Abstract
Spinal muscular atrophy (SMA) is a neuromuscular disorder caused by SMN1 gene deletion/mutation. The drug nusinersen modifies SMN2 mRNA splicing, increasing the production of the full-length SMN protein. Recent studies have demonstrated the beneficial effects of nusinersen in patients with SMA, particularly when treated in early infancy. Because nusinersen treatment can alter disease trajectory, there is a strong rationale for newborn screening. In the current study, we validated the accuracy of a new system for detecting SMN1 deletion (Japanese patent application No. 2017-196967, PCT/JP2018/37732) using dried blood spots (DBS) from 50 patients with genetically confirmed SMA and 50 controls. Our system consists of two steps: (1) targeted pre-amplification of SMN genes by direct polymerase chain reaction (PCR) and (2) detection of SMN1 deletion by real-time modified competitive oligonucleotide priming-PCR (mCOP-PCR) using the pre-amplified products. Compared with PCR analysis results of freshly collected blood samples, our system exhibited a sensitivity of 1.00 (95% confidence interval [CI] 0.96–1.00) and a specificity of 1.00 (95% CI 0.96–1.00). We also conducted a prospective SMA screening study using DBS from 4157 Japanese newborns. All DBS tested negative, and there were no screening failures. Our results indicate that the new system can be reliably used in SMA newborn screening.
Keywords: mCOP-PCR, SMN1, SMN2, spinal muscular atrophy
1. Introduction
Spinal muscular atrophy (SMA) is an autosomal recessive neuromuscular disorder characterized by degeneration of motor neurons in the spinal cord, which results in progressive muscle atrophy and weakness [1]. With a reported incidence of approximately 1 in 6000 to 10,000 live births [2], SMA is the second most common fatal autosomal recessive disorder after cystic fibrosis [3]. SMA is considered to be the leading genetic cause of infant death [1]. Patients with SMA type 1, the severe phenotype, develop symptoms in the first 6 months after birth, never achieve the motor milestone of sitting independently, and have a life expectancy of less than 2 years without respiratory support [1,4,5].
The survival motor neuron (SMN) genes located on chromosome 5q13, SMN1 and SMN2, were identified as the SMA-related genes in 1995 [6]. SMN1 and SMN2 are paralogs and are almost identical except for five nucleotides [6]. SMN1 expresses the full-length transcript and results in the production of the functional, full-length SMN protein [6]. The functional, full-length SMN protein plays a critical role in RNA metabolism and other cellular functions [7]. In contrast, SMN2 expresses two kinds of transcripts, the major one is an exon 7-skipped transcript (Δ7-transcript) due to a C-to-T change in exon 7, producing a non-functional, truncated SMN protein. The minor transcript is a full-length transcript encoding the same protein derived from SMN1. The presence of SMN2 does not fully compensate for the loss of SMN1 because SMN2 can only produce a limited amount of the full-length SMN protein [1,7].
SMN1 is absent (or homozygously deleted) in approximately 95% of patients with SMA and deleteriously mutated in the remaining patients [6]. On the other hand, a higher copy number of SMN2 is associated with a milder phenotype of SMA [8]. SMN1 is proven to be a disease-causing gene, while SMN2 is now considered to be a disease-modifying gene [6,8]. Therefore, the absence of both genes causes embryonic lethality in mice [9,10]. In fact, all patients with homozygous deletion of SMN1 retain at least one copy of the SMN2 gene [1].
SMA was considered to be an incurable disease. Nevertheless, intrathecal administration of an antisense-oligonucleotide drug, nusinersen, has been associated with encouraging clinical efficacy in SMA patients [11,12]. The drug modifies SMN2 splicing and increases the production of the functional, full-length SMN protein [11,13]. Nusinersen has been approved by regulatory agencies in multiple countries [the United States Food and Drug Administration (2016), the European Medicines Agency (2017), the Ministry of Health, Labor and Welfare of Japan (2017), the Ministry of Food and Drug Safety of the Republic of Korea (2018), and the China National Medical Products Association (2019)].
Treatment of SMA patients with nusinersen also appears to result in a better clinical outcome when it is initiated in early infancy [11,12]. Early diagnosis and initiation of treatment, ideally before apparent symptoms develop, may be important for the optimal response to nusinersen [11]. However, without newborn screening for SMA, treatment cannot be initiated until a significant number of motor neurons have been lost [14].
Ideally, implementation of newborn screening programs for SMA would allow pre-symptomatic diagnosis of the disease in many cases and the early initiation of treatment with potential for maximal therapeutic benefit [15]. In the USA, SMA is now included in the recommended uniform screening panel (RUSP), and newborn screening has been implemented in screening programs in a number of states [16,17].
We have developed a rapid, accurate, and high-throughput system for detecting SMN1 deletion using a real-time modified competitive oligonucleotide priming-polymerase chain reaction (mCOP-PCR) technique combined with targeted pre-amplification of the SMN genes from dried blood spots (DBS) [18]. Here, we describe the results of a pilot study of newborn screening for SMA to validate our system and to genotype all DBS collected from newborns in Japan.
2. Materials and Methods
2.1. Objectives and Ethics
The primary objective of this pilot study was to validate the accuracy of our newly developed SMN1-deletion detection system using DBS from individuals with and without genetically confirmed SMA. An additional objective was to investigate the accuracy of the SMN1-deletion detection system for prospective newborn SMA screening using DBS. The study was approved by the institutional review boards at all participating hospitals, as well as the Ethics Committee of the Kobe University Graduate School of Medicine (reference 170165, approved on 27November, 2017), and was conducted in accordance with the World Medical Association Declaration of Helsinki.
2.2. SMA Patients and Non-SMA Controls
The SMN genotype of the patients and controls had been previously analyzed by PCR–restriction fragment length polymorphism (PCR–RFLP) using extracted DNA from freshly collected blood. PCR–RFLP was carried out according to the method of van der Steege and colleagues [19]. DBS from the same patients and controls were stored in the sample library at the Division of Epidemiology, Kobe University Graduate School of Medicine, Japan. DBS from 100 individuals (50 SMA patients and 50 non-SMA controls) were analyzed using the new SMN1-deletion detection system. Written informed consent for the use of all DNA samples was obtained from the patients/controls and/or their parents.
2.3. Newborn Infants
Infants born at 49 hospitals across Japan were eligible to participate in the pilot screening study. Written informed consent for study participation was obtained from parents/guardians of newborn infants. Information about the study was provided, and it was explained that participation was voluntary.
2.4. SMN1-Deletion Detection System
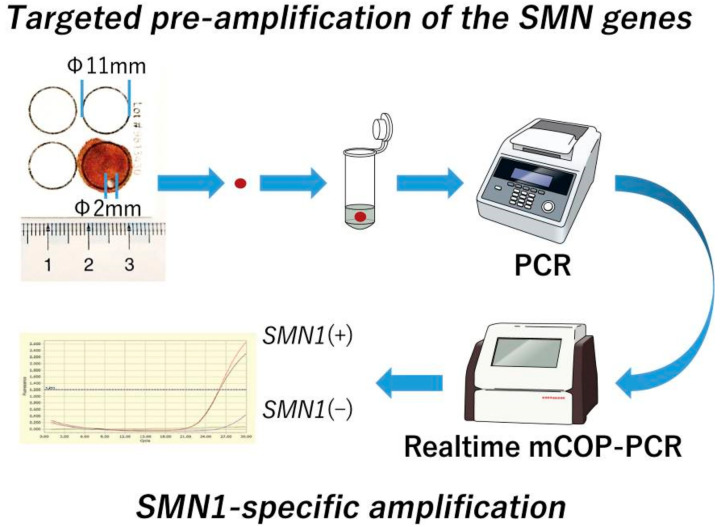
As illustrated in Figure 1, our system consisted of two steps, namely, (1) targeted pre-amplification of the SMN genes: the target sequences of the SMN genes from DBS were pre-amplified by conventional PCR, and (2) gene-specific amplification of SMN1 and SMN2 exon 7: SMN1-deletion was detected by mCOP-PCR with the pre-amplified products (Japanese patent application No. 2017-196967, PCT/JP2018/37732). In the previous version of our system, genomic DNA was extracted from DBS before targeted pre-amplification [18]. However, the new version used in the present study did not include the DNA extraction step. Instead, a punched circle from each DBS was placed directly into the reaction tube of the conventional PCR (direct PCR) [20].
Figure 1.
Scheme of the SMN1-deletion detection system using dried blood spots (DBS). Our system consists of two steps. Step (1) targeted pre-amplification of the SMN genes: the target sequences in the SMN genes from DBS are pre-amplified by conventional polymerase chain reaction (PCR), and Step (2) gene-specific amplification of SMN1 and SMN2 exon 7: SMN1 deletion is detected by real-time modified competitive oligonucleotide priming-PCR (mCOP-PCR) with the pre-amplified products. In the first step, a punched circle from each DBS is placed directly into the reaction tube of conventional PCR (direct PCR).
2.4.1. Targeted Pre-Amplification of the SMN Genes
Targeted pre-amplification of the sequence containing SMN1/SMN2 exon 7 was performed with conventional PCR using the GeneAmp® PCR System 9700 (Applied Biosytems, Foster City, CA, USA). A punched circle 2 mm in diameter (equivalent to ~15 µL of whole blood) from each DBS was added to the reaction mixture with DNA polymerase KOD FX Neo (TOYOBO, Osaka, Japan). The following primers were used to amplify the target sequence containing SMN1/SMN2 exon 7: R111 (5′-AGA CTA TCA ACT TAA TTT CTG ATC A-3′) and 541C770 (5′-TAA GGA ATG TGA GCA CCT TCC TTC-3′) [6]. The PCR conditions for the 50 µL reaction mixture were: (1) initial denaturation at 94 °C for 7 min; (2) 40 cycles of denaturation at 94 °C for 1 min, annealing at 56 °C for 1 min, and extension at 72 °C for 1 min; (3) additional extension at 72 °C for 7 min; and (4) hold at 10 °C. The PCR product (i.e., the pre-amplified SMN gene product) was then subjected to gel electrophoresis and visualized using Midori-Green staining (NIPPON Genetics, Tokyo, Japan).
2.4.2. Gene-Specific Amplification of SMN1 Exon 7
Real-time mCOP-PCR SMN1 and SMN2 exon 7 amplification was performed using the LightCycler® 96 system (Roche Applied Science, Mannheim, Germany). An aliquot of pre-amplified PCR product was added to the reaction mixture with DNA polymerase KOD FX Neo (TOYOBO) and EvaGreen® Dye (Biotium, Hayward, CA, USA). The primer set for SMN1-specific amplification consisted of R111 and SMN1-COP (5′-TGT CTG AAA CC-3′) [18,21]. The PCR conditions for the reaction mixture of 50 µL were: (1) initial denaturation at 94 °C for 7 min; (2) 20 cycles for SMN1 denaturation at 94 °C for 1 min, annealing at 37 °C for 1 min, and extension at 72 °C for 1 min; and (3) melting analysis. Fluorescence signals were detected at the end of each extension procedure.
2.5. Newborn Screening Study Design
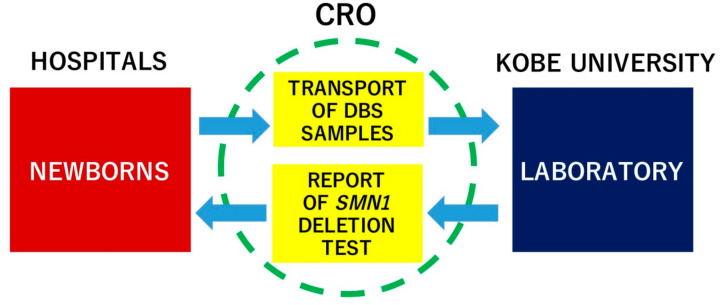
After obtaining written informed consent for SMA screening, approximately 100 µL of blood was collected from newborn infants and spotted onto filter paper (FTA® Elute Cards, GE Healthcare, Boston, MA, USA) during standard blood sampling for tandem mass spectrometry newborn screening. DBS sample collection began at the end of January 2018 and ended at the end of April 2019.
The flow of DBS samples and data collection during the study is illustrated in Figure 2. Filter papers with DBS were transferred to Kobe University within 7 days after blood sampling and stored in the dark at room temperature (20–25 °C) until use. PCR experiments and the final data analysis were completed within 10 ± 4 days after the blood sampling. Any positive screening result (i.e., when SMN1 deletion was detected) was verified by PCR–RFLP using the method of van der Steege et al. [19], and the primary physician of the infant was informed of the result so that the patient could be further examined, definitively diagnosed and, if necessary, treatment could be initiated. Any surplus specimen would be stored at the Division of Epidemiology, Kobe University Graduate School of Medicine, for up to 5 years.
Figure 2.
Data collection and data management flow between hospitals and Kobe University during the pilot study. CRO, contracted research organization.
2.6. Follow-Up Study of the Infants Screened for SMA
To determine the SMA status of the screened infants at 6 and 10 months after DBS collection, a survey was conducted among the physicians participating in the study, with a questionnaire including the question “Are there any patients diagnosed with SMA or SMA-like disease among the infants screened for SMA in this study?”.
2.7. Statistical Analysis
For validation of the screening system, 100 DNA samples (from 50 SMA patients and 50 controls) from the sample library of the Division of Epidemiology, Kobe University Graduate School of Medicine, were analyzed. The sensitivity and specificity of the screening system for the detection of SMN1 exon 7 deletion were calculated using the original results of the PCR–RFLP analysis as the reference. The exact method was used to calculate 95% confidence intervals (CIs) for sensitivity and specificity [22,23]. Statistical analysis was performed using Epi Info (Centers for Disease Control, Atlanta, GA, USA).
3. Results
3.1. Validation Study
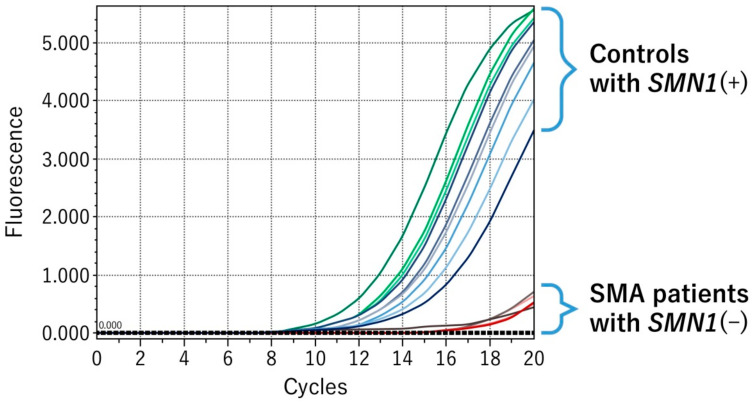
SMN1 exon 7 and SMN2 exon 7 differ in only one nucleotide at position 6, i.e., C in SMN1 exon 7 and T in SMN2. For the detection of SMN1 deletion, gene-specific amplification is essential. Figure 3 shows amplification of SMN1 exon 7 by real-time mCOP-PCR with SMN1-COP primer. DBS samples with the SMN1(+) genotype showed marked amplification by real-time mCOP-PCR with SMN1-COP primer, whereas samples with the SMN1(–) genotype showed no amplification. These results indicated that our system was able to specifically amplify SMN1 exon 7.
Figure 3.
SMN1-specific amplification by real-time mCOP-PCR. SMA patients with SMN1(–) genotype showed no amplification with an SMN1-specific primer (SMN1-COP).
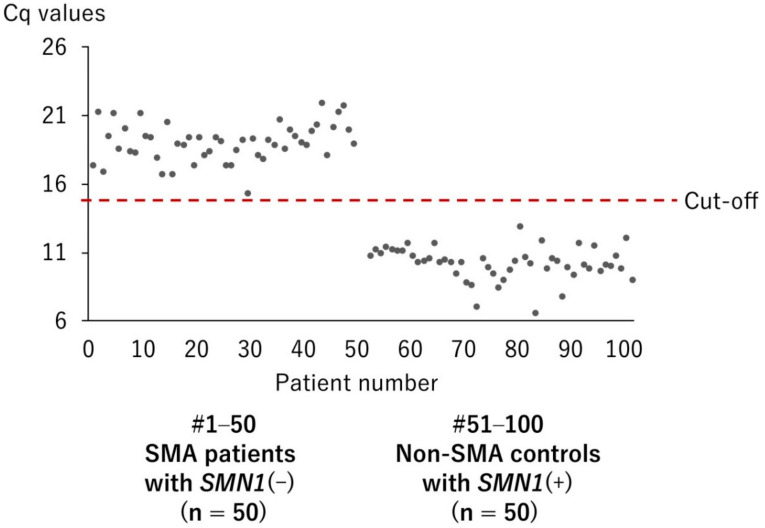
A total of 100 DBS samples from 100 individuals with and without genetically confirmed SMA were analyzed by real-time mCOP-PCR with SMN1-COP primer. They were from 50 SMA patients with SMN1(–) genotype and from 50 controls with SMN1(+) genotype. Figure 4 shows that the quantitation cycle values (Cq values) of 50 SMA patients with SMN1(–) were markedly higher than those of 50 controls with SMN1(+), without overlapping values. The mean ± SD Cq values of SMA patients and controls were 19.0 ± 1.4 and 10.1 ± 1.2, respectively. In the present study, we determined that Cq values ≥14 indicated the absence of SMN1.
Figure 4.
Distribution of quantification cycle number (Cq) values of DBS from 50 patients with SMN1(–) and 50 controls with SMN1(+). A Cq value of 14 was set as the cut-off point for the presence or absence of SMN1.
Compared with the results of PCR–RFLP using DNA from freshly collected blood, the results from real-time mCOP-PCR using DBS for detection of SMN1-deletion showed a sensitivity of 1.00 (95% CI 0.96–1.00) and a specificity of 1.00 (95% CI 0.96–1.00) (Table 1).
Table 1.
Real-time mCOP-PCR versus PCR–restriction fragment length polymorphism (PCR–RFLP) for the detection of SMN1 deletion.
| PCR–RFLP (Fresh Blood) | Total | ||
|---|---|---|---|
| SMN1(–) | SMN1(+) | ||
| Real-time mCOP-PCR (DBS) | |||
| SMN1(–) | 50 | 0 | 50 |
| SMN1(+) | 0 | 50 | 50 |
| Total | 50 | 50 | 100 |
Sensitivity: 1.00 (95% CI 0.96–1.00); specificity: 1.00 (95% CI 0.96–1.00).
3.2. Newborn Screening for SMA
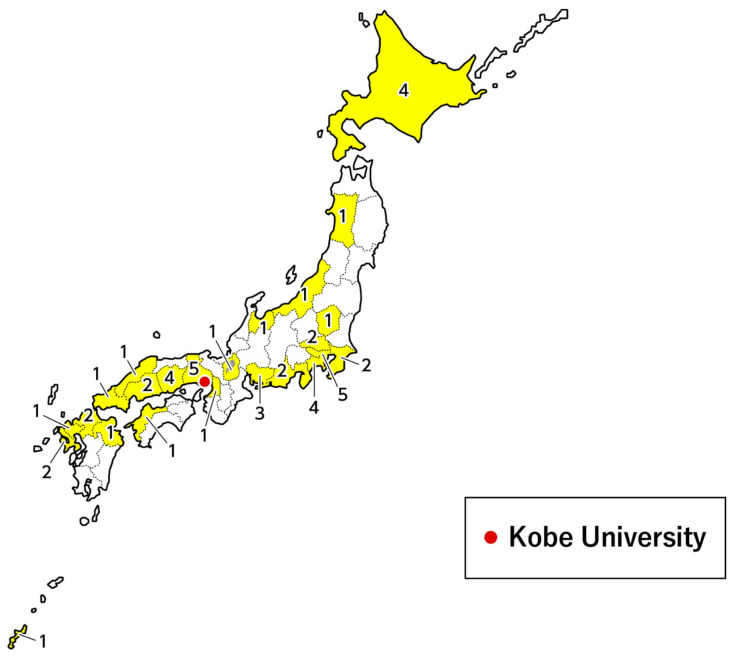
Between January 2018 and April 2019, 4157 DBS samples were collected from newborn infants at 49 hospitals, covering 23 of the 47 prefectures in Japan (Figure 5). Of the 4157 collected samples, all tested negative for SMN1 deletion using our new system described earlier.
Figure 5.
Location of the hospitals that participated in the pilot study. The numbers denote the number of hospitals in each prefecture that participated in the study.
The quantity and quality of the collected DBS were not limiting factors for the detection of SMN1 deletion in our system. Figure 6 shows two examples of good-quantity and -quality DBS (left) and poor-quantity and -quality DBS (right). Both clearly showed the presence of SMN1 exon 7. Pre-amplification guaranteed that a sufficient amount of target sequence of SMN genes required for the identification of SMN1 exon 7 deletion was generated. As a result, there were no samples with screening failure in the current study.
Figure 6.
Two examples of good-quantity and -quality DBS (left) and poor-quantity and -quality DBS (right).
3.3. Follow-Up Study of the Infants Screened for SMA
Extraction survey results were obtained for 2370 babies from 17 hospitals. No infants showed developmental delay in motor milestones indicative of infantile-onset SMA at 6 or 10 months after DBS collection.
4. Discussion
4.1. Targeted Pre-Amplification of SMN1/SMN2 Sequence
Our SMA screening system is an SMN1-deletion detection method that uses real-time mCOP-PCR technique following targeted pre-amplification of the sequence containing SMN1/SMN2 from DBS DNA, which does not require any non-SMN reference genes for validation of PCR quality [18]. The pre-amplification product contains either SMN1 or SMN2, or both. Here, SMN1 and SMN2 can be used as reference genes for each other, because all infants have at least one copy of SMN1 or SMN2 [9,10]. We used the pre-amplification product to confirm the presence of SMN2 in the samples with the SMN1(–) genotype [18].
In the previous version of our system, genomic DNA was extracted from DBS before targeted pre-amplification [18]. However, the new version used in the present study did not include the DNA extraction step [20]. Instead, a punched circle from each DBS was placed directly into the reaction tube of the conventional PCR (direct PCR).
4.2. Modified Competitive Oligonucleotide Priming-PCR (mCOP-PCR)
In the mCOP-PCR, almost identical DNA sequences with one nucleotide difference (SMN1 and SMN2 exon 7 sequences in the present study) compete for annealing of the gene-specific oligonucleotide primer (SMN1-COP), and the better-matched DNA sequence (SMN1 exon 7 sequence) is amplified much more efficiently.
The original COP-PCR is a kind of allele-specific PCR in which two oligonucleotide primers with one nucleotide difference compete for annealing of the target DNA sequence in one PCR tube [24,25]. The core part of the original COP-PCR and our mCOP-PCR is the same: the lengths of the oligonucleotide primer sequences used in PCR amplification are shorter than the usual PCR primers and identical except for a single nucleotide difference that is located in the middle of the primer [24]. Thus, we used the term “modified COP-PCR” (“mCOP-PCR”) to refer to our gene-specific amplification method.
4.3. Accurate Detection System for SMN1 Deletion
In this study, firstly, we confirmed that the new version of our screening system has 100% specificity and sensitivity for the detection of SMN1 deletion. The results were fully consistent with those of PCR–RFLP using DNA from freshly collected blood. Secondly, we successfully genotyped all DBS samples collected from 4157 newborn babies enrolled in the pilot study. There were no screening failures in the present study.
According to the real-time mCOP-PCR analysis in the validation study, the mean Cq values of SMA patients and controls were 19.0 ± 1.4 and 10.1 ± 1.2, respectively. This finding suggested that the amplification of the better-matched sequence (SMN1 exon 7 sequence) was >100-fold more efficient than the amplification of the mismatched sequence (SMN2 exon 7 sequence). Thus, we can claim that our SMA screening system using the real-time mCOP-PCR technique is an accurate method for the detection of SMN1 deletion.
4.4. Robust System for SMA Newborn Screening Using DBS
To date, three other pilot studies have demonstrated the feasibility of population-based screenings of newborn infants for SMA in the USA [16], Taiwan [26], and Germany [27], and another study is underway in Belgium [28].
Of a total of 3826 infants screened in the US study and 120,267 infants in the Taiwan study, the first-pass assay failure rates of false-positive or false-negative were 3.0% and 0.04%, respectively [16,26]. The high first-pass assay failure rate in the US study was attributed to suboptimal DNA quality and quantity [16]. In the German study, in which 213,279 infants were screened, there were no false-positive or false-negative results; however, the authors described some invalid results in initial assessments, due to the DBS collection process or human errors (incorrect pipetting or incomplete sealing of the PCR plate) [27].
Of the 4157 newborn infants screened for SMA in our pilot study, there were no screening failures. Among the DBS collected from the hospitals in the present study, not all showed sufficient quantity and/or dryness of blood (Figure 6). However, we were able to determine the SMN1 genotype of all newborns enrolled in the present study. The key to successful screening with our system may be targeted pre-amplification of SMN genes by direct PCR.
In our system, FTA® Elute Cards could be replaced by other filter papers used for the screening of inborn errors of metabolism, such as phenylketonuria (PKU). We also confirmed that standard DBS filter paper other than FTA® Elute Cards could work in our SMA screening system (data not shown).
4.5. Limitations of SMN1-Deletion Detection as an SMA Screening Strategy
Homozygous deletion of SMN1 has been found in more than 95% of SMA patients, and intragenic mutations of SMN1 have been found in the rest. Therefore, the primary purpose of SMA screening, at this moment, is to determine the presence or absence of SMN1 [18]. None of the SMA screening studies, including ours, has detected any intragenic mutations in SMN1. For the detection of SMA patients with intragenic mutations, the screening system cannot be “simple, rapid, and inexpensive”; instead, a complex, time-consuming, and expensive system with next-generation sequencing may be necessary.
Our real-time mCOP-PCR system is specific for detecting SMN1 deletions and does not provide any information about SMN2 copy number. If required for prognostic purposes, SMN2 copy number should be determined during a subsequent confirmatory assay or a second-tier assay [29,30]. However, SMN2 copy number does not always correspond to disease severity, and factors other than SMN2 copy number are also related to the severity of SMA [31].
5. Conclusions
Our SMN1-deletion detection system consists of a real-time mCOP-PCR technique following targeted pre-amplification of DNA from DBS, which rapidly and accurately detects SMN1 exon 7 deletion even from DNA samples of poor quantity and quality. The current pilot study clearly demonstrated that our system is a useful method for newborn SMA screening, facilitating the early diagnosis of asymptomatic infants and allowing treatment to be started before irreversible motor neuron damage occurs. Thus, our newly developed system is ready to be applied in high-throughput SMA newborn screening.
6. Patents
SMN1-deletion detection system: Japanese patent application No. 2017-196967, PCT/JP2018/37732.
Acknowledgments
We would like to thank the patients and newborn infants who participated in the study and their parents, guardians, and family members; Chiyo Hayashi, Narumi Daikai, and Hiroyuki Yoshida who managed samples and documents of the patients and newborn infants who participated in this study; Madoka Chinen who read drafts of this article and gave suggestions. We also would like to thank Joanne Dalton, on behalf of inScience Communications, Springer Healthcare, who wrote the first draft of this manuscript. This medical writing assistance was funded by Biogen Japan Ltd.
Appendix A
Centres and investigators of the SMA-NBS PILOT STUDY GROUP:
Japanese Red Cross Katsushika Maternity Hospital: Sakae Kumasaka, Chisako Mitsuishi;
Matsuyama Red Cross Hospital: Yoichi Kondo;
Chibune General Hospital: Akihiro Takatera;
St. Marianna University School of Medicine: Isamu Hokuto;
Kurashiki Medical Center: Tetsuro Fujino, Nobuyoshi Mimaki, Noriko Yanazaki;
National Hospital Organization Okayama Medical Center: Makoto Nakamura, Akihito Takeuchi;
Shimane Prefectural Central Hospital: Fumihide Kato;
Department of Community Medicine and Social Healthcare Science, Kobe University Graduate School of Medicine: Emma Tabe Eko Niba, Hisahide Nishio, Masakazu Shinohara, Izumi Takayama, Yogik Onky Silvana Wijaya;
Department of Pediatrics, Kobe University Graduate School of Medicine: Kazumichi Fujioka, Kazumoto Iijima;
Kurashiki Central Hospital: Akihito Takahashi;
Okayama University Hospital: Daisaku Morimoto, Yosuke Washio;
Japan Community Health Care Organization Kyushu Hospital: Yasuhiko Takahashi, Junko Yamamoto;
Fukuoka Children’s Hospital: Yasushi Takahata;
Nihon University School of Medicine: Ichiro Morioka, Nobuhiko Nagano;
Hyogo Prefectural Kobe Children’s Hospital Perinatal Center: Hideto Nakao, Tomoyuki Yokota, Seiji Yoshimoto;
Tosei General Hospital: Kuniko Ieda;
Hiroshima University Graduate School of Biomedical and Health Sciences: Norioki Ohno;
Hokkaido University Hospital: Kazutoshi Cho, Hideaki Shiraishi;
Toyohashi municipal hospital: Norihisa Koyama, Mari Sugimoto;
Saga University: Manabu Iwanaga, Muneaki Matsuo;
Jichi Medical School: Hitoshi Osaka, Hironori Shimozawa, Takanori Yamagata;
Almeida Memorial Hospital: Naoki Fukushima;
Toyooka Hospital: Masaaki Ueda;
Omihachiman Community Medical Center: Shinobu Yoshida;
Hyogo College of Medicine: Hideki Shimomura, Yasuhiro Takeshima;
Nagasaki University Hospital: Fumiko Kinoshita, Tatsuharu Sato;
Juntendo University Nerima Hospital: Shinichi Niijima, Noboru Yoshida;
Tokyo Metropolitan Ohtsuka Hospital: Ken Masunaga;
National Hospital Organization Nagasaki Medical Center: Mikihiro Aoki;
Kakogawa Central City Hospital: Takeshi Morisawa;
Niigata City General Hospital: Yoshihisa Nagayama;
Chiba Children’s Hospital: Kei Murayama, Tomoko Tsuruoka;
Yokosuka General Hospital Uwamachi: Tomoyuki Miyamoto;
Kouseiren Takaoka Hospital: Hiroaki Imamura, Jiro Ogawa;
Hokkaido Medical Center for Child Health and Rehabilitation: Hideomi Asanuma, Shuku Ishikawa;
Sapporo Medical University Hospital: Masaki Kobayashi;
Japanese Red Cross Nagoya Daini Hospital: Taihei Tanaka, Takaharu Yamada;
Akita University Graduate School of Medicine: Hiroyuki Adachi, Atsuko Noguchi, Tsutomu Takahashi;
Japanese Red Cross Saitama Hospital: Yuko Sato;
Graduate School of Medicine, University of the Ryukyus: Koichi Nakanishi, Tomohide Yoshida;
Numazu City Hospital: Masao Murabayashi;
Sapporo City General Hospital: Masato Mizushima, Tatsuo Satomi;
Maternal & Child Health Center Aiiku Hospital: Shinya Hayashida;
Hiroshima City Hiroshima Citizens Hospital: Yutaka Nishimura;
Juntendo University Shizuoka Hospital: Masato Kantake;
Yamaguchi University Hospital: Sasagu Kimura, Kazumasa Takahashi;
National Center for Child Health and Development: Go Tajima;
Kobe Pharmaceutical University: Atsuko Takeuchi
Author Contributions
Conceptualization, H.N. and A.T. (Atsuko Takeuchi); methodology, M.S., K.O., and K.T.; formal analysis, H.N. and M.S.; investigation, E.T.E.N., Y.O.S.W., and I.T.; resources, C.M., S.K., Y.K., A.T. (Akihiro Takatera), I.H., and I.M.; writing—original draft preparation, H.N. and M.S.; writing—review and editing, M.S., E.T.E.N., Y.O.S.W., I.T., C.M., S.K., Y.K., A.T. (Akihiro Takatera), I.H., I.M., K.O., K.T., A.T. (Atsuko Takeuchi), and H.N.
Funding
The present study and article processing charge were funded by Biogen Japan Ltd (Tokyo, Japan).
Conflicts of Interest
H.N. and M.S. have served on the scientific advisory board of Biogen Japan Ltd. and have performed consultancy for Biogen Japan Ltd.
References
- 1.Arnold W.D., Kassar D., Kissel J.T. Spinal muscular atrophy: Diagnosis and management in a new therapeutic era. Muscle Nerve. 2015;51:157–167. doi: 10.1002/mus.24497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verhaart I.E.C., Robertson A., Wilson I.J., Aartsma-Rus A., Cameron S., Jones C.C., Cook S.F., Lochmüller H. Prevalence, incidence and carrier frequency of 5q-linked spinal muscular atrophy—A literature review. Orphanet J. Rare Dis. 2017;12:124. doi: 10.1186/s13023-017-0671-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muralidharan K., Wilson R.B., Ogino S., Nagan N., Curtis C., Schrijver I. Population carrier screening for spinal muscular atrophy a position statement of the association for molecular pathology. J. Mol. Diagn. 2011;13:3–6. doi: 10.1016/j.jmoldx.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finkel R.S., McDermott M.P., Kaufmann P., Darras B.T., Chung W.K., Sproule D.M., Kang P.B., Foley A.R., Yang M.L., Martens W.B., et al. Observational study of spinal muscular atrophy type I and implications for clinical trials. Neurology. 2014;83:810–817. doi: 10.1212/WNL.0000000000000741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zerres K., Rudnik-Schöneborn S. Natural history in proximal spinal muscular atrophy. Clinical analysis of 445 patients and suggestions for a modification of existing classifications. Arch. Neurol. 1995;52:518–523. doi: 10.1001/archneur.1995.00540290108025. [DOI] [PubMed] [Google Scholar]
- 6.Lefebvre S., Bürglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 7.Singh R.N., Howell M.D., Ottesen E.W., Singh N.N. Diverse role of survival motor neuron protein. Biochim. Biophys. Acta Gene Regul. Mech. 2017;1860:299–315. doi: 10.1016/j.bbagrm.2016.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feldkötter M., Schwarzer V., Wirth R., Wienker T.F., Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: Fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am. J. Hum. Genet. 2002;70:358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schrank B., Götz R., Gunnersen J.M., Ure J.M., Toyka K.V., Smith A.G., Sendtner M. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc. Natl. Acad. Sci. USA. 1997;94:9920–9925. doi: 10.1073/pnas.94.18.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsieh-Li H.M., Chang J.G., Jong Y.J., Wu M.H., Wang N.M., Tsai C.H., Li H. A mouse model for spinal muscular atrophy. Nat. Genet. 2000;24:66–70. doi: 10.1038/71709. [DOI] [PubMed] [Google Scholar]
- 11.Finkel R.S., Mercuri E., Darras B.T., Connolly A.M., Kuntz N.L., Kirschner J., Chiriboga C.A., Saito K., Servais L., Tizzano E., et al. Nusinersen versus sham control in infantile-onset spinal muscular atrophy. N. Engl. J. Med. 2017;377:1723–1732. doi: 10.1056/NEJMoa1702752. [DOI] [PubMed] [Google Scholar]
- 12.Mercuri E., Darras B.T., Chiriboga C.A., Day J.W., Campbell C., Connolly A.M., Iannaccone S.T., Kirschner J., Kuntz N.L., Saito K., et al. Nusinersen versus sham control in later-onset spinal muscular atrophy. N. Engl. J. Med. 2018;378:625–635. doi: 10.1056/NEJMoa1710504. [DOI] [PubMed] [Google Scholar]
- 13.Finkel R.S., Chiriboga C.A., Vajsar J., Day J.W., Montes J., De Vivo D.C., Yamashita M., Rigo F., Hung G., Schneider E., et al. Treatment of infantile-onset spinal muscular atrophy with nusinersen: A phase 2, open-label, dose-escalation study. Lancet. 2016;388:3017–3026. doi: 10.1016/S0140-6736(16)31408-8. [DOI] [PubMed] [Google Scholar]
- 14.Swoboda K.J., Prior T.W., Scott C.B., McNaught T.P., Wride M.C., Reyna S.P., Bromberg M.B. Natural history of denervation in SMA: Relation to age, SMN2 copy number, and function. Ann. Neurol. 2005;57:704–712. doi: 10.1002/ana.20473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glascock J., Sampson J., Haidet-Phillips A., Connolly A., Darras B., Day J., Finkel R., Howell R.R., Klinger K., Kuntz N., et al. Treatment algorithm for infants diagnosed with spinal muscular atrophy through newborn screening. J. Neuromuscul Dis. 2018;5:145–158. doi: 10.3233/JND-180304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kraszewski J.N., Kay D.M., Stevens C.F., Koval C., Haser B., Ortiz V., Albertorio A., Cohen L.L., Jain R., Andrew S.P., et al. Pilot study of population-based newborn screening for spinal muscular atrophy in New York state. Genet. Med. 2018;20:608–613. doi: 10.1038/gim.2017.152. [DOI] [PubMed] [Google Scholar]
- 17.Kemper A.R., Lam K.K., Comeau A.M., Kwon J., Green N.S., Ojodu J., Grosse S., Prosser L.A., Jones E., Tanksley S., et al. Evidence-Based Review of Newborn Screening for Spinal Muscular Atrophy (SMA): Final Report (v5.2) [(accessed on 9 August 2019)]; Available online: https://www.hrsa.gov/sites/default/files/hrsa/advisory-committees/heritable-disorders/reports-recommendations/sma-final-report.pdf.
- 18.Ar Rochmah M., Harahap N.I.F., Niba E.T.E., Nakanishi K., Awano H., Morioka I., Iijima K., Saito T., Saito K., Lai P.S., et al. Genetic screening of spinal muscular atrophy using a real-time modified COP-PCR technique with dried blood-spot DNA. Brain Dev. 2017;39:774–782. doi: 10.1016/j.braindev.2017.04.015. [DOI] [PubMed] [Google Scholar]
- 19.Van der Steege G., Grootscholten P.M., van der Vlies P., Draaijers T.G., Osinga J., Cobben J.M., Scheffer H., Buys C.H. PCR-based DNA test to confirm clinical diagnosis of autosomal recessive spinal muscular atrophy. Lancet. 1995;345:985–986. doi: 10.1016/S0140-6736(95)90732-7. [DOI] [PubMed] [Google Scholar]
- 20.Takeuchi A., Tode C., Nishino M., Wijaya Y.O.S., Niba E.T.E., Awano H., Takeshima Y., Saito T., Saito K., Lai P.S., et al. Newborn screening for spinal muscular atrophy: DNA preparation from dried blood spot and DNA polymerase selection in PCR. Kobe J. Med. Sci. 2019;65:E95–E99. [PMC free article] [PubMed] [Google Scholar]
- 21.Kato N., Sa’Adah N., Ar Rochmah M., Harahap N.I., Nurputra D.K., Sato H., Sadewa A.H., Astuti I., Haryana S.M., Saito T., et al. SMA screening system using dried blood spots on filter paper: Application of COP-PCR to the SMN1 deletion test. Kobe J. Med. Sci. 2015;60:E78–E85. [PubMed] [Google Scholar]
- 22.Pezzullo J.C. 2-way Contingency Table Analysis (Japanese) [(accessed on 27 September 2019)]; Available online: http://www.grade-jpn.com/2x2.html.
- 23.Pezzullo J.C. 2-way Contingency Table Analysis (English) [(accessed on 27 September 2019)]; Available online: http://statpages.info/ctab2x2.html.
- 24.Gibbs R.A., Nguyen P.N., Caskey C.T. Detection of single DNA base differences by competitive oligonucleotide priming. Nucleic Acids Res. 1989;17:2437–2448. doi: 10.1093/nar/17.7.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yen T., Nightingale B.N., Burns J.C., Sullivan D.R., Stewart P.M. Butyrylcholinesterase (BCHE) genotyping for post-succinylcholine apnea in an Australian population. Clin. Chem. 2003;49:1297–1308. doi: 10.1373/49.8.1297. [DOI] [PubMed] [Google Scholar]
- 26.Chien Y.H., Chiang S.C., Weng W.C., Lee N.C., Lin C.J., Hsieh W.S., Lee W.T., Jong Y.J., Ko T.M., Hwu W.L. Presymptomatic diagnosis of spinal muscular atrophy through newborn screening. J. Pediatr. 2017;190:124–129. doi: 10.1016/j.jpeds.2017.06.042. [DOI] [PubMed] [Google Scholar]
- 27.Czibere L., Burggraf S., Fleige T., Glück B., Keitel L.M., Landt O., Durner J., Röschinger W., Hohenfellner K., Wirth B., et al. High-throughput genetic newborn screening for spinal muscular atrophy by rapid nucleic acid extraction from dried blood spots and 384-well qPCR. Eur. J. Hum. Genet. 2019 doi: 10.1038/s41431-019-0476-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boemer F., Caberg J.H., Dideberg V., Dardenne D., Bours V., Hiligsmann M., Dangouloff T., Servais L. Newborn screening for SMA in Southern Belgium. Neuromuscul. Disord. 2019;29:343–349. doi: 10.1016/j.nmd.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 29.Taylor J.L., Lee F.K., Yazdanpanah G.K., Staropoli J.F., Liu M., Carulli J.P., Sun C., Dobrowolski S.F., Hannon W.H., Vogt R.F. Newborn blood spot screening test using multiplexed real-time PCR to simultaneously screen for spinal muscular atrophy and severe combined immunodeficiency. Clin. Chem. 2015;61:412–419. doi: 10.1373/clinchem.2014.231019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strunk A., Abbes A., Stuitje A.R., Hettinga C., Sepers E.M., Snetselaar R., Schouten J., Asselman F.-L., Cuppen I., Lemmink H., et al. Validation of a fast, robust, inexpensive, two-tiered neonatal screening test algorithm on dried blood spots for spinal muscular atrophy. Int. J. Neonatal Screen. 2019;5:21. doi: 10.3390/ijns5020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harada Y., Sutomo R., Sadewa A.H., Akutsu T., Takeshima Y., Wada H., Matsuo M., Nishio H. Correlation between SMN2 copy number and clinical phenotype of spinal muscular atrophy: Three SMN2 copies fail to rescue some patients from the disease severity. J. Neurol. 2002;249:1211–1219. doi: 10.1007/s00415-002-0811-4. [DOI] [PubMed] [Google Scholar]