira, Centro de Matemática e Aplicações (CMA-UBI), Universidade da Beira Interior, Covilhã 6200-506, Portugal.
Paula Chaves, Faculty of Health Sciences, University of Beira Interior, Covilhã 6200-506, Portugal; Department of Pathology, Instituto Português de Oncologia de Lisboa Francisco Gentil, EPE, Lisboa 1099-023, Portugal.
Marília Cravo, Department of Gastroenterology, Hospital Beatriz Ângelo, Loures 2674-514, Portugal.
Antonio Dias Pereira, Department of Gastroenterology, Instituto Português de Oncologia de Lisboa Francisco Gentil, EPE, Lisboa 1099-023, Portugal.
Cristina Albuquerque, Unidade de Investigação em Patobiologia Molecular (UIPM), Instituto Português de Oncologia de Lisboa Francisco Gentil, EPE, Lisboa 1099-023, Portugal.
Data sharing statement
No additional data are available.
References
- 1.Stark A, Donahue TR, Reber HA, Hines OJ. Pancreatic Cyst Disease: A Review. JAMA. 2016;315:1882–1893. doi: 10.1001/jama.2016.4690. [DOI] [PubMed] [Google Scholar]
- 2.Singhi AD, Koay EJ, Chari ST, Maitra A. Early Detection of Pancreatic Cancer: Opportunities and Challenges. Gastroenterology. 2019;156:2024–2040. doi: 10.1053/j.gastro.2019.01.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu J, Matthaei H, Maitra A, Dal Molin M, Wood LD, Eshleman JR, Goggins M, Canto MI, Schulick RD, Edil BH, Wolfgang CL, Klein AP, Diaz LA, Jr, Allen PJ, Schmidt CM, Kinzler KW, Papadopoulos N, Hruban RH, Vogelstein B. Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci Transl Med. 2011;3:92ra66. doi: 10.1126/scitranslmed.3002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singhi AD, Nikiforova MN, Fasanella KE, McGrath KM, Pai RK, Ohori NP, Bartholow TL, Brand RE, Chennat JS, Lu X, Papachristou GI, Slivka A, Zeh HJ, Zureikat AH, Lee KK, Tsung A, Mantha GS, Khalid A. Preoperative GNAS and KRAS testing in the diagnosis of pancreatic mucinous cysts. Clin Cancer Res. 2014;20:4381–4389. doi: 10.1158/1078-0432.CCR-14-0513. [DOI] [PubMed] [Google Scholar]
- 5.Singhi AD, Davison JM, Choudry HA, Pingpank JF, Ahrendt SA, Holtzman MP, Zureikat AH, Zeh HJ, Ramalingam L, Mantha G, Nikiforova M, Bartlett DL, Pai RK. GNAS is frequently mutated in both low-grade and high-grade disseminated appendiceal mucinous neoplasms but does not affect survival. Hum Pathol. 2014;45:1737–1743. doi: 10.1016/j.humpath.2014.04.018. [DOI] [PubMed] [Google Scholar]
- 6.Singhi AD, McGrath K, Brand RE, Khalid A, Zeh HJ, Chennat JS, Fasanella KE, Papachristou GI, Slivka A, Bartlett DL, Dasyam AK, Hogg M, Lee KK, Marsh JW, Monaco SE, Ohori NP, Pingpank JF, Tsung A, Zureikat AH, Wald AI, Nikiforova MN. Preoperative next-generation sequencing of pancreatic cyst fluid is highly accurate in cyst classification and detection of advanced neoplasia. Gut. 2018;67:2131–2141. doi: 10.1136/gutjnl-2016-313586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blatt C, Eversole-Cire P, Cohn VH, Zollman S, Fournier RE, Mohandas LT, Nesbitt M, Lugo T, Jones DT, Reed RR. Chromosomal localization of genes encoding guanine nucleotide-binding protein subunits in mouse and human. Proc Natl Acad Sci USA. 1988;85:7642–7646. doi: 10.1073/pnas.85.20.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plagge A, Kelsey G. Imprinting the Gnas locus. Cytogenet Genome Res. 2006;113:178–187. doi: 10.1159/000090830. [DOI] [PubMed] [Google Scholar]
- 9.Bastepe M. The GNAS Locus: Quintessential Complex Gene Encoding Gsalpha, XLalphas, and other Imprinted Transcripts. Curr Genomics. 2007;8:398–414. doi: 10.2174/138920207783406488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turan S, Bastepe M. The GNAS complex locus and human diseases associated with loss-of-function mutations or epimutations within this imprinted gene. Horm Res Paediatr. 2013;80:229–241. doi: 10.1159/000355384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mehta S, Williamson CM, Ball S, Tibbit C, Beechey C, Fray M, Peters J. Transcription driven somatic DNA methylation within the imprinted Gnas cluster. PLoS One. 2015;10:e0117378. doi: 10.1371/journal.pone.0117378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faias S, Duarte M, Albuquerque C, da Silva JP, Fonseca R, Roque R, Dias Pereira A, Chaves P, Cravo M. Clinical Impact of KRAS and GNAS Analysis Added to CEA and Cytology in Pancreatic Cystic Fluid Obtained by EUS-FNA. Dig Dis Sci. 2018;63:2351–2361. doi: 10.1007/s10620-018-5128-y. [DOI] [PubMed] [Google Scholar]
- 13.Singer M, Kosti I, Pachter L, Mandel-Gutfreund Y. A diverse epigenetic landscape at human exons with implication for expression. Nucleic Acids Res. 2015;43:3498–3508. doi: 10.1093/nar/gkv153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hata T, Dal Molin M, Hong SM, Tamura K, Suenaga M, Yu J, Sedogawa H, Weiss MJ, Wolfgang CL, Lennon AM, Hruban RH, Goggins MG. Predicting the Grade of Dysplasia of Pancreatic Cystic Neoplasms Using Cyst Fluid DNA Methylation Markers. Clin Cancer Res. 2017;23:3935–3944. doi: 10.1158/1078-0432.CCR-16-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Innamorati G, Wilkie TM, Kantheti HS, Valenti MT, Dalle Carbonare L, Giacomello L, Parenti M, Melisi D, Bassi C. The curious case of Gαs gain-of-function in neoplasia. BMC Cancer. 2018;18:293. doi: 10.1186/s12885-018-4133-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konukiewitz B, Jesinghaus M, Steiger K, Schlitter AM, Kasajima A, Sipos B, Zamboni G, Weichert W, Pfarr N, Klöppel G. Pancreatic neuroendocrine carcinomas reveal a closer relationship to ductal adenocarcinomas than to neuroendocrine tumors G3. Hum Pathol. 2018;77:70–79. doi: 10.1016/j.humpath.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 17.Ideno N, Yamaguchi H, Ghosh B, Gupta S, Okumura T, Steffen DJ, Fisher CG, Wood LD, Singhi AD, Nakamura M, Gutkind JS, Maitra A. GNASR201C Induces Pancreatic Cystic Neoplasms in Mice That Express Activated KRAS by Inhibiting YAP1 Signaling. Gastroenterology. 2018;155:1593–1607.e12. doi: 10.1053/j.gastro.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bastepe M, Lane AH, Jüppner H. Paternal uniparental isodisomy of chromosome 20q--and the resulting changes in GNAS1 methylation--as a plausible cause of pseudohypoparathyroidism. Am J Hum Genet. 2001;68:1283–1289. doi: 10.1086/320117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Natale F, Vivo M, Falco G, Angrisano T. Deciphering DNA methylation signatures of pancreatic cancer and pancreatitis. Clin Epigenetics. 2019;11:132. doi: 10.1186/s13148-019-0728-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majumder S, Taylor WR, Yab TC, Berger CK, Dukek BA, Cao X, Foote PH, Wu CW, Mahoney DW, Aslanian HR, Fernández-Del Castillo C, Doyle LA, Farrell JJ, Fisher WE, Lee LS, Lee YN, Park W, Rodrigues C, Gould Rothberg BE, Salem RR, Simeone DM, Urs S, Van Buren G, Smyrk TC, Allawi HT, Lidgard GP, Raimondo M, Chari ST, Kendrick ML, Kisiel JB, Topazian MD, Ahlquist DA. Novel Methylated DNA Markers Discriminate Advanced Neoplasia in Pancreatic Cysts: Marker Discovery, Tissue Validation, and Cyst Fluid Testing. Am J Gastroenterol. 2019;114:1539–1549. doi: 10.14309/ajg.0000000000000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No additional data are available.