Abstract
Biogenesis of iron-sulfur (Fe-S) clusters in an essential process in living organisms due to the critical role of Fe-S cluster proteins in myriad cell functions. During biogenesis of Fe-S clusters, multi-protein complexes are used to drive the mobilization and protection of reactive sulfur and iron intermediates, regulate assembly of various Fe-S clusters on an ATPase-dependent, multi-protein scaffold, and target nascent clusters to their downstream protein targets. The evolutionarily ancient sulfur formation (Suf) pathway for Fe-S cluster assembly is found in bacteria and archaea. In Escherichia coli, the Suf pathway functions as an emergency pathway under conditions of iron limitation or oxidative stress. In other pathogenic bacteria, such as Mycobacterium tuberculosis and Enterococcus faecalis, the Suf pathway is the sole source for Fe-S clusters and therefore is a potential target for the development of novel antibacterial compounds. Here we summarize the considerable progress that has been made in characterizing the first step of mobilization and protection of reactive sulfur carried out by the SufS-SufE or SufS-SufU complex, Fe-S cluster assembly on SufBC2D scaffold complexes, and the downstream trafficking of nascent Fe-S clusters to A-type carrier (ATC) proteins.
Keywords: Iron-sulfur cluster, Suf, A-type carrier protein, metallocofactor, iron
1. Introduction to Fe-S cluster biogenesis
Iron-sulfur (Fe-S) clusters are metal cofactors required for essential biological pathways, including respiration, photosynthesis, nitrogen fixation, and DNA repair [1, 2]. Fe-S clusters consist of iron in the Fe2+ or Fe3+ oxidation states bound to sulfide (S2−) in a number of arrangements, including [2Fe-2S], [3Fe-4S], or [4Fe-4S] clusters. Larger, more complex Fe-S clusters are also found in nature, such as the [7Fe-8S] P-cluster and the FeMo cofactor (containing [Mo-3Fe-3S] and [4Fe-3S] clusters) of the nitrogenase complex [3]. Despite their importance, Fe-S clusters can be sensitive to oxidation by reactive oxygen or nitrogen species or to disruption by thiophilic metals, such as copper and cobalt [4–6]. De novo synthesis of Fe-S clusters in cells also requires an adequate iron supply, which can be problematic due to the drop in bioavailable iron that occurs in aerobic environments. These factors necessitate constant assembly and/or repair of Fe-S clusters, especially in cells growing under aerobic conditions.
To meet this need, Fe-S cluster biogenesis systems have emerged. These systems can be relatively simple in evolutionarily ancient anaerobic organisms or highly complex in multicompartmental eukaryotic cells [7–9]. However, there are core components of Fe-S cluster biogenesis pathways found in nearly all organisms examined to date. In most organisms where they have been characterized, Fe-S cluster biogenesis pathways require the enzyme-catalyzed production of sulfide from L-cysteine, a dedicated scaffold protein for the assembly of the nascent Fe-S cluster, and one or more Fe-S cluster trafficking proteins for targeting of the Fe-S cluster to specific apo-proteins in the cell. Here we will discuss the specific biochemical mechanisms elucidated by study of the bacterial Suf systems for Fe-S cluster biogenesis (Figure 1). We will largely focus on the stress-responsive Suf system of E. coli and the housekeeping Suf system of B. subtilis, with some mention of other Suf systems where appropriate.
Figure 1.
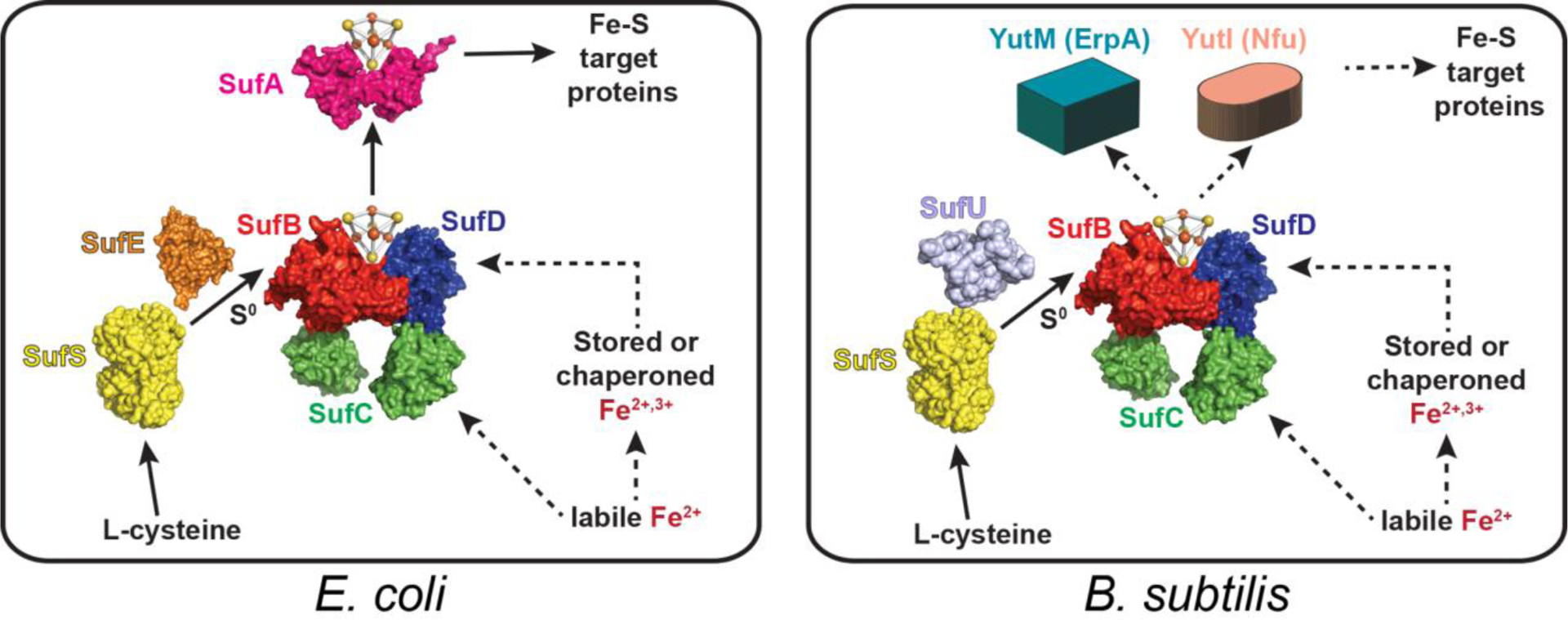
Examples of the Suf pathway for Fe-S cluster biogenesis in bacteria. Solid arrows indicate functional steps that are well-supported by in vitro and in vivo data. Dashed arrows are those steps that are not yet fully characterized. Based on level of homology, we refer to YutM as ErpA, but it has also been annotated as SufA. The yutM gene is separately transcribed from the suf genes in B. subtilis.
2. Sulfur mobilization by SufS and the SufE and SufU accessory proteins
The initiation of Fe-S cluster biogenesis necessitates the acquisition of sulfur from available pools of cysteine. Fe-S cluster biogenesis pathways typically accomplish this feat via a cysteine desulfurase reaction in which free L-cysteine in the cell is converted into an enzyme-bound sulfanylcysteine species (often referred to as a persulfide) as well as alanine. This reaction proceeds to a transpersulfuration step during which the persulfide is transferred to an acceptor protein or metabolites resulting in an overall ping-pong reaction mechanism [10–12]. In the Suf pathway, the SufS protein functions as the cysteine desulfurase for Fe-S cluster biogenesis. This biogenesis step is arguably the most well-understood from the standpoint of mechanistic biochemistry.
2.1. SufS structure and mechanism
Structurally, SufS is a homodimeric protein consisting of two monomers containing the cofactor pyridoxal 5’-phosphate (PLP) that is required for initiating the desulfurase reaction. Analysis of SufS crystal structures from E. coli, B. subtilis, and Synechocystis sp indicates strong structural homology of this protein between organisms [13–15]. In E. coli, SufS requires the PLP cofactor for proper folding and cannot be purified in a soluble form without it, though this may not be true for other organisms. The active site containing PLP is surrounded by several key amino acid residues instrumental in the progression of the cysteine desulfurase reaction. The PLP cofactor in the resting state is bound to Lys226 in an internal aldimine (Schiff base) conformation with the ε amino group of Lys [11]. An active site loop contains a conserved Cys residue where the persulfide species forms during the desulfurase step (Cys364 in E. coli SufS). This loop is shortened in SufS, which limits both solvent accessibility as well as structural flexibility and designates SufS as a group II cysteine desulfurase as opposed to IscS and NifS, which are categorized as group I. Group I desulfurases contain a larger, more flexible active site loop allowing for the interaction of these cysteine desulfurases with multiple other proteins to which they can transfer the persulfide in the transpersulfuration step [13, 16, 17]. Additionally, they do not require a separate shuttling/activating protein, which is the second major distinguishing trait between group I and II desulfurases. Unlike IscS and NifS, SufS requires a shuttle protein (either SufE or SufU depending on the organism) to participate as a co-substrate in the transpersulfuration reaction [18]. Depending on the organism, SufE or SufU operates to remove the persulfide and transfer it to the scaffold protein where de novo Fe-S cluster biogenesis occurs. Both shuttling proteins also enhance the low basal activity of SufS by allosterically altering active site residues around the PLP cofactor prior to or concurrent with desulfuration. Aside from key structural features of SufS, recent studies have elucidated significant enzymatic and physical changes upon SufS interaction with L-cysteine. As shown in Figure 2, PLP begins in an internal aldimine conformation with an absorption feature of 422 nm [11, 19]. After binding substrate cysteine, the PLP-cysteine potentially transits through an unobserved gem-diamine confirmation to subsequently form the enolimine and ketoenamine tautomers of the external aldimine, with absorbance features at 343 nm and 424 nm respectively [19]. The formation of these tautomers is stabilized by π-π stacking interactions between the pyridine ring of PLP and the imidazole ring of His123 in the SufS active site [15]. X-ray crystallography confirmed that the SufS C364A mutation traps the external aldimine species due to the failure of this mutant to carry out the desulfuration step [19].
Figure 2.
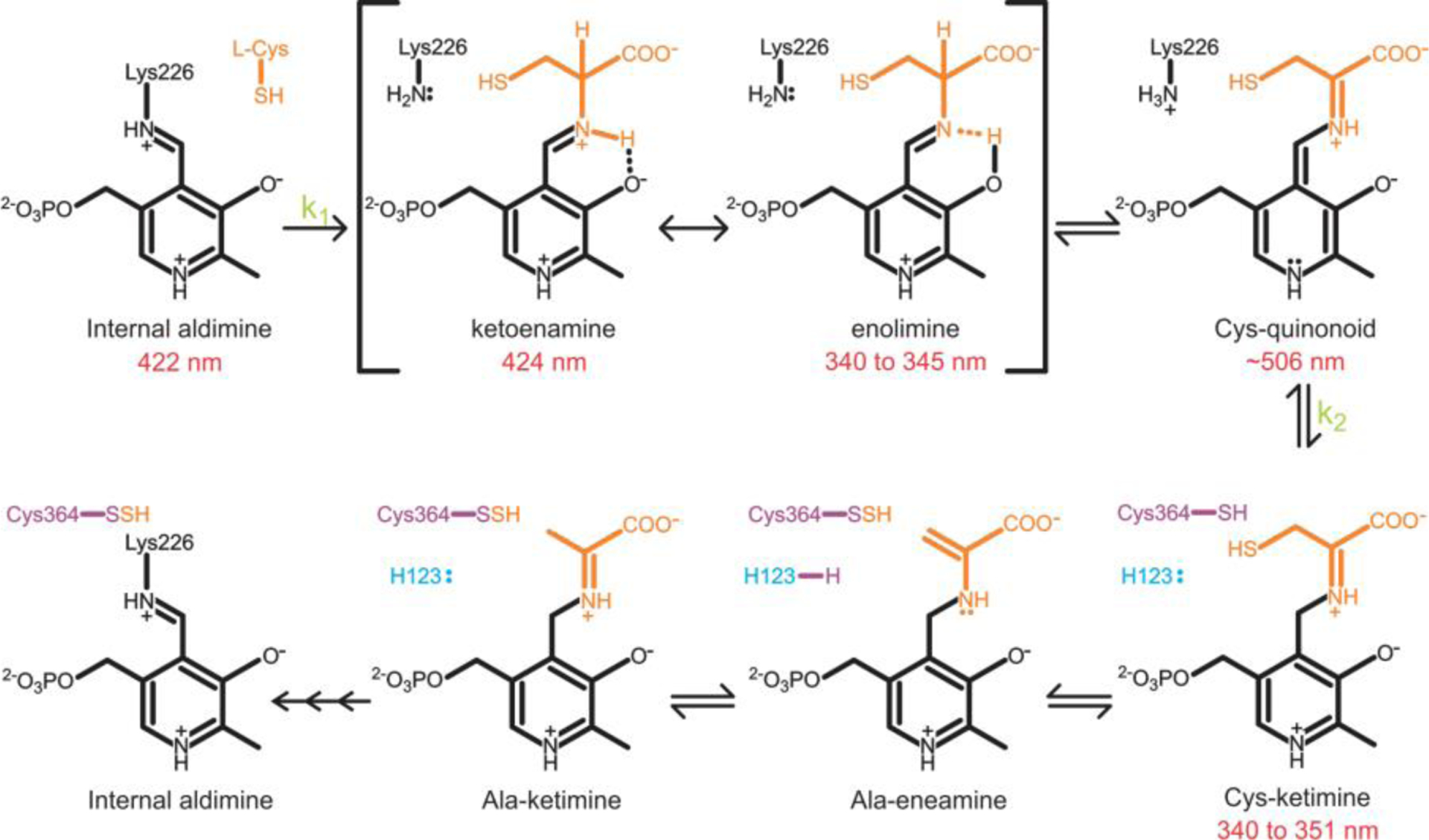
Scheme for the SufS cysteine desulfurase mechanism. PLP is in black, substrate cysteine is orange, active site Cys364 is purple, and His123 acid/base catalyst in blue [19].
Use of stopped-flow spectroscopy on wild-type SufS demonstrated that the shift from the initial 422 nm species to the 343 and the 424 nm intermediates requires a two-summed exponential fit at each wavelength indicating the production of two separate intermediates, although the two species were not spectroscopically distinguishable [19]. Stopped-flow analysis of SufS H123A revealed two distinct species with kinetics matching wild-type SufS, suggesting identical pathways. The H123A mutant lacked noticeable formation of the ketoenamine tautomer of the external aldimine and primarily produced the enolimine tautomer. The two-step transition from the initial internal aldimine species to a final species around 351 nm requires a two-summed exponential fit like wild-type SufS, with formation of the external aldimine corresponding to the initial fast phase of the reaction and absorbing at 345 nm [19]. This Cys-aldimine intermediate was also confirmed for SufS in other organisms by Nakamura et al [20]. The second, slow phase observed with the H123A mutant results in the transformation of the less stable enolimine tautomer at 345 nm into a Cys-ketimine intermediate with absorption at 351 nm. The SufS H123A mutant stalled the desulfurase reaction at the Cys-ketimine state allowing x-ray crystallographic confirmation of the intermediate [19]. In addition to its optimal location for π-π stacking with PLP, His123 is also well-situated to function as an acid-base catalyst during the desulfurase reaction [19]. Although not spectroscopically observed, the transition from Cys-aldimine to Cys-ketimine likely proceeds through a spectroscopically silent quinonoid species, which would absorb around 500 nm.
After generation of the Cys-ketimine, the cysteine desulfurase mechanism likely proceeds by Cys364 attacking the Cys-ketimine sulfhydryl to break the C-S bond, resulting in the formation of an Ala-eneamine species and a persulfide bound to Cys364 [19]. His123 likely deprotonates the thiol group of Cys364 to form the required thiolate anion nucleophile, which is consistent with trapping of the Cys-ketimine intermediate in the H123A mutant [19]. Following formation of Ala-eneamine, the intermediates Ala-ketimine, Ala-quinonoid, and the Ala-aldimine are likely produced based off previous predictions and studies of homologues, although there is a lack of spectroscopic and structural data illustrating these intermediates [21]. Finally, reformation of the PLP internal aldimine species with Lys226 returns the enzyme to its resting state in preparation for another cycle of the cysteine desulfurase mechanism.
2.2. SufE or SufU sulfur shuttling proteins are needed to transfer sulfur from SufS
As previously mentioned, optimum functionality of group II cysteine desulfurases like SufS relies on a sulfur shuttle protein [18, 22]. This protein both enhances the initially lower basal activity of the desulfurase as shown in Table 1 while also transferring the acquired sulfur to the target scaffold protein where Fe-S cluster biogenesis occurs [12, 18]. Two separate proteins, SufE or SufU, can serve this role for SufS but their phylogenetic distribution is distinct, and they do not appear to both occur within the suf operon in the same organism [23]. SufE and SufU are specific to the SufS family members (group II cysteine desulfurases) observed in various organism and have not been seen to function with group I desulfurases. It is worth noting that SufS-SufE homologues are found encoded together in other loci without the rest of the Suf pathway (for example, CsdA-CsdE in E. coli). These homologues can act as multi-copy suppressors of some ΔiscS phenotypes in vivo, but appear to primarily function in other E. coli metabolic pathways [24].
Table 1.
SufS cysteine desulfurase activity enhancement due to sulfur shuttle in various organisms
| Organism | Sulfur shuttling protein | Cysteine desulfurase enhancement | Ref. |
|---|---|---|---|
| Escherichia coli | SufE | (50–100)-fold | [10, 11] |
| Erwinia chrysanthemi | SufE | 40-fold | [22] |
| Plasmodium falciparum | SufE | 17-fold | [25] |
| Bacillus subtilis | SufU | >100-fold | [26] |
| Enterococcus faecalis | SufU | 37-fold | [27] |
| Arabadopsis thaliana | SufE1 | 40-fold | [28] |
| SufE2 | 40-fold | [29] | |
| SufE3 | 70-fold | [29] |
SufE belongs to the E1-like superfamily of proteins, with a monomeric structure consisting primarily of α-helices as well as three contiguous β-strands responsible for constructing the active site of SufE [30]. In E. coli, the active site Cys51 is located on the tip of a loop between two of these strands with the side chain concealed in a hydrophobic cavity isolated from solvent exposure [30]. SufE Cys51 is the site where persulfide is transferred from SufS Cys364 and is required for SufE to function as a substrate for the transpersulfuration reaction [31]. The sequestered nature of the SufE Cys51 active site prevents damage from oxidative stress (such as would hinder the Isc pathway) and renders the Suf pathway more resistant to oxidative stress conditions [10].
The activity of SufS alone pales in comparison to SufS activity after the addition of the SufE shuttle, resulting in a dramatic activity enhancement of up to 50-fold depending on the specific organism [10, 22]. The reaction involving SufE demonstrates a biphasic process where the first, fast phase corresponds to the transfer of sulfur to SufE. The second, slower phase correlates to the accumulation of persulfurated SufE [11]. As determined by Selbach et al., use of a stronger reductant such as TCEP instead of DTT eliminates the biphasic nature of the reaction by promoting the rapid removal of persulfide from SufE. This reaction becomes a single-phase process while maintaining the enhanced rate resulting from SufE [11]. Presumably in vivo, the further transfer of SufE persulfide to the SufB scaffold protein would similarly serve to efficiently recycle SufE.
Despite no current crystal structure of the SufS-SufE complex being available, HDX-MS has revealed that the interaction between SufS and SufE is not limited to simply the transfer of sulfur. SufE binding to SufS results in a notable allosteric effect with structural alterations to the active sites of both proteins in addition to modifications occurring at the SufS dimer interface [32, 33]. Additionally, binding studies of SufE to SufS reveal that the individual protomers of the SufS dimer are not equivalent. The binding to one SufE monomer weakens the affinity of the remaining SufS protomer for a second SufE. This result suggests a two-sites model for the functionality of the SufS cysteine desulfurase when involving SufE [32]. In addition to the solution studies, the crystal structure of the complex formed between the SufS homologue CsdA and the SufE homologue CsdE suggests possible changes that might occur in a SufS-SufE complex. Upon interacting with CsdA, the loop containing the active site cysteine of CsdE becomes exposed and shifts closer and orients itself to the CsdA active site cysteine [34]. This appears true only for the one “activated” CsdE monomer within the CsdA2-CsdE2 structure. The second CsdE monomer that binds to the other CsdA monomer (within the CsdA dimer) does not show positioning of its active site near the CsdA active site [34, 35]. The CsdA protein, like SufS, contains a β-hairpin that blocks the active site and would need to rearrange in order to permit sulfur exchange by moving the β-hairpin from the path [35]. The two cysteines that carry out the transpersulfuration reaction, SufS Cys364 and SufE Cys51 in E. coli, must be oriented proximal to each other. Further support of this allosteric effect is illustrated via the SufE D74R mutation. This mutation enhanced SufE active site loop flexibility, increased solvent accessibility of that loop, and improved the active site Cys51 as a sulfur acceptor. These alterations improved the binding affinity between SufS and SufE without sacrificing sulfur transfer (at least under unstressed conditions) [36].
2.3. SufU as the sulfur transfer protein in some Suf systems
As previously mentioned, not all organisms containing the Suf pathway utilize SufE for sulfur shuttling. Instead, in organisms like B. subtilis and M. tuberculosis, SufU replaces SufE in the operon. SufU is much more similar to the Fe-S scaffold IscU at the primary sequence level than it is to SufE. SufU, like IscU, contains two conserved cysteines near the active site cysteine responsible for persulfide binding (Cys41 in B. subtilis SufU) [37]. Based on analysis of purified SufU and its mutant derivatives, the initial proposed function for these cysteines was to serve as Fe-S cluster ligands as has been observed for the homologous cysteines in the Fe-S scaffold IscU [38]. This proposal was supported by the in vitro ability of SufU to activate Fe-S proteins like Leu1 and by the substoichiometric levels of Fe-S clusters observed in purified Cys mutant forms of SufU [38, 39]. However, Fe-S cluster formation in SufU is much less robust than IscU and it has been suggested the in vitro results stemmed from adventitious iron binding and non-physiological Fe-S cluster reactions. Attempts to characterize the SufU Fe-S cluster by Mössbauer or EPR spectroscopies were unsuccessful and subsequent attempts to reconstitute Fe-S clusters on wild-type SufU were not fruitful [38, 39]. Importantly, it was shown that E. coli sufSE could complement a B. subtilis ΔsufSU mutant strain and that B. subtilis sufSU could rescue the lethality of the ΔiscUA-hscBA ΔsufSE double mutation in E. coli. In contrast, sufU and iscU were not interchangeable between organisms [40]. These results indicate functional redundancy between SufE and SufU for transpersulfuration but argue SufU does not function as the primary Fe-S scaffold.
Functional redundancy between SufE and SufU led to a second proposed function for the SufU as a transpersulfurase in the B. subtilis Suf pathway. SufU improves SufS cysteine desulfurase activity up to 200-fold. SufU Cys41 functions as the site of transpersulfuration from SufS. This enhancement, however, depends on the presence of zinc. As determined by Selbach et al. using EXAFS, a zinc atom binds tightly to SufU via tetrahedral coordination using the three conserved Cys residues (Cys41, Cys66, Cys128) and Asp43 [39]. Mutagenesis of these sites results in disruption of zinc-binding and reduction of sulfur transferase activity as well as secondary structure alterations. SufU binding affinity for zinc is extremely tight with a measured Ka = 1017 M−21 [39]. Mutagenesis of the binding site or removal of zinc using TPEN substantially reduced the cysteine desulfurase enhancement when SufU is added to SufS, demonstrating the importance of zinc to this process [39]. The exact mechanism by which zinc influences SufU interaction with SufS was proposed based on the SufS-SufU crystal structure.
Initially, use of H/DX experiments involving SufS and SufU from B. subtilis revealed structural changes around the active sites when the two proteins interact in order to bring together the active site cysteines Cys361 from SufS and Cys-41 of SufU [14]. X-ray crystallography combined with snapshot analysis of the SufS-SufU complex by Fujishiro et al. illustrated the significant structural alterations experienced by both proteins upon interaction including unusual zinc-ligand swapping shown in Figure 3 [41]. Initially, zinc is bound to SufU in the previously described tetra-coordination [39]. When SufU interacts with SufS, Cys41 is replaced by His342 as the fourth zinc ligand, permitting Cys41 to function in the transpersulfuration reaction with Cys361 of SufS [41]. The conservation of these residues in SufU and SufS indicates the likely occurrence of this exchange in other species containing these proteins. Using snapshot analysis of the reaction revealed secondary structure changes to SufU when interacting with SufS that transferred Cys41 away from zinc and close to Cys361 as well as the PLP [41]. Finally, mutagenesis of the conserved His342 to tyrosine resulted in a major loss of desulfurase function for the SufS-SufU complex [41].
Figure 3.
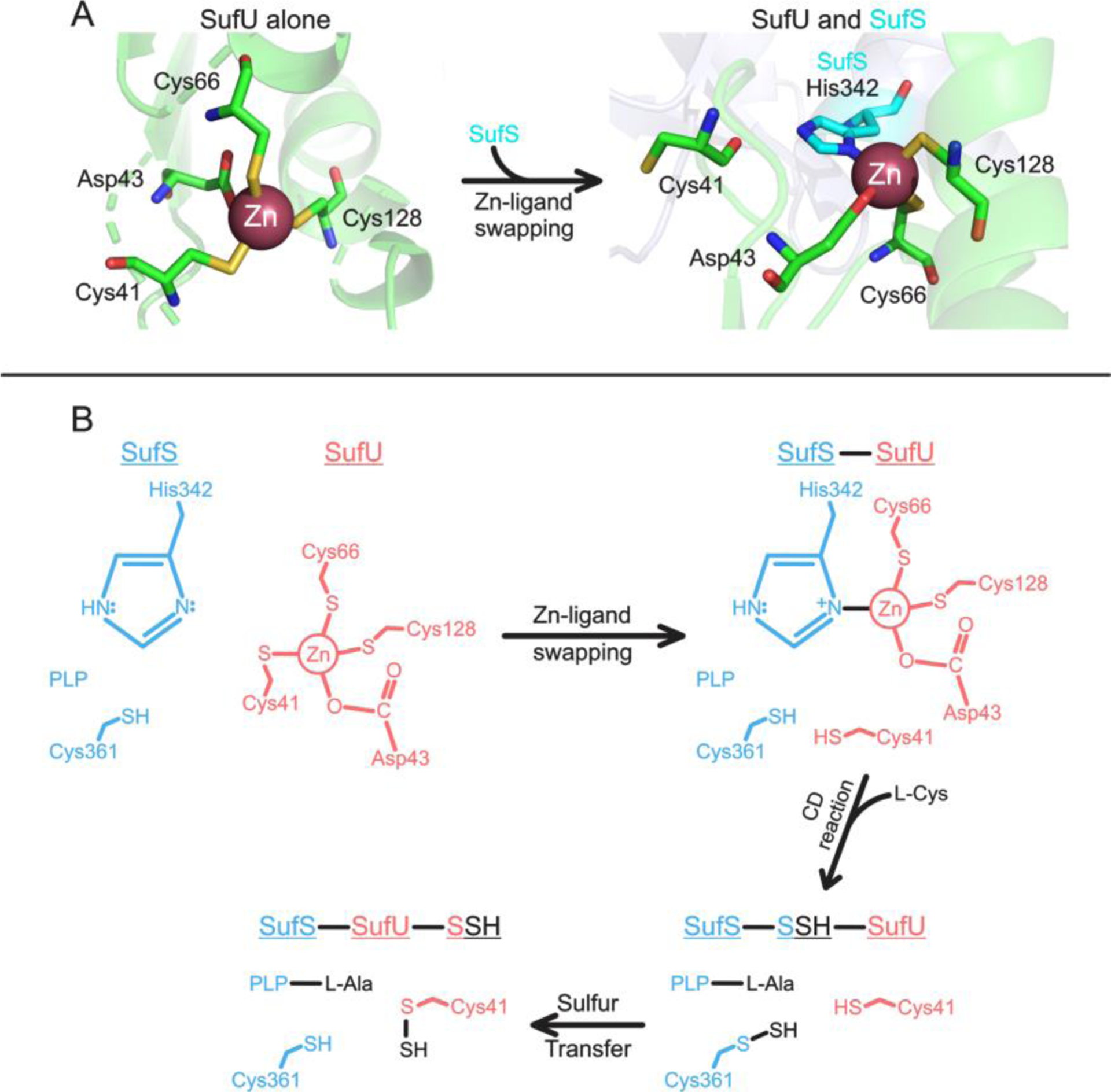
Zinc-ligand swapping between SufU and SufS responsible for cysteine desulfurase enhancement in B. subtilis. A. The starting zinc-coordination structure of SufU alone (left from PDB code 5XT5) as well as the altered zinc-coordination upon the interaction between SufU and SufS (right from PDB code 2AZH). The key event is SufS His342 replacing SufU Cys41 in ligating zinc, making this cysteine available for sulfur transfer. This figure was adapted from Fujishiro et al [41]. B. Mechanism illustrating interaction of SufS and SufU beginning with zinc-ligand swapping and ending with the transfer of sulfur to SufU. This figure was adapted from Fujishiro et al [41].
3. Assembly of the nascent Fe-S cluster on the SufB scaffold complexes
3.1. The role of SufB as site of nascent cluster assembly
SufB is the scaffold protein of the Suf system that can transiently assemble a Fe-S cluster during biogenesis. As a scaffold, SufB can interact with the SufE protein that mobilizes persulfide from the cysteine desulfurase SufS active site. The interaction between SufB and SufE is enhanced if SufC is bound to SufB as SufB2C2 or SufBC2D complexes [42]. When the SufBC2D complex is incubated with SufS, SufE, and L-cysteine in the absence of reducing agent, up to seven sulfur atoms can accumulate onto SufB, as analyzed by ESI mass spectrometry [31]. Through systematic mutational analysis and mapping of critical residues of the β-helix core domain of SufB, Cys254 and Cys405, which are strictly conserved cysteine residues, were identified as potential sulfur acceptor sites from SufE (Table 2) [43]. Cys254 located on the N-terminal side of the helix was shown to be essential for stimulating the cysteine desulfurase activity of SufSE by SufBC2D complex [43].
Table 2.
Functions of critical amino acid residues in the SufBC2D complex in E. coli*
| Protein | Residue | Function | Ref. |
|---|---|---|---|
| SufB | Cys254 | Required for SufBC2D to stimulate the cysteine desulfurase activity of SufS-Suffi Putative initial acceptor site for persulfide transferred from SufE C254A mutation abolished sulfur accumulation on SufB |
[43] |
| Cys405 | Putative final sulfur acceptor and Fe–S cluster ligand (Hg2+ ligand in one SufBC2D structure) Exposed at SufB-SufD interface by SufC dimer formation C405A mutation strongly diminished sulfur binding on SufB in vitro C405A mutation blocked in vivo Fe-S cluster formation by Suf and blocked ability to complement ΔiscΔsuflethality in vivo |
[43, 44] | |
| SufC | Lys40 | Invariant Lys of Walker A motif in ATPase active site K40R mutation reduced in vitro ATPase activity K40R mutation blocked ability to complement ΔiscΔsuf lethality, Suf-mediated resistance to iron starvation, and in vivo Fe-S cluster formation by Suf |
[44, 45] |
| Lys152 | Forms a salt bridge with E171 in monomeric SufC to negatively regulate ATPase activity (salt bridge is broken in SufC complexes) | [44, 46] | |
| Glu171 | Invariant catalytic residue in Walker B motif in ATPase active site E171Q mutation reduced in vitro ATPase activity E171Q mutation blocked ability to complement ΔiscΔsuflethality and in vivo Fe-S cluster formation by Suf |
[44, 46] | |
| His203 | Residue in H-motif in ATPase active site H203A mutation reduced in vitro ATPase activity H203A mutation blocked ability to complement ΔiscΔsuf lethality and in vivo Fe-S cluster formation by Suf |
[44] | |
| SufD | His360 | Invariant residue at SufB-SufD interface H360A mutation blocked in vivo Fe-S cluster formation by Suf and blocked ability to complement ΔiscΔsuflethality in vivo |
[43, 46] |
| Cys358 | Putative Fe-S cluster ligand (Hg2+ ligand in one SufBC2D structure) C358A and C385S mutations had no effect on in vivo Suf function | [43, 44] |
For more extensive mutagenesis results, see Supplementary Table S3 in reference [43]
Also, the Cys254A mutation abolished sulfur accumulation, while the Cys405A mutation strongly diminished sulfur binding on SufB. From these results it was proposed that Cys254 is the initial acceptor site for persulfide transferred from SufE. The persulfide species is then transferred along a “Cys relay” to Cys405 on the C-terminal side of the helix for cluster assembly (Figure 4). E. coli SufB also contains a putative Fe-S cluster motif consisting of Cys96, Cys99, and Cys103 in its N-terminal region that could serve as a binding site for the Fe-S cluster [31]. However, mutagenesis studies refuted this hypothesis since the cysteine triple mutant lacking this motif still assembled a Fe-S cluster after in vitro reconstitution, suggesting that these cysteines are not cluster ligands. A mutated SufB protein lacking this motif was still able to complement the synthetic lethality of a Δisc Δsuf double mutant strain, suggesting the N-terminal Cys motif is not essential for SufB function [43].
Figure 4.
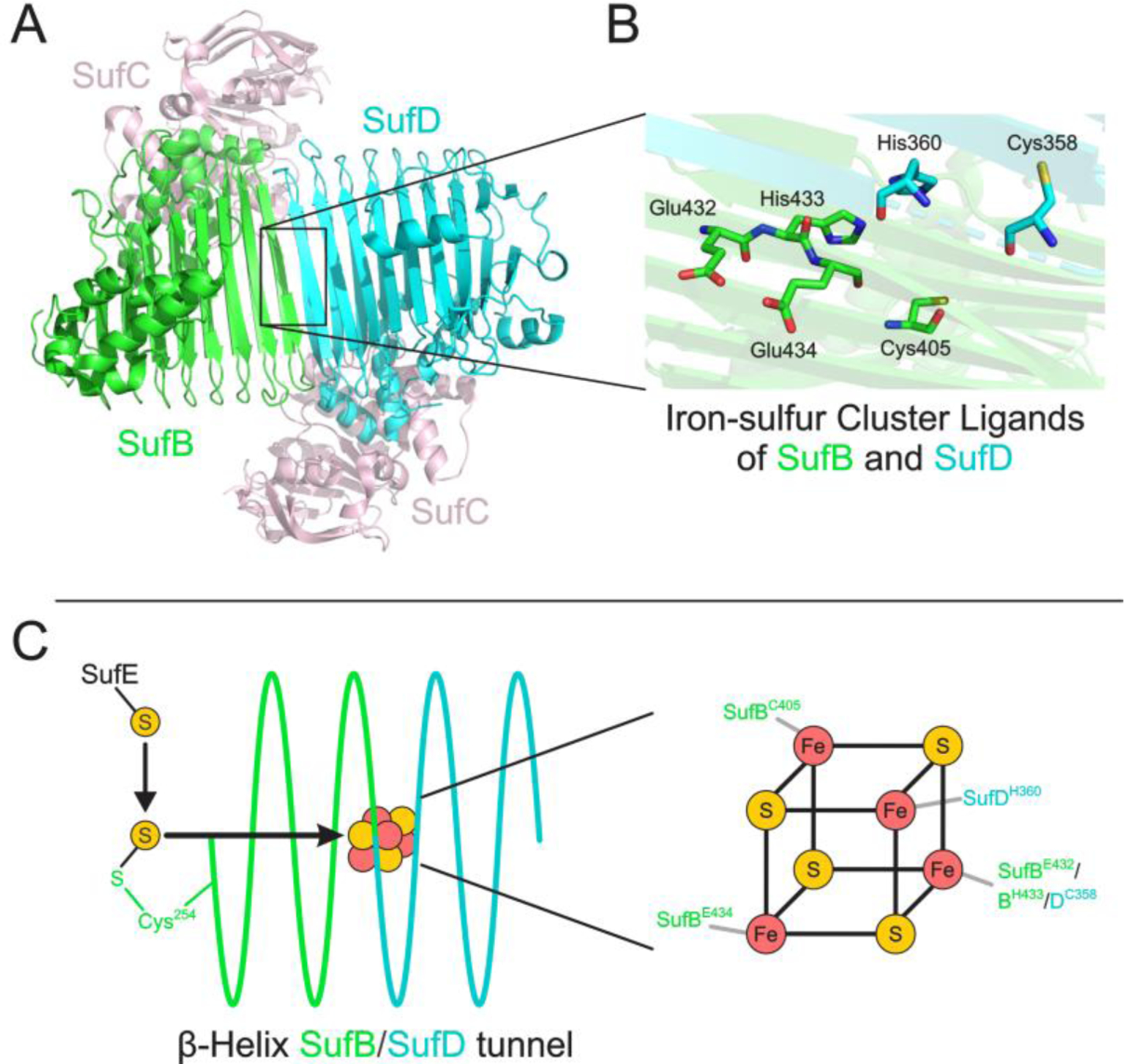
A. Structure of the SufBC2D protein complex (PDB code: 5AWG) with the putative Fe-S binding site boxed [44]. B. The specific residues from SufB, as determined by Takahashi et al. and SufD proposed to ligate the cluster are illustrated [43]. C. The proposed path through which sulfur may travel, starting with the sulfur shuttling protein SufE and proceeding into the β-helical structure of SufB and SufD, is illustrated (left) as are the proposed Fe-S ligands (right).
E. coli SufB is a difficult protein to manipulate in vitro as it tends to be insoluble, and it exists in different oligomerization states (Outten FW, unpublished data and [31]). While there are no structures of E. coli SufB alone, a homologue of SufB from Methanosarcina mazei Go1 has been crystallized (PDB 4DN7). The two proteins are 23% identical and 44% over a shared 381 residue region. Structural alignment of the M. mazei SufB homodimer with the E. coli SufB-SufD heterodimer (from the SufBC2D structure, see below) shows remarkable structural similarities between the two complexes (data not shown). This is consistent with phylogenetic analysis suggesting that sufD arose from a gene duplication of sufB [7]. Phylogenetic distribution of Suf components also indicates that SufB can likely act as the sole cluster scaffold in some organisms that lack SufD. This hypothesis is consistent with biochemical experiments in E. coli showing that a SufB oligomer can form a [4Fe-4S] cluster similar to SufBC2D in vitro [31].
The stepwise mechanism of Fe-S cluster assembly on SufB remains unclear. Cluster synthesis by IscU proceeds through initial formation of two [2Fe-2S]2+ clusters that can be reductively coupled to form a single [4Fe-4S]2+ cluster. In vitro Fe-S reconstitution experiments initially reported assembly of a [4Fe-4S] cluster on SufB without any observable accumulation of [2Fe-2S] clusters during the process [31]. However, the [2Fe-2S] SufB form is obtained by anaerobic incubation of apo-SufB in the absence of DTT with a 3-fold molar excess of ferric iron and sulfide followed by purification with an anion exchange column. These conditions may abort cluster assembly at the [2Fe-2S] cluster stage due to the lower molar ratio of iron and sulfide and the absence of a reductant. Adding DTT to the SufB reconstitution along with increasing the ratio of iron and sulfide resulted in a [4Fe-4S] cluster form of SufB [47]. One interpretation of these results is that the addition of a reductant allows reductive coupling of [2Fe-2S] clusters to form [4Fe-4S] SufB, although this has not been confirmed by monitoring stepwise cluster assembly over time on SufB. Finally, after in vivo co-expression with the sufCDSE genes, both [4Fe-4S] and linear [3Fe-4S] clusters were detected on purified His6-SufB as part of the SufBC2D complex [45]. Interestingly, the SufB [2Fe-2S] cluster is more stable and more resistant to H2O2, O2, and iron chelators than the [2Fe-2S] cluster of IscU. This resistance is further increased if SufB is part of the SufBC2D complex, supporting a specific role for SufBC2D as a stress-resistant scaffold complex in E. coli [47]. In vitro, both the [2Fe-2S] and [4Fe-4S] holo forms of SufB can transfer their cluster to [2Fe-2S] and [4Fe-4S] client proteins such as SufA, ferredoxin and aconitase [48–50].
3.2. The SufBC2D scaffold complex
In E. coli, the scaffolding complex is the ternary SufBC2D complex with a 1:2:1 stoichiometry as indicated by mass spectrometry analysis and biochemical analyses [48]. While the SufBC2D complex can be obtained in vivo in strains expressing the entire suf operon, it cannot be reconstituted in vitro starting from the purified components. Whether this reflects some cotranslational folding or assembly constraint remains to be investigated. There are several structures of this complex and the associated SufC2D2 sub-complex but, thus far none of them contain an Fe-S cluster [48]. Negative-stain electron microscopy and small angle X-ray scattering (SAXS) data from the as-isolated SufBC2D complex in solution are in agreement with the crystal structure [44]. The SufBC2D complex shares a common architecture with a separately isolated SufC2D2 sub-complex, where each of the SufC subunits is bound to a subunit of the SufB-SufD heterodimer or the SufD homodimer [46].
E. coli SufC, a monomer in solution, was found to possess ATPase activity. Additionally, the crystal structure of monomeric E. coli SufC exhibits many traits typical of ABC ATPases such as Walker A and B motifs as well as D- and Q-loops [51–53] (PDB accession number:2D3W) [54]. Mutation of SufC Lys40 in the Walker A box, Lys152 or Glu171 in the Walker B, or Asp173 and His203 in the H-motif, abolishes SufC ATPase activity [44]. The ATPase activity of SufC is not required for Fe-S assembly on SufBC2D in vitro. However, in vivo function of the Suf pathway is abolished by deleting sufC or mutating residues in the ATPase active site (Table 2) [45, 53, 55].
The structural fold of the SufC protomers in the SufC2D2 and SufBC2D complexes shows significant physical changes around the ATP-binding pocket compared to monomeric SufC. In the SufC monomer, the conserved glutamate residue (Glu171 in E. coli) is rotated away from the ATP binding site and forms a salt-bridge with Lys152, leading to nearly undetectable ATPase activity in SufC alone. By contrast, in the SufC protomers of the SufC2D2 and SufBC2D complexes, the Glu171-Lys152 salt bridge is broken leading to rotation of Glu171 towards the ATP-binding pocket [46]. A dramatic shift of Q-loop residue Gln85 towards the ATP binding site accompanies the Glu171 rotation. His203, another key residue for the activity of ABC ATPases, is shifted 5 Å toward Glu171. These structural changes remodel the catalytic pocket of SufC to be suitable for ATP binding and hydrolysis, resulting in a SufC local structure that more closely resembles that of other activated ABC-ATPases. Due to these changes, the ATPase activity of SufC is significantly improved upon interaction with SufB or SufD in Thermus thermophilus (180-fold as SufBC and 5-fold with SufCD) [56]. The ATPase activity of SufC is thought to induce conformational changes when binding to SufBD. This permits SufBC2D to form Fe-S clusters because such complexes containing SufC variants (mutations at Lys40, Glu171, His203) that lacked any ATPase activity could not perform Fe-S cluster assembly in vivo [44].
Despite the active site rearrangement, however, the SufC protomers in the complexes are widely separated (>40 Å) in the structure. Since ABC ATPases must dimerize to form two composite active sites for ATP hydrolysis, the current SufC2D2 and SufBC2D structures may represent a resting state of the complex. Fluorescence labelling and cross-linking experiments in solution revealed that, during the catalytic step of ATP binding and hydrolysis, SufC does form a transient head-to-tail dimer within the SufBC2D complex [44]. Furthermore, SufC dimerization induces conformational changes in its two partners, SufB and SufD, notably in SufB Cys405; the residue reputed to be a Fe-S cluster ligand when exposed at the complex’s surface [44]. Most likely, the conformational changes also allow SufD His360, another postulated ligand for cluster coordination, to migrate adjacently to Cys405 of SufB [44, 46]. The association between SufC and SufB/SufD is made via the so-called “transmission interface” mode observed in homologous ABC transporters, which was proposed to allow transmission of the dimerization motion within the ABC ATPase to its binding partners during ATP binding and hydrolysis [51]. This structural analysis shows that the ATPase activity of SufC likely drives conformational change of its SufB-SufD binding partners resulting in ATP-dependent conformational changes similar to those arising within other ABC transporters (even though it is not a membrane-bound transport complex) [44]. It seems clear that ATP hydrolysis must be essential to drive conformational changes that suitably rearranges the scaffold complex to facilitate Fe-S cluster biogenesis, but it is not clear at exactly which step(s) it is required. The SufC K40R mutation blocks in vivo Fe-S cluster formation in the SufBC2D complex primarily by reducing its iron content with only mild effects on sulfide content [45]. This result may indicate a role for SufC ATPase activity for in vivo iron acquisition.
The structure of the E. coli SufD homodimer (PDB number: 1VH4) demonstrates a flattened righthanded β-helix of nine turns with two strands per turn, which can form homodimers [57]. Two highly conserved residues, Pro347 and His360, interact at the dimer interface [57]. The structure of the SufD subunit in SufC2D2 and SufBC2D is almost identical to that of SufD protomers in the SufD homodimer [57]. SufB and SufD share a similar structural organization, with an N-terminal helical domain, a core domain consisting of a right-handed parallel β-helix, and a C-terminal helical domain that contains the SufC binding site (Figure 4). The C-terminal subdomain is well-conserved between SufD and SufB. The β-helix architecture of the SufB-SufD protomers was speculated to be specialized for the Fe-S cluster biogenesis family of proteins [44]. Deletion of sufD reduces the iron content of the subcomplex SufB2C2 and for this reason SufD is thought to play a role in iron acquisition [45]. Early studies reported a link between SufD and iron metabolism [45, 53, 58, 59]. However, in vitro evidence for a SufD iron interaction is still incomplete.
At present it is unclear exactly where the nascent Fe-S cluster binds within SufB oligomers or SufBC2D. A holo form of SufBC2D has been crystallized containing two Hg2+ ions coordinated at the SufB/SufD dimer interface. Cys405 of SufB and Cys358 of SufD in the heterodimer interface each bind one Hg2+ ion while His360 of SufD is also in the vicinity of the Hg2+ binding site near the interface of SufB and SufD (Table 2) [44]. It is likely that these mercury binding sites may overlap with those of iron ions in the complex. However, as pointed out by the authors, this “closed interface” conformation of SufB-SufD does not allow sufficient space for coordination of an Fe-S cluster in that region. As noted above, it is hypothesized that ATP binding and/or hydrolysis leads to SufC dimerization, which in turn leads to movements that expose or expand the SufB-SufD dimer interface, possibly allowing for formation of an Fe-S cluster in that region. An in vivo assay to monitor Fe-S cluster biogenesis by Suf showed that the SufB C405A and SufD H360A mutations blocked Fe-S cluster synthesis, although the SufD C358A variant retained function [44]. Similarly, the SufB C405A mutant was unable to complement the synthetic lethality phenotype of a ΔiscΔsuf double mutant strain. SufC mutations that abolish ATPase activity (K40R, E171Q, or H203A) also blocked in vivo Fe-S cluster formation, further supporting that SufC ATPase activity is essential for Fe-S cluster biogenesis [44].
In addition to binding Fe-S clusters, SufBC2D also binds one equivalent of FADH2 with a dissociation constant (Kd) of 12μM [48]. E. coli SufBC2D can be purified anaerobically bound to FADH2, however, upon oxygen exposure the flavin is oxidized to FAD and released from the complex [45, 48]. A binding site for the FADH2 has been proposed in SufB based on the presence of a R(x)6ExxY(x)5G(x)8Y motif with sequence homology to the p-cresol methylhydroxylase family [48]. However, subsequent structural analysis of SufB in the SufBC2D complex suggested its conformation in that region is distinct from flavin-binding proteins [43]. Furthermore, SufB mutant proteins containing point mutations in the proposed flavin-binding motif are still able to complement the ΔsufABCDSE ΔiscUA-hscBA double mutant strain, indicating that motif is not absolutely required for Suf function [43]. Since it was shown that all three SufBCD proteins are required for stoichiometric binding of FADH2 by the complex, it is likely that there is a composite site that only forms when the active SufBC2D complex is assembled [45, 48].
The differential affinity of SufBC2D for reduced versus oxidized flavin suggests a reaction cycle where FADH2 is bound, undergoes a controlled oxidation to release electrons to SufBC2D, and then is released as oxidized FAD. The functional significance of the flavin oxidation is unclear. Unlike Isc, the Suf system does not contain a dedicated [2Fe-2S] ferredoxin protein to provide electrons during cluster biogenesis for steps such as persulfide (S0) reduction to sulfide (S2−). The FADH2 could be used for this process. In vitro, SufBC2D-FADH2 also was shown to release iron from ferric citrate or the ferric-loaded form of bacterial frataxin (CyaY), most likely by reducing Fe3+ to Fe2+ [48]. This result led to the proposal that flavin is used by Suf to reduce ferric iron in an iron storage or iron chaperone protein during Fe-S cluster biogenesis in vivo. Since the in vivo iron donor for the Suf pathway is unknown, this hypothesis has been hard to test rigorously. The use of FADH2 as a cellular reductant in place of Isc Fdx would be logical in E. coli when one considers that an Fe-S ferredoxin may not function adequately under oxidative stress and iron starvation conditions where Suf must carry out Fe-S cluster biogenesis.
4. Fe-S cluster trafficking by SufA and SufT
4.1. Biochemical features of A-type carrier (ATC) proteins in E. coli
A-type carrier (ATC) proteins can be divided into three families, type-I, type-II, and type-III [60]. Genes for type-I ATC proteins such as ErpA, reside on separate loci in E. coli. This organism also expresses type-II ATC proteins IscA and SufA, which are encoded by the isc and suf operons respectively [60]. Type-III ATC proteins, which include NifIscA and are not present in E. coli, are a smaller subfamily found within the operon of Fe-S cluster biogenesis genes in nitrogen fixing bacteria [60]. The exact function of ATC proteins has been a subject of some debate. Biochemical analyses have clearly demonstrated that IscA, SufA, ErpA, and NifIscA homodimers can coordinate either a [2Fe-2S]2+ or a [4Fe-4S]2+ cluster [49, 50, 61–64]. Further, evidence suggests that these enzymes can deliver the bound Fe-S clusters to apo-proteins in vitro [49, 50, 61–64]. NifIscA from Azotobacter vinelandii has been shown to bind a [2Fe-2S] cluster in the presence of oxygen, with reversible conversion to a [4Fe-4S] cluster occurring under anaerobic and reducing conditions [65]. While the biochemical details of the transformation have not been fully elucidated, it seems likely that NifIscA and other ATCs may be able to modify the trafficked cluster type depending on environmental factors.
It is also clear that IscA and NifIscA homodimers can bind a single Fe2+ or Fe3+ in a mononuclear site with an apparent association constant of 3.0×1019 M−1 for ferric iron [65, 66]. Iron is at least partially coordinated by the conserved Cys residues, although a Tyr residue has also been implicated in iron binding for E. coli IscA possibly via an oxygen ligand [66]. The Y40F mutation in IscA significantly decreased the iron-binding activity of this ATC protein but did not alter Fe-S cluster binding activity in vitro and was found to be inactive in vivo. The results suggest that IscA can efficiently bind iron and that this process is crucial for Fe-S cluster biogenesis [66]. For IscA, the ferric iron can be released in the presence of L-cysteine, presumably by reduction of Fe3+ to Fe2+, allowing the iron to be incorporated into nascent cluster assembly on the IscU scaffold in vitro [67]. There is still some disagreement about whether E. coli SufA also coordinates ferrous and ferric iron in the same manner, but it seems a distinct possibility. Iron-binding properties of ErpA have not been thoroughly investigated. In summary, it is possible that ATC proteins have multiple distinct biochemical functions such as Fe-S cluster trafficking and iron donation for biogenesis or repair of Fe-S clusters. To date, it has been difficult to directly assess the possible multifunctional nature of ATCs in vivo.
The crystal structure of ~13 kDa ATC protein apo-SufA reported by Wada et al. in 2005 (PDB 2D2A) shows a dimeric form in which the two monomers (α1 and α2) are stabilized by H-bonding and extensive hydrophobic interactions [68]. The C-terminal region of the α1 monomer interacts with the C-terminus of the α2 monomer. Similar to other ATC proteins, SufA possesses three highly conserved cysteine residues at positions 50, 114, and 116 which are arranged in a C50XC114XC116 motif near the C-terminus. Cys50 is positioned in a way that is much farther than the other conserved cysteines, Cys114 and Cys116 (Figure 5). Arrangement of these cysteines in close proximity to one another would ensure efficient binding of iron, [2Fe-2S], and/or [4Fe-4S] [68]. However, in the α1 monomer of the crystal structure, Cys50 is oriented further away from Cys114 and Cys116 (Figure 5). Though clearly visible in the α1 monomer, these residues are disordered and unresolved within the α2 monomer [68]. Previous work has demonstrated that these conserved cysteine residues are crucial in homologous ATC proteins. In the S. cerevisiae ATC homologues, Isa1p and Isa2p, the three conserved cysteines were individually replaced with a serine residue [69]. The Isa1p C178S, C242S, and C244S mutant proteins each failed to complement the severe growth defect of the Δisa1 mutant strain. Similarly, Isa2p C89S, C175S, and C177S mutations blocked the ability of those proteins to complement the Δisa2 mutant. Protein stability of each of these same mutants was comparable to the wild-type Isa1p and Isa2p, demonstrating that protein stability remained unaffected in the cysteine mutants. This work supports the idea that each of these conserved cysteines is crucial for ATC protein activity [69].
Figure 5.
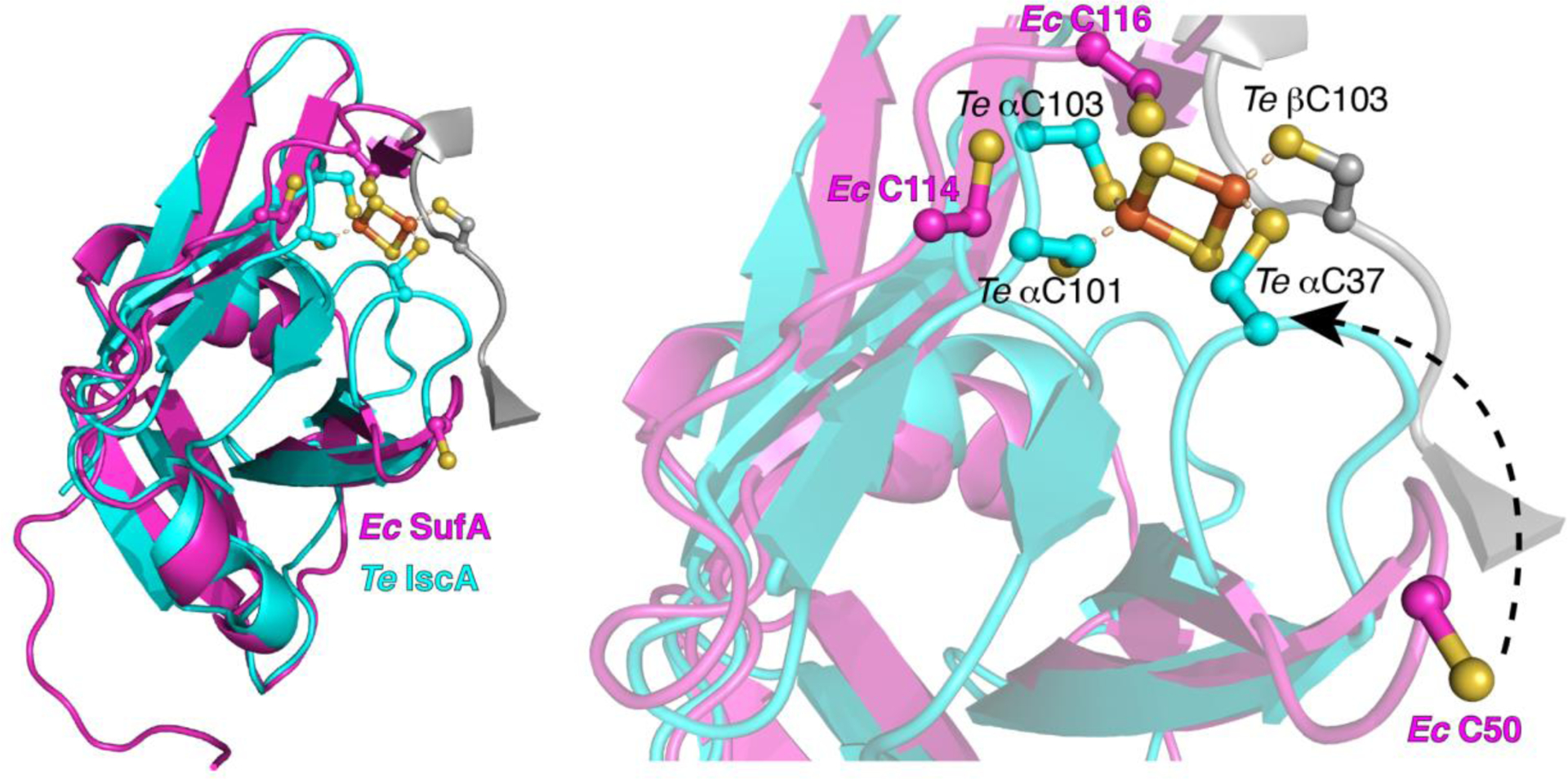
Structural overlay of the α (magenta) subunit of apo-SufA (PDB 2D2A, Ec SufA) with the α (cyan) and β (grey) subunits of [2Fe-2S] IscA (PDB 1X0G, Te IscA). Left, full protein structure. Right, enlargement of the Fe-S cluster binding site. For clarity, only the portion of the β subunit containing C103 is shown (in grey).
Alignment of the E. coli SufA apo-protein (PDB 2D2A) with the [2Fe-2S]-bound form of IscA from Thermosynechococcus elongatus (PDB 1X0G) suggests a possible Fe-S cluster binding conformation of SufA [70]. The T. elongatus holo-IscA is in an unusual asymmetric tetramer conformation containing two [2Fe-2S] clusters. Each [2Fe-2S] cluster is coordinated by Cys37, Cys101, Cys103 from the α (or α’) protomer and Cys103 from the β (or β’) protomer (see partial structure in Figure 5). In the alignment, SufA Cys114 and Cys116 are on a flexible loop in the vicinity of the IscA α Cys101 and Cys103 ligands, although the SufA loop would have to rotate slightly to bring those two ligands into the correct orientation for cluster coordination (Figure 5). By contrast, the SufA Cys50 loop is flipped out and away from the cluster binding site relative to the IscA α Cys37 ligand and would have to move approximately 14 Å to be in the same orientation. The [2Fe-2S] IscA structure suggests that some significant reordering of the flexible Cys loops within SufA would create a similar Fe-S cluster binding site. Based on the homology with IscA, it is logical to suggest that Cys116 from the other SufA monomer would provide the fourth cluster ligand. It is worth noting, however, that only the α or α’ protomers of IscA could be aligned with the SufA monomers. The unusual β and β’ protomers from the IscA tetramer are conformationally distinct from apo-SufA. They form an intertwined domain-swapped dimer where their central domains are exchanged [70]. It is possible the asymmetric IscA tetramer does not fully represent the structural changes that actually occur in holo-SufA, at least in regard to the β and β’ protomers.
4.2. In vivo roles for SufA and other ATC proteins in Fe-S cluster trafficking
While this review is focused primarily on the Suf proteins (including SufA), it is necessary to briefly consider all ATCs in parallel when evaluating in vivo roles for these proteins. In vitro and in vivo evidence suggests that SufA and IscA interface with their respective scaffolds (SufBC2D and IscU). IscA and SufA have partially overlapping functions since an ΔiscA ΔsufA double mutant in E. coli grows very poorly under aerobic conditions whereas the single deletion strains showed almost no phenotypes [71]. The ΔiscA ΔsufA mutant shows defects in the maturation of multiple [4Fe-4S] enzymes (Figure 6) [60, 71, 72]. With the exception of the NsrR nitric oxide response regulator, the [2Fe-2S] proteins tested were not defective in the double mutant strain (although there is some suggestion that E. coli NsrR may be a [4Fe-4S] protein in vivo). These results suggest the ATC proteins have a specific role in delivering [4Fe-4S] clusters. In support of this hypothesis, the eukaryotic ATC I and II homologues found in the mitochondrion have also been clearly linked to maturation of mitochondrial [4Fe-4S] clusters in vivo [69].
Figure 6.
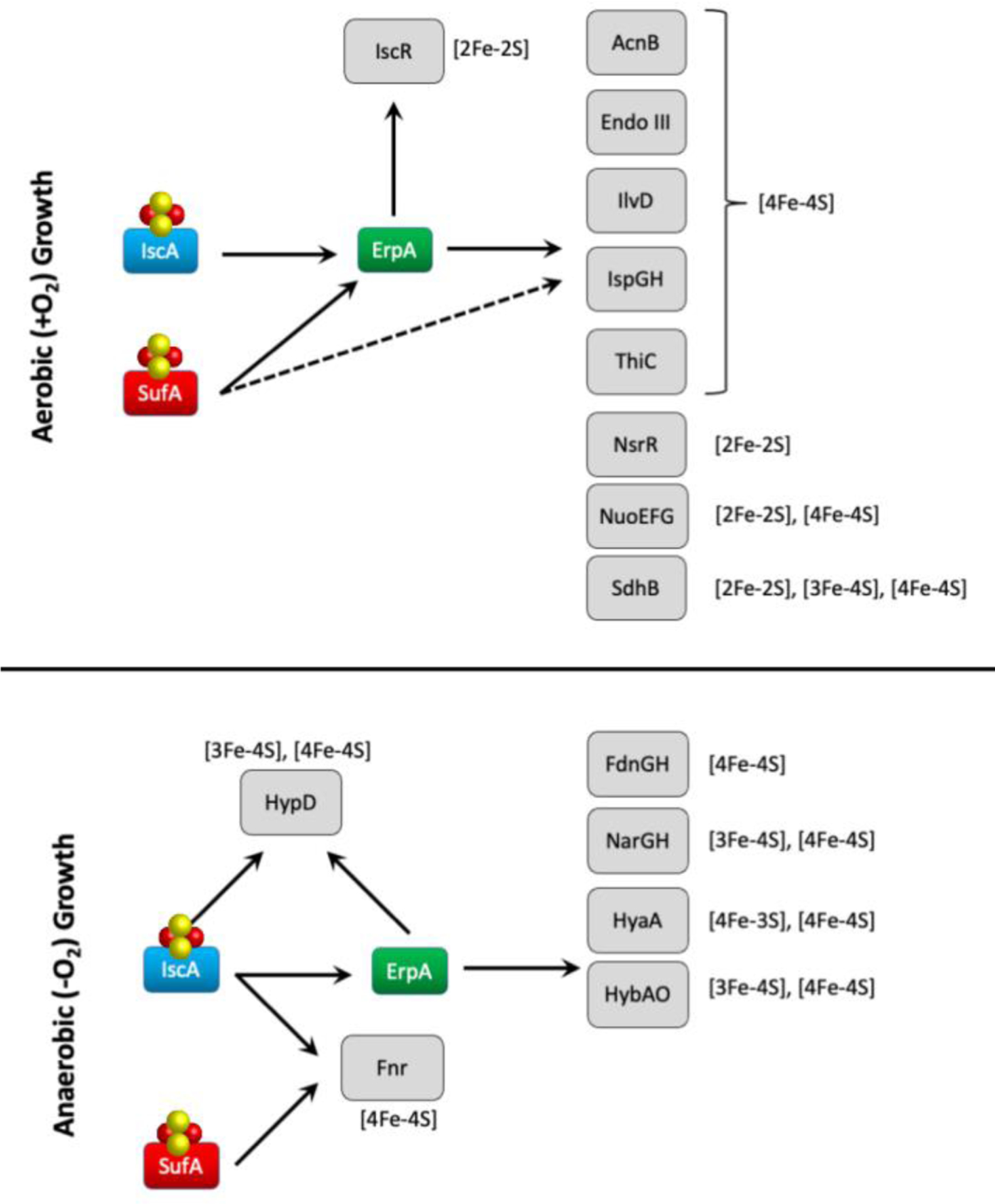
Fe-S cluster trafficking interactions between the ATC proteins and target enzymes in E. coli under aerobic (top) and anaerobic (bottom) growth conditions. Dashed lines indicate interactions that occur when SufA is highly expressed. See text for details.
In contrast to the relatively mild phenotypes of either an ΔiscA or a ΔsufA single mutation, an ΔerpA mutation was found to be lethal under both aerobic and anaerobic growth conditions that require respiration [61]. The ΔerpA phenotype appears to result from a decreased quinone pool caused by disruption of Fe-S cluster maturation of IspG, an enzyme involved in the isopentenyl diphosphate (IPP) biosynthesis pathway [61]. Since respiration using oxygen, nitrate, or fumarate as terminal electron acceptors requires ubiquinone, demethylmenaquinone, and/or menaquinone as intermediate electron carriers, the IspG deficiency is lethal under those growth conditions. The ΔerpA strain, however, is viable under anaerobic fermentative growth using glucose as the carbon source since that allows E. coli to bypass the need for the quinone pool.
Multicopy suppressor analysis of strains with combinations of ATC deletions provided a model of Fe-S cluster trafficking where the Type II ATCs IscA and SufA transfer Fe-S clusters to the Type I ErpA carrier, which then delivers the Fe-S cluster to the target metalloprotein (Figure 6) [60]. This model is supported by recent biochemical experiments showing that the ErpA protein can physically interact with both IscA and SufA with Kd values of 40±3 and 902 μM respectively [63]. These dissociation constants were measured with apo-proteins and the affinities may increase if the IscA or SufA proteins were in their Fe-S cluster forms due to the logical promotion of interactions with the downstream cluster acceptor, ErpA. While this model suggests some redundancy between IscA and SufA, the in vivo results also indicate that SufA may provide additional functionality under oxidative stress and/or be able to completely bypass ErpA for some target enzymes. All three ATC proteins show similar Fe-S cluster stability when exposed to oxygen in vitro (within experimental error) [63]. Interaction with other partner or client proteins may also influence cluster stability, as observed for the ErpA-NfuA interaction that makes the ErpA Fe-S cluster more resistant to oxygen. Therefore, it is not clear if SufA itself is more resistant to oxidative stress than IscA or if it is simply a better transfer partner for the SufS-SufE-SufBC2D cluster assembly complexes, which have been shown to be more stress resistant than Isc in vitro.
4.3. SufT, a new trafficking protein for the Suf pathway.
Many bacterial and archaeal genomes show an additional open reading frame associated with the suf operon, especially retained with sufBC genes, although it is lacking in the E. coli and B. subtilis model bacteria where Suf has been most well-studied [73]. The protein encoded by this accessory ORF has been annotated as SufT [74]. SufT is nearly entirely composed of a single DUF59 domain. DUF59 domain proteins include several members of eukaryotic Fe-S cluster biogenesis systems, including the CIA2B protein of the cytosolic iron-sulfur machinery in eukaryotes, and the HCF101 protein utilized for maturation of the photosystem I complex in the chloroplast [75–77]. The DUF59 domain contains a “DPE-X26–31-T-X2/3-C” motif where the cysteine is strictly conserved. This conserved Cys residue is highly reactive and appears to be required for function of DUF59 proteins that are involved in Fe-S cluster biogenesis [78]. Readers are referred to a recent review for more comprehensive information on the DUF59 family of proteins [79]. Recently it was demonstrated in Staphylococcus aureus and Sinorhizobium meliloti that the sufT gene is involved in Fe-S cluster biogenesis in bacteria [73, 80].
In S. aureus, deletion of sufT resulted in decreased activity of multiple Fe-S cluster metalloenzymes, including AcnA, LeuCD, and IlvD [73]. Multiple conditions which increased cellular demand for de novo Fe-S cluster biogenesis exacerbated the ΔsufT phenotype [73, 81]. While a ΔsufA ΔsufT mutant did not show any additive phenotypes, Fe-S phenotypes of the Δnfu ΔsufT strain were more severe than those observed in single deletion strains. In addition, multicopy expression of nfu was able to partially rescue the ΔsufT phenotypes in vivo [73, 81]. There is strong in vivo evidence that the Fe-S enzyme LipA, which is used for lipoic acid synthesis, is a bona fide target of SufT, especially during growth conditions that require high levels of lipoamide production [81]. These findings suggest a role for this DUF59 protein SufT in Fe-S cluster trafficking, perhaps as a part of a network of trafficking proteins in S. aureus that includes Nfu and SufA. Such a role would be consistent with the known function of DUF59 proteins in the eukaryotic cytosol and chloroplast.
In Sinorhizobium meliloti, the sufT gene is located just downstream of the sufBCDS genes and can be expressed as part of a sufBCDSTA polycistronic mRNA [82]. However, sufTA also is under the control of an internal RpoH1-dependent promoter, leading to transcriptional induction under heat shock stress [80]. Unlike deletions within sufBCDS, which are lethal, the ΔsufT mutant of S. meliloti is viable. The ΔsufT strain did show a slow growth phenotype linked to disruption of Fe-S dependent enzymes and was more sensitive to elevated temperature, acidic pH, and iron starvation stress [80]. Similarly, a recently published study shows that a ΔsufT strain of Mycobacterium smegmatis is sensitive to iron limitation, showing both reduced planktonic growth and reduced biofilm formation under those conditions [83]. Surprisingly, neither of the ΔsufT mutant strains in M. smegmatis or S. meliloti showed increased sensitivity to oxidative stress. In contrast, the S. aureus ΔsufT strain was mildly sensitive to paraquat exposure and to the shift from anaerobic fermentative to aerobic respiratory growth conditions [73]. The sum of the in vivo studies on sufT in bacteria points to a specific role for this accessory suf gene under a number of different stress conditions where Fe-S cluster demand is increased.
5. Outlook.
Clearly great strides have been made in characterizing stepwise Fe-S cluster biogenesis by the Suf pathway in bacteria. In particular, the detailed structural characterization of SufS reaction intermediates and SufS interactions with its transpersulfuration partner proteins SufE and SufU have provided us with a nearly complete picture of the sulfur mobilization step. We also now have three dimensional structures of all Suf components in E. coli, albeit the most interesting conformations still elude us (such as the Fe-S holo-forms). Despite these achievements, much remains to be learned. We have only an incomplete picture of the exact role of SufD or the SufC ATPase reaction cycle during Fe-S cluster assembly on SufB. It is feasible that SufD and/or SufC are somehow involved in iron acquisition by the SufBC2D complex, but the in vivo iron donor is not known so it is difficult to ascertain how they could assist in this step. Possibly SufD and/or SufC also assists in the transfer of the Fe-S cluster to SufA or other unknown Fe-S trafficking proteins. Once the Fe-S cluster has formed there are a number of mechanistic details still missing for the stepwise process of cluster transfer and the targeting of the Fe-S cluster to downstream target proteins. Eukaryotic Fe-S cluster trafficking pathways have a plethora of accessory targeting factors that assist with cluster delivery [8]. Possibly E. coli and other bacteria have novel targeting factors that have not yet been identified. A broader question about the Suf system arises from its phylogenetic distribution and physiological roles. It seems clear there is a stress-response role for Suf in some bacteria and a house-keeping role for Suf in other bacteria. Understanding what differentiates these two types of Suf systems is a critical question. For example, elegant genetic studies have demonstrated that the E. coli stress-response sufCDSE and the B. subtilis housekeeping sufCDSU are at least partially interchangeable whereas sufB was not [40]. Point mutations that allowed B. subtilis SufB to function in E. coli were identified, but their biochemical significance has not been determined
[40]. Clearly much work remains to determine the biochemical characteristics that differentiate stress-response Suf from housekeeping Suf in bacteria. As the interest in the Suf pathway across the phylogenetic spectrum has increased, it is likely many of the answers to these questions will be revealed in the near future.
HIGHLIGHTS.
SufS sulfur transfer requires SufE or SufS
Structural analysis of SufS reaction intermediates elucidates its mechanism
SufB scaffold function requires SufC and SufD
Iron-sulfur carrier proteins for Suf include SufA and SufT
Acknowledgements
The authors thank their lab members, past and present, for many insightful discussions. They also are thankful for the scientifically rigorous yet collegial community of researchers who have contributed to the field of Fe-S cluster biogenesis.
Funding
This work was supported by the United States National Institutes of Health (grant GM112919 to F.W.O.).
Abbreviations
- ATC
A-type carrier
- EXAFS
Extended X-ray Absorption Fine Structure
- Fe-S
iron-sulfur
- PLP
pyridoxal 5’-phosphate
- SAXS
small angle X-ray scattering
- Suf
sulfur formation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Przybyla-Toscano J, Roland M, Gaymard F, Couturier J, Rouhier N, Roles and maturation of iron-sulfur proteins in plastids, J Biol Inorg Chem, 23(4) (2018) 545–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rouault TA, The indispensable role of mammalian iron sulfur proteins in function and regulation of multiple diverse metabolic pathways, Biometals, 32(3) (2019) 343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ribbe MW, Hu Y, Hodgson KO, Hedman B, Biosynthesis of nitrogenase metalloclusters, Chem Rev, 114(8) (2014) 4063–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Brancaccio D, Gallo A, Piccioli M, Novellino E, Ciofi-Baffoni S, Banci L, [4Fe-4S] Cluster Assembly in Mitochondria and Its Impairment by Copper, J Am Chem Soc, 139(2) (2017) 719–730. [DOI] [PubMed] [Google Scholar]
- [5].Garcia-Santamarina S, Uzarska MA, Festa RA, Lill R, Thiele DJ, Cryptococcus neoformans Iron-Sulfur Protein Biogenesis Machinery Is a Novel Layer of Protection against Cu Stress, mBio, 8(5) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Imlay JA, The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium, Nat Rev Microbiol, 11(7) (2013) 443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Boyd ES, Thomas KM, Dai Y, Boyd JM, Outten FW, Interplay between oxygen and Fe-S cluster biogenesis: insights from the Suf pathway, Biochemistry, 53(37) (2014) 5834–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lill R, Freibert SA, Mechanisms of Mitochondrial Iron-Sulfur Protein Biogenesis, Annu Rev Biochem, 89 (2020) 471–499. [DOI] [PubMed] [Google Scholar]
- [9].Tsaousis AD, On the Origin of Iron/Sulfur Cluster Biosynthesis in Eukaryotes, Front Microbiol, 10 (2019) 2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Dai Y, Outten FW, The E coli SufS-SufE sulfur transfer system is more resistant to oxidative stress than IscS-IscU, FEBS Lett, 586(22) (2012) 4016–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Selbach BP, Pradhan PK, Dos Santos PC, Protected sulfur transfer reactions by the Escherichia coli Suf system, Biochemistry, 52(23) (2013) 4089–96. [DOI] [PubMed] [Google Scholar]
- [12].Albrecht AG, Peuckert F, Landmann H, Miethke M, Seubert A, Marahiel MA, Mechanistic characterization of sulfur transfer from cysteine desulfurase SufS to the iron-sulfur scaffold SufU in Bacillus subtilis, FEBS Lett, 585(3) (2011) 465–70. [DOI] [PubMed] [Google Scholar]
- [13].Tirupati B, Vey JL, Drennan CL, Bollinger JM Jr., Kinetic and structural characterization of Slr0077/SufS, the essential cysteine desulfurase from Synechocystis sp. PCC 6803, Biochemistry, 43(38) (2004) 12210–9. [DOI] [PubMed] [Google Scholar]
- [14].Blauenburg B, Mielcarek A, Altegoer F, Fage CD, Linne U, Bange G, Marahiel MA, Crystal Structure of Bacillus subtilis Cysteine Desulfurase SufS and Its Dynamic Interaction with Frataxin and Scaffold Protein SufU, PLoS One, 11(7) (2016) e0158749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mihara H, Fujii T, Kato S, Kurihara T, Hata Y, Esaki N, Structure of external aldimine of Escherichia coli CsdB, an IscS/NifS homolog: implications for its specificity toward selenocysteine, J Biochem, 131(5) (2002) 679–85. [DOI] [PubMed] [Google Scholar]
- [16].Cupp-Vickery JR, Urbina H, Vickery LE, Crystal structure of IscS, a cysteine desulfurase from Escherichia coli, J Mol Biol, 330(5) (2003) 1049–59. [DOI] [PubMed] [Google Scholar]
- [17].Fujii T, Maeda M, Mihara H, Kurihara T, Esaki N, Hata Y, Structure of a NifS homologue: X-ray structure analysis of CsdB, an Escherichia coli counterpart of mammalian selenocysteine lyase, Biochemistry, 39(6) (2000) 1263–73. [DOI] [PubMed] [Google Scholar]
- [18].Outten FW, Wood MJ, Munoz FM, Storz G, The SufE protein and the SufBCD complex enhance SufS cysteine desulfurase activity as part of a sulfur transfer pathway for Fe-S cluster assembly in Escherichia coli, J Biol Chem, 278(46) (2003) 45713–9. [DOI] [PubMed] [Google Scholar]
- [19].Blahut M, Wise CE, Bruno MR, Dong G, Makris TM, Frantom PA, Dunkle JA, Outten FW, Direct observation of intermediates in the SufS cysteine desulfurase reaction reveals functional roles of conserved active-site residues, J Biol Chem, 294(33) (2019) 12444–12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nakamura R, Hikita M, Ogawa S, Takahashi Y, Fujishiro T, Snapshots of PLP-substrate and PLP-product external aldimines as intermediates in two types of cysteine desulfurase enzymes, FEBS J, 287(6) (2020) 1138–1154. [DOI] [PubMed] [Google Scholar]
- [21].Behshad E, Bollinger JM Jr., Kinetic analysis of cysteine desulfurase CD0387 from Synechocystis sp. PCC 6803: formation of the persulfide intermediate, Biochemistry, 48(50) (2009) 12014–23. [DOI] [PubMed] [Google Scholar]
- [22].Loiseau L, Ollagnier-de-Choudens S, Nachin L, Fontecave M, Barras F, Biogenesis of Fe-S cluster by the bacterial Suf system: SufS and SufE form a new type of cysteine desulfurase, J Biol Chem, 278(40) (2003) 38352–9. [DOI] [PubMed] [Google Scholar]
- [23].Perard J, Ollagnier de Choudens S, Iron-sulfur clusters biogenesis by the SUF machinery: close to the molecular mechanism understanding, J Biol Inorg Chem, 23(4) (2018) 581–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Loiseau L, Ollagnier-de Choudens S, Lascoux D, Forest E, Fontecave M, Barras F, Analysis of the heteromeric CsdA-CsdE cysteine desulfurase, assisting Fe-S cluster biogenesis in Escherichia coli, J Biol Chem, 280(29) (2005) 26760–9. [DOI] [PubMed] [Google Scholar]
- [25].Charan M, Singh N, Kumar B, Srivastava K, Siddiqi MI, Habib S, Sulfur mobilization for Fe-S cluster assembly by the essential SUF pathway in the Plasmodium falciparum apicoplast and its inhibition, Antimicrob Agents Chemother, 58(6) (2014) 3389–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Selbach B, Earles E, Dos Santos PC, Kinetic analysis of the bisubstrate cysteine desulfurase SufS from Bacillus subtilis, Biochemistry, 49(40) (2010) 8794–802. [DOI] [PubMed] [Google Scholar]
- [27].Riboldi GP, de Oliveira JS, Frazzon J, Enterococcus faecalis SufU scaffold protein enhances SufS desulfurase activity by acquiring sulfur from its cysteine-153, Biochim Biophys Acta, 1814(12) (2011) 1910–8. [DOI] [PubMed] [Google Scholar]
- [28].Ye H, Abdel-Ghany SE, Anderson TD, Pilon-Smits EA, Pilon M, CpSufE activates the cysteine desulfurase CpNifS for chloroplastic Fe-S cluster formation, J Biol Chem, 281(13) (2006) 8958–69. [DOI] [PubMed] [Google Scholar]
- [29].Ollagnier-de-Choudens NMM,S, Sanakis Y Abdel-Ghany SE, Rousset C, Ye H, Fontecave M, Pilon-Smits EA, Pilon M, Characterization of Arabidopsis thaliana SufE2 and SufE3: functions in chloroplast iron-sulfur cluster assembly and Nad synthesis, J Biol Chem, 282(25) (2007) 18254–64. [DOI] [PubMed] [Google Scholar]
- [30].Goldsmith-Fischman S, Kuzin A, Edstrom WC, Benach J, Shastry R, Xiao R, Acton TB, Honig B, Montelione GT, Hunt JF, The SufE sulfur-acceptor protein contains a conserved core structure that mediates interdomain interactions in a variety of redox protein complexes, J Mol Biol, 344(2) (2004) 549–65. [DOI] [PubMed] [Google Scholar]
- [31].Layer G, Gaddam SA, Ayala-Castro CN, Ollagnier-de Choudens S, Lascoux D, Fontecave M, Outten FW, SufE transfers sulfur from SufS to SufB for iron-sulfur cluster assembly, J Biol Chem, 282(18) (2007) 13342–50. [DOI] [PubMed] [Google Scholar]
- [32].Kim D, Singh H, Dai Y, Dong G, Busenlehner LS, Outten FW, Frantom PA, Changes in Protein Dynamics in Escherichia coli SufS Reveal a Possible Conserved Regulatory Mechanism in Type II Cysteine Desulfurase Systems, Biochemistry, 57(35) (2018) 5210–5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Singh H, Dai Y, Outten FW, Busenlehner LS, Escherichia coli SufE sulfur transfer protein modulates the SufS cysteine desulfurase through allosteric conformational dynamics, J Biol Chem, 288(51) (2013) 36189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kim S, Park S, Structural changes during cysteine desulfurase CsdA and sulfur acceptor CsdE interactions provide insight into the trans-persulfuration, J Biol Chem, 288(38) (2013) 27172–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Dunkle JA, Bruno MR, Outten FW, Frantom PA, Structural Evidence for Dimer-Interface-Driven Regulation of the Type II Cysteine Desulfurase, SufS, Biochemistry, 58(6) (2019) 687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Dai Y, Kim D, Dong G, Busenlehner LS, Frantom PA, Outten FW, SufE D74R Substitution Alters Active Site Loop Dynamics To Further Enhance SufE Interaction with the SufS Cysteine Desulfurase, Biochemistry, 54(31) (2015) 4824–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Riboldi GP, Verli H, Frazzon J, Structural studies of the Enterococcus faecalis SufU [Fe-S] cluster protein, BMC Biochem, 10 (2009) 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Albrecht AG, Netz DJ, Miethke M, Pierik AJ, Burghaus O, Peuckert F, Lill R, Marahiel MA, SufU is an essential iron-sulfur cluster scaffold protein in Bacillus subtilis, J Bacteriol, 192(6) (2010) 1643–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Selbach BP, Chung AH, Scott AD, George SJ, Cramer SP, Dos Santos PC, Fe-S cluster biogenesis in Gram-positive bacteria: SufU is a zinc-dependent sulfur transfer protein, Biochemistry, 53(1) (2014) 152–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yokoyama N, Nonaka C, Ohashi Y, Shioda M, Terahata T, Chen W, Sakamoto K, Maruyama C, Saito T, Yuda E, Tanaka N, Fujishiro T, Kuzuyama T, Asai K, Takahashi Y, Distinct roles for U-type proteins in iron-sulfur cluster biosynthesis revealed by genetic analysis of the Bacillus subtilis sufCDSUB operon, Mol Microbiol, 107(6) (2018) 688–703. [DOI] [PubMed] [Google Scholar]
- [41].Fujishiro T, Terahata T, Kunichika K, Yokoyama N, Maruyama C, Asai K, Takahashi Y, Zinc-Ligand Swapping Mediated Complex Formation and Sulfur Transfer between SufS and SufU for Iron-Sulfur Cluster Biogenesis in Bacillus subtilis, J Am Chem Soc, 139(51) (2017) 18464–18467. [DOI] [PubMed] [Google Scholar]
- [42].Garcia PS, Gribaldo S, Py B, Barras F, The SUF system: an ABC ATPase-dependent protein complex with a role in Fe-S cluster biogenesis, Res Microbiol, 170(8) (2019) 426–434. [DOI] [PubMed] [Google Scholar]
- [43].Yuda E, Tanaka N, Fujishiro T, Yokoyama N, Hirabayashi K, Fukuyama K, Wada K, Takahashi Y, Mapping the key residues of SufB and SufD essential for biosynthesis of iron-sulfur clusters, Sci Rep, 7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Hirabayashi K, Yuda E, Tanaka N, Katayama S, Iwasaki K, Matsumoto T, Kurisu G, Outten FW, Fukuyama K, Takahashi Y, Wada K, Functional Dynamics Revealed by the Structure of the SufBCD Complex, a Novel ATP-binding Cassette (ABC) Protein That Serves as a Scaffold for Iron-Sulfur Cluster Biogenesis, J Biol Chem, 290(50) (2015) 29717–29731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Saini A, Mapolelo DT, Chahal HK, Johnson MK, Outten FW, SufD and SufC ATPase Activity Are Required for Iron Acquisition during in Vivo Fe-S Cluster Formation on SufB, Biochemistry, 49(43) (2010) 9402–9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wada K, Sumi N, Nagai R, Iwasaki K, Sato T, Suzuki K, Hasegawa Y, Kitaoka S, Minami Y, Outten FW, Takahashi Y, Fukuyama K, Molecular Dynamism of Fe-S Cluster Biosynthesis Implicated by the Structure of the SufC2-SufD2 Complex, J Mol Biol, 387(1) (2009) 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Blanc B, Clemancey M, Latour JM, Fontecave M, de Choudens SO, Molecular Investigation of Iron Sulfur Cluster Assembly Scaffolds under Stress, Biochemistry, 53(50) (2014) 7867–7869. [DOI] [PubMed] [Google Scholar]
- [48].Wollers S, Layer G, Garcia-Serres R, Signor L, Clemancey M, Latour JM, Fontecave M, de Choudens SO, Iron-Sulfur (Fe-S) Cluster Assembly: the SufBCD complex is a new type of Fe-S scaffold with a flavin redox cofactor, J Biol Chem, 285(30) (2010) 23329–23339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Chahal HK, Dai YY, Saini A, Ayala-Castro C, Outten FW, The SufBCD Fe-S Scaffold Complex Interacts with SufA for Fe-S Cluster Transfer, Biochemistry, 48(44) (2009) 10644–10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Chahal HK, Outten FW, Separate Fe - S scaffold and carrier functions for SufB(2)C(2) and SufA during in vitro maturation of 2Fe-2S Fdx, J Biol Inorg Chem, 116 (2012) 126–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hollenstein K, Dawson RJP, Locher KP, Structure and mechanism of ABC transporter proteins, Curr. Opin. Struct. Biol, 17(4) (2007) 412–418. [DOI] [PubMed] [Google Scholar]
- [52].Nachin L, El Hassouni M, Loiseau L, Expert D, Barras F, SoxR-dependent response to oxidative stress and virulence of Erwinia chrysanthemi: the key role of SufC, an orphan ABC ATPase, Mol Microbiol, 39(4) (2001) 960–972. [DOI] [PubMed] [Google Scholar]
- [53].Nachin L, Loiseau L, Expert D, Barras F, SufC: an unorthodox cytoplasmic ABC/ATPase required for Fe-S biogenesis under oxidative stress, EMBO J, 22(3) (2003) 427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kitaoka S, Wada K, Hasegawa Y, Minami Y, Fukuyama K, Takahashi Y, Crystal structure of Escherichia coli SufC, an ABC-type ATPase component of the SUF iron-sulfur cluster assembly machinery, FEBS Lett, 580(1) (2006) 137–143. [DOI] [PubMed] [Google Scholar]
- [55].Outten FW, Djaman O, Storz G, A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli, Mol Microbiol, 52(3) (2004) 861–872. [DOI] [PubMed] [Google Scholar]
- [56].Petrovic A, Davis CT, Rangachari K, Clough B, Wilson RJM, Eccleston JF, Hydrodynamic characterization of the SufBC and SufCD complexes and their interaction with fluorescent adenosine nucleotides, Protein Sci, 17(7) (2008) 1264–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Badger J, Sauder JM, Adams JM, Antonysamy S, Bain K, Bergseid MG, Buchanan SG, Buchanan MD, Batiyenko Y, Christopher JA, Emtage S, Eroshkina A, Feil I, Furlong EB, Gajiwala KS, Gao X, He D, Hendle J, Huber A, Hoda K, Kearins P, Kissinger C, Laubert B, Lewis HA, Lin J, Loomis K, Lorimer D, Louie G, Maletic M, Marsh CD, Miller I, Molinari J, Muller-Dieckmann HJ, Newman JM, Noland BW, Pagarigan B, Park F, Peat TS, Post KW, Radojicic S, Ramos A, Romero R, Rutter ME, Sanderson WE, Schwinn KD, Tresser J, Winhoven J, Wright TA, Wu L, Xu J, Harris TJR, Structural analysis of a set of proteins resulting from a bacterial genomics project, Proteins, 60(4) (2005) 787–796. [DOI] [PubMed] [Google Scholar]
- [58].Patzer SI, Hantke K, SufS is a NifS-like protein, and SufD is necessary for stability of the 2Fe-2S FhuF protein in Escherichia coli, J Bacteriol, 181(10) (1999) 3307–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Expert D, Boughammoura A, Franza T, Siderophore-controlled Iron Assimilation in the Enterobacterium Erwinia chrysanthemi evidence for the involvement of bacterioferritin and the suf iron-sulfur cluster assembly machinery, J Biol Chem, 283(52) (2008) 36564–36572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Vinella D, Brochier-Armanet C, Loiseau L, Talla E, Barras F, Iron-sulfur (Fe/S) protein biogenesis: phylogenomic and genetic studies of A-type carriers, PLoS Genet, 5(5) (2009) e1000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Loiseau L, Gerez C, Bekker M, Ollagnier-de Choudens S, Py B, Sanakis Y, Teixeira de Mattos J, Fontecave M, Barras F, ErpA, an iron-sulfur (Fe-S) protein of the A-type essential for respiratory metabolism in Escherichia coli, Proc Natl Acad Sci U S A, 104(34) (2007) 13626–13631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Gupta V, Sendra M, Naik SG, Chahal HK, Huynh BH, Outten FW, Fontecave M, Ollagnier de Choudens S, Native Escherichia coli SufA, coexpressed with SufBCDSE, purifies as a [2Fe-2S] protein and acts as an Fe-S transporter to Fe-S target enzymes, Journal of the American Chemical Society, 131(17) (2009) 6149–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Py B, Gerez C, Huguenot A, Vidaud C, Fontecave M, Ollagnier de Choudens S, Barras F, The ErpA/NfuA complex builds an oxidation-resistant Fe-S cluster delivery pathway, J Biol Chem, 293(20) (2018) 7689–7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Mapolelo DT, Zhang B, Randeniya S, Albetel AN, Li H, Couturier J, Outten CE, Rouhier N, Johnson MK, Monothiol glutaredoxins and A-type proteins: partners in Fe-S cluster trafficking, Dalton Trans, 42(9) (2013) 3107–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Mapolelo DT, Zhang B, Naik SG, Huynh BH, Johnson MK, Spectroscopic and Functional Characterization of Iron-Sulfur Cluster-Bound Forms of Azotobacter vinelandii NifIscA, Biochemistry, 51(41) (2012) 8071–8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Landry AP, Cheng Z, Ding H, Iron binding activity is essential for the function of IscA in iron-sulphur cluster biogenesis, Dalton transactions (Cambridge, England : 2003), 42(9) (2013) 3100–3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Ding B, Smith ES, Ding H, Mobilization of the iron centre in IscA for the iron-sulphur cluster assembly in IscU, Biochem J, 389(Pt 3) (2005) 797–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Wada K, Hasegawa Y, Gong Z, Minami Y, Fukuyama K, Takahashi Y, Crystal structure of Escherichia coli SufA involved in biosynthesis of iron-sulfur clusters: Implications for a functional dimer, FEBS Lett, 579(29) (2005) 6543–6548. [DOI] [PubMed] [Google Scholar]
- [69].Jensen LT, Culotta VC, Role of Saccharomyces cerevisiae ISA1 and ISA2 in iron homeostasis, Mol Cell Biol, 20(11) (2000) 3918–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Morimoto K, Yamashita E, Kondou Y, Lee SJ, Arisaka F, Tsukihara T, Nakai M, The asymmetric IscA homodimer with an exposed [2Fe-2S] cluster suggests the structural basis of the Fe-S cluster biosynthetic scaffold, J Mol Biol, 360(1) (2006) 117–32. [DOI] [PubMed] [Google Scholar]
- [71].Lu J, Yang J, Tan G, Ding H, Complementary roles of SufA and IscA in the biogenesis of iron-sulfur clusters in Escherichia coli, Biochem. J, 409(2) (2008) 535–543. [DOI] [PubMed] [Google Scholar]
- [72].Vinella D, Loiseau L, Ollagnier de Choudens S, Fontecave M, Barras F, In vivo [Fe-S] cluster acquisition by IscR and NsrR, two stress regulators in Escherichia coli, Mol Microbiol, 87(3) (2013) 493–508. [DOI] [PubMed] [Google Scholar]
- [73].Mashruwala AA, Bhatt S, Poudel S, Boyd ES, Boyd JM, The DUF59 Containing Protein SufT Is Involved in the Maturation of Iron-Sulfur (FeS) Proteins during Conditions of High FeS Cofactor Demand in Staphylococcus aureus, PLoS Genet, 12(8) (2016) e1006233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Tsaousis AD, Gentekaki E, Eme L, Gaston D, Roger AJ, Evolution of the cytosolic iron-sulfur cluster assembly machinery in Blastocystis species and other microbial eukaryotes, Eukaryot Cell, 13(1) (2014) 143–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Stehling O, Vashisht AA, Mascarenhas J, Jonsson ZO, Sharma T, Netz DJ, Pierik AJ, Wohlschlegel JA, Lill R, MMS19 assembles iron-sulfur proteins required for DNA metabolism and genomic integrity, Science, 337(6091) (2012) 195–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Stehling O, Mascarenhas J, Vashisht AA, Sheftel AD, Niggemeyer B, Rosser R, Pierik AJ, Wohlschlegel JA, Lill R, Human CIA2A-FAM96A and CIA2B-FAM96B integrate iron homeostasis and maturation of different subsets of cytosolic-nuclear iron-sulfur proteins, Cell Metab, 18(2) (2013) 187–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Schwenkert S, Netz DJ, Frazzon J, Pierik AJ, Bill E, Gross J, Lill R, Meurer J, Chloroplast HCF101 is a scaffold protein for [4Fe-4S] cluster assembly, Biochem J, 425(1) (2009) 207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, Bachovchin DA, Mowen K, Baker D, Cravatt BF, Quantitative reactivity profiling predicts functional cysteines in proteomes, Nature, 468(7325) (2010) 790–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Mashruwala AA, Boyd JM, Investigating the role(s) of SufT and the domain of unknown function 59 (DUF59) in the maturation of iron-sulfur proteins, Curr Genet, 64(1) (2018) 9–16. [DOI] [PubMed] [Google Scholar]
- [80].Sasaki S, Minamisawa K, Mitsui H, A Sinorhizobium meliloti RpoH-Regulated Gene Is Involved in Iron-Sulfur Protein Metabolism and Effective Plant Symbiosis under Intrinsic Iron Limitation, J Bacteriol, 198(17) (2016) 2297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Mashruwala AA, Roberts CA, Bhatt S, May KL, Carroll RK, Shaw LN, Boyd JM, Staphylococcus aureus SufT: an essential iron-sulphur cluster assembly factor in cells experiencing a high-demand for lipoic acid, Mol Microbiol, 102(6) (2016) 1099–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Chao TC, Buhrmester J, Hansmeier N, Puhler A, Weidner S, Role of the regulatory gene rirA in the transcriptional response of Sinorhizobium meliloti to iron limitation, Appl Environ Microbiol, 71(10) (2005) 5969–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Tamuhla T, Joubert L, Willemse D, Williams MJ, SufT is required for growth of Mycobacterium smegmatis under iron limiting conditions, Microbiology, 166(3) (2020) 296–305. [DOI] [PubMed] [Google Scholar]


