Abstract
Background
The therapeutic role of mesenchymal stem cells (MSCs) has been widely confirmed in several animal models of premature infant diseases. Micromolecule peptides have shown promise for the treatment of premature infant diseases. However, the potential role of peptides secreted from MSCs has not been studied. The purpose of this study is to help to broaden the knowledge of the hUC-MSC secretome at the peptide level through peptidomic profile analysis.
Methods
We used tandem mass tag (TMT) labeling technology followed by tandem mass spectrometry to compare the peptidomic profile of preterm and term umbilical cord MSC (hUC-MSC) conditioned medium (CM). Gene Ontology (GO) enrichment analysis and ingenuity pathway analysis (IPA) were conducted to explore the differentially expressed peptides by predicting the functions of their precursor proteins. To evaluate the effect of candidate peptides on human lung epithelial cells stimulated by hydrogen peroxide (H2O2), quantitative real-time PCR (qRT-PCR), western blot analysis, and enzyme-linked immunosorbent assay (ELISA) were, respectively, adopted to detect inflammatory cytokines (TNF-α, IL-1β, and IL-6) expression levels at the mRNA and protein levels.
Results
A total of 131 peptides derived from 106 precursor proteins were differentially expressed in the preterm hUC-MSC CM compared with the term group, comprising 37 upregulated peptides and 94 downregulated peptides. Bioinformatics analysis showed that these differentially expressed peptides may be associated with developmental disorders, inflammatory response, and organismal injury. We also found that peptides 7118TGAKIKLVGT7127 derived from MUC19 and 508AAAAGPANVH517 derived from SIX5 reduced the expression levels of TNF-α, IL-1β, and IL-6 in H2O2-treated human lung epithelial cells.
Conclusions
In summary, this study provides further secretomics information on hUC-MSCs and provides a series of peptides that might have antiinflammatory effects on pulmonary epithelial cells and contribute to the prevention and treatment of respiratory diseases in premature infants.
Keywords: hUC-MSCs, Peptidomics, LC-MS/MS, TMT labeling, Infant diseases
Introduction
The incidence of preterm birth has increased over the past 20 years in most countries [1, 2]. Despite recent advances in perinatal medicine, severe diseases related to premature birth, including periventricular leukomalacia (PVL), bronchopulmonary dysplasia (BPD), necrotizing enterocolitis (NEC), and retinopathy of prematurity (ROP), remain major causes of mortality and morbidity, which represent a heavy burden for families and society [3]. Therefore, it is an urgent and significant task to develop new safe and effective treatments to improve the prognosis of these diseases in premature infants.
In the past several decades, the development of mesenchymal stem cell (MSC) therapy and its continuous advancement have gained extensive attention. MSCs are multipotent progenitor cells, which can be raised from different tissues, for instance, adipose tissue, umbilical cord, and bone marrow [4, 5]. Human umbilical cord MSCs (hUC-MSCs) are easily accessible and can be harvested from donors without risks or damage [6]. Additionally, the therapeutic application of MSCs is not limited by the aging-like nature of adult tissues such as bone marrow and adipose tissue [7, 8]. Mechanically, MSCs function in vivo via direct differentiation or paracrine action. The therapeutic potential of MSC engraftment has been proved in premature infant diseases, and early clinical trials in preterm neonates with BPD (NCT01297205 [9], NCT01632475 [10]) and severe intraventricular hemorrhage (NCT02274428 [11]) have been conducted. A myriad of bioactive factors are readily available in the conditioned medium (CM) of MSCs and the medium can mediate multiple known functions of MSCs, such as angiogenesis, anti-fibrosis, and antiinflammatory effects [12]. Extracellular vesicles such as exosomes have been isolated from CM, and they have been shown to contain microRNAs and proteins, which partially mediated the effects of MSC [13–15]. Many studies have established that the secretome from MSCs can reduce organ damage in animal models of PVL, BPD, NEC, and ROP [16–20]. Other studies have also shown that soluble factors such as heme oxygenase-1 (HO-1) and erythropoietin (EPO) may be principally responsible for the ability of MSC-CM to ameliorate inflammation, angiogenesis, fibrosis, and so on [21, 22]. More types of MSC secreted factors and regulatory mechanisms still need to be established.
Peptides, a type of compound with two or more amino acids connected by peptide bonds, have been shown to play important roles in the treatment of diseases. Glucagon-like peptide-1 (GLP-1), a well-known peptide hormone secreted from the L cells of the duodenum, colon, terminal ileum, and rectal mucosa, has been used in the clinical treatment of type 2 diabetes [23]. Extrinsic calcitonin gene-related peptide (CGRP) could suppress apoptosis, oxidative stress, and ROS production in hyperoxia-induced alveolar epithelium type II (AECII) cells [24]. WKYMVm hexapeptide could attenuate hyperoxia-induced lung injuries in newborn mice [25]. Additionally, peptides from human milk such as PDC213, β-casein 197, and Casein201 exhibited obvious antimicrobial effects on the common pathogenic bacterial species S. aureus and Y. enterocolitica in neonatal intensive care units [26–28]. These studies indicated that peptides may hold great promise for the treatment of premature infant diseases. However, the secreted peptidomic profile from hUC-MSCs has not been fully characterized.
In the present study, we compared the secreted peptides from preterm and term hUC-MSCs using the tandem mass tag (TMT) labeling method with liquid chromatography (LC)-tandem mass spectrometry (MS/MS) analysis. Moreover, using ingenuity pathway analysis (IPA) software, we predicted that the differentially expressed peptides are associated with developmental disorder, inflammatory response, and organismal injury. And we preliminarily investigated the antiinflammatory effect of differentially expressed peptides on human lung epithelial cells. This study helps to broaden the knowledge of the hUC-MSC secretome at the peptide level and provided potential clues for the treatment of respiratory diseases in premature infants.
Materials and methods
Patients and samples
Umbilical cords were obtained from six infants without genetic or structural anomalies delivered at 27–41 weeks of gestation with parental written consent. Cases involving maternal diabetes, pre-eclampsia, eclampsia, intrauterine growth retardation (IUGR), or infectious diseases were excluded, because these factors may influence cell proliferation, cytokine expression, and other functions [29–33]. This study was approved by the Ethics Committee of Changzhou Maternal and Child Health Care Hospital (approval number 2019126) and conducted in accordance with the approved guidelines.
Preparation of hUC-MSCs and CM
Human umbilical cords were collected after preterm (n = 3) or full term (n = 3) deliveries hUC-MSCs were obtained by the tissue explants adherent method, as previously reported [17, 34]. Each umbilical cord (about 10 cm) was washed in phosphate-buffered saline (PBS; Gibco, Grand Island, CA, USA) and with 1% penicillin/streptomycin (P/S; Gibco) to remove residual blood from the vein and arteries. After the cord was cut longitudinally, and the arteries and vein were removed, Wharton’s jelly was finely dissected into small pieces. The pieces were individually placed on 100-mm2 tissue culture dishes with Dulbecco’s modified Eagle medium/nutrient mixture F-12 (DMEM/F-12; Gibco) containing 10% fetal bovine serum (FBS; Gibco), and 1% P/S (Gibco) and incubated for 10–12 days at 37 °C with 5% CO2. The medium was subsequently exchanged every 2–3 days. The cultures were passaged when they reached 80–90% confluency after carefully removing the umbilical cord tissues. hUC-MSCs at passage 3 were cultured to 80–90% confluence in a T75 culture flask (about 106 cells). The complete medium was replaced with serum-free DMEM/F-12 medium (5 ml) to avoid peptides contamination from FBS. The collected serum-free medium was centrifuged for 10 min at 300×g at 4 °C to remove cell debris, and protease inhibitor (Roche, Basel, Switzerland) was added. Lastly, the mixture was snap-frozen in liquid nitrogen and stored at − 80 °C until used.
Peptide extraction and purification
Before peptide extraction, the protein integrity of the CM samples was appraised. The collected CM samples were concentrated by centrifugation under vacuum (LaboGene, Allerød, Denmark), boiled in sodium dodecyl sulfate (SDS)-sample buffer at 95 °C for 10 min and then subjected to 12% SDS-polyacrylamide gel electrophoresis (PAGE). The SDS-PAGE gel was stained using a Pierce Silver Stain Kit (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s protocol. Thereafter, the samples were filtered through an ultrafiltration tube (Amicon Ultra-15, Millipore, MA, USA) with a molecular weight cutoff (MWCO) of 10 kDa to acquire the filtered liquid containing the peptides. The protein concentration of the supernatant was also measured using the Bradford protein assay [35].
TMT labeling and LC-MS/MS analysis
The peptides from preterm and term hUC-MSC CM were reduced with 10 mM DL-dithiothreitol (DTT; Promega, WI, USA) for 1 h at 56 °C and alkylated with 55 mM iodoacetamide (Promega) for 1 h in the dark at RT. Thereafter, precooled acetone was added, and the peptides were precipitated over 3 h at − 20 °C. After centrifuging for 20 min at 20,000×g and 4 °C, the precipitate was dissolved in 300 μl of the following buffer: 50% triethylamine borane (Sigma) and 0.1% SDS (Sigma). Next, the peptide solution was desalted using a Strata-X C18 column (Phenomenex, Torrance, CA, USA) and dried and labeled with TMT reagent (TMT 6-plex Label Reagent; Thermo Fisher Scientific) for 1 h [36]. Next, the preterm and term samples were mixed at a 1:1 ratio on the basis of the total peptide amount. Analysis of labeled peptides was performed on a Q Exactive Orbitrap LC-MS/MS system (Thermo Fisher Scientific). Qualitative and relative quantitative analyses of the detected peptides were performed using the SWISSPROT_human database and Mascot software (version 2.3.01). Peptides with absolute fold change ≥ 1.5 and P value < 0.05 were considered differentially expressed.
Bioinformatics analysis
The molecular weight (MW) and isoelectric point (PI) of the identified peptides were calculated using the online tool PI/MW (http://web.expasy.org/compute.pi/). A Gene Ontology (GO) analysis (http://www.blast2go.com/b2ghome) was carried out to explore the possible cellular components, biological processes, and molecular functions related to the precursor proteins. Diseases and regulator effects networks analysis of the differentially expressed peptides and their precursors were performed using ingenuity pathway analysis (IPA) software v7.1 (Ingenuity Systems, Mountain View, CA, USA) [37]. The UniProt database (http://www.uniprot.org/) was used to detect the predominant subcellular locations of the precursors of the differentially expressed peptides. The Open Targets Platform database (http://www.targetvalidation.org/) was applied to study the diseases associated with the protein precursors [38].
Synthetic peptides
All the peptides used in this study were synthesized by GenScript (Nanjing, Jiangsu, China) through the solid-phase method. The purity of each peptide was 95% detected by HPLC-MS method. All the used peptides were preserved in freeze-drying at − 20 °C until immediately dissolved in aseptic water for cell treatment in vitro.
Cell culture
Two peptides TGAKIKLVGT and AAAAGPANVH were selected with high fold change, and we investigated the effects on human lung epithelial cells A549 (Meiyan, Shanghai, China) stimulated by hydrogen peroxide (H2O2; Kelong, Chengdu, Sichuan, China). A549 cell is a human lung carcinoma cell line from an adult with similar characteristics of human alveolar basal epithelial cells, and was often used for researches in BPD. A549 cells were cultured in Dulbecco’s modified Eagle medium (Gibco) with 10% FBS and 1% P/S. The A549 cells were exposed to 1 mM H2O2 with or without peptides (1 μM, 10 μM, and 100 μM) for 24 h in serum-free DMEM with 1% P/S. The sample size was 3 biological independent samples per group.
Quantitative real-time polymerase chain reaction (QRT-PCR)
Total RNA was extracted with Trizol reagent (Invitrogen; Thermo Fisher Scientific Inc., Shanghai, China) according to the manufacturer’s instructions. The RNA concentration was detected by ultraviolet spectrophotometer and the appropriate OD value at 260 to 280 nm was 1.8 to 2.0. Then, qRT-PCR was carried out to measure the gene expression levels of inflammatory factors: tumor necrosis factor-α (TNF-α), interleukin-1β (IL-1β), and interleukin-6 (IL-6) mRNAs in A549 cells using SYBR Green qPCR method (Thermo Fisher Scientific, Waltham, MA). Primer sequences were the following: human TNF-α: forward, 5′ CCTCTCTCTAATCAGCCCTCTG 3′, reverse, 5′ GAGGACCTGGGAGTAGATGAG 3′; human IL-1β: forward, 5′ AGCTACGAATCTCCGACCAC 3′, reverse, 5′ CGTTATCCCATGTGTCGAAGAA 3′; human IL-6: forward, 5′ ACTCACCTCTTCAGAACGAATTG 3′, reverse, 5′ CCATCTTTGGAAGGTTCAGGTTG 3′; and human β-actin: forward, 5′ AGCGAGCATCCCCCAAAGTT 3′, reverse, 5′ GGGCACGAAGGCTCATCATT 3′. To calculate fold change in the expression of these genes, ΔCt = Ct of individual genes − Ct of β-actin was first obtained. ΔΔCt = ΔCt of treated groups − ΔCt of control groups was then obtained. Fold change was calculated as 2−ΔΔCt, with control groups as 1.0-fold.
Western blot analysis
Lysates from cultured cells were similarly prepared using a cell scraper. Homogenates were clarified by centrifugation (10,000×g, 4 °C, 10 min). Protein concentration was quantified with a BCA protein assay (Beyotime, Shanghai, China). Cell extracts resolved on a 10% or 12% reducing SDS-PAGE gel were transferred to a nitrocellulose membrane according to molecular weight. Blots were probed with the following antibodies: rabbit anti-TNF-α (1:2000; Proteintech, Wuhan, China), anti-IL-1β (1:1000; Proteintech, Wuhan, China), and mouse anti-IL-6 (1:5000; Proteintech, Wuhan, China), whereas mouse GAPDH (1:5000; Proteintech, Wuhan, China) served as a loading control.
Enzyme-linked immunosorbent assay (ELISA)
ELISA was also performed to detect the expression levels of inflammatory factors in A549 cells by ELISA kits for human TNF-α, IL-1β, and IL-6 (4A Biotech, Beijing, China), according to the manufacturer’s instructions. Optical density was measured at 450 nm using an ELISA microplate reader. No significant cross-reactivity or interference was observed.
Statistical analysis
Student’s t test or one-way ANOVA was employed for statistical comparisons. The results of the bioinformatics analysis were visualized using GraphPad Prism 5/7 software. The statistical significances were calculated as P values, and P < 0.05 was considered statistically significant.
Results
Isolation and characterization of MSCs derived from human umbilical cords
The range of gestational ages (GA) was 31–32 weeks of gestation for the preterm umbilical cords and 40–41 weeks for the term cords (Table 1). The hUC-MSCs (obtained by the tissue explants adherent method) had a typical fibroblast phenotype (Fig. S1A). Both preterm and term hUC-MSCs were positive for CD29, CD73, and CD105 staining and negative for CD31, CD34, and HLA-DR staining (Fig. S1B). Furthermore, these isolated cells had the potential to differentiate into adipocytes and chondrocytes (Fig. S1A). These results confirmed that MSCs from human umbilical cords were successfully isolated, without significant differences in morphology, expression of cell surface markers or differentiation capacities between the preterm and term groups.
Table 1.
Cohort clinical date of hUC-MSC samples
| Sample | Gestational age (weeks) | Birth weight (g) | Sex | Apgar score 1 min | Apgar score 5 min | Maternal age (years) | Gravidity | Parity | Perinatal history |
|---|---|---|---|---|---|---|---|---|---|
| P 1 | 32 + 5 | 1630 | Female | 10 | 10 | 28 | 2 | 1 | Cesarean section due to placental abruption |
| P 2 | 32 + 5 | 1850 | Male | 10 | 10 | 29 | 2 | 1 | Vaginal delivery due to premature rupture of fetal membranes |
| P 3 | 31 | 1600 | Female | 8 | 8 | 33 | 5 | 3 | Cesarean section due to active premature labor |
| T 1 | 40 + 3 | 4670 | Male | 10 | 10 | 23 | 1 | 1 | Cesarean section due to primary uterine atony |
| T 2 | 41 + 1 | 3360 | Female | 10 | 10 | 29 | 2 | 2 | Repeated cesarean section |
| T 3 | 40 + 6 | 4250 | Female | 10 | 10 | 27 | 2 | 1 | Cesarean section due to non-reassuring fetal status |
Identification of differentially expressed peptides in hUC-MSC CM from preterm and term infants
We verified the protein integrity of the hUC-MSC CM by silver staining (Fig. S2). The peptides from preterm and term hUC-MSC CM were directly analyzed by the TMT labeling method combined with LC-MS/MS. We identified a total of 3099 peptides in hUC-MSC CM from both groups. A total of 131 peptides were observed to be significantly differentially expressed (absolute fold change ≥ 1.5, P value < 0.05) in the hUC-MSC CM from the preterm group compared with the term group, comprising 37 upregulated peptides (Fig. 1a) and 94 downregulated peptides (Fig. 1b). The top 20 upregulated and top 20 downregulated peptides are shown with their precursor proteins in Fig. 1c and d. All the differentially expressed peptides are shown in Table 2.
Fig. 1.
Differentially expressed peptides in hUC-MSC conditioned medium (CM) from preterm infants compared with term infants. a, b Upregulated and downregulated peptides visualized using heatmaps (n = 3 per group, P1–3 represent preterm infants and T1–3 represent term infants). c, d Top 20 upregulated and top 20 downregulated peptides visualized using heatmaps
Table 2.
Differentially expressed peptides in hUC-MSC conditioned medium from preterm and term infants
| Accession | Gene | Protein | Peptide | MW (kD) | Fold change | − 10lgP |
|---|---|---|---|---|---|---|
| Upregulated peptides | ||||||
| Q6PEZ8-3 | PODNL1 | Podocan-like protein 1 | PSLERLHLQNNLISKVPR | 5.94 | ∞ | 6.96 |
| A0A0A0MTS7 | TTN | Titin | ESDSG | 2.92 | 13.62 | 9.66 |
| Q9BYJ1-2 | ALOXE3 | Hydroperoxide isomerase ALOXE3 | LNGRQQY | 0.54 | 7.95 | 10.08 |
| Q15772-1 | SPEG | Striated muscle preferentially expressed protein kinase | SCTVAVARVPGKLAPPEVPQ | 0.76 | 5.96 | 10.05 |
| Q8N196 | SIX5 | Homeobox protein SIX5 | AAAAGPANVH | 1.00 | 5.93 | 11.82 |
| P49815-3 | TSC2 | Tuberin | PAGPAVRL | 0.57 | 4.52 | 10.67 |
| A0A0A6YYA3 | CDHR1 | Cadherin-related family member 1 | RVLRKRPSPAPRTIRIE | 0.69 | 3.82 | 9.34 |
| Q7Z5P9-2 | MUC19 | Mucin-19 | DDFMSSQN | 2.05 | 3.46 | 5.01 |
| P54259 | ATN1 | Atrophin-1 | GPARPYHP | 0.67 | 3.28 | 11.27 |
| A0A140TA73 | SNTB2 | Beta-2-syntrophin | NGLPNGGGAGDS | 0.88 | 2.98 | 7.03 |
| Q9UQD0-2 | SCN8A | Sodium channel protein type 8 subunit alpha | EAGID | 1.81 | 2.67 | 16.18 |
| Q5TZA2-2 | CROCC | Rootletin | RLLKGEASLEV | 0.68 | 2.67 | 9.78 |
| E7EMZ9 | TACC2 | Transforming acidic coiled-coil-containing protein 2 | RMSESPTPC | 1.29 | 2.57 | 8.92 |
| A0A075B756 | KLF14 | Krueppel-like factor 14 | TKHARRHP | 0.94 | 2.52 | 17.50 |
| A0A0A0MTS7 | TTN | Titin | LEDGG | 3.06 | 2.50 | 12.31 |
| O15027-2 | SEC16A | Protein transport protein Sec16A | KSILTQ | 2.31 | 2.49 | 13.78 |
| Q9NRA0-3 | SPHK2 | Sphingosine kinase 2 | EWDGIVTVSGDGLLHEVLN | 0.56 | 2.46 | 10.14 |
| Q8WXG9 | ADGRV1 | Adhesion G-protein coupled receptor V1 | EAGLD | 2.02 | 2.23 | 19.57 |
| Q9Y4D8 | HECTD4 | Probable E3 ubiquitin-protein ligase HECTD4 | KLAKLQRIARQAVAALCALGG | 1.02 | 2.21 | 6.58 |
| A0A087WVF8 | PDE4DIP | Myomegalin | QSMMAV | 1.01 | 2.18 | 21.26 |
| Q01167 | FOXK2 | Forkhead box protein K2 | QTVHVVH | 0.67 | 2.18 | 12.99 |
| Q5IJ48 | CRB2 | Protein crumbs homolog 2 | LLEVAVPAACACLLLLLLGLLSGILAARK | 0.78 | 2.18 | 5.08 |
| Q9ULE3 | DENND2A | DENN domain-containing protein 2A | FLHKK | 1.70 | 2.17 | 21.46 |
| P53420 | COL4A4 | Collagen alpha-4(IV) chain | PGEPGLVGPPGQPGRPG | 0.84 | 2.14 | 7.51 |
| Q7Z5P9-2 | MUC19 | Mucin-19 | FLGGS | 2.11 | 2.11 | 13.79 |
| P01833 | PIGR | Polymeric immunoglobulin receptor | QADGSRASVD | 0.49 | 2.10 | 5.11 |
| Q09666 | AHNAK | Neuroblast differentiation-associated protein AHNAK | KLKGDI | 1.57 | 2.08 | 15.35 |
| P36776 | LONP1 | Lon protease homolog, mitochondrial | KHKPR | 0.99 | 2.07 | 8.69 |
| A0A0A0MTS7 | TTN | Titin | VPEAPKEVVPEKKVPVTPPKK | 2.94 | 1.98 | 10.42 |
| Q8NEZ4 | KMT2C | Histone-lysine N-methyltransferase 2C | QQNNLSNP | 1.03 | 1.96 | 10.44 |
| A0A0A0MTS7 | TTN | Titin | SPPSP | 2.94 | 1.93 | 11.70 |
| Q7Z5P9-2 | MUC19 | Mucin-19 | AGTSI | 2.24 | 1.89 | 23.15 |
| O75592-2 | MYCBP2 | E3 ubiquitin-protein ligase MYCBP2 | QLLYR | 2.05 | 1.79 | 11.10 |
| Q9H6K5-2 | PRR36 | Proline-rich protein 36 | PPSLQTLPSPPATPPSQVPPTQ | 0.81 | 1.78 | 9.94 |
| P08913 | ADRA2A | Alpha-2A adrenergic receptor | ISAVISFPPLISIEKKGGGG | 0.93 | 1.71 | 9.59 |
| Q9UJ55 | MAGEL2 | MAGE-like protein 2 | PPPIRPGP | 1.10 | 1.65 | 15.90 |
| Q4V328-4 | GRIPAP1 | GRIP1-associated protein 1 | LCSQMEQLE | 0.63 | 1.60 | 6.91 |
| Downregulated peptides | ||||||
| O14526-3 | FCHO1 | F-BAR domain only protein 1 | AGIVRVF | 0.53 | −1.56 | 9.90 |
| Q7Z5P9-2 | MUC19 | Mucin-19 | KTLAAGS | 2.15 | −1.57 | 14.06 |
| Q99814 | EPAS1 | Endothelial PAS domain-containing protein 1 | TPLSSMGGRS | 1.00 | −1.58 | 18.34 |
| Q6ZNL6 | FGD5 | FYVE, RhoGEF and PH domain-containing protein 5 | EDHAQ | 0.77 | −1.58 | 13.24 |
| Q9BW04-2 | SARG | Specifically androgen-regulated gene protein | LTTPKPRKLPPN | 0.61 | −1.60 | 5.40 |
| O15027-2 | SEC16A | Protein transport protein Sec16A | QACAASGS | 2.41 | −1.61 | 17.59 |
| S4R393 | ZSWIM8 | Zinc finger SWIM domain-containing protein 8 | QTHKPQT | 0.99 | −1.64 | 13.03 |
| Q2VWA4 | SKOR2 | SKI family transcriptional corepressor 2 | GGSGGDCSAG | 0.50 | −1.65 | 8.13 |
| Q9Y6V0-6 | PCLO | Protein piccolo | QQPGPAKPPP | 1.00 | −1.69 | 6.21 |
| P28329 | CHAT | Choline O-acetyltransferase | GLPKLPVPPLQQ | 0.66 | −1.70 | 5.12 |
| Q8WXH0 | SYNE2 | Nesprin-2 | KIYKKFLKKAQDLTSLLKEL | 2.04 | −1.71 | 5.55 |
| P13611-5 | VCAN | Versican core protein | QPEFSS | 1.97 | −1.74 | 29.72 |
| Q8TE85 | GRHL3 | Grainyhead-like protein 3 homolog | LFIPNVHFSSLQRSG | 0.54 | −1.78 | 9.85 |
| A0A1B0GUF7 | IQCM | IQ domain-containing protein M | KTFKT | 0.88 | −1.79 | 14.67 |
| A0A087WXW9 | COL5A1 | Collagen alpha-1(V) chain | PPGEV | 2.70 | −1.83 | 8.75 |
| A0A0A0MTS7 | TTN | Titin | KACDPVF | 2.92 | −1.87 | 8.46 |
| A0A0A0MTS7 | TTN | Titin | IVASDVTKRLIKANLLANN | 2.78 | −1.87 | 5.62 |
| H0Y5I7 | SFI1 | Protein SFI1 homolog | QQLAARRQEQRATVRALW | 0.82 | −1.88 | 6.99 |
| A6NMZ7 | COL6A6 | Collagen alpha-6(VI) chain | RRAIN | 0.91 | −1.89 | 13.43 |
| Q8NEZ4 | KMT2C | Histone-lysine N-methyltransferase 2C | EGCVK | 1.08 | −1.89 | 12.10 |
| A0A087WXW9 | COL5A1 | Collagen alpha-1(V) chain | GPRGITGKPGPK | 2.70 | −1.90 | 10.79 |
| Q9Y6W6 | DUSP10 | Dual specificity protein phosphatase 10 | DNQAQT | 1.21 | −1.91 | 9.85 |
| A0A0J9YXV3 | N/A | Uncharacterized protein | KIGLGY | 0.94 | −1.91 | 14.05 |
| A0A0A0MTS7 | TTN | Titin | EGNKDD | 3.08 | −1.91 | 12.34 |
| Q92616 | GCN1 | eIF-2-alpha kinase activator GCN1 | ILDVASLEVLN | 0.66 | −1.92 | 5.74 |
| Q9NR09 | BIRC6 | Baculoviral IAP repeat-containing protein 6 | DNESCTN | 1.47 | −1.95 | 6.90 |
| Q8TEP8 | CEP192 | Centrosomal protein of 192 kDa | LLSTTK | 1.70 | −1.96 | 17.83 |
| A0A0A0MTS7 | TTN | Titin | DPPGKPVPLN | 3.22 | −2.08 | 14.35 |
| Q8TE73 | DNAH5 | Dynein heavy chain 5, axonemal | QRVKSKIPAAIEQLIVPHLAKVDEALQPGLAAL | 1.84 | −2.10 | 7.15 |
| P10827-4 | THRA | Thyroid hormone receptor alpha | LHARAV | 0.51 | −2.11 | 9.87 |
| Q9P2D3 | HEATR5B | HEAT repeat-containing protein 5B | HAKGK | 0.83 | −2.12 | 12.50 |
| O60423-3 | ATP8B3 | Phospholipid-transporting ATPase IK | YGLVI | 0.98 | − 2.13 | 11.77 |
| P06401-2 | PGR | Progesterone receptor | GPLLKGKPRALGGAAAGGG | 0.77 | − 2.14 | 5.47 |
| A0A140T8Y3 | TNXB | Tenascin-X | HGRGRCEEGRCLCDPGYTGPTCATRMCPADCRGRGRCVQGVCLCHVGYGGEDCGQ | 1.71 | −2.14 | 5.92 |
| P27658 | COL8A1 | Collagen alpha-1(VIII) chain | GIDGVKPPHAYGAKKGKN | 0.65 | −2.14 | 6.82 |
| Q9C093 | SPEF2 | Sperm flagellar protein 2 | ESLCEKVKEILTTEIAKKKN | 0.69 | −2.14 | 7.26 |
| Q8NAC3-3 | IL17RC | Interleukin-17 receptor C | AAALSLILLLKKDHAKGWLRLLKQ | 0.48 | −2.15 | 5.00 |
| Q92771 | DDX12P | Putative ATP-dependent RNA helicase DDX12 | KGGLLGRLAARKKIFQEPK | 0.67 | −2.16 | 6.09 |
| Q6PJG9 | LRFN4 | Leucine-rich repeat and fibronectin type-III domain-containing protein 4 | VAVGGVLVAALLVFTVALLVRGRGAGNGRL | 0.69 | −2.17 | 5.32 |
| Q9HD67 | MYO10 | Unconventional myosin-X | KTSCVE | 0.82 | −2.23 | 14.68 |
| Q96DN2 | VWCE | von Willebrand factor C and EGF domain-containing protein | RPVLHLLQLLLRTNLMKTQTL | 0.50 | −2.26 | 9.34 |
| Q96QD8 | SLC38A2 | Sodium-coupled neutral amino acid transporter 2 | VFNLSNAIVGSGILGLS | 0.60 | −2.31 | 12.68 |
| A0A0C4DGG6 | NPC1L1 | NPC1-like intracellular cholesterol transporter 1 | VFAVVTILLVGFRVAPARDKSKMVDPKK | 0.68 | −2.32 | 12.53 |
| Q9UQD0-2 | SCN8A | Sodium channel protein type 8 subunit alpha | VSLVSLIAN | 1.71 | −2.33 | 12.11 |
| Q9UPA5 | BSN | Protein bassoon | KGGPRPR | 2.05 | −2.34 | 10.76 |
| Q9UKV8 | AGO2 | Protein argonaute-2 | KLQAN | 1.17 | −2.36 | 13.86 |
| Q9UQD0-2 | SCN8A | Sodium channel protein type 8 subunit alpha | YLALL | 1.72 | −2.36 | 14.90 |
| I6L894 | ANK2 | Ankyrin-2 | KHKLNVP | 0.89 | −2.38 | 11.67 |
| O95996 | APC2 | Adenomatous polyposis coli protein 2 | PAAEAATKKPLPPLRH | 0.78 | −2.39 | 5.35 |
| P36776 | LONP1 | Lon protease homolog, mitochondrial | TIAAKRAGVT | 0.99 | −2.46 | 12.42 |
| H7BXZ5 | KALRN | Kalirin | VKKCIHKATRKDVAVKFVSKKMKKKEQA | 0.91 | −2.46 | 9.58 |
| Q8NF91-4 | SYNE1 | Nesprin-1 | QKAVDHRKAIILSIN | 1.02 | −2.56 | 8.45 |
| J3KQC6 | TMPRSS13 | Transmembrane protease serine 13 | LPLIGCVLLLIALVVSLIILFQFW | 0.45 | −2.60 | 5.00 |
| Q8TE73 | DNAH5 | Dynein heavy chain 5, axonemal | AQTKRLVGDVLLATAFLSYSGP | 1.82 | −2.62 | 5.48 |
| I3L2R4 | SLC2A4 | Solute carrier family 2 (Facilitated glucose transporter), member 4, isoform CRA_b | IGAGVVNTVFTLVSVLLVERAGRRTLHLLGLA | 0.62 | −2.63 | 7.43 |
| P42167 | TMPO | Lamina-associated polypeptide 2, isoforms beta/gamma | KSEKTKKGRSIPVWIKILLFVVVAV | 0.49 | −2.66 | 6.27 |
| Q9BW11-4 | MXD3 | Max dimerization protein 3 | GPIHRRK | 0.50 | −2.66 | 7.43 |
| E7EPG1 | MMRN1 | Multimerin-1 | LPDIQLLQKGLTEFV | 0.66 | −2.70 | 6.46 |
| G5EA42 | TMOD2 | Tropomodulin 2 (Neuronal), isoform CRA_a | HVKKF | 0.67 | −2.76 | 12.93 |
| A0A0A0MTS7 | TTN | Titin | SSRLECKI | 2.84 | −2.77 | 7.13 |
| Q7Z5P9-2 | MUC19 | Mucin-19 | QNGIIVI | 2.18 | −2.78 | 18.23 |
| Q5H8A4 | PIGG | GPI ethanolamine phosphate transferase 2 | WLAAGGVMVLASALLCVIVSVLTNVLVGGN | 1.18 | −2.80 | 9.72 |
| Q9NR09 | BIRC6 | Baculoviral IAP repeat-containing protein 6 | TRIGLKLIDILLRNCAAS | 1.36 | −2.82 | 5.14 |
| A0A0A0MTS7 | TTN | Titin | VVHAGGVIRIIAYV | 2.83 | −2.89 | 5.92 |
| Q8NG04 | SLC26A10 | Solute carrier family 26 member 10 | EPVVKALTSGAALHVLLSQLPSLLGLSL | 0.59 | −2.91 | 11.40 |
| J3KNF3 | TET3 | Methylcytosine dioxygenase TET3 | GPEGCSA | 1.65 | −2.94 | 15.44 |
| Q92835-2 | INPP5D | Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | PLPVKSPA | 1.01 | −2.97 | 22.11 |
| Q9Y6R1 | SLC4A4 | Electrogenic sodium bicarbonate cotransporter 1 | HHTIYIGVHVPKSYR | 0.78 | −2.99 | 8.24 |
| Q99707 | MTR | Methionine synthase | KSARVMKKAVG | 1.66 | −3.12 | 21.33 |
| Q8WXG9 | ADGRV1 | Adhesion G-protein coupled receptor V1 | RFLQSIYLVPEEDHILIIPVVRGKDN | 1.97 | −3.13 | 8.73 |
| P43243-2 | MATR3 | Matrin-3 | HLILN | 0.67 | −3.21 | 8.71 |
| O75970-3 | MPDZ | Multiple PDZ domain protein | FISLLKT | 0.58 | −3.22 | 11.08 |
| Q96N23-2 | CFAP54 | Cilia- and flagella-associated protein 54 | HLKKPKIKISGSPLTLKPPLRRSSSVKET | 0.62 | −3.25 | 6.96 |
| F8WBW8 | BAHCC1 | BAH and coiled-coil domain-containing protein 1 | RSVKAKVGTTL | 1.16 | −3.27 | 15.77 |
| F8VZY0 | MYBPC1 | Myosin-binding protein C, slow-type | TDAKIFVRVKAVNAAGAS | 0.76 | −3.39 | 9.25 |
| G0XQ39 | STIM1 | STIM1L | GVHPGSLVEKLPDSPALAKKALLALNHGL | 0.59 | −3.56 | 12.45 |
| P0CJ78 | ZNF865 | Zinc finger protein 865 | MEANPAGSGAGGGGSSGIGGEDGVHFQSYPFDFLEFLNHQRFEPMELYGEHAKAVAA | 0.86 | −3.84 | 11.26 |
| Q7LBC6-2 | KDM3B | Lysine-specific demethylase 3B | VKSKASLPN | 0.69 | −3.87 | 7.94 |
| Q9UI33-3 | SCN11A | Sodium channel protein type 11 subunit alpha | IGAIPAILNV | 0.83 | −4.13 | 15.16 |
| Q5XPI4 | RNF123 | E3 ubiquitin-protein ligase RNF123 | HYLRLTIAI | 0.67 | −4.16 | 8.06 |
| O15027-2 | SEC16A | Protein transport protein Sec16A | AGSLCQALLPGPSNEAAGDVWGDTASTGVPDASGSQYE | 2.30 | −4.48 | 5.51 |
| B7ZLJ5 | MPIG6B | C6orf25 protein | YPQLLIPLLGAGLVLGLGALG | 0.48 | −4.66 | 5.03 |
| Q86WI1 | PKHD1L1 | Fibrocystin-L | LFVGR | 1.16 | −4.97 | 17.10 |
| Q7Z3U7 | MON2 | Protein MON2 homolog | KPPQYGQLETKHIAN | 1.01 | −5.28 | 21.73 |
| F1T0I1 | SEC16A | Protein transport protein sec16 | RRRAN | 2.30 | −5.79 | 11.96 |
| A0A087WXW9 | COL5A1 | Collagen alpha-1(V) chain | HPGLI | 2.50 | −5.82 | 5.85 |
| Q9H3S7 | PTPN23 | Tyrosine-protein phosphatase non-receptor type 23 | KLELLRQN | 0.65 | −6.11 | 5.79 |
| Q7Z5P9-2 | MUC19 | Mucin-19 | TGAKIKLVGT | 2.11 | −6.37 | 6.06 |
| P52746 | ZNF142 | Zinc finger protein 142 | TGLKP | 4.94 | -∞ | 13.46 |
| P43088-7 | PTGFR | Prostaglandin F2-alpha receptor | QKSKASFLL | 3.34 | -∞ | 9.46 |
| A0A1C7CYW7 | TTC34 | Tetratricopeptide repeat protein 34 | TGGQRLLAAL | 3.68 | -∞ | 12.39 |
| Q9Y219 | JAG2 | Protein jagged-2 | CGSDAGPGMPGTAASGVCGPHGRCVSQPGGN | 3.52 | -∞ | 10.83 |
| E7ERG8 | LRP1B | Low-density lipoprotein receptor-related protein 1B | QVDQFSCGNGRCIPRAWLCDREDDCGDQTDEMASCEFPTCEPLT | 3.26 | -∞ | 5.58 |
| O75129-2 | ASTN2 | Astrotactin-2 | TCHLC | 5.80 | -∞ | 12.93 |
Basic characteristics of the differentially expressed peptides in hUC-MSC CM from preterm and term infants
The MW and PI of the differentially expressed peptides were analyzed. The MW of most peptides ranged from 500 to 700 Da (Fig. 2a), and the PI ranged from 3 to 11 (Fig. 2b). We also investigated the distribution of the MW relative to the PI (Fig. 2c). Peptides are cleaved from their precursor proteins by specific enzymes [37], so we analyzed the cleavage sites at the N- and C-terminals of the identified peptides. Lysine (K) was the most common N-terminal amino acid (accounting for 13.7% of the peptides), while asparagine (N) was the most common C-terminal amino acid (accounting for 16.0% of the peptides) (Fig. 2d).
Fig. 2.
Basic features of the differentially expressed peptides in hUC-MSC CM from preterm and term infants. a, b Molecular weights (MW) and isoelectric points (PI) of the differentially expressed peptides. c Scatter plot of MW versus PI of the differentially expressed peptides. d Distributions of the N- and C-terminals of the differentially expressed peptides
GO analysis and subcellular location analysis of the differentially expressed peptide precursors
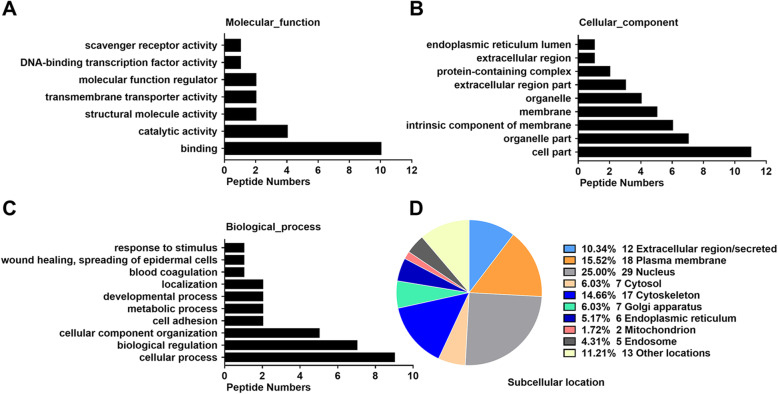
Next, molecular functions, cellular components and biological processes of the corresponding precursor proteins were determined by GO analysis to predict the latent functions of the differentially expressed peptides. Binding and catalytic activity were the most highly enriched molecular functions (Fig. 3a). Cell part, organelle part, and intrinsic component of membrane were the most highly enriched cellular components (Fig. 3b). Cellular process, biological regulation, and cellular component organization were the most highly enriched biological processes (Fig. 3c). Furthermore, we categorized the subcellular locations of the precursor proteins of all 131 peptides in accordance with their annotations in the UniProt database. The analysis revealed that the nucleus (25%), plasma membrane (16%), and cytoskeleton (15%) were the predominant subcellular locations of the differentially expressed peptide precursors. About 10% of the precursors were types of proteins that are located in the extracellular region of hUC-MSCs (Fig. 3d).
Fig. 3.
Gene Ontology (GO) and subcellular location analysis of the differentially expressed peptide precursors. a Molecular functions. b Cellular components. c Biological processes. d Subcellular locations
Diseases and regulator effects networks associated with the differentially expressed peptide precursors
We further evaluated the diseases and regulator effects networks associated with the differentially expressed peptide precursors using IPA software. Disease and functional protein network analysis indicated that several precursor proteins were involved in developmental disorders and inflammatory responses (Fig. 4a, b). More precisely, precursor proteins including Alpha-2A adrenergic receptor (ADRA2A), Protein argonaute-2 (AGO2), Baculoviral IAP repeat-containing protein 6 (BIRC6), Kalirin (KALRN), and Histone-lysine N-methyltransferase 2C (KMT2C) were involved in developmental disorders, and KMT2C, Solute carrier family 2 (SLC2A4), Electrogenic sodium bicarbonate cotransporter 1 (SLC4A4), STIM1L (STIM1), and Rootletin (CROCC) were involved in inflammatory responses. All the putative precursor proteins associated with diseases are shown in Table S1. Furthermore, the regulator effects network analysis showed that some of the protein precursors participated in the networks of cellular development, embryonic development, organismal development, and organismal injury and abnormalities (Fig. 4c, d). For example, AGO2, CROCC, DENN domain-containing protein 2A (DENND2A), Krueppel-like factor 14 (KLF14), and Lon protease homolog (LONP1) were involved in the networks of cellular development, embryonic development, and organismal development. Additionally, Atrophin-1 (ATN1), Collagen alpha-1 (VIII) chain (COL8A1), Protein jagged-2 (JAG2), KMT2C, and Mucin-19 (MUC19) were related to the network of organismal injury and abnormalities. All the precursor proteins involved in regulator effects networks are shown in Table S2.
Fig. 4.
Diseases and regulator effects networks associated with the differentially expressed peptide precursors. Functional proteins related to a developmental disorder and b inflammatory response. Regulator effects network related to c cellular development, embryonic development, and organismal development and d organismal injury and abnormalities. Precursor proteins, diseases, and functions are shown as nodes, and the biological relationships between nodes are represented as lines with arrows. All lines are supported by at least one literature reference from the ingenuity pathway analysis (IPA) analysis. The intensity of the node color indicates the degree of upregulation (red) or downregulation (green)
Putative bioactive peptides associated with respiratory diseases
It is well known that peptides with biological functions have functions that are related to the functions of their precursor proteins, with domains playing key roles in the biological functions [39, 40]. The UniProt database was used to analyze the peptides and their precursors and the results showed that 25 peptides were located in the functional domains of their corresponding precursors (Table S3). The preceding results suggested that these precursors are mainly associated with inflammatory responses and abnormal organ development, which contribute to premature infant respiratory diseases. Therefore, we focused on peptides and their precursors related to respiratory diseases. Using the Open Targets Platform database, we investigated whether these peptides might play potential roles in respiratory diseases. All told, 17 precursor proteins were found to be closely related to respiratory diseases (association score ≥ 0.5) (Table S4).
TNF-α, IL-1β, and IL-6 in H2O2-treated lung epithelial cells with differentially expressed peptides
Previous researches have reported that lung epithelial cells A549 can be stimulated by H2O2 to induce inflammatory response [41, 42]. Therefore, we explored the effect of differentially expressed peptides on inflammatory response of H2O2-treated A549 cells. As observed, TNF-α, IL-1β, and IL-6 mRNA and protein expression levels were higher in A549 cells from the H2O2 (1 mM) group compared with the control group (Fig. 5), respectively, measured by qRT-PCR, ELISA, and western blot. According to the high fold change, we selected two peptides 508AAAAGPANVH517 derived from Homeobox protein SIX5 (SIX5, absolute fold change: 5.9) and 7118TGAKIKLVGT7127 derived from MUC19 (absolute fold change, 6.3) for future study. Peptide 7118TGAKIKLVGT7127 (MUC19) at 10 and 100 μM significantly attenuated the H2O2-induced increase of TNF-α, IL-1β, and IL-6. Moreover, we observed that the levels of inflammatory cytokines decreased more obviously with the 7118TGAKIKLVGT7127 at the concentration of 100 μM than 10 μM (Fig. 5). Meanwhile TNF-α, IL-1β, and IL-6 were significantly reduced in H2O2-treated A549 cells with peptide 508AAAAGPANVH517 derived from SIX5 at concentration of 100 μM, but not at 1 μM or 10 μM (Fig. 5). Both the two peptides help to suppress inflammatory response in H2O2-treated A549 cells.
Fig. 5.
Function analysis of differentially expressed peptides in vitro. (a) TNF-α, IL-1β, and IL-6 mRNA expression assessed by qRT-PCR in A549 cells stimulated by H2O2 (1 mM) with or without peptides 7118TGAKIKLVGT7127 (MUC19) and 508AAAAGPANVH517 (SIX5) at concentrations of 1, 10, and 100 μM. b The protein levels of TNF-α, IL-1β, and IL-6 measured by ELISA analyze in H2O2-treated A549 cells with or without peptides. c, d The protein levels of TNF-α, IL-1β, and IL-6 measured by western blot in H2O2-treated A549 cells with or without peptides (n = 3 biological independent samples per group in qRT-PCR and WB, technical replication = 3 in ELISA. *P < 0.05, **P < 0.01, ***P < 0.001)
Discussion
The MSC secretome and its therapeutic effects have been extensively demonstrated in preterm diseases, such as BPD [43] and NEC [19]. Previously, most researchers considered soluble factors and extracellular vesicles as the primary components of the secretome derived from MSCs [44–46]. However, the paracrine substances from MSCs are not limited to these biomolecules. Therefore, further studies are required to explore more types of components derived from MSCs and investigate their potential functions.
With the advance of detection technologies, mounting evidence has confirmed the differences in hUC-MSCs between preterm and term groups, which may help to identify the possible regulators or mechanisms underlying MSC function. To compare the global gene expression patterns in hUC-MSCs between these two groups, Iwatani et al. used microarray analysis and revealed that upregulated WNT2B in preterm hUC-MSCs was involved in the control of hUC-MSC proliferation [47]. A very recent study comparing hUC-MSC transcriptomics and proteomics profiles from term and preterm groups showed that Frizzled-2 (FZD2) protein and mRNA expression levels were both higher in preterm hUC-MSCs [48]. Importantly, FZD2 is the receptor of Wnt5a/b and FZD2 mutations influence Wnt signaling, which mediates the epithelial to mesenchymal transition (EMT) during lung development [49]. In addition, by comparing the proteome of microvesicles collected from hUC-MSC CM between preterm and term groups, Bruschi et al. found that 173 proteins were significantly changed, 163 of which were increased in the preterm group [50]. However, there have been no comprehensive comparisons of hUC-MSC CM peptidomic profiles between preterm and term infants. In the present study, we found that 131 peptides derived from 106 precursor proteins were differentially expressed in the preterm hUC-MSC CM compared with the term group by TMT labeling quantification (Fig. 1). Our study provides hUC-MSC CM polypeptide profiles for preterm and term infants. Secreted peptides have been shown to have important biological functions. Neuropeptide Y, a 36-amino acid peptide secreted by the hypothalamus, was found to play key roles in neurodegenerative diseases including modulation of neurogenesis, food intake, and thermogenesis [51]. Mao et al. showed that peptides derived from human beta-defensins are secreted by viable human cryopreserved amniotic membrane and exhibited direct antimicrobial effects against P. aeruginosa [52]. Further studies are needed to understand and explore the functions of secreted peptides from hUC-MSCs. In our study, we used p value to screen differentially expressed peptides between term and preterm groups. Although the false discovery rate (FDR) can detect differentially expressed peptides more accurately than p value, we would get very few peptides by FDR. Additionally, in order to identify more differentially expressed peptides, some studies also applied the p value to the analysis of peptides [53, 54]. Thus, in some condition, it is also appropriate to use the p value.
Subcellular location analysis of precursor proteins can help to better understand the source and potential functions of peptides. As shown in Fig. 3d, most precursor proteins were annotated as being part of organelles or membranes (56%). Notably, a small fraction of peptides were annotated as being derived from proteins located in the extracellular region, termed secreted proteins from hUC-MSCs. Classically, a large proportion of peptides (such as peptide neurotransmitters) are generated by the proteolysis of macromolecular proteins followed by release into the space outside of cells [55]. However, some other peptides containing one or more cleavage sites do not derive from endosomal processing [56]. These peptides are the N- or C-terminal peptides of their precursor proteins, rather than internal fragments [57]. In addition, the identification in our study of several peptides arising from secreted proteins raised the possibility that some bioactive peptides may be produced by enzymatic hydrolysis of extracellular proteins. These observations provide us with more methods to evaluate the potential functions of differentially expressed peptides in our study; additional studies are needed to ascertain whether these peptides are actually secreted from hUC-MSCs and have biological activities. From the results of IPA analysis, we found that a series of precursor proteins were involved in networks of developmental disorders, inflammatory responses, and organismal injury and abnormalities associated with premature infant diseases and most of these precursor proteins participated in the course of premature infant diseases according to the Open Targets Platform database. These observations raised the possibility that the peptides secreted by hUC-MSCs may have beneficial effects with a similar role of precursor proteins in neonatal respiratory diseases, which are worthy of in-depth functional studies.
As the functions of secretory peptides derived from hUC-MSCs were unclear, we assessed whether the peptides were located in the functional domains of their precursor proteins to analyze their functionality. Using the UniProt database, we discovered that 25 peptides were situated in the functional domains of their corresponding precursors. Peptides derived from Titin (8990ESDSG8994, 26941EGNKDD26946, 6684SSRLECKI6691, and 19103VVHAGGVIRIIAYV19116) and one peptide derived from striated muscle preferentially expressed protein kinase (2669SCTVAVARVPGKLAPPEVPQ2688) were located in Ig-like domains. The Ig-like domain was initially characterized as a structure composed of two sheets of antiparallel β strands [58]. Okano et al. reported that the Ig-like domain contributed to the maintenance of the structure, activity, and stability of metagenome-derived glycoside hydrolase family 9 endoglucanase [59]. Interestingly, the Ig-like domain made endoglucanase Cel9A from Alicyclobacillus acidocaldarius dependent on calcium [60]. Additionally, Ig-like domains have been shown to play various roles in functions [61, 62]. We also found that two peptides derived from MUC19 (961DDFMSSQN968 and 1382QNGIIVI1388) were located in the type D von Willebrand factor (VWFD) domain. This domain plays a substantial role in reducing soluble VWF binding to platelet GpIba and regulates platelet activation and adhesion [63, 64]. It also participates in fertilization, as it binds to sperm proteases [65]. Like the Ig-like domain, the VWFD domain is partially responsible for biological processes and functions. The above motif analysis may provide a new perspective for clarifying the possible roles of these newly identified peptides.
Based on the above results, we focused on exploring the potential role of differentially expressed peptides in developmental disorders and inflammatory responses. BPD is clearly one of the most common respiratory morbidity in preterm infants, and it may not simply be a consequence of lung immaturity [66]. And inflammation, one key contributor, often initiated by a pulmonary fetal inflammatory response, is aggravated by invasive or non-invasive mechanical ventilation and exposure to hyperoxia [67]. Given the large predominance of inflammation in the pathogenesis of BPD, we focused on the effect of peptides on inflammation response. A549 cells, a human lung carcinoma cell line, are often used for researches in pathological mechanism and treatment of BPD by A549 cells [68, 69]. And in our study, H2O2, known as a strong two-electron oxidant, could induce the production for TNF-α, IL-1β, and IL-6 in human lung epithelial cell line A549 in agreement with previous studies [70]. As known to all, primary human AECII are difficult to isolate and culture and also are mutable and unsuitable for experimental intervention, while A549 cells have similar biological characteristics to AECII cells [69], although embryonic alveolar epithelial cells are more in line with the purpose of the project and can be better used to evaluate the effects of peptides on the characteristics of lung development. However, this cell is difficult to obtain and easy to mutate [69]. Thus, we chose human lung epithelial cells A549 to explore the effects of differentially expressed peptides in vitro. Accordingly, to make a full understanding of these peptides, animal experiments will be carried out to support our researches in the following experiments.
We also observed that 7118TGAKIKLVGT7127 derived from MUC19 reduced the levels of TNF-α, IL-1β, and IL-6 in H2O2-treated A549 cells and the degree of decrease is related to the concentration of peptide as shown in Fig. 5. As a secreted mucin, MUC19 is released to the extracellular medium and has been identified in respiratory, digestive, and reproductive tracts [71]. It has been reported that MUC19 was differentially regulated after exposure to inflammatory cytokines [72]. And one recent study found that MUC19 peptides may enhance vaginal mucous immunity against infections [73]. These results indicated that 7118TGAKIKLVGT7127 derived from MUC19 may also play a role in protecting from inflammatory response stimulated by H2O2 in human lung epithelial cells. In addition, the other peptide 508AAAAGPANVH517 derived from SIX5 also reduced the expression levels of TNF-α, IL-1β, and IL-6 in H2O2-treated A549 cells only at the maximum concentration of 100 μM. It has been acknowledged that SIX5 was correlated with eye development [74] and myotonic dystrophy [75], which are related to embryonic and organismal development. Thus, we put forward one hypothesis that the hUC-MSCs from preterm infants may secrete protective substances such as peptides under stress. These findings indicated that two peptides may play an antiinflammatory role in the process of BPD. And previous studies have shown that the regulation of inflammation-related signal pathways such as p38 mitogen activated protein kinases (p38MAPK) signal pathways and nuclear factor-κB (NF-κB) signal pathway can reduce the level of pro-inflammatory cytokines, thus relieving the pulmonary inflammation of BPD [76, 77]. Combined with previous studies, we consider that the two peptides secreted from hUC-MSCs may decrease the production of pro-inflammatory cytokines through inflammation-related signal pathways and further researches need to be conducted to validate the possible pathway.
Conclusion
As far as we know, no large-scale quantitative peptidomic analysis has been carried out on the secretory components of hUC-MSCs. Our study identified the differentially expressed peptides secreted by preterm and term hUC-MSCs using TMT-based LC-MS/MS technology. Furthermore, bioinformatics analysis of precursors predicted the possible functions of peptides that may be useful in the treatment of premature respiratory diseases in connection with inflammatory responses and developmental disorders. And we first investigated the antiinflammatory effect of the peptides 7118TGAKIKLVGT7127 derived from MUC19 and 508AAAAGPANVH517 derived from SIX5 on human lung epithelial cells. This study expands our knowledge of the hUC-MSC secretome and may provide insights into new therapy for premature respiratory diseases.
Supplementary information
Additional file 1: Figure S1. Characterization of hUC-MSCs from preterm and term umbilical cords. (A) hUC-MSCs from preterm and term umbilical cords exhibited a fibroblast phenotype in culture. Adipogenesis capacity was confirmed by oil red O staining, and chondrocytes were evaluated by Alcian blue staining after differentiation. (B) Flow cytometry was used to detect the expression of positive markers (CD29, CD73, CD105) and negative markers (CD31, CD34 and HLA-DR) on hUC-MSCs.
Additional file 2: Figure S2. Identification of protein integrity of hUC-MSC CM from preterm and term infants by silver staining. The protein integrity of hUC-MSC CM was visualized by SDS-PAGE and silver staining (n=3 per group, P1-3 represent preterm infants and T1-3 term represent term infants).
Additional file 3: Table S1. Putative precursor proteins associated with diseases.
Additional file 4: Table S2. Precursor proteins involved in networks.
Additional file 5: Table S3. Differentially peptides located in functional domain based on Uniprot database.
Additional file 6: Table S4. Protein precursors and identified peptides related to respiratory system diseases.
Acknowledgements
This study was supported by contract grant sponsors and numbers: Jiangsu Provincial Women and Children Health Research Project (grant no. F201816 and F201744), National Natural Science Foundation of China (grant no. 81600687 and 81701491), Nanjing Medical Science and Technique Development Foundation (grant no. YKK18155), Jiangsu Provincial Medical Youth Talent (grant no. QNRC2016111), and Six Talent Peaks Project of Jiangsu Province (grant no. YY-112). The TMT method followed by mass spectrometry analysis was supported by the Analysis and Testing Center of Nanjing Medical University.
Abbreviations
- UC-MSC
Umbilical cord mesenchymal stem cell
- TMT
Tandem mass tag
- CM
Conditioned medium
- GO
Gene Ontology
- IPA
Ingenuity pathway analysis
- PVL
Periventricular leukomalacia
- BPD
Bronchopulmonary dysplasia
- NEC
Necrotizing enterocolitis
- ROP
Retinopathy of prematurity
- Ang-1
Angiopoietin-1
- HO-1
Heme oxygenase-1
- EPO
Erythropoietin
- GLP-1
Glucagon-like peptide-1
- CGRP
Calcitonin gene-related peptide
- AECII
Alveolar epithelium type II cells
- LC-MS/MS
Liquid chromatography-tandem mass spectrometry
- DMEM/F-12
Dulbecco’s modified Eagle medium/nutrient mixture F-12
- RT
Room temperature
- SDS
Sodium dodecyl sulfate
- MWCO
Molecular weight cutoff
- MW
Molecular weight
- PI
Isoelectric point
- H2O2
Hydrogen peroxide
- TNF-α
Tumor necrosis factor α
- IL-1β
Interleukin 1β
- IL-6
Interleukin 6
- GA
Gestational ages
- ADRA2A
Alpha-2A adrenergic receptor
- AGO2
Protein argonaute-2
- BIRC6
Baculoviral IAP repeat-containing protein 6
- KALRN
Kalirin
- KMT2C
Histone-lysine N-methyltransferase 2C
- SLC2A4
Solute carrier family 2
- SLC4A4
Electrogenic sodium bicarbonate cotransporter 1
- CROCC
Rootletin
- DENND2A
DENN domain-containing protein 2A
- KLF14
Krueppel-like factor 14
- LONP1
Lon protease homolog
- ATN1
Atrophin-1
- COL8A1
Collagen alpha-1 (VIII) chain
- JAG2
Protein jagged-2
- MUC19
Mucin-19
- MPDZ
Multiple PDZ domain protein
- SIX5
Homeobox protein SIX5
- FZD2
Frizzled-2
- EMT
Epithelial to mesenchymal transition
- HCAECs
Human coronary artery endothelial cells
- HUVECs
Human umbilical vein endothelial cells
- VWFD
Type D von Willebrand factor
Authors’ contributions
YuW and LZ performed the experiments, interpreted the results of the experiments, and drafted the manuscript. YunW and RPZ prepared the figures. YanW and YC analyzed the data. WL and CBJ participated in the discussion. HYW and LHY conceived and designed the experiments and provided funding to regents. All authors read and approved the final manuscript.
Funding
This study was supported by contract grant sponsors and numbers: Jiangsu Provincial Women and Children Health Research Project (grant no. F201816 and F201744), National Natural Science Foundation of China (grant no. 81600687 and 81701491), Nanjing Medical Science and Technique Development Foundation (grant no. YKK18155), Jiangsu Provincial Medical Youth Talent (grant no. QNRC2016111), and Six Talent Peaks Project of Jiangsu Province (grant no. YY-112).
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
This study has been approved by the Ethics Committee of Changzhou Maternal and Child Health Care Hospital (approval number 2019126). Written consent to participate was obtained from the parents of the patients.
Consent for publication
Written informed consent for publication of their clinical details was obtained from the parents of the patients.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yu Wang and Lin Zhang contributed equally to this work and should be regarded as joint first authors.
Contributor Information
Huaiyan Wang, Email: huaiyanwang@njmu.edu.cn.
Lianghui You, Email: y_lianghui@126.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13287-020-01931-0.
References
- 1.Blencowe H, Cousens S, Oestergaard MZ, Chou D, Moller AB, Narwal R, et al. National, regional, and worldwide estimates of preterm birth rates in the year 2010 with time trends since 1990 for selected countries: a systematic analysis and implications. Lancet. 2012;379:2162–2172. doi: 10.1016/S0140-6736(12)60820-4. [DOI] [PubMed] [Google Scholar]
- 2.Li C, Liang Z, Bloom MS, Wang Q, Shen X, Zhang H, et al. Temporal trends of preterm birth in Shenzhen, China: a retrospective study. Reprod Health. 2018;15:47. doi: 10.1186/s12978-018-0477-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brink LT, Gebhardt GS, Mason D, Groenewald CA, Odendaal HJ. The association between preterm labour, perinatal mortality and infant death (during the first year) in Bishop Lavis, Cape Town, South Africa. S Afr Med J. 2019;109:102–106. doi: 10.7196/SAMJ.2019.v109i2.13438. [DOI] [PubMed] [Google Scholar]
- 4.Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 5.Kern S, Eichler H, Stoeve J, Kluter H, Bieback K. Comparative analysis of mesenchymal stem cells from bone marrow, umbilical cord blood, or adipose tissue. Stem Cells. 2006;24:1294–1301. doi: 10.1634/stemcells.2005-0342. [DOI] [PubMed] [Google Scholar]
- 6.Pu L, Meng M, Wu J, Zhang J, Hou Z, Gao H, et al. Compared to the amniotic membrane, Wharton’s jelly may be a more suitable source of mesenchymal stem cells for cardiovascular tissue engineering and clinical regeneration. Stem Cell Res Ther. 2017;8:72. doi: 10.1186/s13287-017-0501-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma RR, Pollock K, Hubel A, McKenna D. Mesenchymal stem or stromal cells: a review of clinical applications and manufacturing practices. Transfusion. 2014;54:1418–1437. doi: 10.1111/trf.12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bongso A, Fong CY. The therapeutic potential, challenges and future clinical directions of stem cells from the Wharton's jelly of the human umbilical cord. Stem Cell Rev Rep. 2013;9:226–240. doi: 10.1007/s12015-012-9418-z. [DOI] [PubMed] [Google Scholar]
- 9.Chang YS, Ahn SY, Yoo HS, Sung SI, Choi SJ, Oh WI, et al. Mesenchymal stem cells for bronchopulmonary dysplasia: phase 1 dose-escalation clinical trial. J Pediatr. 2014;164:966–972. doi: 10.1016/j.jpeds.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 10.Ahn SY, Chang YS, Kim JH, Sung SI, Park WS. Two-year follow-up outcomes of premature infants enrolled in the phase I trial of mesenchymal stem cells transplantation for bronchopulmonary dysplasia. J Pediatr. 2017;185:49–54. doi: 10.1016/j.jpeds.2017.02.061. [DOI] [PubMed] [Google Scholar]
- 11.Ahn SY, Chang YS, Sung SI, Park WS. Mesenchymal stem cells for severe intraventricular hemorrhage in preterm infants: phase I dose-escalation clinical trial. Stem Cells Transl Med. 2018;7:847–856. doi: 10.1002/sctm.17-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohammadipoor A, Antebi B, Batchinsky AI, Cancio LC. Therapeutic potential of products derived from mesenchymal stem/stromal cells in pulmonary disease. Respir Res. 2018;19:218. doi: 10.1186/s12931-018-0921-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan Y, Jiang W, Tan Y, Zou S, Zhang H, Mao F, et al. hucMSC exosome-derived GPX1 is required for the recovery of hepatic oxidant injury. Mol Ther. 2017;25:465–479. doi: 10.1016/j.ymthe.2016.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang N, Chen C, Yang D, Liao Q, Luo H, Wang X, et al. Mesenchymal stem cells-derived extracellular vesicles, via miR-210, improve infarcted cardiac function by promotion of angiogenesis. Biochim Biophys Acta Mol basis Dis. 1863;2017:2085–2092. doi: 10.1016/j.bbadis.2017.02.023. [DOI] [PubMed] [Google Scholar]
- 15.Shi B, Wang Y, Zhao R, Long X, Deng W, Wang Z. Bone marrow mesenchymal stem cell-derived exosomal miR-21 protects C-kit+ cardiac stem cells from oxidative injury through the PTEN/PI3K/Akt axis. PLoS One. 2018;13:e191616. doi: 10.1371/journal.pone.0191616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morioka C, Komaki M, Taki A, Honda I, Yokoyama N, Iwasaki K, et al. Neuroprotective effects of human umbilical cord-derived mesenchymal stem cells on periventricular leukomalacia-like brain injury in neonatal rats. Inflamm Regen. 2017;37:1. doi: 10.1186/s41232-016-0032-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willis GR, Fernandez-Gonzalez A, Anastas J, Vitali SH, Liu X, Ericsson M, et al. Mesenchymal stromal cell Exosomes ameliorate experimental bronchopulmonary dysplasia and restore lung function through macrophage immunomodulation. Am J Respir Crit Care Med. 2018;197:104–116. doi: 10.1164/rccm.201705-0925OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCulloh CJ, Olson JK, Wang Y, Zhou Y, Tengberg NH, Deshpande S, et al. Treatment of experimental necrotizing enterocolitis with stem cell-derived exosomes. J Pediatr Surg. 2018;53:1215–1220. doi: 10.1016/j.jpedsurg.2018.02.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rager TM, Olson JK, Zhou Y, Wang Y, Besner GE. Exosomes secreted from bone marrow-derived mesenchymal stem cells protect the intestines from experimental necrotizing enterocolitis. J Pediatr Surg. 2016;51:942–947. doi: 10.1016/j.jpedsurg.2016.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moisseiev E, Anderson JD, Oltjen S, Goswami M, Zawadzki RJ, Nolta JA, et al. Protective effect of intravitreal administration of exosomes derived from mesenchymal stem cells on retinal ischemia. Curr Eye Res. 2017;42:1358–1367. doi: 10.1080/02713683.2017.1319491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han XP, Zhang FQ, Tan XS, Liu L, Ma WX, Ou-Yang HF, et al. EPO modified MSCs can inhibit asthmatic airway remodeling in an animal model. J Cell Biochem. 2018;119:1008–1016. doi: 10.1002/jcb.26268. [DOI] [PubMed] [Google Scholar]
- 22.Liu YT, Lin ZM, He SJ, Zuo JP. Heme oxygenase-1 as a potential therapeutic target in rheumatic diseases. Life Sci. 2019;218:205–212. doi: 10.1016/j.lfs.2018.12.033. [DOI] [PubMed] [Google Scholar]
- 23.Abd EAM, Kahle M, Meier JJ, Nauck MA. A meta-analysis comparing clinical effects of short- or long-acting GLP-1 receptor agonists versus insulin treatment from head-to-head studies in type 2 diabetic patients. Diabetes Obes Metab. 2017;19:216–227. doi: 10.1111/dom.12804. [DOI] [PubMed] [Google Scholar]
- 24.Bai YX, Fang F, Jiang JL, Xu F. Extrinsic calcitonin gene-related peptide inhibits hyperoxia-induced alveolar epithelial type II cells apoptosis, oxidative stress, and reactive oxygen species (ROS) production by enhancing notch 1 and homocysteine-induced endoplasmic reticulum protein (HERP) expression. Med Sci Monit. 2017;23:5774–5782. doi: 10.12659/MSM.904549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim YE, Park WS, Ahn SY, Sung DK, Sung SI, Kim JH, et al. WKYMVm hexapeptide, a strong formyl peptide receptor 2 agonist, attenuates hyperoxia-induced lung injuries in newborn mice. Sci Rep. 2019;9:6815. doi: 10.1038/s41598-019-43321-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun Y, Zhou Y, Liu X, Zhang F, Yan L, Chen L, et al. Antimicrobial activity and mechanism of PDC213, an endogenous peptide from human milk. Biochem Biophys Res Commun. 2017;484:132–137. doi: 10.1016/j.bbrc.2017.01.059. [DOI] [PubMed] [Google Scholar]
- 27.Zhang F, Cui X, Fu Y, Zhang J, Zhou Y, Sun Y, et al. Antimicrobial activity and mechanism of the human milk-sourced peptide Casein201. Biochem Biophys Res Commun. 2017;485:698–704. doi: 10.1016/j.bbrc.2017.02.108. [DOI] [PubMed] [Google Scholar]
- 28.Fu Y, Ji C, Chen X, Cui X, Wang X, Feng J, et al. Investigation into the antimicrobial action and mechanism of a novel endogenous peptide beta-casein 197 from human milk. AMB Express. 2017;7:119. doi: 10.1186/s13568-017-0409-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tozour JN, Delahaye F, Suzuki M, Praiss A, Zhao Y, Cai L, et al. Intrauterine hyperglycemia is associated with an impaired postnatal response to oxidative damage. Stem Cells Dev. 2018;27:683–691. doi: 10.1089/scd.2017.0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amrithraj AI, Kodali A, Nguyen L, Teo A, Chang CW, Karnani N, et al. Gestational diabetes alters functions in offspring's umbilical cord cells with implications for cardiovascular health. Endocrinology. 2017;158:2102–2112. doi: 10.1210/en.2016-1889. [DOI] [PubMed] [Google Scholar]
- 31.Kim J, Piao Y, Pak YK, Chung D, Han YM, Hong JS, et al. Umbilical cord mesenchymal stromal cells affected by gestational diabetes mellitus display premature aging and mitochondrial dysfunction. Stem Cells Dev. 2015;24:575–586. doi: 10.1089/scd.2014.0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan PY, Chang CW, Duan K, Poidinger M, Ng KL, Chong YS, et al. E2F1 orchestrates transcriptomics and oxidative metabolism in Wharton's jelly-derived mesenchymal stem cells from growth-restricted infants. PLoS One. 2016;11:e163035. doi: 10.1371/journal.pone.0163035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hwang JH, Lee MJ, Seok OS, Paek YC, Cho GJ, Seol HJ, et al. Cytokine expression in placenta-derived mesenchymal stem cells in patients with pre-eclampsia and normal pregnancies. Cytokine. 2010;49:95–101. doi: 10.1016/j.cyto.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 34.Nagamura-Inoue T, He H. Umbilical cord-derived mesenchymal stem cells: their advantages and potential clinical utility. World J Stem Cells. 2014;6:195–202. doi: 10.4252/wjsc.v6.i2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li GC, Zhang L, Yu M, Jia H, Tian T, Wang J, et al. Identification of novel biomarker and therapeutic target candidates for acute intracerebral hemorrhage by quantitative plasma proteomics. Clin Proteomics. 2017;14:14. doi: 10.1186/s12014-017-9149-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao W, Xu J, Wang F, Zhang L, Peng R, Shu Y, et al. Plasma membrane proteomic analysis of human gastric cancer tissues: revealing flotillin 1 as a marker for gastric cancer. BMC Cancer. 2015;15:367. doi: 10.1186/s12885-015-1343-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, Liang D, Cheng Q, Cao L, Wu Y, Wang Y, et al. Peptidomic analysis of fetal heart tissue for identification of endogenous peptides involved in tetralogy of Fallot. DNA Cell Biol. 2017;36:451–461. doi: 10.1089/dna.2017.3647. [DOI] [PubMed] [Google Scholar]
- 38.Yin J, Wang X, Zhang L, Wang X, Liu H, Hu Y, et al. Peptidome analysis of lung tissues from a hyperoxia-induced bronchopulmonary dysplasia mouse model: insights into the pathophysiological process of bronchopulmonary dysplasia. J Cell Physiol. 2018;233:7101–7112. doi: 10.1002/jcp.26633. [DOI] [PubMed] [Google Scholar]
- 39.Hatano H, Shaw J, Marquardt K, Zhang Z, Gauthier L, Chanteux S, et al. The D0 Ig-like domain plays a central role in the stronger binding of KIR3DL2 to B27 free H chain dimers. J Immunol. 2015;194:1591–1601. doi: 10.4049/jimmunol.1402214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koide A, Bailey CW, Huang X, Koide S. The fibronectin type III domain as a scaffold for novel binding proteins. J Mol Biol. 1998;284:1141–1151. doi: 10.1006/jmbi.1998.2238. [DOI] [PubMed] [Google Scholar]
- 41.Rahman I, Gilmour PS, Jimenez LA, Biswas SK, Antonicelli F, Aruoma OI. Ergothioneine inhibits oxidative stress- and TNF-alpha-induced NF-kappa B activation and interleukin-8 release in alveolar epithelial cells. Biochem Biophys Res Commun. 2003;302:860–864. doi: 10.1016/S0006-291X(03)00224-9. [DOI] [PubMed] [Google Scholar]
- 42.Rahman I, Gilmour PS, Jimenez LA, MacNee W. Oxidative stress and TNF-alpha induce histone acetylation and NF-kappaB/AP-1 activation in alveolar epithelial cells: potential mechanism in gene transcription in lung inflammation. Mol Cell Biochem. 2002;234–235:239–48. [PubMed]
- 43.Monsel A, Zhu YG, Gudapati V, Lim H, Lee JW. Mesenchymal stem cell derived secretome and extracellular vesicles for acute lung injury and other inflammatory lung diseases. Expert Opin Biol Ther. 2016;16:859–871. doi: 10.1517/14712598.2016.1170804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haraszti RA, Miller R, Stoppato M, Sere YY, Coles A, Didiot MC, et al. Exosomes produced from 3D cultures of MSCs by tangential flow filtration show higher yield and improved activity. Mol Ther. 2018;26:2838–2847. doi: 10.1016/j.ymthe.2018.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lai RC, Tan SS, Yeo RW, Choo AB, Reiner AT, Su Y, et al. MSC secretes at least 3 EV types each with a unique permutation of membrane lipid, protein and RNA. J Extracell Vesicles. 2016;5:29828. doi: 10.3402/jev.v5.29828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang YS, Ahn SY, Jeon HB, Sung DK, Kim ES, Sung SI, et al. Critical role of vascular endothelial growth factor secreted by mesenchymal stem cells in hyperoxic lung injury. Am J Respir Cell Mol Biol. 2014;51:391–399. doi: 10.1165/rcmb.2013-0385OC. [DOI] [PubMed] [Google Scholar]
- 47.Iwatani S, Shono A, Yoshida M, Yamana K, Thwin K, Kuroda J, et al. Involvement of WNT signaling in the regulation of gestational age-dependent umbilical cord-derived mesenchymal stem cell proliferation. Stem Cells Int. 2017;2017:8749751. doi: 10.1155/2017/8749751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chien C, Lo Y, Wu H, Hsuan Y, Lin C, Chen Y, Lin W, Han C. Transcriptomic and proteomic profiling of human mesenchymal stem cell derived from umbilical cord in the study of preterm birth. Prot Clin Appl. 2019;14:1900024. doi: 10.1002/prca.201900024. [DOI] [PubMed] [Google Scholar]
- 49.Kadzik RS, Cohen ED, Morley MP, Stewart KM, Lu MM, Morrisey EE. Wnt ligand/frizzled 2 receptor signaling regulates tube shape and branch-point formation in the lung through control of epithelial cell shape. Proc Natl Acad Sci U S A. 2014;111:12444–12449. doi: 10.1073/pnas.1406639111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bruschi M, Santucci L, Ravera S, Bartolucci M, Petretto A, Calzia D, et al. Metabolic signature of microvesicles from umbilical cord mesenchymal stem cells of preterm and term infants. Proteomics Clin Appl. 2018;12:e1700082. doi: 10.1002/prca.201700082. [DOI] [PubMed] [Google Scholar]
- 51.Reichmann F, Holzer P. Neuropeptide Y: A stressful review. Neuropeptides. 2016;55:99–109. [DOI] [PMC free article] [PubMed]
- 52.Mao Y, Hoffman T, Singh-Varma A, Duan-Arnold Y, Moorman M, Danilkovitch A, et al. Antimicrobial peptides secreted from human cryopreserved viable amniotic membrane contribute to its antibacterial activity. Sci Rep. 2017;7:13722. doi: 10.1038/s41598-017-13310-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xue Y, Xu P, Xu S, Xue K, Xu L, Chen J, et al. Peptidomic analysis of endometrial tissue from patients with ovarian endometriosis. Cell Physiol Biochem. 2018;47:107–118. doi: 10.1159/000489753. [DOI] [PubMed] [Google Scholar]
- 54.Wu L, Li H, Li X, Chen Y, Zhang Q, Cheng Z, et al. Peptidomic analysis of cultured cardiomyocytes exposed to acute ischemic-hypoxia. Cell Physiol Biochem. 2017;41:358–368. doi: 10.1159/000456282. [DOI] [PubMed] [Google Scholar]
- 55.Hook V, Bark S, Gupta N, Lortie M, Lu WD, Bandeira N, et al. Neuropeptidomic components generated by proteomic functions in secretory vesicles for cell-cell communication. AAPS J. 2010;12:635–645. doi: 10.1208/s12248-010-9223-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clement CC, Becerra A, Yin L, Zolla V, Huang L, Merlin S, et al. The dendritic cell major histocompatibility complex II (MHC II) peptidome derives from a variety of processing pathways and includes peptides with a broad spectrum of HLA-DM sensitivity. J Biol Chem. 2016;291:5576–5595. doi: 10.1074/jbc.M115.655738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fricker LD. Analysis of mouse brain peptides using mass spectrometry-based peptidomics: implications for novel functions ranging from non-classical neuropeptides to microproteins. Mol BioSyst. 2010;6:1355–1365. doi: 10.1039/c003317k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sato K, Kakuda S, Yukitake H, Kondo Y, Shoji M, Takebe K, et al. Immunoglobulin-like domains of the cargo proteins are essential for protein stability during secretion by the type IX secretion system. Mol Microbiol. 2018;110:64–81. doi: 10.1111/mmi.14083. [DOI] [PubMed] [Google Scholar]
- 59.Okano H, Kanaya E, Ozaki M, Angkawidjaja C, Kanaya S. Structure, activity, and stability of metagenome-derived glycoside hydrolase family 9 endoglucanase with an N-terminal Ig-like domain. Protein Sci. 2015;24:408–419. doi: 10.1002/pro.2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pazhang M, Younesi FS, Mehrnejad F, Najavand S, Tarinejad A, Haghi M, et al. Ig-like domain in endoglucanase Cel9A from alicyclobacillus acidocaldarius makes dependent the enzyme stability on calcium. Mol Biotechnol. 2018;60:698–711. doi: 10.1007/s12033-018-0105-4. [DOI] [PubMed] [Google Scholar]
- 61.Centa A, Rodriguez-Barrueco R, Montero JC, Pandiella A. The immunoglobulin-like domain of neuregulins potentiates ErbB3/HER3 activation and cellular proliferation. Mol Oncol. 2018;12:1061–1076. doi: 10.1002/1878-0261.12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim TK, Park CS, Na HJ, Lee K, Yoon A, Chung J, et al. Ig-like domain 6 of VCAM-1 is a potential therapeutic target in TNFalpha-induced angiogenesis. Exp Mol Med. 2017;49:e294. doi: 10.1038/emm.2016.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Madabhushi SR, Zhang C, Kelkar A, Dayananda KM, Neelamegham S. Platelet GpIba binding to von Willebrand factor under fluid shear:contributions of the D'D3-domain, A1-domain flanking peptide and O-linked glycans. J Am Heart Assoc. 2014;3:e1420. doi: 10.1161/JAHA.114.001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Madabhushi SR, Shang C, Dayananda KM, Rittenhouse-Olson K, Murphy M, Ryan TE, et al. von Willebrand factor (VWF) propeptide binding to VWF D'D3 domain attenuates platelet activation and adhesion. Blood. 2012;119:4769–4778. doi: 10.1182/blood-2011-10-387548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Akasaka M, Harada Y, Sawada H. Vitellogenin C-terminal fragments participate in fertilization as egg-coat binding partners of sperm trypsin-like proteases in the ascidian Halocynthia roretzi. Biochem Biophys Res Commun. 2010;392:479–484. doi: 10.1016/j.bbrc.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 66.Kalikkot TR, Guaman MC, Shivanna B. Bronchopulmonary dysplasia: a review of pathogenesis and pathophysiology. Respir Med. 2017;132:170–177. doi: 10.1016/j.rmed.2017.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Speer CP. Pulmonary inflammation and bronchopulmonary dysplasia. J Perinatol. 2006;26:S57–S62. doi: 10.1038/sj.jp.7211476. [DOI] [PubMed] [Google Scholar]
- 68.Kunzmann S, Ottensmeier B, Speer CP, Fehrholz M. Effect of progesterone on Smad signaling and TGF-beta/Smad-regulated genes in lung epithelial cells. PLoS One. 2018;13:e200661. doi: 10.1371/journal.pone.0200661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang X, Chu X, Gong X, Zhou H, Cai C. The expression of miR-125b in Nrf2-silenced A549 cells exposed to hyperoxia and its relationship with apoptosis. J Cell Mol Med. 2020;24:965–972. doi: 10.1111/jcmm.14808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang W, Zheng JP, Zhu SX, Guan WJ, Chen M, Zhong NS. Carbocisteine attenuates hydrogen peroxide-induced inflammatory injury in A549 cells via NF-kappaB and ERK1/2 MAPK pathways. Int Immunopharmacol. 2015;24:306–313. doi: 10.1016/j.intimp.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 71.Ballester B, Milara J, Cortijo J. Mucins as a new frontier in pulmonary fibrosis. J Clin Med. 2019;8(9):1447. [DOI] [PMC free article] [PubMed]
- 72.Kerschner JE, Khampang P, Erbe CB, Kolker A, Cioffi JA. Mucin gene 19 (MUC19) expression and response to inflammatory cytokines in middle ear epithelium. Glycoconj J. 2009;26:1275–1284. doi: 10.1007/s10719-009-9245-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yan L, Luan T, Hua Q, Gu Y, Fu Z, Liu X, et al. Peptidomic analysis of female reproductive tract secretion to identify putative anti-infection peptides in the female genital system via nanotechnologies. J Biomed Nanotechnol. 2018;14:215–226. doi: 10.1166/jbn.2018.2500. [DOI] [PubMed] [Google Scholar]
- 74.Pham YC, Man N, Holt I, Sewry CA, Pall G, Johnson K, et al. Characterisation of the transcription factor, SIX5, using a new panel of monoclonal antibodies. J Cell Biochem. 2005;95:990–1001. doi: 10.1002/jcb.20454. [DOI] [PubMed] [Google Scholar]
- 75.Sarkar PS, Paul S, Han J, Reddy S. Six5 is required for spermatogenic cell survival and spermiogenesis. Hum Mol Genet. 2004;13:1421–1431. doi: 10.1093/hmg/ddh161. [DOI] [PubMed] [Google Scholar]
- 76.Li K, Zhang F, Wei L, Han Z, Liu X, Pan Y, et al. Recombinant human elafin ameliorates chronic hyperoxia-induced lung injury by inhibiting nuclear factor-kappa B signaling in neonatal mice. J Interf Cytokine Res. 2020;40:320–330. doi: 10.1089/jir.2019.0241. [DOI] [PubMed] [Google Scholar]
- 77.Zhang Z, Sun C, Wang J, Jiang W, Xin Q, Luan Y. Timing of erythropoietin modified mesenchymal stromal cell transplantation for the treatment of experimental bronchopulmonary dysplasia. J Cell Mol Med. 2018;22:5759–5763. doi: 10.1111/jcmm.13843. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Characterization of hUC-MSCs from preterm and term umbilical cords. (A) hUC-MSCs from preterm and term umbilical cords exhibited a fibroblast phenotype in culture. Adipogenesis capacity was confirmed by oil red O staining, and chondrocytes were evaluated by Alcian blue staining after differentiation. (B) Flow cytometry was used to detect the expression of positive markers (CD29, CD73, CD105) and negative markers (CD31, CD34 and HLA-DR) on hUC-MSCs.
Additional file 2: Figure S2. Identification of protein integrity of hUC-MSC CM from preterm and term infants by silver staining. The protein integrity of hUC-MSC CM was visualized by SDS-PAGE and silver staining (n=3 per group, P1-3 represent preterm infants and T1-3 term represent term infants).
Additional file 3: Table S1. Putative precursor proteins associated with diseases.
Additional file 4: Table S2. Precursor proteins involved in networks.
Additional file 5: Table S3. Differentially peptides located in functional domain based on Uniprot database.
Additional file 6: Table S4. Protein precursors and identified peptides related to respiratory system diseases.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.