Abstract
5-flurouracil (5-FU)-based chemotherapy is the main pharmacological therapy for advanced colorectal cancer (CRC). Despite significant progress in the treatment of CRC during the last decades, 5-FU drug resistance remains the most important cause of failure in CRC therapy. Resistance to 5-FU is a complex and multistep process. Different mechanisms including microsatellite instability, increased expression level of key enzyme thymidylate synthase and its polymorphism, increased level of 5-FU-activating enzymes and mutation of TP53 are proposed as the main determinants of resistance to 5-FU in CRC cells. Recently, micro-ribonucleic acids (miRNA) and their alterations were found to have a crucial role in 5-FU resistance. In this regard, the miRNA-mediated mechanisms of 5-FU drug resistance reside among the new fields of pharmacogenetics of CRC drug response that has not been completely discovered. Identification of the biological markers that are related to response to 5-FU-based chemotherapy is an emerging field of precision medicine. This approach will have an important role in defining those patients who are most likely to benefit from 5-FU-based chemotherapy in the future. Thereby, the identification of 5-FU drug resistance mechanisms is an essential step to predict and eventually overcome resistance. In the present comprehensive review, we will summarize the latest knowledge regarding the molecular determinants of response to 5-FU-based chemotherapy in CRC by emphasizing the role of miRNAs.
Keywords: 5-flurouracil; Colorectal cancer; Chemotherapy resistance, Thymidylate synthase; Microsatellite instability; Micro-ribonucleic acid; TP53
Core Tip: Resistance to the main chemotherapy drug, 5-flurouracil (5-FU), is an important cause of failure in clinical colorectal cancer therapy. Microsatellite instability, increased activity and expression level of thymidylate synthase and dihydropyridine dehydrogenase, mutation of TP53 as well as micro-ribonucleic acids alterations are among the main molecular determinants of response to 5-FU-based chemotherapy in colorectal cancer. The identification of potential molecular determinants of response to 5-FU could be an important clinical tool for developing treatment strategies and selecting colorectal cancer patients who are most likely to benefit from 5-FU chemotherapy.
INTRODUCTION
Colorectal cancer (CRC) is the third most common malignancy and the second leading cause of cancer related-death globally[1,2]. Surgical procedures are the primary treatment choice for CRC, and other strategies such as chemotherapy are also used as complementary therapy to decrease the risk of local recurrences and to increase quality of life for patients afflicted with metastatic CRC (mCRC). 5-fluorouracil (5-FU), leucovorin, capecitabine, oxaliplatin and irinotecan chemotherapy drugs are the standard neoadjuvant and adjunctive therapeutic options[3]. 5-FU is the main component in most of the gastrointestinal malignancies chemotherapy regimens[4,5]. Although combined treatment approaches have contributed to the current improvements in response rates, drug resistance and tumor recurrence occur in most colon cancer patients[6,7].
A great proportion of clinical data highlights the need for addressing the interindividual differences in 5-FU-based chemotherapy responses. Different mechanisms including increased expression of the target thymidylate synthase (TS) and a decreased level of 5-FU-activating enzymes are the cause for resistance of CRC cells to the cytotoxic effect of 5-FU[8,9]. On the other hand, micro RNAs (miRNAs) and their alterations play a crucial role in cancer initiation, progression and drug resistance[10,11]. The miRNA-mediated mechanisms of drug resistance reside among the new fields of pharmacogenetics of cancer, yet they have not been discovered thoroughly[10]. Therefore, investigating the involvement of miRNAs in anticancer drug resistance is important in overcoming the barriers in human cancer therapy applications. In this review, we summarized the latest knowledge regarding the molecular determinants of response to 5-FU-based chemotherapy in CRC with more emphasis on the role of miRNAs.
FLUOROURACIL-BASED CHEMOTHERAPY AND CRC
Cytotoxic chemotherapy has long been used as the backbone of treatment for CRC patients and evolved significantly over the last decades. 5-FU-based chemotherapy is a primary chemotherapeutic option for advanced CRC. Although the response rate to 5-FU as a single antitumor medicine is typically below 20%, there are interindividual differences regarding the clinical effectiveness of 5-FU-based chemotherapy[12,13]. 5-FU is a synthetic pyrimidine antagonist (uracil analog) and belongs to the group of anticancer medicine known as antimetabolites[14,15].
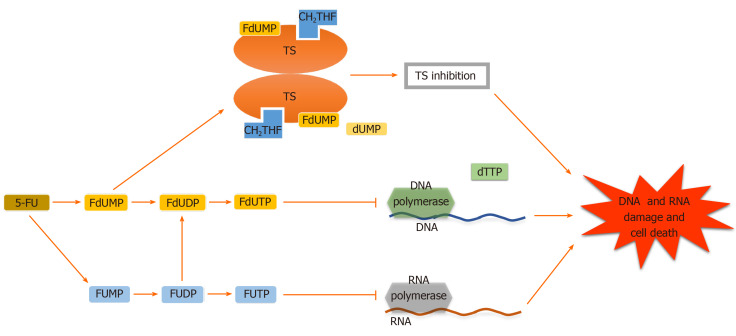
Adequate 5-FU transformation to 5-fluorouridine-5’-monophosphate fluorouridine monophosphate is necessary for its function[16]. Nevertheless, 5-FU is converted intracellularly to several other active metabolites. Each of them has a distinctive role in cytotoxic effects. These metabolites include (1) 5-fluorodeoxyuridine-5’- monophosphate fluorodeoxyuridine monophosphate, which interferes with TS, a key enzyme in the creation of the DNA nucleotide deoxythymidine-5 monophosphate deoxythymidine monophosphate, in the ternary complex; (2) 5-fluorodeoxyuridine-5’-triphosphate fluorodeoxyuridine triphosphate, which acts as a substrate for DNA polymerases and is incorporated into DNA in place of deoxythymidine-5 triphosphate deoxythymidine triphosphate and inhibits DNA elongation thus leading to DNA fragmentation; and (3) 5-fluorouridine-5 triphosphate fluorouridine triphosphate, an eligible substrate for RNA polymerases, which is integrated into RNA instead of uridine triphosphate. These active metabolites disrupt RNA synthesis, making single and double-strand DNA breaks and inhibit TS action, resulting in cellular apoptosis (Figures 1-3)[16-19].
Figure 1.
Different mechanisms of 5-fluorouracil action. 5-Fluorouracil (5_FU) and its derivative active metabolites exert their antitumor function at the levels of enzyme thymidylate synthase, DNA and RNA, leading to DNA and RNA damage and cell death. TS: Thymidylate synthase; FdUMP: Fluorodeoxyuridine’ monophosphate; FdUDP: Fluorodeoxyuridine diphosphate; FdUTP: Fluorodeoxyuridine triphosphate; FUMP: Fluorouridine monophosphate; FUDP: Fluorouridine diphosphate; FUTP: Fluorouridine triphosphate; CH2THF: 5,10-methylenetetrahydrofolate; dTTP: Deoxythymidine triphosphate; dUMP: Deoxyuridine monophosphate.
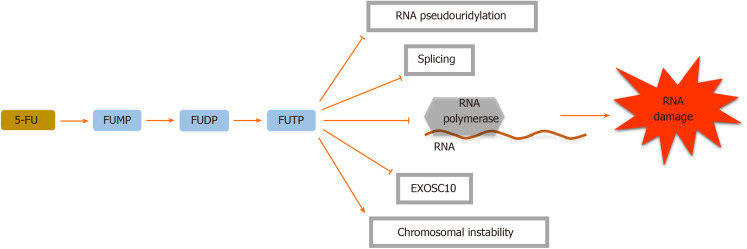
Figure 3.
5-Fluorouracil inhibits ribonucleic acid pseudouridylation including the conversion of uridine to pseudouridine and formation of stable ribonucleoprotein complexes. These actions will interrupt cellular tRNA, pre-mRNA and rRNA production and also post-transcriptional modification, leading to RNA biosynthesis cessation. Moreover, 5-fluorouracil (5-FU) disrupts the assembly and activity of snRNA and protein complexes through its effect on the pseudouridylation of U2 snRNA, thereby inhibiting the splicing of pre-mRNA[21]. Because of the similarity between the 5-FU metabolite fluorouridine triphosphate (FUTP) with uridine triphosphate, FUTP can be identified by RNA polymerases and incorporated into both nuclear and cytoplasmic RNA molecules, interrupting normal RNA processing and function[22]. RNA-based 5-FU toxicity decreases the cellular levels of the nuclear exosome Rrp6 or exosome component 10, a breakdown complex for RNA, banning the effectual turn-over of aberrant RNA transcripts[21]. FUMP: Fluorouridine monophosphate; FUDP: Fluorouridine diphosphate.
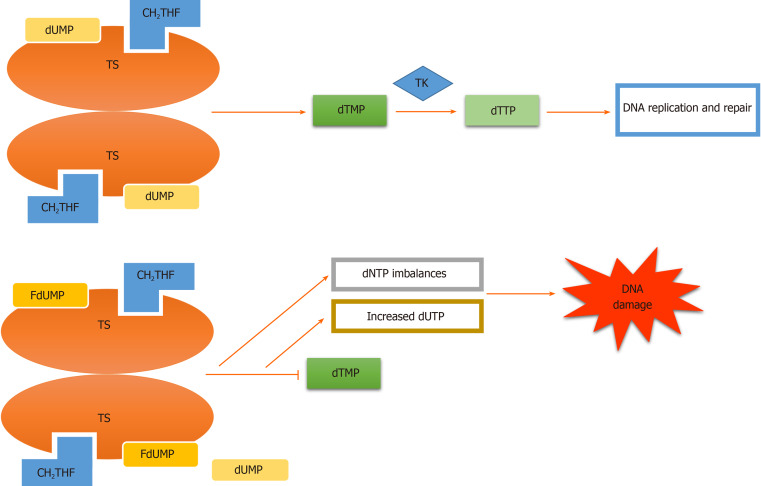
Figure 2.
Thymidylate synthase inhibition. Fluorodeoxyuridine monophosphate (FdUMP) incorporation into thymidylate synthase in place of deoxyuridine monophosphate (dUMP) results in suppression of thymidylate synthase (TS). Consequently, the synthesis of thymidine monophosphate required for DNA replication and repair is diminished, which leads to deoxyuridine triphosphate (dUTP) imbalances, increased dUTP, DNA damage and finally apoptosis of actively dividing cancerous cells[20]. TK: Thymidylate kinase; CH2THF: 5,10-methylene tetrahydrofolate; dTMP: Deoxythymidine monophosphate; dTTP: Deoxythymidine triphosphate; dNTP: Deoxynucleotide triphosphate.
RESISTANCE TO 5-FU
Malignant tumors may show either intrinsic or acquired resistance that requires special lines of treatment. Innate resistance is generally attributed to the capacity of tumoral cells to escape from drug effects spontaneously during the early phase of drug administration[23]. Acquired resistance has different mechanisms dedicated to specific cytotoxic and targeted therapy. However, different mechanisms of drug resistance interact with each other, and acquired resistance to one drug may result in resistance to other drugs. This phenomenon is known as multidrug resistance[23,24]. Different molecular mechanisms are involved in drug resistance. Decreased drug delivery rates to the cancerous cells through excessive efflux, reduced influx, the increment of drug inactivation, mutation of the drug target genes, defective drug transportation and overexpression of p170 (protein of multidrug resistance) are the main mechanisms for drug resistance. Moreover, alterations in the enzyme activity involved in the metabolism of drugs are considered the other main pathways of drug resistance during cancer management[23,24].
One of the greatest challenges for CRC management is drug resistance to 5-FU that can be intrinsic or acquired during treatment and is believed to occur in nearly half of patients with metastatic cancer[25]. Identification of the biological rules that are related to 5-FU-based chemotherapy response is an emerging field of precision medicine. This approach will have an important role in defining those patients who are most likely to benefit from 5-FU-based chemotherapy in the future. Thereby, comprehending the mechanisms by which tumors become resistant to 5-FU is a prerequisite for predicting or overcoming resistance.
MIRNAS FUNCTION IN CANCER PROGRESSION AND 5-FU DRUG RESISTANCE IN CRC
miRNAs are short RNA molecules with various regulatory effects. These small RNA molecules are transcribed from DNA sequences within exons or introns. miRNA genes transcribed by RNA polymerase undergo a maturation process by different enzymes before gaining their ability to perform their action[25,26].
During cancer, miRNAs may become dysregulated[25-27]. In some cancers including leukemia, some DNA chromosomal regions become deleted, and therefore specific miRNAs in these locations may be lost. A decrease or complete loss of specific miRNA expression in cancers may upregulate the expression of oncogenes[25]. Also, the upregulation of specific transcription factors may result in the upregulation of oncogenic miRNAs and therefore dysregulate further cellular pathways[25]. Despite the role of miRNAs in the development and progression of cancers, these small RNA molecules are also important in the development of chemotherapy resistance. Each chemotherapy drug performs its action from a specific cellular pathway. For example, 5-FU induces apoptosis and cell cycle arrest as the main antitumor action[28]. However, overexpressing miR-21 can induce 5-FU chemoresistance through inhibition of apoptosis and cell cycle arrest in CRC cells[28,29]. miR-10b can also induce 5-FU resistance by downregulating proapoptotic proteins[28-30].
Various studies evaluated the expression of different miRNAs in CRC tissue and discussed their effect on 5-FU response. Each of these miRNAs summarized in Table 1 has their unique targets that affect specific cellular functions in CRC cells including apoptosis, proliferation, colony formation or metastasis as well as resistance to 5-FU[31-35].
Table 1.
Summary of studies evaluating the association of different micro-ribonucleic acids with 5-fluorouracil sensitivity or resistance in colorectal cancer
| Ref. | miRNA | CRC source | miRNA target | miRNA effect on 5-FU treatment | miRNA-related pathways (KEGG Identifier)[89] | Other findings |
| Zhang et al[82] | miR-24 | HCT116, RKO, SW480, and SW48 cell lines | Downregulation of RNA binding protein DND1 expression | Overexpression enhanced the chemosensitivity of SW48 cell lines | Fatty acid biosynthesis (00061). Proteoglycans in cancer (05205). Cell cycle (04110). Glycosaminoglycan biosynthesis-keratan sulfate (00533). Pancreatic cancer (05212) | miR-24 acted as a tumor suppressor |
| Zhang et al[83] | miR-361 | HCT116 and HT29 | Forkhead box M1 (FOXM1) | Overexpression increased chemosensitivity via modulation of FOXM1-ABCC5/10 | Adherens junction (04520). Proteoglycans in cancer (05205). Protein processing in the endoplasmic reticulum (04141). miRNAs in cancer (05206). ECM-receptor interaction (04512) | ATP binding cassette subfamily C members 5 and 10 (ABCC5/10) were the downstream effectors of miR-361 |
| Xu et al[84] | miR-375-3p | Tissue and cells | YAP1 and SP1 | Overexpression enhanced the chemosensitivity | Hippo signaling pathway (04390). Lysine degradation (00310). Protein processing in the endoplasmic reticulum (04141). Proteoglycans in cancer (05205). Viral carcinogenesis (05203) | Low miR-375 expression was strongly correlated with poor overall survival in CRC patients |
| Lv et al[85] | miR-133b | Tissue and cells | H3K79me2 | Overexpression enhanced the chemosensitivity and reduced CRC stemness | Sulfur relay system (04122). ECM-receptor interaction (04512) | miR-133b overexpression suppressed DOT1L-mediated H3K79me2 modification of stem cell genes |
| Zhao et al[41] | miR-552 | 5-FU-resistant tissues and cells | SMAD2 cascade | Overexpression decreased chemoresistance and miR-552 inhibition led to increased chemoresistance | Fatty acid degradation (00071). Fatty acid metabolism (01212). Fatty acid elongation (00062). Biosynthesis of unsaturated fatty acids (01040). TGF-beta signaling pathway (04350) | Expression of miR-552 downregulated in 5-FU-resistant tissues and cells that were regulated by dMMR |
| Liu et al[88] | miR-543 | HCT8/FU | Phosphatase and tensin homolog/ PI3K/ protein kinase B (PTEN/PI3K/AKT) | Downregulation enhanced chemosensitivity | TGF-beta signaling pathway (04350). Signaling pathways regulating pluripotency of stem cells (04550). FoxO signaling pathway (04068). Transcriptional misregulation in cancer (05202). Hippo signaling pathway (04390) | miR-543 enhanced chemoresistance by downregulating expression of PTEN, which negatively regulated AKT activation |
| Zhao et al[89] | miR-1260b | HCT116 | Cell death 4 was a direct target of miR-1260b inhibitor | Inhibition of miR-1260b enhanced chemosensitivity | Wnt signaling pathway (04310). Hippo signaling pathway (04390). Central carbon metabolism in cancer (05230). Adherens junction (04520).Lysine degradation (00310) | miR-1260b inhibitor reduced proliferation and increased apoptosis while downregulating cell death 4, phosphorylated-Akt, and phosphorylated-extracellular-signal-regulated kinase (p-ERK) expression |
| Guo et al[90] | miR-191 | Beta-elemene on colorectal carcinoma HCT116 and HT29 | Expression of kinases, including Wnt3a and β-catenin | Downregulation of miR-191 improved chemoresistance | Colorectal cancer (05210). Lysine degradation (00310). Steroid biosynthesis (00100). Hippo signaling pathway (04390). Transcriptional misregulation in cancer (05202) | Beta-elemene downregulated miR-191 and thereby inhibited the Wnt/β-catenin pathway |
| Cao et al[31] | miR-761 | HT29 | FOXM1 | Overexpression increased chemosensitivity | Glycosphingolipid biosynthesis. Lacto and neolacto series (00601). Mucin Type O-Glycan biosynthesis (00512). Biosynthesis of unsaturated fatty acids (01040) | MiR-761 expression negatively associated with FOXM1 expression and elevated FOXM1 expression suppressed cell proliferation, colony formation and invasion |
| Kong et al[91] | miR-195 | 5-FU-resistant HCT116 and SW480 | PI3K/AKT and NF-κB pathways | Overexpression enhanced chemosensitivity | Viral carcinogenesis (05203). Hippo signaling pathway (04390). Proteoglycans in cancer (05205). Adherens junction (04520). Pathways in cancer (05200). Fatty acid degradation (00071) | Schizandrin A (SchA) sensitized 5-FU-resistant cells by upregulating miR-195, which inhibited PI3K/AKT and NF-κB pathways |
| Que et al[92] | miR-874-3p | Tissue and cells | Transcriptional co-activators YAP and TAZ of the Hippo signaling pathway | Overexpression enhanced chemosensitivity | miR-874-3p indirectly inactivated TEAD transcription | |
| Liu et al[79] | miR-135b and miR-182 | 5-FU resistant Cell lines | ST6GALNAC2 via PI3K/AKT pathway | Overexpression of these two miRNAs increased drug resistance | miR-135b: Hippo signaling pathway (04390). Thyroid hormone signaling pathway (04919). Steroid hormone biosynthesis (00140). Signaling pathways regulating pluripotency of stem cells (04550) cGMP-PKG signaling pathway (04022) miR-182. Fatty acid biosynthesis (00061). Viral carcinogenesis (05203). Adherens junction (04520). Cell cycle (04110). Pancreatic cancer (05212) | miR-135b and miR-182 overexpression were seen in 5-FU resistance cell lines |
| Zhao et al[32] | miR-15b-5p | Tissue and cells | NF-κB pathway and XIAP | Overexpression decreased chemoresistance | Fatty acid biosynthesis (00061). Fatty acid metabolism (01212). Viral carcinogenesis (05203). Hippo signaling pathway (04390). Adherens junction (04520) | miR-15b mediated apoptosis regulation by negative regulation of NF-κB1 and kinase complexes IKK-α and also targeting anti-apoptosis protein XIAP |
| Ye et al[40] | miR-1290 | Tissue and cells | hMSH2 | Inhibition decreased chemoresistance | Proteoglycans in cancer (05205). Fatty acid degradation (00071). ErbB signaling pathway (04012). Signaling pathways regulating pluripotency of stem cells (04550). Estrogen signaling pathway (04915) | miR-1290 was positively correlated with dMMR status and predicted poor prognosis in stage II and III colon cancer patients who received 5-FU |
| Ren et al[34] | miR-196b-5p | Tissue and cells | SOCS1 and SOCS3 of STAT3 signaling pathway | Downregulation increased chemosensitivity and ectopic expression yielded the opposite effect | Adherens junction (04520). TGF-beta signaling pathway (04350). Proteoglycans in cancer (05205). Hippo signaling pathway (04390). Glycosaminoglycan biosynthesis. Keratan sulfate (00533) | Silencing miR-196b-5p suppressed spheroids formation ability and expression of stem cell factors and enhanced the apoptosis induced by 5-FU. Overexpression of this microRNA correlated with poor survival |
| Xu et al[60] | miR-330 | Tissues and cell | TYMS | Ectopic expression decreased cell proliferation and enhanced chemosensitivity | Adherens junction (hsa04520). ECM-receptor interaction (04512). Thyroid hormone signaling pathway (04919). Proteoglycans in cancer (05205). Hippo signaling pathway (04390) | miR-330 affected thymidylate synthase (TYMS) in the cell apoptosis pathway |
| Yu et al[34] | miR-125b | Tissues and cell | CXCL12/CXCR4 and Wnt/β-catenin signaling | Expression interfered with chemoresistance | Fatty acid biosynthesis (00061). Fatty acid metabolism (01212). Hippo signaling pathway (04390). Other types of O-glycan biosynthesis (00514). Lysine degradation (00310) | Overexpression of miR-125b triggered epidermal mesenchymal transition and invasion |
| Jiang et al[93] | miR-577 | 5-FU resistant SW480 cells | Heat shock protein 27 (HSP27) | Ectopic expression enhanced chemosensitivity | Lysine degradation (00310). Mucin Type O-Glycan biosynthesis (00512). Viral carcinogenesis (05203). p53 signaling pathway (04115). Colorectal cancer (05210) | Restoration of miR-577 suppressed proliferation and induced a G0/G1 cell cycle arrest |
| Fu et al[94] | miR-20b | 5-FU-sensitive (HCT116) and -resistant (HCT116-R) | ADAM9/EGFR | Expression reduced chemoresistance and induced apoptosis | Prolactin signaling pathway (04917). TGF-beta signaling pathway (04350). FoxO signaling pathway (04068). Pancreatic cancer (05212). Bladder cancer (05219) | miR-20b expressed at lower levels in the 5-FU resistant tissues |
| Liu et al[95] | miR302a | HCT116 and HT29 | Insulin-like growth factor1 receptor (IGF1R) | Expression induced chemosensitivity | Lysine degradation (00310). Wnt signaling pathway (04310). Proteoglycans in cancer (05205). Signaling pathways regulating pluripotency of stem cells (04550). Transcriptional misregulation in cancer (05202) | miR302a enhanced 5FUinduced cell death |
| Dong et al[96] | miR-429 | Tissues and cells | - | Decreased expression correlated with better response | Fatty acid elongation (00062). Fatty acid degradation (00071). Adherens junction (04520). Fatty acid metabolism (01212). Ras signaling pathway (04014) | An increase in miR-429 level was associated with tumor size, metastasis, lymph node invasion and TNM stage |
| Liu et al[35] | miR-149 | 5-FU-resistant CRC cells (HCT-8/5-FU and LoVo/5-FU) | FOXM1 | Re-expression enhanced chemosensitivity | RNA transport (03013). Spliceosome (03040). Gap junction (04540). Sulfur relay system (04122). Thyroid hormone synthesis (04918) | miR-149 enhanced 5-FU sensitivity by increasing apoptosis |
| Han et al[97] | miR-874 | Tissues and cell line | XIAP | Re-expression enhanced chemosensitivity, inhibited proliferation and enhanced apoptosis | Tyrosine metabolism (00350). Thyroid hormone signaling pathway (04919). Glycosphingolipid biosynthesis. Globo series (00603). Thyroid cancer (05216). Glycosphingolipid biosynthesis. ganglio series (00604) | miR-874 expression negatively correlated with TNM stage and lymph node metastasis |
| Liu et al[98] | miR-139-5p | HCT-8/5-FU and HCT-116/5-FU | NOTCH-1 and MDR-associated genes | Ectopic expression enhanced chemosensitivity and induced apoptosis | Hippo signaling pathway (04390). Neurotrophin signaling pathway (04722). Glioma (05214). Prostate cancer (05215). Estrogen signaling pathway (04915) | Upregulation of NOTCH-1 repealed miR-139-5p-mediated sensitization to 5-FU |
| Wu et al[99] | miR-204 | HCT116 and SW480 | High mobility group protein A2 | Overexpression decreased chemoresistance | Steroid biosynthesis (00100). Estrogen signaling pathway (04915). ECM-receptor interaction (04512). Metabolism of xenobiotics by cytochrome P450 (00980). Apoptosis (04210) | miR-204 took action by activation of high mobility group protein A2 and the PI3K/AKT pathway |
| Li et al[61] | miR-218 | HCT116 and HT29 cells | Thymidylate synthase (TS) | Overexpression increased chemosensitivity | Fatty acid biosynthesis (00061). Proteoglycans in cancer (05205). Viral carcinogenesis (05203). Pathways in cancer (05200). p53 signaling pathway (04115) | miR-218 suppressed BIRC5 and TS and promoted apoptosis |
| Zhang et al[81] | miR-587 | Tissues and cell | PPP2R1B | Inhibition decreased chemoresistance | GABAergic synapse (04727). Valine, leucine and isoleucine degradation (00280). TGF-beta signaling pathway (04350). Lysine degradation (00310). Signaling pathways regulating pluripotency of stem cells (04550) | miR-587 inhibited AKT activation |
| Amankwatia et al[80] | miR-224 | isogenic KRAS WT and mutant HCT116 cells | ERK and AKT | Decreased expression increased chemosensitivity | Lysine degradation (hsa00310). Fatty acid metabolism (01212). Glioma (05214). Biosynthesis of unsaturated fatty acids (01040). Fatty acid elongation (00062) | miR-224 target genes altered cell proliferation and invasion as well as epithelial-mesenchymal transition |
| Zhang et al[75] | miR-520g-3p | Tissues and cell | p21 | Inhibition of miR-520g in p53 negative cells increased chemosensitivity. | TGF-beta signaling pathway (04350). Folate biosynthesis (00790). Transcriptional misregulation in cancer (05202). Signaling pathways regulating pluripotency of stem cells (04550) | p53 suppressed the miR-520g expression |
| Kim et al[74] | miR-96 | Tissues and cell | XIAP and UBE2N | Expression modulated chemosensitivity and promoted apoptosis | Adherens junction (04520). ECM-receptor interaction (04512). Prostate cancer (05215). Viral carcinogenesis (05203). Protein processing in the endoplasmic reticulum (04141) | Following 5-FU exposure, the expression of the antiapoptotic regulator (XIAP) and p53 stability regulator (UBE2N) decreased |
| Li et al[59] | miR-203 | 5-FU-resistant cell line LoVo/5-Fu | Thymidylate synthase (TYMS) | Inhibition of expression increased chemoresistance and the overexpression increased chemosensitivity | Sulfur metabolism (00920). MAPK signaling pathway (04010). Thyroid hormone signaling pathway (04919) | miR-203 increased the inhibitory effect of 5-FU on tumor growth and suppressed TYMS protein levels |
| Boni et al[58] | miR-192 and miR-215 | Tissue and cells | TYMS | The expression did not affect chemoresistance but overexpression reduced cell proliferation | miR-192: Folate biosynthesis (00790). Wnt signaling pathway (04310). Steroid biosynthesis (00100). Lysine degradation (00310). Signaling pathways regulating pluripotency of stem cells (04550). miR-215: Folate biosynthesis (00790). Estrogen signaling pathway (04915). One carbon pool by folate (00670) | Ectopic expression of these miRNAs was a strong predictor of 5-FU response. The expression did not affect chemoresistance but overexpression reduced cell proliferation |
miRNA: Micro-ribonucleic acid; 5-FU: 5-fluorouracil; CRC: Colorectal cancer; TS: Thymidylate synthase; TK: Thymidylate kinase; MMR: Mismatch repair; MDR: Multidrug resistance; DOT1L: Disruptor of telomeric silencing 1-like; ECM: Extracellular matrix; KEGG: Kyoto Encyclopedia of Genes and Genomes; TNM: Tumor, node, metastasis.
In the following sections, we reviewed the latest and most important molecular mechanisms in 5-FU resistance including microsatellite instability (MSI), altered expression of TS and dihydropyrimidine dehydrogenase (DPD) enzymes and loss of p53, along with the roles of related miRNAs.
MICROSATELLITE INSTABILITY AND RESISTANCE TO 5-FU
MSI is an inherent mechanism of resistance to 5-FU. Microsatellites or simple sequence repeats are tracts of 15-30 repeated DNA units composed of 1-6 nucleotides distributed among all DNA regions (coding and noncoding). Instability of microsatellites result from sporadic or germline mutations in DNA mismatch repair (MMR) machinery genes including PMS1, PMS6, MSH2, MSH6, MGMT and MLH1, which are responsible for correcting errors occurring during DNA replication[36-38].
About 10%-15% of all CRCs and more than 90% of cases of hereditary nonpolyposis colon cancer show levels of DNA MMR deficiencies (dMMR). MMR mutant genes are responsible for aberrant scanning and recognizing errors during DNA replication that lead to aberrant insertion or deletion of repeated units in microsatellites[24]. Loss of detection of mismatched and unpaired bases in cancerous cells make DNA damage and apoptosis caused by 5-FU tolerable, leading to 5-FU resistance[23]. Meyers et al[39] demonstrated that restoring MMR efficiency through inserting a corrected clone of the hMLH1 gene in MMR deficient cell lines, gave rise to an increased response to 5-FU treatment, suggesting that MMR-deficient cells are more resistant to 5-FU. Nonetheless, some conflicting results indicated that the MSI phenotype correlated with considerable survival of patients who benefit from adjuvant 5-FU-based chemotherapy. The finding can be explained by the fact that MSI-positive tumors may have intrinsic biological characteristics such as wild-type tumor suppressor p53, which aids 5-FU-mediated cytotoxicity in contrast to MSI-negative tumors. When wild-type p53 in MSI-positive tumors is compared to MSI-negative (TP53-mutant) tumors, it highlights the MSI phenotype influence[20].
The role of different miRNAs targeting the MMR system is summarized in Table 1. Ye et al[40] evaluated the effect of miR-1290 on CRC cells, which targets HMSH2. They reported that this miRNA is positively correlated with dMMR status. Moreover, the expression of miR-1290 was associated with poor prognosis in stage II and III CRC patients who received 5-FU. They also demonstrated that inhibition of miR-1290 decreased 5-FU chemoresistance[40]. miR-552 is another miRNA that targets the SMAD2 cascade. Zhao et al[41] evaluated the effect of different expression levels of miR-522 on 5-FU chemoresistance. They demonstrated that expression of miR-552 that was downregulated in 5-FU-resistant tissues and cells was regulated by dMMR. Moreover, the expression of this miRNA was associated with poor post-chemotherapy prognosis. In terms of 5-FU resistance, overexpression of miR-552 reduced chemoresistance, and its inhibition may lead to increased 5-FU chemoresistance (Table 1)[41].
THYMIDYLATE SYNTHASE AND RESISTANCE TO 5-FU
Abundant research regarding 5-FU resistance in mCRC have highlighted that various genes were involved in 5-FU pharmacokinetic and pharmacodynamic metabolic pathways. One of the major key enzymes in 5-FU metabolism is TS. TS expression is the main determinant of 5-FU sensitivity. Disturbed methylenetetrahydrofolate or one of its polyglutamate pools that results in an unstable ternary complex of fluorodeoxyuridine’ monophosphate and TS (which leads to poor inhibition of TS) along with a high level of TS enzyme before treatment are two main factors of intrinsic resistance to 5-FU[42]. 5-FU chemotherapy nonrespondents have higher TS enzyme activity in comparison to 5-FU respondents. TS has a negative autoregulatory role in the translational level by binding to its own TS mRNA, thereby it inhibits the production of functional TS enzyme. However, the inhibition becomes interrupted by TS enzyme interaction with 5-FU metabolite. As a result, the 5-FU treatment causes acquired resistance by affecting TS stability. Besides, acquired resistance to chemotherapy that is emanating from the gene amplification of TS with consequent increases in TS mRNA and protein is observed in 5-FU and 5-fluorouridine deoxyribose (5-FUDR) treated cell lines[43-46]. Moreover, TS overexpression may result in oncogenic phenotypes by decreasing the translational efficiency of the TP53 transcript[47]. Manifold clinical and preclinical investigations showed that colorectal tumors are more sensitive to 5-FU-based therapy in patients with low tumoral TS expression[48-50]. In line with these findings, Abdallah et al[51] reported that analyzing TS expression in circulating tumor cells of mCRC patients would be a promising tool as a 5-FU resistance predictor biomarker.
Genotyping of the TYMS promoter shows interindividual differences among patients with variable sensitivity to 5-FU treatment and divides CRC patients into those who receive a survival benefit from 5-FU chemotherapy and those who do not[52,53]. The 5’-region of the TYMS gene promoter has a variable number of tandem repeats, and this is composed of usually either two (TSER*2 or 2R) or three (TSER*3 or 3R) 28-base-pair tandem-repeat sequences[54]. Preliminary studies indicated that TSER*3/TSER*3 homozygous patients are less likely to respond to 5-FU-based chemotherapy than TSER*2/TSER*2 homozygous or TSER*2/TSER*3 heterozygous patients[55,56]. In vitro studies showed that TYMS promoter variants observed in tissue tumors with the TSER*3 alleles produce nearly four times higher mRNA in comparison to patients with mCRC who carry TSER*2 alleles (P < 0.004)[57]. Homozygous TSER*2/TSER*2 alleles have a significantly higher percentage of a favorable response to 5-FU treatment compared to those who were homozygous for TSER*3/TSER*3 (50% vs 9%, P = 0.04)[20].
TYMS is the target of miRNAs, and it is demonstrated in Table 1. Boni et al[58] demonstrated that ectopic expression of miR-192 and miR-215 are stronger predictors of 5-FU response than TYMS inhibition. In their study, miR-192 and miR-215 expression do not affect chemoresistance, although their overexpression reduced cell proliferation. Li et al[59] reported that miR-203 increased the inhibitory effect of 5-FU on tumor growth and suppressed TYMS protein levels. Inhibition of miR-203 expression increased chemoresistance, and the miR-203 overexpression increased chemosensitivity[59]. Another miRNA affecting 5-FU response is miR-330. Ectopic expression of miR-330 reduced cell proliferation and enhanced chemosensitivity[60]. In HCT116 and HT29 cell lines, miR-218 expression suppressed BIRC5 and TS, promoting apoptosis. Overexpression of this miRNA showed an increase in 5-FU chemosensitivity[61] (Table 1).
DIHYDROPYRIMIDINE DEHYDROGENASE AND RESISTANCE TO 5-FU
Patients with decreased 5-FU catabolize capacity due to partial or complete DPD deficiency are prone to severe systemic toxicity in response to 5-FU[12]. Increased drug concentration and longer half-life due to the decreased drug catabolism may be the possible explanation for this finding. The assessment of more than 60 human cancer cell lines illustrated a highly significant inverse relationship between the 5-FU response and both DPD mRNA expression and DPD activity. Low DPD mRNA levels and DPD activity in human tumor xenografts are in strong correlation with better response to 5-FU in comparison with tumors expressing a higher level of DPD mRNA[12]. Furthermore, in vitro studies have also shown that high levels of DPD mRNA expression in colorectal tumors and the corresponding catabolism of 5-FU correlate with resistance to 5-FU[62]. Importantly, previous studies showed that DPD, TS and thymidine phosphorylase are independent prognostic markers of 5-FU response, and measurement of all three markers significantly enhanced the ability to predict tumor response to 5-FU-based chemotherapy[63-65].
As alterations in DPD is correlated with 5-FU chemoresistance, miRNAs regulating its expression can interfere with chemotherapy resistance. When miR-494 is bound to the 3’UTR of DPD’s gene, it can decrease its expression[66]. Similarly, miR-27a and miR-27b decrease 5-FU resistance by targeting DPD[67].
P53 AND RESISTANCE TO 5-FU
Some in vitro and in vivo studies reported that loss of p53 function (due to mutant or inactive TP53) reduced cellular sensitivity to cytotoxic agents including 5-FU, which is accompanied by poor prognosis and poor survival rates[68-70]. One of the main outcomes of 5-FU treatment is the induction of apoptosis in normal and tumoral intestinal cells. Thereby, any alterations in genes involved in apoptotic pathways may have extensive impacts on chemotherapy and can take part in the induction of 5-FU resistance by cancerous cells[71]. Disrupting both alleles of TP53 in a colon cancer cell line made the cells strikingly resistant to apoptosis induced by 5-FU compared with the parental line[69]. Remarkably, several clinical studies reported that p53 overexpression correlates with resistance to 5-FU[72,73].
Some studies evaluated the effect of the expression of different miRNAs targeting p53. Kim et al[74] studied the p53 stability regulator (UBE2N) in CRC tissue. miR-96 targets UBE2N. Kim et al[74] demonstrated that exposing CRC cells to 5-FU decreased the expression of an antiapoptotic regulator (XIAP) and UBE2N[74]. Also, in another study, the miR-96 expression was reported to be a modulator of 5-FU chemosensitivity and promoted apoptosis of CRC cells[74]. Zhang et al[75] investigated the miR-520g expression and its target, p21, in CRC cells. p53 suppressed miR-520g expression, and inhibition of miR-520g in p53 negative cells increased chemosensitivity to 5-FU[75] (Table 1).
CIRCULAR RNAS AND RESISTANCE TO 5-FU
One of the new research fields of transcriptome studies performed on drug resistance in CRC is circular RNAs (circRNAs). Recently, these noncoding closely looped RNAs were found to be involved in 5-FU-based chemotherapy resistance in CRC mainly because of their ability to act as a miRNA sponge. Using microarray analysis, Xiong et al[76] for the first time revealed that circRNAs were expressed differently in 5-FU chemoradiation resistant CRC cells than parental control cells, suggesting the new potential role of these biomolecules in 5-FU drug resistance. This study introduced hsa circ 0007031 as the most upregulated circRNA (116 fold) in 5-FU chemoradiation resistant CRC cells[76]. In line with the work of Xiong et al[76], Abu et al[77] compared the expression profile of circRNAs between chemosensitive and chemoresistant FOLFOX (5-FU + oxaliplatin) HCT-116 colon cancer cells. They concluded that up to 773 and 732 circRNAs were upregulated and downregulated in these two cell lines, respectively. Among the investigated circRNAs, hsa circ 32883 that encodes for the EML5 gene was believed to be a potential molecular target demanding further research[77].
MIRNAS PROMOTE 5-FU DRUG RESISTANCE IN CRC
Among miRNAs presented in Table 1, some miRNAs can induce 5-FU resistance. Upregulation of ST6GALNAC2 (which further activates the PI3K/AKT pathway) is one of the main mechanisms resulting in chemoresistance caused by miR-182 and miR-135b[78]. Based on the DIANA-miRPath database, these two miRNAs play a role in signaling pathways regulating the pluripotency of stem cells, adherent junctions and cell cycle[79]. In a study by Amankwatia et al[80], it has been shown that miR-224 knockdown significantly increased sensitivity of HCT116 KRAS wild-type cells to 5-FU but did not affect sensitivity to oxaliplatin or irinotecan. Additionally, they demonstrated that miR-224 silencing markedly enhanced KRAS activity and ERK and AKT phosphorylation, suggesting a direct effect of miR-224 on prosurvival RAS/AKT/PI3K pathway[80]. miR-587 is another miRNA that can downregulate activation of the AKT/XIAP axis and induce 5-FU resistance[81]. miR-587 inhibited AKT activation; when this miRNA was inhibited, it decreased chemoresistance to 5-FU[81]. Regardless of the cancer cells, expression of some specific miRNAs including miR-196b-5p in cancer stem cells is also related to 5-FU resistance, and it activates JAK/STAT3 signaling[33].
MIRNAS REVERSE 5-FU DRUG RESISTANCE IN CRC
In contrast to the previous section, other miRNAs are involved in reducing 5-FU chemoresistance in CRC patients. miR-24 is a tumor suppressor and increases the chemosensitivity of SW48 cells to 5-FU, probably due to targeting and downregulating DND1, which is upregulated in CRC cell lines up to 5.3-fold[82]. Moreover, according to the DIANA-miRPath database, miR-24-3p is involved in proteoglycans in cancer, cell cycle and pancreatic cancer pathways. Likewise, miR-361 overexpression increased cell apoptosis in 5-FU-resistant HCT116 and HT29 cells through targeting and modulation of FOXM1 and ABCC5/10, respectively[83]. miR-361 is also involved in adherent junction as well as extracellular matrix receptor interaction cellular pathways (DIANA-miRPath database). miR-375-3p targets YAP1 and SP1. Inhibition of YAP1 downstream genes including survivin, CTGF, and cyclin D1 promoted CRC cell sensitivity to 5-FU[84]. miR-133b also increased the sensitivity of cancer stem cells within colorectal spheroids to 5-FU by targeting disruptor of telomeric silencing 1-like, which is responsible for H3K79 methylation[85]. Similar to miRNAs promoting 5-FU drug resistance, some miRNAs reverse 5-FU drug resistance in CRC and are listed in Table 1 along with detailed explanations.
CONCLUSION
The latest and most important molecular mechanisms of 5-FU resistance including MSI, altered expression of TS and DPD enzymes and loss of p53 were introduced in detail. As discussed above, MSI induced by sporadic or germline mutations in DNA mismatch repair machinery genes increased the tolerance of CRC cells to DNA damage and apoptosis caused by 5-FU, leading to 5-FU resistance. High expression levels of TS and DPD before treatment are two important factors for intrinsic resistance to 5-FU in CRC. Polymorphism of the 5’-region of the TS gene promoter and loss of p53 function due to mutation or TP53 overexpression are other molecular determinants of response to 5-FU. miRNAs and their alterations are other important factors in 5-FU drug resistance in CRC. New emerging evidence indicated that the consideration of miRNAs not only confer great assurance for the diagnosis, prognosis and clinical follow-up of CRC patients but also can be considered as a potential tool for anticancer therapies. Moreover, miRNAs can be utilized as predictive markers for CRC patients[86,87]. As it is shown in Table 1, some miRNAs promote 5-FU drug resistance in CRC, while some reverse it. The identification of new potential molecular determinants of response to 5-FU could be an important clinical tool to develop treatment strategies and select CRC patients who would most likely benefit from 5-FU-based chemotherapy.
Footnotes
Conflict-of-interest statement: The authors declare no potential financial interests.
Manuscript source: Invited manuscript
Peer-review started: March 21, 2020
First decision: April 26, 2020
Article in press: July 19, 2020
Specialty type: Oncology
Country/Territory of origin: Iran
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Fiorentini G, Zhao W S-Editor: Zhang L L-Editor: Filipodia P-Editor: Li JH
Contributor Information
Amirsaeed Sabeti Aghabozorgi, Medical Genetics Research Center, Mashhad University of Medical Sciences, Mashhad 1394491388, Iran.
Mostafa Moradi Sarabi, Department of Biochemistry and Genetics, School of Medicine, Lorestan University of Medical Sciences, Khorramabad 381251698, Iran.
Reza Jafarzadeh-Esfehani, Department of Medical Genetics, Faculty of Medicine, Mashhad University of Medical Sciences, Mashhad 1394491388, Iran.
Shabnaz Koochakkhani, Molecular Medicine Research Center, Hormozgan Health Institute, Hormozgan University of Medical Sciences, Bandar Abbas 7919915519, Iran.
Marziyeh Hassanzadeh, Molecular Medicine Research Center, Hormozgan Health Institute, Hormozgan University of Medical Sciences, Bandar Abbas 7919915519, Iran.
Soudabeh Kavousipour, Molecular Medicine Research Center, Hormozgan Health Institute, Hormozgan University of Medical Sciences, Bandar Abbas 7919915519, Iran.
Ebrahim Eftekhar, Molecular Medicine Research Center, Hormozgan Health Institute, Hormozgan University of Medical Sciences, Bandar Abbas 7919915519, Iran. eftekhar19@gmail.com.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Moradi Sarabi M, Mohammadrezaei Khorramabadi R, Zare Z, Eftekhar E. Polyunsaturated fatty acids and DNA methylation in colorectal cancer. World J Clin Cases. 2019;7:4172–4185. doi: 10.12998/wjcc.v7.i24.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y, Chen Z, Li J. The current status of treatment for colorectal cancer in China: A systematic review. Medicine (Baltimore) 2017;96:e8242. doi: 10.1097/MD.0000000000008242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malet-Martino M, Jolimaitre P, Martino R. The prodrugs of 5-fluorouracil. Curr Med Chem Anticancer Agents. 2002;2:267–310. doi: 10.2174/1568011023354146. [DOI] [PubMed] [Google Scholar]
- 5.Eftekhar E, Naghibalhossaini F. Carcinoembryonic antigen expression level as a predictive factor for response to 5-fluorouracil in colorectal cancer. Mol Biol Rep. 2014;41:459–466. doi: 10.1007/s11033-013-2880-0. [DOI] [PubMed] [Google Scholar]
- 6.Hu T, Li Z, Gao CY, Cho CH. Mechanisms of drug resistance in colon cancer and its therapeutic strategies. World J Gastroenterol. 2016;22:6876–6889. doi: 10.3748/wjg.v22.i30.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van der Jeught K, Xu HC, Li YJ, Lu XB, Ji G. Drug resistance and new therapies in colorectal cancer. World J Gastroenterol. 2018;24:3834–3848. doi: 10.3748/wjg.v24.i34.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furuta T. Pharmacogenomics in chemotherapy for GI tract cancer. J Gastroenterol. 2009;44:1016–1025. doi: 10.1007/s00535-009-0124-9. [DOI] [PubMed] [Google Scholar]
- 9.Peters GJ, Backus HH, Freemantle S, van Triest B, Codacci-Pisanelli G, van der Wilt CL, Smid K, Lunec J, Calvert AH, Marsh S, McLeod HL, Bloemena E, Meijer S, Jansen G, van Groeningen CJ, Pinedo HM. Induction of thymidylate synthase as a 5-fluorouracil resistance mechanism. Biochim Biophys Acta. 2002;1587:194–205. doi: 10.1016/s0925-4439(02)00082-0. [DOI] [PubMed] [Google Scholar]
- 10.Zheng T, Wang J, Chen X, Liu L. Role of microRNA in anticancer drug resistance. Int J Cancer. 2010;126:2–10. doi: 10.1002/ijc.24782. [DOI] [PubMed] [Google Scholar]
- 11.Moradi Sarabi M, Zahedi SA, Pajouhi N, Khosravi P, Bagheri S, Ahmadvand H, Shahryarhesami S. The effects of dietary polyunsaturated fatty acids on miR-126 promoter DNA methylation status and VEGF protein expression in the colorectal cancer cells. Genes Nutr. 2018;13:32. doi: 10.1186/s12263-018-0623-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Che J, Pan L, Yang X, Liu Z, Huang L, Wen C, Lin A, Liu H. Thymidine phosphorylase expression and prognosis in colorectal cancer treated with 5-fluorouracil-based chemotherapy: A meta-analysis. Mol Clin Oncol. 2017;7:943–952. doi: 10.3892/mco.2017.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gan Z, Zou Q, Lin Y, Xu Z, Huang Z, Chen Z, Lv Y. Identification of a 13-gene-based classifier as a potential biomarker to predict the effects of fluorouracil-based chemotherapy in colorectal cancer. Oncol Lett. 2019;17:5057–5063. doi: 10.3892/ol.2019.10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luqmani YA. Mechanisms of drug resistance in cancer chemotherapy. Med Princ Pract. 2005;14 Suppl 1:35–48. doi: 10.1159/000086183. [DOI] [PubMed] [Google Scholar]
- 15.Chen P, Ni W, Xie T, Sui X. Meta-Analysis of 5-Fluorouracil-Based Chemotherapy Combined With Traditional Chinese Medicines for Colorectal Cancer Treatment. Integr Cancer Ther. 2019;18:1534735419828824. doi: 10.1177/1534735419828824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miura K, Kinouchi M, Ishida K, Fujibuchi W, Naitoh T, Ogawa H, Ando T, Yazaki N, Watanabe K, Haneda S, Shibata C, Sasaki I. 5-fu metabolism in cancer and orally-administrable 5-fu drugs. Cancers (Basel) 2010;2:1717–1730. doi: 10.3390/cancers2031717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pietrantonio F, De Braud F, Da Prat V, Perrone F, Pierotti MA, Gariboldi M, Fanetti G, Biondani P, Pellegrinelli A, Bossi I, Di Bartolomeo M. A review on biomarkers for prediction of treatment outcome in gastric cancer. Anticancer Res. 2013;33:1257–1266. [PubMed] [Google Scholar]
- 18.Zargar P, Ghani E, Mashayekhi FJ, Ramezani A, Eftekhar E. Acriflavine enhances the antitumor activity of the chemotherapeutic drug 5-fluorouracil in colorectal cancer cells. Oncol Lett. 2018;15:10084–10090. doi: 10.3892/ol.2018.8569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eftekhar E, Jaberie H, Naghibalhossaini F. Carcinoembryonic Antigen Expression and Resistance to Radiation and 5-Fluorouracil-Induced Apoptosis and Autophagy. Int J Mol Cell Med. 2016;5:80–89. [PMC free article] [PubMed] [Google Scholar]
- 20.Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 21.Chon J, Stover PJ, Field MS. Targeting nuclear thymidylate biosynthesis. Mol Aspects Med. 2017;53:48–56. doi: 10.1016/j.mam.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grem JL. 5-Fluorouracil: forty-plus and still ticking. A review of its preclinical and clinical development. Invest New Drugs. 2000;18:299–313. doi: 10.1023/a:1006416410198. [DOI] [PubMed] [Google Scholar]
- 23.Hammond WA, Swaika A, Mody K. Pharmacologic resistance in colorectal cancer: a review. Ther Adv Med Oncol. 2016;8:57–84. doi: 10.1177/1758834015614530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moradi Sarabi M, Doosti M, Einollahi N, Hesami SS, Dashti N. Effect of eicosapentaenoic acid on the expression of ABCG1 gene in the human monocyte THP-1 cells. Acta Med Iran. 2014;52:176–181. [PubMed] [Google Scholar]
- 25.Longley DB, Allen WL, Johnston PG. Drug resistance, predictive markers and pharmacogenomics in colorectal cancer. Biochim Biophys Acta. 2006;1766:184–196. doi: 10.1016/j.bbcan.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 26.Tan W, Liu B, Qu S, Liang G, Luo W, Gong C. MicroRNAs and cancer: Key paradigms in molecular therapy. Oncol Lett. 2018;15:2735–2742. doi: 10.3892/ol.2017.7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Negrini M, Ferracin M, Sabbioni S, Croce CM. MicroRNAs in human cancer: from research to therapy. J Cell Sci. 2007;120:1833–1840. doi: 10.1242/jcs.03450. [DOI] [PubMed] [Google Scholar]
- 28.Si W, Shen J, Zheng H, Fan W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin Epigenetics. 2019;11:25. doi: 10.1186/s13148-018-0587-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valeri N, Gasparini P, Braconi C, Paone A, Lovat F, Fabbri M, Sumani KM, Alder H, Amadori D, Patel T, Nuovo GJ, Fishel R, Croce CM. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2) Proc Natl Acad Sci USA. 2010;107:21098–21103. doi: 10.1073/pnas.1015541107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chai H, Liu M, Tian R, Li X, Tang H. miR-20a targets BNIP2 and contributes chemotherapeutic resistance in colorectal adenocarcinoma SW480 and SW620 cell lines. Acta Biochim Biophys Sin (Shanghai) 2011;43:217–225. doi: 10.1093/abbs/gmq125. [DOI] [PubMed] [Google Scholar]
- 31.Cao S, Lin L, Xia X, Wu H. MicroRNA-761 promotes the sensitivity of colorectal cancer cells to 5-Fluorouracil through targeting FOXM1. Oncotarget. 2018;9:321–331. doi: 10.18632/oncotarget.20109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao C, Zhao Q, Zhang C, Wang G, Yao Y, Huang X, Zhan F, Zhu Y, Shi J, Chen J, Yan F, Zhang Y. miR-15b-5p resensitizes colon cancer cells to 5-fluorouracil by promoting apoptosis via the NF-κB/XIAP axis. Sci Rep. 2017;7:4194. doi: 10.1038/s41598-017-04172-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren D, Lin B, Zhang X, Peng Y, Ye Z, Ma Y, Liang Y, Cao L, Li X, Li R, Sun L, Liu Q, Wu J, Zhou K, Zeng J. Maintenance of cancer stemness by miR-196b-5p contributes to chemoresistance of colorectal cancer cells via activating STAT3 signaling pathway. Oncotarget. 2017;8:49807–49823. doi: 10.18632/oncotarget.17971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu X, Shi W, Zhang Y, Wang X, Sun S, Song Z, Liu M, Zeng Q, Cui S, Qu X. CXCL12/CXCR4 axis induced miR-125b promotes invasion and confers 5-fluorouracil resistance through enhancing autophagy in colorectal cancer. Sci Rep. 2017;7:42226. doi: 10.1038/srep42226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X, Xie T, Mao X, Xue L, Chu X, Chen L. MicroRNA-149 Increases the Sensitivity of Colorectal Cancer Cells to 5-Fluorouracil by Targeting Forkhead Box Transcription Factor FOXM1. Cell Physiol Biochem. 2016;39:617–629. doi: 10.1159/000445653. [DOI] [PubMed] [Google Scholar]
- 36.Sarabi MM, Naghibalhossaini F. The impact of polyunsaturated fatty acids on DNA methylation and expression of DNMTs in human colorectal cancer cells. Biomed Pharmacother. 2018;101:94–99. doi: 10.1016/j.biopha.2018.02.077. [DOI] [PubMed] [Google Scholar]
- 37.Mokarram P, Kavousipour S, Sarabi MM, Mehrabani G, Fahmidehkar MA, Shamsdin SA, Alipour A, Naini MA. MGMT-B gene promoter hypermethylation in patients with inflammatory bowel disease - a novel finding. Asian Pac J Cancer Prev. 2015;16:1945–1952. doi: 10.7314/apjcp.2015.16.5.1945. [DOI] [PubMed] [Google Scholar]
- 38.Mokarram P, Zamani M, Kavousipour S, Naghibalhossaini F, Irajie C, Moradi Sarabi M, Hosseini SV. Different patterns of DNA methylation of the two distinct O6-methylguanine-DNA methyltransferase (O6-MGMT) promoter regions in colorectal cancer. Mol Biol Rep. 2013;40:3851–3857. doi: 10.1007/s11033-012-2465-3. [DOI] [PubMed] [Google Scholar]
- 39.Meyers M, Wagner MW, Hwang HS, Kinsella TJ, Boothman DA. Role of the hMLH1 DNA mismatch repair protein in fluoropyrimidine-mediated cell death and cell cycle responses. Cancer Res. 2001;61:5193–5201. [PubMed] [Google Scholar]
- 40.Ye L, Jiang T, Shao H, Zhong L, Wang Z, Liu Y, Tang H, Qin B, Zhang X, Fan J. miR-1290 Is a Biomarker in DNA-Mismatch-Repair-Deficient Colon Cancer and Promotes Resistance to 5-Fluorouracil by Directly Targeting hMSH2. Mol Ther Nucleic Acids. 2017;7:453–464. doi: 10.1016/j.omtn.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao P, Ma YG, Zhao Y, Liu D, Dai ZJ, Yan CY, Guan HT. MicroRNA-552 deficiency mediates 5-fluorouracil resistance by targeting SMAD2 signaling in DNA-mismatch-repair-deficient colorectal cancer. Cancer Chemother Pharmacol. 2019;84:427–439. doi: 10.1007/s00280-019-03866-7. [DOI] [PubMed] [Google Scholar]
- 42.Zhang N, Yin Y, Xu SJ, Chen WS. 5-Fluorouracil: mechanisms of resistance and reversal strategies. Molecules. 2008;13:1551–1569. doi: 10.3390/molecules13081551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watson RG, Muhale F, Thorne LB, Yu J, O'Neil BH, Hoskins JM, Meyers MO, Deal AM, Ibrahim JG, Hudson ML, Walko CM, McLeod HL, Auman JT. Amplification of thymidylate synthetase in metastatic colorectal cancer patients pretreated with 5-fluorouracil-based chemotherapy. Eur J Cancer. 2010;46:3358–3364. doi: 10.1016/j.ejca.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnston PG, Drake JC, Trepel J, Allegra CJ. Immunological quantitation of thymidylate synthase using the monoclonal antibody TS 106 in 5-fluorouracil-sensitive and -resistant human cancer cell lines. Cancer Res. 1992;52:4306–4312. [PubMed] [Google Scholar]
- 45.Copur S, Aiba K, Drake JC, Allegra CJ, Chu E. Thymidylate synthase gene amplification in human colon cancer cell lines resistant to 5-fluorouracil. Biochem Pharmacol. 1995;49:1419–1426. doi: 10.1016/0006-2952(95)00067-a. [DOI] [PubMed] [Google Scholar]
- 46.Imam AM, Crossley PH, Jackman AL, Little PF. Analysis of thymidylate synthase gene amplification and of mRNA levels in the cell cycle. J Biol Chem. 1987;262:7368–7373. [PubMed] [Google Scholar]
- 47.Wilson PM, Danenberg PV, Johnston PG, Lenz HJ, Ladner RD. Standing the test of time: targeting thymidylate biosynthesis in cancer therapy. Nat Rev Clin Oncol. 2014;11:282–298. doi: 10.1038/nrclinonc.2014.51. [DOI] [PubMed] [Google Scholar]
- 48.Lenz HJ, Hayashi K, Salonga D, Danenberg KD, Danenberg PV, Metzger R, Banerjee D, Bertino JR, Groshen S, Leichman LP, Leichman CG. p53 point mutations and thymidylate synthase messenger RNA levels in disseminated colorectal cancer: an analysis of response and survival. Clin Cancer Res. 1998;4:1243–1250. [PubMed] [Google Scholar]
- 49.Johnston PG, Lenz HJ, Leichman CG, Danenberg KD, Allegra CJ, Danenberg PV, Leichman L. Thymidylate synthase gene and protein expression correlate and are associated with response to 5-fluorouracil in human colorectal and gastric tumors. Cancer Res. 1995;55:1407–1412. [PubMed] [Google Scholar]
- 50.Edler D, Blomgren H, Allegra CJ, Johnston PG, Lagerstedt U, Magnusson I, Ragnhammar P. Immunohistochemical determination of thymidylate synthase in colorectal cancer--methodological studies. Eur J Cancer. 1997;33:2278–2281. doi: 10.1016/s0959-8049(97)00306-7. [DOI] [PubMed] [Google Scholar]
- 51.Abdallah EA, Fanelli MF, Buim ME, Machado Netto MC, Gasparini Junior JL, Souza E Silva V, Dettino AL, Mingues NB, Romero JV, Ocea LM, Rocha BM, Alves VS, Araújo DV, Chinen LT. Thymidylate synthase expression in circulating tumor cells: a new tool to predict 5-fluorouracil resistance in metastatic colorectal cancer patients. Int J Cancer. 2015;137:1397–1405. doi: 10.1002/ijc.29495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marsh S, McLeod HL. Thymidylate synthase pharmacogenetics in colorectal cancer. Clin Colorectal Cancer. 2001;1:175–8; discussion 179-81. doi: 10.3816/CCC.2001.n.018. [DOI] [PubMed] [Google Scholar]
- 53.Naghibalhossaini F, Shefaghat M, Mansouri A, Jaberi H, Tatar M, Eftekhar E. The Impact of Thymidylate Synthase and Methylenetetrahydrofolate Reductase Genotypes on Sensitivity to 5-Fluorouracil Treatment in Colorectal Cancer Cells. Acta Med Iran. 2017;55:751–758. [PubMed] [Google Scholar]
- 54.Horie N, Aiba H, Oguro K, Hojo H, Takeishi K. Functional analysis and DNA polymorphism of the tandemly repeated sequences in the 5'-terminal regulatory region of the human gene for thymidylate synthase. Cell Struct Funct. 1995;20:191–197. doi: 10.1247/csf.20.191. [DOI] [PubMed] [Google Scholar]
- 55.Marsh S, McKay JA, Cassidy J, McLeod HL. Polymorphism in the thymidylate synthase promoter enhancer region in colorectal cancer. Int J Oncol. 2001;19:383–386. doi: 10.3892/ijo.19.2.383. [DOI] [PubMed] [Google Scholar]
- 56.Pullarkat ST, Stoehlmacher J, Ghaderi V, Xiong YP, Ingles SA, Sherrod A, Warren R, Tsao-Wei D, Groshen S, Lenz HJ. Thymidylate synthase gene polymorphism determines response and toxicity of 5-FU chemotherapy. Pharmacogenomics J. 2001;1:65–70. doi: 10.1038/sj.tpj.6500012. [DOI] [PubMed] [Google Scholar]
- 57.Kawakami K, Omura K, Kanehira E, Watanabe Y. Polymorphic tandem repeats in the thymidylate synthase gene is associated with its protein expression in human gastrointestinal cancers. Anticancer Res. 1999;19:3249–3252. [PubMed] [Google Scholar]
- 58.Boni V, Bitarte N, Cristobal I, Zarate R, Rodriguez J, Maiello E, Garcia-Foncillas J, Bandres E. miR-192/miR-215 influence 5-fluorouracil resistance through cell cycle-mediated mechanisms complementary to its post-transcriptional thymidilate synthase regulation. Mol Cancer Ther. 2010;9:2265–2275. doi: 10.1158/1535-7163.MCT-10-0061. [DOI] [PubMed] [Google Scholar]
- 59.Li T, Gao F, Zhang XP. miR-203 enhances chemosensitivity to 5-fluorouracil by targeting thymidylate synthase in colorectal cancer. Oncol Rep. 2015;33:607–614. doi: 10.3892/or.2014.3646. [DOI] [PubMed] [Google Scholar]
- 60.Xu W, Jiang H, Zhang F, Gao J, Hou J. MicroRNA-330 inhibited cell proliferation and enhanced chemosensitivity to 5-fluorouracil in colorectal cancer by directly targeting thymidylate synthase. Oncol Lett. 2017;13:3387–3394. doi: 10.3892/ol.2017.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li PL, Zhang X, Wang LL, Du LT, Yang YM, Li J, Wang CX. MicroRNA-218 is a prognostic indicator in colorectal cancer and enhances 5-fluorouracil-induced apoptosis by targeting BIRC5. Carcinogenesis. 2015;36:1484–1493. doi: 10.1093/carcin/bgv145. [DOI] [PubMed] [Google Scholar]
- 62.Panczyk M. Pharmacogenetics research on chemotherapy resistance in colorectal cancer over the last 20 years. World J Gastroenterol. 2014;20:9775–9827. doi: 10.3748/wjg.v20.i29.9775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Salonga D, Danenberg KD, Johnson M, Metzger R, Groshen S, Tsao-Wei DD, Lenz HJ, Leichman CG, Leichman L, Diasio RB, Danenberg PV. Colorectal tumors responding to 5-fluorouracil have low gene expression levels of dihydropyrimidine dehydrogenase, thymidylate synthase, and thymidine phosphorylase. Clin Cancer Res. 2000;6:1322–1327. [PubMed] [Google Scholar]
- 64.Bai W, Wu Y, Zhang P, Xi Y. Correlations between expression levels of thymidylate synthase, thymidine phosphorylase and dihydropyrimidine dehydrogenase, and efficacy of 5-fluorouracil-based chemotherapy for advanced colorectal cancer. Int J Clin Exp Pathol. 2015;8:12333–12345. [PMC free article] [PubMed] [Google Scholar]
- 65.Taddia L, D'Arca D, Ferrari S, Marraccini C, Severi L, Ponterini G, Assaraf YG, Marverti G, Costi MP. Inside the biochemical pathways of thymidylate synthase perturbed by anticancer drugs: Novel strategies to overcome cancer chemoresistance. Drug Resist Updat. 2015;23:20–54. doi: 10.1016/j.drup.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 66.Chai J, Dong W, Xie C, Wang L, Han DL, Wang S, Guo HL, Zhang ZL. MicroRNA-494 sensitizes colon cancer cells to fluorouracil through regulation of DPYD. IUBMB Life. 2015;67:191–201. doi: 10.1002/iub.1361. [DOI] [PubMed] [Google Scholar]
- 67.Offer SM, Butterfield GL, Jerde CR, Fossum CC, Wegner NJ, Diasio RB. microRNAs miR-27a and miR-27b directly regulate liver dihydropyrimidine dehydrogenase expression through two conserved binding sites. Mol Cancer Ther. 2014;13:742–751. doi: 10.1158/1535-7163.MCT-13-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Elsaleh H, Powell B, McCaul K, Grieu F, Grant R, Joseph D, Iacopetta B. P53 alteration and microsatellite instability have predictive value for survival benefit from chemotherapy in stage III colorectal carcinoma. Clin Cancer Res. 2001;7:1343–1349. [PubMed] [Google Scholar]
- 69.Bunz F, Hwang PM, Torrance C, Waldman T, Zhang Y, Dillehay L, Williams J, Lengauer C, Kinzler KW, Vogelstein B. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J Clin Invest. 1999;104:263–269. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Longley DB, Boyer J, Allen WL, Latif T, Ferguson PR, Maxwell PJ, McDermott U, Lynch M, Harkin DP, Johnston PG. The role of thymidylate synthase induction in modulating p53-regulated gene expression in response to 5-fluorouracil and antifolates. Cancer Res. 2002;62:2644–2649. [PubMed] [Google Scholar]
- 71.Hector S, Prehn JH. Apoptosis signaling proteins as prognostic biomarkers in colorectal cancer: a review. Biochim Biophys Acta. 2009;1795:117–129. doi: 10.1016/j.bbcan.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 72.Liang JT, Huang KC, Cheng YM, Hsu HC, Cheng AL, Hsu CH, Yeh KH, Wang SM, Chang KJ. P53 overexpression predicts poor chemosensitivity to high-dose 5-fluorouracil plus leucovorin chemotherapy for stage IV colorectal cancers after palliative bowel resection. Int J Cancer. 2002;97:451–457. doi: 10.1002/ijc.1637. [DOI] [PubMed] [Google Scholar]
- 73.Ahnen DJ, Feigl P, Quan G, Fenoglio-Preiser C, Lovato LC, Bunn PA, Jr, Stemmerman G, Wells JD, Macdonald JS, Meyskens FL., Jr Ki-ras mutation and p53 overexpression predict the clinical behavior of colorectal cancer: a Southwest Oncology Group study. Cancer Res. 1998;58:1149–1158. [PubMed] [Google Scholar]
- 74.Kim SA, Kim I, Yoon SK, Lee EK, Kuh HJ. Indirect modulation of sensitivity to 5-fluorouracil by microRNA-96 in human colorectal cancer cells. Arch Pharm Res. 2015;38:239–248. doi: 10.1007/s12272-014-0528-9. [DOI] [PubMed] [Google Scholar]
- 75.Zhang Y, Geng L, Talmon G, Wang J. MicroRNA-520g confers drug resistance by regulating p21 expression in colorectal cancer. J Biol Chem. 2015;290:6215–6225. doi: 10.1074/jbc.M114.620252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xiong W, Ai YQ, Li YF, Ye Q, Chen ZT, Qin JY, Liu QY, Wang H, Ju YH, Li WH, Li YF. Microarray Analysis of Circular RNA Expression Profile Associated with 5-Fluorouracil-Based Chemoradiation Resistance in Colorectal Cancer Cells. Biomed Res Int. 2017;2017:8421614. doi: 10.1155/2017/8421614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Abu N, Hon KW, Jeyaraman S, Yahaya A, Abdullah NM, Mustangin M, Sulaiman SA, Jamal R, Ab-Mutalib NS. Identification of differentially expressed circular RNAs in chemoresistant colorectal cancer. Epigenomics. 2019;11:875–884. doi: 10.2217/epi-2019-0042. [DOI] [PubMed] [Google Scholar]
- 78.Wang X, Hu H, Hua R, Yang J, Zheng P, Niu X, Cheng H, Dai G, Liu X, Zhang Z, An Y. Effect of umbilical cord mesenchymal stromal cells on motor functions of identical twins with cerebral palsy: pilot study on the correlation of efficacy and hereditary factors. Cytotherapy. 2015;17:224–231. doi: 10.1016/j.jcyt.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 79.Liu B, Liu Y, Zhao L, Pan Y, Shan Y, Li Y, Jia L. Upregulation of microRNA-135b and microRNA-182 promotes chemoresistance of colorectal cancer by targeting ST6GALNAC2 via PI3K/AKT pathway. Mol Carcinog. 2017;56:2669–2680. doi: 10.1002/mc.22710. [DOI] [PubMed] [Google Scholar]
- 80.Amankwatia EB, Chakravarty P, Carey FA, Weidlich S, Steele RJ, Munro AJ, Wolf CR, Smith G. MicroRNA-224 is associated with colorectal cancer progression and response to 5-fluorouracil-based chemotherapy by KRAS-dependent and -independent mechanisms. Br J Cancer. 2015;112:1480–1490. doi: 10.1038/bjc.2015.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Y, Talmon G, Wang J. MicroRNA-587 antagonizes 5-FU-induced apoptosis and confers drug resistance by regulating PPP2R1B expression in colorectal cancer. Cell Death Dis. 2015;6:e1845. doi: 10.1038/cddis.2015.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Q, Li W, Liu G, Tang W. MicroRNA-24 regulates the growth and chemosensitivity of the human colorectal cancer cells by targeting RNA-binding protein DND1. J BUON. 2019;24:1476–1481. [PubMed] [Google Scholar]
- 83.Zhang L, Li B, Zhang B, Zhang H, Suo J. miR-361 enhances sensitivity to 5-fluorouracil by targeting the FOXM1-ABCC5/10 signaling pathway in colorectal cancer. Oncol Lett. 2019;18:4064–4073. doi: 10.3892/ol.2019.10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu X, Chen X, Xu M, Liu X, Pan B, Qin J, Xu T, Zeng K, Pan Y, He B, Sun H, Sun L, Wang S. miR-375-3p suppresses tumorigenesis and partially reverses chemoresistance by targeting YAP1 and SP1 in colorectal cancer cells. Aging (Albany NY) 2019;11:7357–7385. doi: 10.18632/aging.102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lv L, Li Q, Chen S, Zhang X, Tao X, Tang X, Wang S, Che G, Yu Y, He L. miR-133b suppresses colorectal cancer cell stemness and chemoresistance by targeting methyltransferase DOT1L. Exp Cell Res. 2019;385:111597. doi: 10.1016/j.yexcr.2019.111597. [DOI] [PubMed] [Google Scholar]
- 86.Jarry J, Schadendorf D, Greenwood C, Spatz A, van Kempen LC. The validity of circulating microRNAs in oncology: five years of challenges and contradictions. Mol Oncol. 2014;8:819–829. doi: 10.1016/j.molonc.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Iorio MV, Palmieri D. Editorial: From "Junk DNA" to Clinically Relevant Tools for Cancer Diagnosis, Staging, and Tailored Therapies: The Incredible Case of Non-Coding RNAs. Front Oncol. 2019;9:389. doi: 10.3389/fonc.2019.00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu G, Zhou J, Dong M. Down-regulation of miR-543 expression increases the sensitivity of colorectal cancer cells to 5-Fluorouracil through the PTEN/PI3K/AKT pathway. Biosci Rep. 2019;39 doi: 10.1042/BSR20190249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhao J, Cao J, Zhou L, Du Y, Zhang X, Yang B, Gao Y, Wang Y, Ma N, Yang W. MiR-1260b inhibitor enhances the chemosensitivity of colorectal cancer cells to fluorouracil by targeting PDCD4/IGF1. Oncol Lett. 2018;16:5131–5139. doi: 10.3892/ol.2018.9307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guo Z, Liu Z, Yue H, Wang J. Beta-elemene increases chemosensitivity to 5-fluorouracil through down-regulating microRNA-191 expression in colorectal carcinoma cells. J Cell Biochem. 2018;119:7032–7039. doi: 10.1002/jcb.26914. [DOI] [PubMed] [Google Scholar]
- 91.Kong D, Zhang D, Chu X, Wang J. Schizandrin A enhances chemosensitivity of colon carcinoma cells to 5-fluorouracil through up-regulation of miR-195. Biomed Pharmacother. 2018;99:176–183. doi: 10.1016/j.biopha.2018.01.035. [DOI] [PubMed] [Google Scholar]
- 92.Que K, Tong Y, Que G, Li L, Lin H, Huang S, Wang R, Tang L. Downregulation of miR-874-3p promotes chemotherapeutic resistance in colorectal cancer via inactivation of the Hippo signaling pathway. Oncol Rep. 2017;38:3376–3386. doi: 10.3892/or.2017.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jiang H, Ju H, Zhang L, Lu H, Jie K. microRNA-577 suppresses tumor growth and enhances chemosensitivity in colorectal cancer. J Biochem Mol Toxicol. 2017;31 doi: 10.1002/jbt.21888. [DOI] [PubMed] [Google Scholar]
- 94.Fu Q, Cheng J, Zhang J, Zhang Y, Chen X, Luo S, Xie J. miR-20b reduces 5-FU resistance by suppressing the ADAM9/EGFR signaling pathway in colon cancer. Oncol Rep. 2017;37:123–130. doi: 10.3892/or.2016.5259. [DOI] [PubMed] [Google Scholar]
- 95.Liu N, Li J, Zhao Z, Han J, Jiang T, Chen Y, Hou N, Huang C. MicroRNA-302a enhances 5-fluorouracil-induced cell death in human colon cancer cells. Oncol Rep. 2017;37:631–639. doi: 10.3892/or.2016.5237. [DOI] [PubMed] [Google Scholar]
- 96.Dong SJ, Cai XJ, Li SJ. The Clinical Significance of MiR-429 as a Predictive Biomarker in Colorectal Cancer Patients Receiving 5-Fluorouracil Treatment. Med Sci Monit. 2016;22:3352–3361. doi: 10.12659/MSM.900674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Han J, Liu Z, Wang N, Pan W. MicroRNA-874 inhibits growth, induces apoptosis and reverses chemoresistance in colorectal cancer by targeting X-linked inhibitor of apoptosis protein. Oncol Rep. 2016;36:542–550. doi: 10.3892/or.2016.4810. [DOI] [PubMed] [Google Scholar]
- 98.Liu H, Yin Y, Hu Y, Feng Y, Bian Z, Yao S, Li M, You Q, Huang Z. miR-139-5p sensitizes colorectal cancer cells to 5-fluorouracil by targeting NOTCH-1. Pathol Res Pract. 2016;212:643–649. doi: 10.1016/j.prp.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 99.Wu H, Liang Y, Shen L, Shen L. MicroRNA-204 modulates colorectal cancer cell sensitivity in response to 5-fluorouracil-based treatment by targeting high mobility group protein A2. Biol Open. 2016;5:563–570. doi: 10.1242/bio.015008. [DOI] [PMC free article] [PubMed] [Google Scholar]