Abstract
microRNAs are powerful regulators of growth, development, and stress responses in plants. The Arabidopsis thaliana microRNA miR167 was previously found to regulate diverse processes including flower development, root development, and response to osmotic stress by controlling the patterns of expression of its target genes AUXIN RESPONSE FACTOR 6 (ARF6), ARF8, and IAA‐Ala RESISTANT 3. Here, we report that miR167 also modulates defense against pathogens through ARF6 and ARF8. miR167 is differentially expressed in response to the bacterial pathogen Pseudomonas syringae, and overexpression of miR167 confers very high levels of resistance. This resistance appears to be due to suppression of auxin responses and is partially dependent upon salicylic acid signaling, and also depends upon altered stomatal behavior in these plants. Closure of stomata upon the detection of P. syringae is an important aspect of the basal defense response, as it prevents bacterial cells from entering the leaf interior and causing infection. Plants overexpressing miR167 constitutively maintain small stomatal apertures, resulting in very high resistance when the pathogen is inoculated onto the leaf surface. Additionally, the systemic acquired resistance (SAR) response is severely compromised in plants overexpressing miR167, in agreement with previous work showing that the activation of SAR requires intact auxin signaling responses. This work highlights a new role for miR167, and also emphasizes the importance of hormonal balance in short‐ and long‐term defense and of stomata as an initial barrier to pathogen entry.
Keywords: auxin (AUX), biotic stress, hormone signaling, microRNA, plant defense response, salicylic acid (SA)
1. INTRODUCTION
Plants are constantly exposed to microbes and have evolved complex and multilayered defense strategies to prevent infection. These include the basal defense mechanisms triggered by plant cell recognition of microbe‐associated molecular patterns (MAMPs), resulting in responses such as the accumulation of salicylic acid (SA) and reactive oxygen species (ROS), deposition of callose to strengthen cell walls, activation of pathogenesis‐related (PR), and other defense genes, and production of antimicrobial compounds (Chisholm et al., 2006; Nimchuk et al., 2003). Alternatively, plant resistance (R) proteins may recognize the activity of specific pathogenic virulence effectors, and this “gene‐for‐gene” interaction between R proteins and their cognate virulence effectors triggers a second (much stronger) defense system called effector‐triggered immunity (ETI) which includes the hypersensitive response (HR), a form of programmed cell death induced at the site of infection which leads to death of infected host cells and the pathogen (Chisholm et al., 2006; Nimchuk et al., 2003). Cell death that results from HR or disease leads to the activation of systemic acquired resistance (SAR), a broad‐spectrum and long‐lasting resistance that is characterized by the accumulation of SA and priming of defense genes in distal, uninfected tissues (Durrant & Dong, 2004).
Many of the defense mechanisms employed during basal defense and ETI are controlled by phytohormones, particularly SA, jasmonic acid (JA), and ethylene (ET). SA is a master regulator of defense against biotrophic and hemibiotrophic pathogens and is also required for the establishment of SAR (Durrant & Dong, 2004; Kunkel & Brooks, 2002). In contrast to SA, JA and ET mainly regulate resistance against necrotrophic pathogens which kill the host plant, and herbivorous insects. Because the different strategies of biotrophic versus necrotrophic pathogens require different approaches for defense, SA and JA/ET responses are largely mutually antagonistic and the balance between different hormone signaling pathways plays a large role in the strength of the defense response (Kunkel & Brooks, 2002; Robert‐Seilaniantz et al., 2011). Bacterial pathogens such as P. syringae have evolved ways to take advantage of the antagonistic effects of JA on SA, and use virulence effectors such as coronatine, a JA‐Ile mimic, to suppress SA responses (Nomura et al., 2005; Zheng et al., 2012).
Studies in recent years have also revealed an important role for the hormone auxin in pathogenesis. In general, auxin responses are detrimental to defense against biotrophic pathogens and tend to modify host physiology in ways that aid pathogen growth (Kazan & Manners, 2009). As with JA, mutual antagonism has been observed between SA and auxin signaling pathways, and auxin also promotes disease through a pathway independent of its effect on SA signaling (Kazan & Manners, 2009; Mutka et al., 2013; Park et al., 2007; Wang et al., 2007). P. syringae has evolved mechanisms to exploit the effects of auxin to cause disease; for example, the P. syringae type III effector AvrRpt2 enhances auxin production and signaling in infected plants to promote virulence (Chen et al., 2007; Cui et al., 2013).
Numerous studies have shown that various types of small noncoding RNAs play important roles in disease resistance (reviewed in Ruiz‐Ferrer & Voinnet, 2009; Padmanabhan et al., 2009; Katiyar‐Agarwal & Jin, 2010; Seo et al., 2013; Staiger et al., 2013; Gupta et al., 2014, and Yang & Huang, 2014, and Rose et al., 2019). One type of small RNAs, microRNAs (miRNA), are short, 21 to 24‐nucleotide RNAs that regulate gene expression at the post‐transcriptional level by binding to their target mRNAs by complementary base pairing and directing cleavage or preventing the translation of those targets (Bartel, 2004). The first miRNA shown to play a role in plant defense was miR393, which is induced by the MAMP flagellin and silences auxin receptor genes, thereby suppressing auxin responses during infection and preventing them from antagonizing SA signaling (Navarro et al., 2006). miR393 also triggers a second response by which metabolic pathways are redirected to produce antimicrobial compounds that are most effective against biotrophic pathogens (Robert‐Seilaniantz et al., 2011). Other defense‐associated small RNAs such as nat‐siRNAATGB2, AtlsiRNA‐1 miR472, miR400, and miR398 regulate components of defense signaling pathways or aid in the management of responses such as the burst of ROS (Katiyar‐Agarwal et al., 2006, Katiyar‐Agarwal et al., 2007, Jagadeeswaran et al., 2009, Li et al., 2010, Boccara et al., 2014, Park et al., 2014). These examples serve to illustrate the variety in small RNA species and their target genes that contribute to defense against P. syringae by affecting not only specific defense functions but also hormone responses and metabolic pathways.
The microRNA miR167 is an important regulator of auxin‐mediated development. It has been shown to target the mRNAs for Auxin Response Factor6 (ARF6) and Auxin Response Factor8 (ARF8), two members of a large family of transcription factors that direct gene expression in response to auxin (Guilfoyle & Hagen, 2007; Wu et al., 2006). ARF6 and ARF8 regulate the maturation of both male and female floral organs, and removal of these proteins either in arf6 arf8 double mutants or in plants overexpressing miR167 results in sterility (Nagpal et al., 2005; Wu et al., 2006). arf6 arf8 plants have impaired responses to exogenous auxin and do not produce detectable levels of JA in flowers (which is critical for pollen release) (Nagpal et al., 2005). Ectopic expression of ARF6 and ARF8 transcripts that are immune to regulation by miR167 also causes floral defects and sterility (Wu et al., 2006). Therefore, miR167 is essential for directing the pattern of ARF6 and ARF8 expression in each floral organ, thereby ensuring that the correct auxin and JA responses occur. ARF8 has also been shown to affect hypocotyl and root growth in seedlings through the regulation of auxin levels, and miR167 regulation of ARF6 and ARF8 is involved in adventitious rooting (a process regulated by auxin) and lateral root growth in response to nitrogen (Gifford et al., 2008; Gutierrez et al., 2009; Tian et al., 2004).
More recently, IAA‐Ala‐Resistant3 (IAR3) was identified as a third target gene of miR167 (Kinoshita et al., 2012). IAR3 encodes an indole‐3‐acetic acid (IAA)‐Ala hydrolase, which releases bioactive auxin (IAA) from the inactive IAA‐Ala storage conjugate (Davies et al., 1999). Under conditions of osmotic stress, miR167 expression is reduced, allowing higher expression of IAR3 and increased levels of free IAA which then drives adaptive changes in root architecture (Kinoshita et al., 2012).
Given the importance of hormone signaling networks during pathogen defense, it is unsurprising that miR167 has been shown to be differentially expressed in response to a number of pathogens, including bacteria, viruses, and cyst nematodes (Fahlgren et al., 2007; Hewezi et al., 2008; Feng et al., 2009; Zhang et al., 2011; Gupta et al., 2012; Gao et al., 2013). In this study, we present evidence that miR167 is differentially expressed in response to P. syringae and modulates resistance based on its function in regulating auxin signaling. Plants overexpressing miR167 are highly resistant to P. syringae, possibly due to reduced auxin responses and to their tendency to maintain stomata in a relatively closed state, thus preventing pathogens from entering the leaf interior and establishing an infection.
2. MATERIALS AND METHODS
2.1. Plant materials and growth conditions
All Arabidopsis thaliana plants used were of the Col‐0 ecotype. Plants were grown in soil (Metro‐Mix 360, Sun Gro Horticulture) or on plates containing Murashige and Skoog media supplemented with 1% sucrose and 0.8% agar. Growth chambers were kept at 25/23°C (day/night), 50%–60% relative humidity, and a photosynthetic photon flux density (PPFD) of 100–150 µmol/m2 s–1 with a 10 hr photoperiod. The P35S::MIR167a, mARF6, and mARF8 constructs (described previously in Wu et al., 2006) were transformed into Col‐0, DR5::GUS, eds16‐1, or gdg1 plants via the floral dip method (Clough & Bent, 1998). T1 generation transformants were selected on soil via resistance to BASTA (P35S::MIR167a and mARF6) or on plates supplemented with 50 mg/ml kanamycin (mARF8). Due to defects in floral development caused by the P35S::MIR167a, mARF6, and mARF8 transgenes, the resulting transgenic plants are sterile and thus independent transgenic lines could not be established. Instead, all experiments were performed on populations of independent T1 transformants that were confirmed by selection as described above as well as the confirmation of sterility after plants reached the flowering stage. arf6‐2 arf8‐3, eds16‐1, and gdg1 mutants were isolated and described previously (Jagadeeswaran et al., 2007; Nagpal et al., 2005; Wildermuth et al., 2001).
2.2. Plant inoculation and measurement of in planta bacterial growth
Growth of Pseudomonas syringae strains and infiltration of plants was performed essentially as described (Devadas et al., 2002). For spray inoculation, a bacterial suspension of 5 × 108 cfu/mL was added to a small aerosol spray bottle and Silwet L‐77 (OSi Specialties) was added to a concentration of 0.02%. After spraying, trays were covered with a clear dome for 24 hr to maintain high humidity, then domes were cracked open to allow air circulation for the remainder of the experiment. Three leaf punches (approximately 0.3 cm2 each) were taken per plant using a standard paper hole punch and bacteria were quantified as described (Tornero & Dangl, 2001). For growth assays including auxin cotreatment, the auxin analog NAA (1‐Naphthaleneacetic acid) was added into the inoculation solution at the indicated final concentration. All experiments were performed for a minimum of three times.
Data from pathogen growth assays performed using spray inoculation were not normally distributed and often had unequal variances, therefore, standard parametric statistical analyses could not be used. The nonparametric Kruskal‐Wallis and Wilcoxon rank‐sum tests were, therefore, used to test for statistically significant differences in pathogen growth among genotypes. All statistical analysis was performed using R software.
2.3. Northern blot analysis and RT‐PCR
Tissue samples were flash‐frozen in liquid nitrogen upon collection. Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Small RNA gel blot analysis was performed as described (Dhar et al., 2020; Wu et al., 2006). Antisense probes used for hybridization were 5′‐TAGATCATGCTGGCAGCTTCA‐3′ for miR167 and 5′‐CTCGATTTATGCGTGTCATCCTTGC‐3′ for U6 snRNA. Differences in expression were quantified using a phosphorimager and ImageQuant software (GE Healthcare).
For semi‐quantitative RT‐PCR, 2 µg of total RNA for each sample was reverse transcribed with Superscript III Reverse Transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Resulting cDNA was diluted 1:2 with ddH2O and 1 µl cDNA was used in PCR reactions. PCR was performed by amplifying samples for the appropriate number of cycles ranging from 26 to 30 before reaching the stationary phase. Primers used for amplification are listed in Table S1.
2.4. Histochemical assay for β‐glucuronidase activity
After appropriate treatments, tissue samples were incubated overnight at 37°C in GUS assay buffer (50 mM Phosphate buffer (Na3PO4) pH 7.2, 0.5 mM K3[Fe(CN)6], 0.5 mM K4[Fe(CN)6], 2 mM 5‐bromo‐4‐chloro‐3‐indoyl‐beta‐D‐glucuronic acid (X‐gluc)). After overnight staining, samples were incubated at 37°C in 70% ethanol for 2–3 days, changing to fresh 70% ethanol each day, to remove chlorophyll for better visualization of blue color resulting from β‐glucuronidase (GUS) activity.
2.5. Stomatal aperture measurement
Treatment of whole leaves of Col‐0 and P35S::MIR167 plants and measurement of stomatal apertures were performed as described previously (Chitrakar & Melotto, 2010). Briefly, plants were kept under light for at least 3 hr to induce the opening of stomata prior to the start of the experiment. Whole leaves of each genotype were detached and placed into Petri plates, and water or Pst DC3000 at a titer of 5 × 108 cfu/ml (suspended in water) was pipetted under the leaves so that the entire underside of each leaf was in contact with liquid. Plates were then placed back under normal growth conditions. At 1 and 4 hr after the start of the experiment, leaves were photographed for the measurement of stomatal apertures.
For measurement of guard cell length, dark‐adapted plants were used to ensure that stomata were uniformly closed. For the measurement of stomatal density, the number of stomata was counted in an area of 0.050 mm2 for each sample.
Stomata were viewed and photographed using a Nikon Eclipse E400 compound microscope and SPOT 5.0 software. Aperture and length measurements were taken using ImageJ Software (NIH, USA), and data were analyzed using Student's t test in Microsoft Excel to test for statistically significant differences between WT and transgenic plants.
3. RESULTS
3.1. miR167 is differentially expressed in response to bacterial pathogens
As part of a previous project, we performed whole‐genome microarray analysis to identify transcriptome changes in response to the avirulent bacterial pathogen Pseudomonas syringae pv. tomato DC3000 expressing avrRpm1 [Pst DC3000 (avrRpm1)]. Interestingly, miR167 was suppressed in response to the pathogen treatment (S. Maqbool, personal communication). To confirm the results of our microarray experiment, we performed small RNA northern blot analysis to examine the expression levels of miR167 in response to several bacterial pathogens. Five‐week‐old wild‐type (WT) Col‐0 plants were treated with 10mM MgSO4 (infiltration control) and Pst DC3000 (avrRpm1), as well as other avirulent strains expressing avrRpt2 or avrRps4 and the virulent strain Pst DC3000. We observed suppression of miR167 by 6 hr post‐infiltration (hpi) in response to Pst DC3000 (avrRpm1), and by 12 hpi in response to Pst DC3000 (avrRpt2) and Pst DC3000 (avrRps4) (Figure 1a). In general, suppression of miR167 was correlated with the timing and intensity of the visible HR response: leaves treated with Pst DC3000 (avrRpm1) are fully collapsed by 6 hpi, while visible HR does not occur until 8 hpi with Pst DC3000 (avrRpt2) or 10–12 hpi with Pst DC3000 (avrRps4). We did not observe changes in miR167 expression in response to virulent Pst DC3000 (Figure 1b).
FIGURE 1.
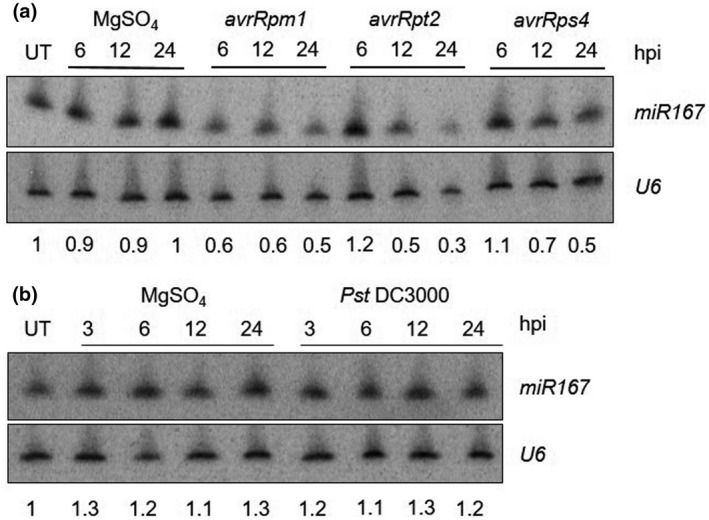
miR167 is differentially expressed in response to P. syringae. Northern blot analyses of miR167 expression in response to Pst DC3000 carrying different avirulence factors (a) or virulent Pst DC3000 (b). Five‐week‐old wild‐type Col‐0 plants were left untreated (UT) or infiltrated with the indicated P. syringae strains at a titer of 5 × 107 cfu/ml or MgSO4 (control). Tissue samples were collected from three independent plants and pooled for RNA isolation at the indicated hours post infiltration (hpi) and used for small RNA northern blotting. U6 snRNA is included as a loading control. Signal intensity relative to untreated control was quantified using a phosphorimager. Numbers beneath lanes indicate relative transcript levels normalized to loading control. These experiments were repeated four times with similar results
3.2. miR167 promotes resistance against P. syringae
miR167 is a conserved microRNA that regulates flower and root development and response to osmotic stress (Gifford et al., 2008; Kinoshita et al., 2012; Wu et al., 2006), but to date, the specific function of miR167a in pathogen defense has not been experimentally determined. To determine whether it plays a role in pathogen defense, we tested the growth of Pst DC3000 in transgenic plants overexpressing MIR167a under the control of the Cauliflower Mosaic Virus 35S promoter (Wu et al., 2006). These plants produce high levels of miR167 and have morphological phenotypes very similar to those of arf6‐2 arf8‐3 double mutant plants. As overexpression of miR167a causes floral defects leading to sterility, stable transgenic lines for P35S::MIR167a could not be generated and all experiments were performed on T1 generation transformants. For each experiment, populations of transgenic T1 plants were first identified via selection for resistance to BASTA (selection marker linked to the P35S:MIR167a T‐DNA). T1 plants overexpressing miR167 could also easily be identified based on their slightly smaller size, curled leaves, and broad petioles (Figure 2a), and this enabled us to perform pathogen growth assays before plants reached the flowering stage. Small RNA northern blots of RNA from representative individual plants confirmed that this was a reliable method to identify plants overexpressing miR167 (Figure S1a), as all plants selected for their curled leaves and leafy petioles did, indeed, overexpress miR167. In addition, we kept all plants until the flowering stage to confirm that they also displayed arrested flower development and sterility known to be caused by overexpression of miR167.
FIGURE 2.

Growth of P. syringae after infiltration into plants overexpressing miR167a. (a) Phenotype of P35S::MIR167a plants. Five‐week‐old Col‐0 and P35S::MIR167a plants were photographed. White arrows indicate leaves with curled shape and leafy petioles that were used to identify P35S::MIR167a plants during vegetative growth. Scale bars indicate 2 cm. (b) and (c) Five‐week‐old Col‐0 or P35S::MIR167 plants were infiltrated with Pst DC3000 (b) or Pst DC3000 carrying avirulence factors (avrRpm1) or (avrRpt2) (c) at a titer of 5 x 105 cfu/ml and bacterial growth was determined after 0 and 3 days (b) or 3 days only (c). Bars represent mean + SEM of pathogen growth. For all graphs, asterisks indicate significantly different growth from Col‐0 at p < .05 using the Wilcoxon rank‐sum test. Experiments were performed on eight to ten plants per genotype and each experiment was repeated three to five times with similar results. Results of one such experiment are shown. As P35S::MIR167a plants are sterile, a population of independent T1 transgenic plants was used rather than stable transgenic lines
When infiltrated with virulent Pst DC3000, P35S::MIR167a plants were two to fivefold less susceptible to infection than wild‐type plants (Figure 2b). We also tested the growth of two avirulent strains in these plants. We did not observe a difference in susceptibility to Pst DC3000 expressing avrRpm1, but P35S::MIR167a plants were an average of fivefold more resistant to the pathogen expressing avrRpt2 (Figure 2c).
We also tested pathogen growth in plants that were treated by spraying virulent Pst DC3000 onto the surface of leaves rather than by infiltration. In contrast to the modest difference observed after infiltration, P35S::MIR167a plants developed dramatically weaker disease symptoms (chlorosis and water‐soaked lesions) and were far less susceptible to Pst DC3000 as compared to wild‐type plants (Figure 3a). We generally recovered only 1–10 bacterial colony forming units (cfu) per leaf disc (and often none at all) from most P35S::MIR167a plants, despite recovering 105 to 106 cfu/leaf disc from wild‐type plants. Surprisingly, the variance within each T1 population of P35S::MIR167a plants was unusually high and in most trials, mean colony counts were skewed by high numbers in just a few individuals (Figure 3b). We hypothesize that this variation may be due to the altered stomatal behavior of these plants and their tendency to keep most stomata closed (see Section 3.9), creating a situation in which a rare few stomata are more open and allow bacterial entry into the leaf interior and subsequent multiplication to levels far greater than is typical for the plant population. Thus, while the mean colony number for plants overexpressing miR167 was usually around 50‐fold less than in wild‐type plants, the true level of resistance of P35S::MIR167a plants may be even greater. We could not test the growth of avirulent strains of P. syringae by spray inoculation, as the naturally low growth of these strains due to HR requires a much higher initial inoculum than we could achieve by spraying to produce detectable growth after four days.
FIGURE 3.

Growth of P. syringae after spray inoculation onto plants overexpressing miR167a and arf6 arf8 mutants. (a) Disease symptom development in Col‐0 and P35S::MIR167 plants sprayed with 5 × 108 cfu/ml Pst DC3000. Plants were photographed 4 days after inoculation. Lower panel, close‐up photograph showing chlorosis and water‐soaked lesions in wild‐type Col‐0 (left) and P35S::MIR167 (right) leaves. (b) and (c) Growth of Pst DC3000 in P35S::MIR167 (b) and arf6 arf8 double mutants (c). Plants were sprayed with Pst DC3000 at a titer of 5 × 108 cfu/ml. After spraying (unlike infiltration), the mean values for P35S::MIR167 and arf6 arf8 plants were very severely skewed by a few individuals. Therefore, data points from individual plants are plotted to better illustrate growth in each genotype. Horizontal bars represent treatment means. For all graphs, asterisks indicate significantly different growth from Col‐0 at p < .05 using the Wilcoxon rank‐sum test. Experiments were performed on eight to ten plants per genotype and each experiment was repeated three to five times with similar results. Results of one such experiment are shown. As P35S::MIR167a plants are sterile, a population of independent T1 transgenic plants was used rather than stable transgenic lines
3.3. miR167 modulates pathogen defense through its targets ARF6 and ARF8
To confirm the above results, we also tested the growth of Pst DC3000 in arf6‐2 arf8‐3 double mutant plants, which have the same developmental phenotypes as P35S:MIR167a plants (Wu et al., 2006). As with the P35S:MIR167a overexpression plants, arf6‐2 arf8‐3 plants were extremely resistant to Pst DC3000 (Figure 3c) and we saw high variation in colony count in these plants, with many zeros and very small counts making up the majority of data points and high counts observed for only a few individuals. Again, this variation could be due to altered stomatal behavior. As arf6‐2 arf8‐3 mutants recapitulate the developmental and pathogen‐responsive phenotypes of P35S:MIR167a plants, it is likely that miR167 influences physiology, and thereby pathogen defense, mainly through targeting the transcripts of ARF6 and ARF8 transcription factors.
3.4. Ectopic expression of ARF6 and ARF8 does not affect defense
As miR167 arises from four different precursor genes in Arabidopsis, obtaining complete loss‐of‐function lines for miR167 is relatively difficult and time‐consuming. Instead, Wu et al., produced constructs in which eight translationally silent mutations were introduced into the miR167 target sites of the ARF6 and ARF8 coding sequences. These new transcripts, called mARF6 and mARF8, are under the control of their normal 5′ and 3′ flanking sequences and produce functional proteins but are immune to cleavage directed by miR167 (Wu et al., 2006). mARF6 and mARF8 transgenic plants are dwarfed with small, rounded leaves, short petioles, and have sterile flowers, and the strength of these phenotypes is directly correlated with the level of mARF transcript expressed (Figure S1b) (Wu et al., 2006).
To test the effect of loss of miR167‐directed cleavage of ARF6 and ARF8, we tested the growth of Pst DC3000 in mARF6 and mARF8 plants. As with the overexpression of miR167a, sterility of mARF6 and mARF8 plants meant that stable transgenic lines could not be generated and all experiments were performed on populations of individual T1 generation transformants. We did not observe a significant difference in pathogen growth in plants expressing either the mARF6 or mARF8 transgenes (Figure 4). This indicates that while wild‐type levels of ARF6 and ARF8 contribute to susceptibility to Pst DC3000, excess ARF6 or ARF8 does not further increase susceptibility.
FIGURE 4.

Growth of P. syringae in mARF plants. Five‐week‐old Col‐0, mARF6, and mARF8 plants were sprayed with Pst DC3000 at a titer of 5 × 108 cfu/ml and bacterial growth was determined after 3 days. Bars represent mean + SEM of pathogen growth. No significant differences in pathogen growth were detected using the Wilcoxon rank‐sum test. This experiment was performed on eight to ten plants per genotype and was repeated three times with similar results. Results of one such experiment are shown. As mARF6 and mARF8 plants are sterile, a population of independent T1 transgenic plants was used rather than stable transgenic lines
3.5. Overexpression of miR167a does not affect the expression of hormone response genes
As discussed above, cross‐talk between hormones is a major component of the defense response, and previous studies performed on arf6‐2 arf8‐3 knockouts have demonstrated that multiple hormone responses are altered in flowers or seedlings of these plants (Nagpal et al., 2005; Reeves et al., 2012; Tabata et al., 2010; Tian et al., 2004; Wu et al., 2006). Therefore, we wanted to determine whether the high resistance in P35S:MIR167a plants might be due to altered hormone responses.
We used semi‐quantitative reverse transcriptase‐PCR to test the expression of several genes related to hormone responses in pathogen‐infiltrated P35S::MIR167a plants. We first tested the PATHOGENESIS‐RELATED GENE1 (PR1) gene as a marker of SA‐responsive defense activation, but we did not observe any significant change in its expression in P35S:MIR167a plants as compared to Col‐0. We also did not observe differences in the expression of two genes required for SA accumulation, ISOCHORISMATE SYNTHASE1 (ICS1) or GH3‐LIKE DEFENSE GENE1 (GDG1) (Wildermuth et al., 2001; Jagadeeswaran et al., 2007) (Figure S2).
ARF8 has previously been reported to regulate auxin levels in seedlings by inducing genes for auxin‐conjugating enzymes in the GH3 family (Tian et al., 2004). One of these genes is GH3.5, which is induced by auxin, SA, and P. syringae and has been proposed to have a dual role in the modulation of both SA and auxin signaling during infection (Zhang et al., 2007). As with PR1, expression of GH3.5 was not different in P35S::MIR167a plants compared to wild‐type after infection by virulent or avirulent P. syringae (Figure S2).
Several genes associated with JA biosynthesis are dependent on the expression of ARF6 and ARF8 in flowers, including LOX2 (a lipoxygenase which is also often used as a marker for JA‐pathway activation), AOS (allene oxide synthase), and OPR3 (OPDA reductase) (Nagpal et al., 2005). None of these genes showed expression patterns different from wild‐type in P35S::MIR167a plants, nor did PDF1.2 (plant defensin 1.2) or VSP2 (vegetative storage protein 2), downstream marker genes for the JA/ethylene‐mediated defense pathway.
The absence of major effects on the expression levels of the tested response genes was not entirely unexpected. In fact, our results are consistent with results observed for miR393, which targets auxin receptor transcripts for degradation. While overexpression of miR393 leads to increased resistance to P. syringae, it does not cause changes in expression levels of SA‐ or JA‐regulated defense genes (Navarro et al., 2006; Robert‐Seilaniantz et al., 2011).
3.6. Plants overexpressing miR167a are less responsive to auxin
Although we did not observe changes in the expression of the tested subset of SA‐, auxin‐ and JA‐related genes, we wanted to further investigate the possibility that altered auxin levels or responses were responsible for the resistance of P35S::MIR167a plants. Responses to auxin are reduced in arf6‐2 arf8‐3 mutant flowers, and arf8‐1 seedlings have reduced levels of auxin (though sensitivity to auxin is not affected) (Nagpal et al., 2005; Tian et al., 2004). We tested responses to auxin in P35S::MIR167a plants using the synthetic DR5::GUS construct, which is a widely used reporter for auxin‐responsive transcriptional activity. This construct consists of seven tandem repeats of a modified auxin response element plus a minimal cauliflower mosaic virus (CaMV) 35S promoter, fused upstream of the β‐glucuronidase (GUS) reporter gene (Ulmasov et al., 1997). We transformed the P35S:MIR167a construct into transgenic DR5::GUS plants and assayed GUS activity in populations of independent T1 transgenic plants. At 3.5 weeks of age (when phenotypes associated with overexpression of miR167a could be clearly identified), DR5::GUS plants overexpressing miR167a displayed lower basal levels of GUS expression than the DR5::GUS parent line (Figure 5). Treatment of these plants with the synthetic auxin 1‐napthaleneacetic acid (NAA) induced strong GUS activity in DR5::GUS plants, but overexpression of miR167 largely prevented this response (Figure 5). Thus, plants overexpressing miR167a have reduced auxin‐responsive transcriptional activity, which may at least partially account for their resistance to Pst DC3000.
FIGURE 5.

Overexpression of miR167 suppresses auxin responses. GUS staining in 3.5‐week‐old DR5::GUS and P35S::MIR167a DR5::GUS plants after overnight incubation in water or in 2 μM NAA. Scale bar = 1 cm. This experiment was repeated three times with 3–5 plants per treatment, with similar results. Results of one such experiment are shown. As P35S::MIR167a plants are sterile, a population of independent T1 transgenic plants was used rather than stable transgenic lines
Co‐treatment of plants with auxin at the time of infection has been shown to enhance disease symptom severity and in one report, the overall growth of P. syringae (Chen et al., 2007; Navarro et al., 2006; Wang et al., 2007). We hypothesized that if the defect in auxin responses in P35S:MIR167a plants is at the level of signaling rather than due to decreased auxin production, treatment of these plants with exogenous auxin should not restore susceptibility. To test this hypothesis, we treated wild‐type and P35S::MIR167a plants with either NAA or Pst DC3000 alone or Pst DC3000 plus 50 μM NAA by both infiltration and spray inoculation. Treatment with NAA alone caused leaves to become epinastic but did not have any other visible effect, as expected based on other researchers’ work (data not shown; see Chen et al., 2007; Navarro et al., 2006; Wang et al., 2007). NAA cotreatment with Pst DC3000 caused leaves of both genotypes to become epinastic and to develop enhanced disease symptoms (chlorosis and water‐soaked lesions) (Figure 6a); however, it did not have any effect on overall bacterial growth for either genotype or either infection method (Figure 6b,c). Thus, increased resistance in P35S::MIR167a plants must be at least partially at the level of auxin signaling. It also appears that the enhancement of disease symptom development caused by NAA occurs via a mechanism that is equally effective in both genotypes, as the relative increase in symptoms appeared similar between wild‐type and P35S::MIR167a plants. This would indicate that this mechanism is independent of ARF6 and ARF8 function.
FIGURE 6.

Effect of auxin treatment on pathogen growth in P35S::MIR167a plants. (a) Five‐week‐old Col‐0 and P35S::MIR167a plants were sprayed either with Pst DC3000 at a titer of 5 × 108 cfu/ml alone (top row) or Pst DC3000 plus 50 μM NAA (bottom row). Plants were photographed four days after treatment (b) and (c) Growth of Pst DC3000 in plants infiltrated (b) or sprayed (c) with pathogen alone or pathogen + 50 μM NAA. Means + SEM are displayed in (a) and counts from individual plants are shown in (b), with horizontal bars indicating population means. Different letters indicate statistically significant differences in pathogen counts (p < .05, Kruskal‐Wallis test followed by pairwise Wilcoxon rank‐sum tests using Hochberg p‐value adjustment). All experiments were performed on seven to ten plants of each genotype and were repeated three to five times with similar results. Results of one such experiment are shown. As P35S::MIR167a plants are sterile, a population of independent T1 transgenic plants was used rather than stable transgenic lines
3.7. MIR167a‐mediated resistance is dependent on salicylic acid
Because of the strong mutual antagonism between SA‐ and auxin/JA‐mediated defense pathways, we reasoned that plants overexpressing miR167 might be resistant because of the derepression of SA responses. To test whether resistance in P35S::MIR167a plants was dependent on SA, we transformed the overexpression construct into the eds16 mutant. eds16 plants do not accumulate SA due to a mutation in the ISOCHORISMATE SYNTHASE1 (ICS1) gene, a key enzyme in the SA biosynthesis pathway, and are, therefore, very susceptible to P. syringae (Wildermuth et al., 2001). When infiltrated with Pst DC3000, eds16 plants were highly susceptible (roughly 100‐fold more) compared to wild‐type, and pathogen growth was equally high in eds16 P35S:MIR167a plants (Figure 7a). Therefore, resistance conferred by overexpression of MIR167a is at least partially dependent on SA.
FIGURE 7.

Resistance in P35S::MIR167a plants is dependent on Salicylic Acid. Col‐0, P35S::MIR167a, eds16, and eds16 P35S::MIR167a plants were infiltrated (a) or sprayed (b) with Pst DC3000 at a titer of 5 × 105 or 5 × 108 cfu/ml, respectively. Means + SEM are displayed in (a) and counts from individual plants are shown in (b), with horizontal bars indicating population means. Different letters indicate statistically significant differences in pathogen counts (p < .05, Kruskal‐Wallis test followed by pairwise Wilcoxon rank‐sum tests using Hochberg p‐value adjustment). All experiments were performed on seven to ten plants of each genotype and were repeated three to five times with similar results. Results of one such experiment are shown. As P35S::MIR167a and eds16 P35S::MIR167a plants are sterile, populations of independent T1 transgenic plants were used rather than stable transgenic lines
Interestingly, when we performed infection by spray inoculation we did not observe any increase in susceptibility in eds16 mutant plants. This was an unexpected result but has also been reported by others when SA‐deficient plants were surface‐inoculated rather than infiltrated (Brooks et al., 2005). We have observed a similar phenomenon in the gdg1 (GH3‐LIKE DEFENSE GENE1) mutant, which is also deficient in SA accumulation and is highly susceptible to P. syringae when inoculated by infiltration (Jagadeeswaran et al., 2007). When spray inoculated, gdg1 mutant plants are not more susceptible to P. syringae than wild‐type (J. Caruana and N. Dhar, unpublished). After spraying, both eds16 P35S::MIR167a and gdg1 P35S::MIR167a plants displayed resistance intermediate between Col‐0 and P35S::MIR167a (Figure 7b and Figure S3). This suggests that resistance of P35S::MIR167a plants is partly dependent on SA when they are surface‐inoculated.
3.8. Overexpression of miR167a compromises the activation of SAR
As defenses activated during effector‐triggered immunity (ETI) are generally faster and stronger compared to basal defense mechanisms, we found it very surprising that miR167 was suppressed in response to avirulent pathogens when its overexpression aids defense. One key difference between ETI and basal defense is that the activation of ETI by avirulent pathogens leads to systemic acquired resistance (SAR), a long‐lasting, broad‐spectrum resistance that prevents further infections (Durrant & Dong, 2004). Accumulation of SA in distal tissues has long been known to be required for SAR, but studies have also shown that the early stages of SAR establishment also require auxin and JA signaling (Truman et al., 2007, 2010). As overexpression of miR167 represses auxin responses, we hypothesized that high levels of miR167 may compromise SAR activation.
To test the ability of P35S::MIR167a plants to establish SAR, we first treated older (primary) leaves of wild‐type and P35S::MIR167a plants with 10 mM MgSO4 (infiltration control) or Pst DC3000 (avrRpm1). Two days later, systemic leaves located above the primary leaves were infiltrated with Pst DC3000. Bacterial growth was quantified three days after this challenge infection. Pretreatment of wild‐type plants with Pst DC3000 (avrRpm1) caused a strong reduction in the subsequent growth of virulent Pst DC3000, but we observed no such effect in P35S::MIR167a plants (Figure 8). Thus, the SAR response is severely compromised in plants overexpressing miR167, suggesting that the activation of SAR in response to avirulent pathogens is suppressed by miR167.
FIGURE 8.

P35s::MIR167 plants are unable to activate systemic acquired resistance. Three lower leaves of Col‐0 and P35S::MIR167 plants were infiltrated with either 10 mM MgSO4 (mock) or Pst DC3000 (avrRpm1) at 1 × 108 cfu/ml to induce SAR. Two days later, secondary leaves were infiltrated with 5 × 105 cfu/ml Pst DC3000. Bacterial growth was measured after 3 days. Bars represent means + SEM; different letters indicate statistically significant differences in pathogen counts (p < .05, Kruskal‐Wallis test followed by pairwise Wilcoxon rank‐sum tests using Hochberg p‐value adjustment). This experiment was performed on seven to ten plants of each genotype and was repeated three times with similar results. Results of one such experiment are shown. As P35S::MIR167a plants are sterile, a population of independent T1 transgenic plants was used rather than stable transgenic lines
3.9. Stomatal behavior is altered in P35S::MIR167a plants
The difference in the level of resistance of P35S::MIR167a plants when sprayed versus infiltrated with P. syringae is very striking, and suggests that one of the major effects of miR167 overexpression is to impair the ability of bacterial cells to gain entry to the interior of the leaf. In order to multiply effectively and cause disease, bacterial cells that are inoculated onto the leaf surface must enter leaf tissues through wounds or natural openings, usually the stomata. Detection of pathogen‐associated molecular patterns (MAMPs) such as flagellin and lipopolysaccharide by guard cells results in the rapid closure of stomata. However, virulent pathogens such as P. syringae produce effector molecules that can force stomata to reopen (Melotto et al., 2006; Zeng & He, 2010). To test whether stomatal responses are altered in plants overexpressing miR167, we measured stomatal apertures of P35S::MIR167a plants after treatment with water or Pst DC3000 using the whole‐leaf method described previously (Chitrakar & Melotto, 2010). We found that the baseline stomatal aperture in P35S::MIR167a plants treated with water was smaller than that of wild‐type plants (Figure 9a). In response to Pst DC3000, both wild‐type and P35S::MIR167a stomata closed within 1 hpt and reopened by 4 hpt, but P35S::MIR167a plants always maintained significantly smaller apertures than wild‐type plants (Figure 9a). Even in their “reopened” state, the average stomatal aperture of P35S::MIR167a plants was smaller than the average aperture of “closed” wild‐type stomata. To confirm that the smaller baseline stomatal aperture of P35S::MIR167a plants was not a response to incubation in water, we also measured apertures of freshly detached leaves hourly throughout the ten‐hour photoperiod. The average stomatal aperture of P35S::MIR167a plants changed very little over the course of the day, and without incubation in water, the difference in stomatal aperture between wild‐type and P35S::MIR167a plants was even more pronounced (Figure 9b).
FIGURE 9.

Plants overexpressing miR167 maintain smaller stomatal apertures. (a) Whole leaves of five‐week‐old Col‐0 and P35S::MIR167a plants were detached and incubated in water alone or in 5 × 108 cfu/ml Pst DC3000 suspended in water and stomatal apertures were measured at the indicated time points. Asterisks indicate significant differences between wild‐type and P35S::MIR167a at p < .001 (ANOVA followed by Tukey's HSD post‐hoc tests). (b) Fresh whole leaves of five‐week‐old Col‐0 and P35S::MIR167a plants were detached and stomatal apertures were immediately measured each hour from lights‐on until lights‐out. Differences between Col‐0 and P35S::MIR167a were statistically significant at all time points (p < .001, Student's t test). All bars indicate means + SEM for 85 stomata per sample. These experiments were repeated three times with similar results. Results of one such experiment are shown. As P35S::MIR167a plants are sterile, a population of independent T1 transgenic plants was used rather than stable transgenic lines
P35S::MIR167a plants are slightly smaller than wild‐type plants, so it is possible that they have smaller stomatal apertures due to the overall smaller cell size. To address this possibility, we measured guard cell length in wild‐type and P35S::MIR167a plants. We did not observe any significant difference between the two genotypes, indicating that the guard cell size is not likely to account for the observed difference in the stomatal aperture in P35S::MIR167a plants (Figure S4a). We also did not observe any significant difference in stomatal density between wild‐type and P35S::MIR167a plants (Figure S4b). Therefore, overexpression of miR167a does not appear to affect the stomatal size or density, but alters overall stomatal behavior to constitutively maintain small apertures. This may prevent pathogen entry and account for the very high resistance of P35S::MIR167a plants.
4. DISCUSSION
In this study, we have demonstrated a role for the microRNA miR167 in defense against P. syringae. miR167 has been shown to regulate floral organ development by controlling the expression of ARF6 and ARF8, transcription factors that regulate auxin responses (Gifford et al., 2008; Wu et al., 2006). Our results demonstrate that miR167 also modulates pathogen defense through the degradation of ARF6 and ARF8 transcripts, as arf6 arf8 double mutants recapitulated the defense phenotype of P35S::MIR167a plants. Overexpression of miR167a results in enhanced resistance to Pst DC3000; however, the level of resistance is variable depending on the inoculation method used. When the virulent pathogen is infiltrated into the leaf interior, resistance due to the overexpression of miR167 is only modest. This is similar to the level of resistance conferred by the overexpression of miR393, which represses auxin responses by targeting the transcript for the auxin receptor for degradation (Navarro et al., 2006). When we tested for an effect of overexpression of miR167 on resistance to avirulent pathogens, there was no difference in susceptibility to Pst DC3000 expressing avrRpm1, but P35S::MIR167a plants were slightly more resistant to the Pst DC3000 expressing avrRpt2. This may be due to the fact that the virulence effector AvrRpt2 manipulates host auxin responses to promote bacterial growth in susceptible genotypes, and auxin responses are be repressed in plants overexpressing miR167 (Chen et al., 2007; Cui et al., 2013; Wu et al., 2006).
In contrast to the results observed for bacterial infiltration, high resistance is seen when the pathogen is sprayed onto the surface of leaves. This resistance appears mainly to be due to the fact that P35S::MIR167a plants constitutively maintain small stomatal apertures, which prevents bacterial cells on the leaf surface from gaining access to the leaf interior where they can effectively multiply. The more modest resistance of P35S::MIR167a plants after pathogen infiltration indicates an additional (secondary) mechanism, and genetic experiments suggest that this is at least partially dependent on SA. Finally, miR167 overexpression also eliminated SAR against secondary infections, despite causing an increase in resistance during local infection by Pst DC3000. This result indicates distinct requirements for auxin response in local and systemic immunity.
Auxin can promote stomatal opening, in part by affecting potassium uptake into guard cells (Acharya & Assmann, 2009). As responses to auxin are repressed in P35S::MIR167a plants, our results suggest that ARF6 and ARF8 may activate genes encoding downstream effectors of stomatal opening or inhibitors of stomatal closing. Closure of stomata in response to the detection of pathogens is a critical part of the plant's innate immune system. As it represents a barrier to entry into the interior of the leaf where pathogens can establish infection, the mechanism regulating stomatal closure is a target of bacterial virulence factors. While the detection of MAMPs such as flagellin and LPS causes rapid closure of stomata, virulence factors such as the JA mimic coronatine (COR) can reverse this effect and force stomata to reopen (Melotto et al., 2006).
Mutants with altered stomatal responses often display differential susceptibility depending on whether the pathogen is inoculated onto the leaf surface or infiltrated directly into the leaf interior (bypassing the stomatal layer of basal defense) (Liu et al., 2009; Melotto et al., 2006; Zeng et al., 2011; Zeng & He, 2010). We observed this effect in P35S::MIR167a plants, as resistance was fairly minimal (fivefold) when the pathogen was infiltrated but was very strong (sometimes greater than 500‐fold) when bacteria were sprayed onto leaves. Stomata of P35S::MIR167a plants do close in response to Pst DC3000 and also reopen at the same time as wild‐type stomata, so overexpression of miR167 does not eliminate response to the pathogen. However, P35S::MIR167a plants maintain a smaller average stomatal aperture than wild‐type plants and even when “reopened,” apertures remain much smaller than in wild‐type. Thus, overexpression of miR167 has a significant effect on the physiology of guard cells, and this suggests that P35S::MIR167a plants prevent bacterial entry through maintenance of stomata in a closed state. This may explain the extremely high level of resistance in P35S::MIR167a plants when the pathogen is sprayed onto the leaf surface, and it may also account for the higher pathogen growth observed in a small number of P35S::MIR167a and arf6‐2 arf8‐3 individuals, as small variations in stomatal aperture among leaves can have a large effect on pathogen entry and eventual growth.
The exact mechanism by which miR167 controls the stomatal aperture is unclear, though it is likely to be due to altered hormonal responses in P35S::MIR167a plants. Like other aspects of defense, the behavior of guard cells is dependent on multiple hormones. Abscisic acid (ABA) is the master regulatory hormone that drives stomatal closure, but other hormones including SA, JA, and auxin also contribute to stomatal behavior. Auxin generally promotes stomatal opening (Acharya & Assmann, 2009), but auxin‐induced transcriptional activity is reduced in P35S::MIR167 plants, indicating that they are less responsive to auxin than wild‐type plants. SA is required in combination with ABA for stomatal closure in response to P. syringae, and it was recently demonstrated that high levels of SA in the siz2, cpr5, and acd6 mutants result in constitutive stomatal closure and drought tolerance (Melotto et al., 2006; Miura et al., 2013). Given the strong antagonism between auxin and SA responses (Chen et al., 2007; Robert‐Seilaniantz et al., 2011; Wang et al., 2007), it seems likely that one of the major reasons for increased resistance in P35S::MIR167 plants is that SA responses, including stomatal closure, may be more effective when auxin signaling is weakened. This is supported by our evidence that resistance to spray‐inoculated Pst DC3000 conferred by the overexpression of miR167 is lessened in the eds16 and gdg1 mutant backgrounds, which are unable to accumulate SA. A model for miR167 involvement in defense through its modulation of hormone balance and stomatal behavior is shown in Figure 10.
FIGURE 10.

Proposed model for the effect of miR167 on defense responses. In response to pathogen detection, SA is produced and directs closure of stomata and activation of defense responses. At the same time, pathogen effectors activate auxin signaling to reopen stomata and repress defense. Overexpression of miR167 represses auxin (and possibly JA) signaling, thereby shifting the balance between hormones to favor closure of stomata and stronger defense induced by SA. Not shown but also present: mutual antagonism between SA and JA/auxin
Surprisingly, while overexpression of miR167 has a positive effect on defense during local infection, it prevents the establishment of SAR. Usually, activation of the HR at the site of a local infection results in the priming of defense genes in distal tissues to allow faster, stronger activation of immune responses upon subsequent pathogen infection. Establishment of SAR has long been known to require the accumulation of SA in distal tissues, but it was recently reported that the first systemic responses to avirulent pathogen challenge are associated with JA and auxin signaling (Truman et al., 2007, 2010). Mutants defective in either JA or auxin biosynthesis, signaling, or transport are impaired in the establishment of SAR, and a model has been proposed in which SAR is activated by temporally spaced phases of JA, then auxin, then SA signaling (Truman et al., 2007, 2010).
We hypothesize that this requirement of all three hormones for the establishment of SAR may account for the differential expression of miR167 in response to P. syringae. miR167 is suppressed during the HR against avirulent strains of P. syringae, with the degree of suppression of miR167 correlated with the strength of the HR. When it is present, miR167 suppression of its targets ARF6 and ARF8 might prevent critical aspects of auxin response that are necessary for the first stages of SAR. Therefore, it might be more advantageous for plants to sacrifice the potential benefits provided by miR167 during a local infection in order to enable the longer‐lasting protection of SAR.
Our studies of miR167 expression patterns complement several previous studies in which global profiling methods were used to study changes in microRNA expression in response to biotic and abiotic stresses. Fahlgren et al. used deep sequencing to study miRNA expression in response to the non‐host pathogen Pst DC3000 hrcC, and found miR167 to be induced fivefold by 3 hr post‐infiltration (Fahlgren et al., 2007). Zhang et al. also used deep sequencing and found miR167 to be induced both by Pst DC3000 and Pst DC3000 hrcC (Zhang et al., 2011). We did not observe any change in miR167 levels in response to Pst DC3000 by northern blot analysis (Figure 1b). This difference from previous results is likely due to the lower sensitivity of our method (small RNA northern blot) versus their method (next‐generation RNA sequencing) for detecting small changes in expression.
Zhang et al. also found miR167 to be slightly induced by avirulent Pst DC3000 expressing avrRpt2, rather than suppressed as our data indicate. They report induction of three to fourfold by 6 hpi with expression returning to basal levels by 14 hpi, while we observed suppression beginning at 12 hpi and continuing through 24 hpi with Pst DC3000 (avrRpt2). One possible explanation for this difference in results is that Zhang et al. used a lower titer of pathogen than we did for our assays (2 × 107 cfu/ml vs. 5 × 107 cfu/ml), and specify that they did not observe visible HR symptoms at 14 hpi when samples were collected. In contrast, we observed visible HR beginning at 8 hpi, and leaves were fully collapsed by 12 hpi. If the strength of suppression of miR167 is in fact correlated with the strength of the HR, then this difference in the extent of visible HR (and perhaps by extension, the rapidity of SAR induction) may explain the discrepancy between the two datasets.
The work described here illustrates a new role for miR167, a microRNA previously studied for its role in the growth and developmental processes. Our results demonstrate that miR167 is also involved in defense against bacterial pathogens such as P. syringae and build on a growing body of evidence that miR167 plays a role in responses to multiple biotic and abiotic stresses. miR167 was recently shown to mediate responses to osmotic stress (Kinoshita et al., 2012). It is also differentially regulated by high salinity, drought, cold, hypoxia, UV‐B radiation, and in response to changes in nutrient concentrations, as well as by bacterial and viral pathogens and nematodes (Feng et al., 2009; Gao et al., 2013; Gifford et al., 2008; Gupta et al., 2012; Hewezi et al., 2008; Huang et al., 2010; Jia et al., 2009; Liu et al., 2008; Zhang et al., 2008).
Taken together, these results indicate that the role of miR167 extends far beyond growth and developmental processes and reinforce the importance of small RNA‐mediated mechanisms in the regulation of all aspects of plant biology.
CONFLICT OF INTEREST
The authors declare that this research was conducted in the absence of any commercial or financial relationships that would cause any potential conflicts of interest.
AUTHOR CONTRIBUTIONS
JC, ND, and RR conceived the study and designed all experiments; JC and ND conducted experiments; JC conducted statistical analyses; JC wrote the main manuscript text; JC and ND prepared figures. All authors approved the final version of the manuscript.
Supporting information
Fig S1‐S4‐Table S1
ACKNOWLEDGMENTS
We thank Dr. Jason Reed (University of North Carolina at Chapel Hill) for helpful discussions on the manuscript as well as for providing P35S::MIR167a, mARF6, and mARF8 contruct and arf6‐2 arf8‐3 seeds. Pseudomonas syringae strains were provided by Dr. Barbara Kunkel (Washington University). eds16‐1 mutant seeds were provided by Dr. Mary Wildermuth (Wildermuth et al., 2001), and DR5::GUS seeds were provided by Dr. Tom Guilfoyle (Ulmasov et al., 1997). This work was supported by an NSF Arabidopsis 2010 grant (DEB0313492) and funds from Syracuse University to RR. JC was supported in part by a Syracuse University Scholarship.
Caruana JC, Dhar N, Raina R. Overexpression of Arabidopsis microRNA167 induces salicylic acid‐dependent defense against Pseudomonas syringae through the regulation of its targets ARF6 and ARF8 . Plant Direct. 2020;4:1–16. 10.1002/pld3.270
Julie C. Caruana and Nikhilesh Dhar have equal contribution.
Funding information
Work in the laboratory of R.R. was supported by a grant from the National Science Foundation (IOS‐1146128). J.C. was supported in part by a Syracuse University Fellowship.
REFERENCES
- Acharya, B. R. , & Assmann, S. M. (2009). Hormone interactions in stomatal function. Plant Molecular Biology, 69, 451–462. 10.1007/s11103-008-9427-0 [DOI] [PubMed] [Google Scholar]
- Bartel, D. P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell, 116, 281–297. 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Boccara, M. , Sarazin, A. , Thiébeauld, O. , Jay, F. , Voinnet, O. , Navarro, L. , & Colot, V. (2014). The Arabidopsis miR472‐RDR6 silencing pathway modulates PAMP‐ and effector‐triggered immunity through the post‐transcriptional control of disease resistance genes. PLoS Path, 10(1), e1003883 10.1371/journal.ppat.1003883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks, D. M. , Bender, C. L. , & Kunkel, B. N. (2005). The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid‐dependent defenses in Arabidopsis thaliana . Molecular Plant Pathology, 6, 629–639. [DOI] [PubMed] [Google Scholar]
- Chen, Z. , Agnew, J. L. , Cohen, J. D. , He, P. , Shan, L. , Sheen, J. , & Kunkel, B. N. (2007). Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proceedings of the National Academy of Sciences of the United States of America, 104, 20131–20136. 10.1073/pnas.0704901104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm, S. T. , Coaker, G. , Day, B. , & Staskawicz, B. J. (2006). Host‐microbe interactions: Shaping the evolution of the plant immune response. Cell, 124, 803–814. 10.1016/j.cell.2006.02.008 [DOI] [PubMed] [Google Scholar]
- Chitrakar, R. , & Melotto, M. (2010). Assessing stomatal response to live bacterial cells using whole leaf imaging. Journal of Visualized Experiments, 44, e2185 10.3791/2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S. J. , & Bent, A. F. (1998). Floral dip: A simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . The Plant Journal, 16, 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Cui, F. , Wu, S. , Sun, W. , Coaker, G. , Kunkel, B. , He, P. , & Shan, L. (2013). The Pseudomonas syringae type III effector AvrRpt2 promotes pathogen virulence via stimulating Arabidopsis auxin/indole acetic acid protein turnover. Plant Physiology, 162, 1018–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, R. T. , Goetz, D. H. , Lasswell, J. , Anderson, M. N. , & Bartel, B. (1999). IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. The Plant Cell, 11, 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devadas, S. K. , Enyedi, A. , & Raina, R. (2002). The Arabidopsis hrl1 mutation reveals novel overlapping roles for salicylic acid, jasmonic acid and ethylene signalling in cell death and defence against pathogens. The Plant Journal, 30, 467–480. 10.1046/j.1365-313X.2002.01300.x [DOI] [PubMed] [Google Scholar]
- Dhar, N. , Caruana, J. , Erdem, I. , & Raina, R. (2020). An Arabidopsis DISEASE RELATED NONSPECIFIC LIPID TRANSFER PROTEIN 1 is required for resistance against various phytopathogens and tolerance to salt stress. Gene, 753, 144802 10.1016/j.gene.2020.144802 [DOI] [PubMed] [Google Scholar]
- Durrant, W. E. , & Dong, X. (2004). Systemic acquired resistance. Annual Review of Phytopathology, 42, 185–209. 10.1146/annurev.phyto.42.040803.140421 [DOI] [PubMed] [Google Scholar]
- Fahlgren, N. , Howell, M. D. , Kasschau, K. D. , Chapman, E. J. , Sullivan, C. M. , Cumbie, J. S. , Givan, S. A. , Law, T. F. , Grant, S. R. , Dangl, J. L. , & Carrington, J. C. (2007). High‐throughput sequencing of Arabidopsis microRNAs: Evidence for frequent birth and death of MIRNA genes. PLoS One, 2, e219 10.1371/journal.pone.0000219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, J. , Wang, K. , Liu, X. , Chen, S. , & Chen, J. (2009). The quantification of tomato microRNAs response to viral infection by stem‐loop real‐time RT‐PCR. Gene, 437, 14–21. 10.1016/j.gene.2009.01.017 [DOI] [PubMed] [Google Scholar]
- Gao, R. , Wan, Z. Y. , & Wong, S.‐M. (2013). Plant growth retardation and conserved miRNAs are correlated to hibiscus chlorotic ringspot virus infection. PLoS One, 8, e85476 10.1371/journal.pone.0085476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford, M. L. , Dean, A. , Gutierrez, R. A. , Coruzzi, G. M. , & Birnbaum, K. D. (2008). Cell‐specific nitrogen responses mediate developmental plasticity. Proceedings of the National Academy of Sciences of the United States of America, 105, 803–808. 10.1073/pnas.0709559105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle, T. J. , & Hagen, G. (2007). Auxin response factors. Current Opinion in Plant Biology, 10, 453–460. 10.1016/j.pbi.2007.08.014 [DOI] [PubMed] [Google Scholar]
- Gupta, O. P. , Permar, V. , Koundal, V. , Sing, U. D. , & Praveen, S. (2012). MicroRNA regulated defense responses in Tricitum aestivum L. during Puccinia graminis f.sp. triciti infection. Molecular Biology Reports, 39, 817–824. [DOI] [PubMed] [Google Scholar]
- Gupta, O. P. , Sharma, P. , Gupta, R. K. , & Sharma, I. (2014). Current status on role of miRNAs during plant‐fungus interaction. Physiological and Molecular Plant Pathology, 85, 1–7. 10.1016/j.pmpp.2013.10.002 [DOI] [Google Scholar]
- Gutierrez, L. , Bussell, J. D. , Pacurar, D. I. , Schwambach, J. , Pacurar, M. , & Bellini, C. (2009). Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. The Plant Cell, 21, 3119–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewezi, T. , Howe, P. , Maier, T. R. , & Baum, T. J. (2008). Arabidopsis small RNAs and their targets during cyst nematode parasitism. Molecular Plant‐Microbe Interactions, 21, 1622–1634. [DOI] [PubMed] [Google Scholar]
- Huang, S. Q. , Xiang, A. L. , Che, L. L. , Chen, S. , Li, H. , Song, J. B. , & Yang, Z. M. (2010). A set of miRNAs from Brassica napus in response to sulphate deficiency and cadmium stress. Plant Biotechnology Journal, 8, 887–899. 10.1111/j.1467-7652.2010.00517.x [DOI] [PubMed] [Google Scholar]
- Jagadeeswaran, G. , Raina, S. , Acharya, B. R. , Maqbool, S. B. , Appel, H. M. , Schultz, J. C. , Klessig, D. F. , & Raina, R. (2007). Arabidopsis GH3‐LIKE DEFENSE GENE 1 is required for accumulation of salicylic acid, activation of defense responses and resistance to Pseudomonas syringae . The Plant Journal, 51, 234–246. [DOI] [PubMed] [Google Scholar]
- Jagadeeswaran, G. , Saini, A. , & Sunkar, R. (2009). Biotic and abiotic stress down‐regulate miR398 expression in Arabidopsis . Planta, 229, 1009–1014. 10.1007/s00425-009-0889-3 [DOI] [PubMed] [Google Scholar]
- Jia, X. , Ren, L. , Chen, Q. J. , Li, R. , & Tang, G. (2009). UV‐B‐responsive microRNAs in Populus tremula . Journal of Plant Physiology, 166, 2046–2057. 10.1016/j.jplph.2009.06.011 [DOI] [PubMed] [Google Scholar]
- Katiyar‐Agarwal, S. , Gao, S. , Vivian‐Smith, A. , & Jin, H. (2007). A novel class of bacteria‐induced small RNAs in Arabidopsis . Genes & Development, 21, 3123–3134. 10.1101/gad.1595107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar‐Agarwal, S. , & Jin, H. (2010). Role of small RNAs in host‐microbe interactions. Annual Review of Phytopathology, 48, 225–246. 10.1146/annurev-phyto-073009-114457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar‐Agarwal, S. , Morgan, R. , Dahlbeck, D. , Borsani, O. , Villegas, A. Jr , Zhu, J. K. , Staskawicz, B. J. , & Jin, H. (2006). A pathogen‐inducible endogenous siRNA in plant immunity. Proceedings of the National Academy of Sciences of the United States of America, 103, 18002–18007. 10.1073/pnas.0608258103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan, K. , & Manners, J. M. (2009). Linking development to defense: Auxin in plant‐pathogen interactions. Trends in Plant Science, 14, 373–382. 10.1016/j.tplants.2009.04.005 [DOI] [PubMed] [Google Scholar]
- Kinoshita, N. , Wang, H. , Kasahara, H. , Liu, J. , MacPherson, C. , Machida, Y. , Kamiya, Y. , Hannah, M. , & Chua, N. H. (2012). IAA‐Ala‐Resistant3, an evolutionarily conserved target of miR167, mediates Arabidopsis root architecture changes during high osmotic stress. The Plant Cell, 24, 3590–3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel, B. N. , & Brooks, D. M. (2002). Cross talk between signaling pathways in pathogen defense. Current Opinion in Plant Biology, 5, 325–331. 10.1016/S1369-5266(02)00275-3 [DOI] [PubMed] [Google Scholar]
- Li, Y. , Zhang, Q. , Zhang, J. , Wu, L. , Qi, Y. , & Zhou, J. M. (2010). Identification of microRNAs involved in pathogen‐associated molecular pattern‐triggered plant innate immunity. Plant Physiology, 152, 2222–2231. 10.1104/pp.109.151803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. H. , Tian, X. , Li, Y. J. , Wu, C. A. , & Zheng, C. C. (2008). Microarray‐based analysis of stress‐regulated microRNAs in Arabidopsis thaliana . RNA, 14, 836–843. 10.1261/rna.895308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Elmore, J. M. , Fuglsang, A. T. , Palmgren, M. G. , Staskawicz, B. J. , & Coaker, G. (2009). RIN4 functions with plasma membrane H+‐ATPases to regulate stomatal apertures during pathogen attack. PLoS Biology, 7, e1000139 10.1371/journal.pbio.1000139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto, M. , Underwood, W. , Koczan, J. , Nomura, K. , & He, S. Y. (2006). Plant stomata function in innate immunity against bacterial invasion. Cell, 126, 969–980. 10.1016/j.cell.2006.06.054 [DOI] [PubMed] [Google Scholar]
- Miura, K. , Okamoto, H. , Okuma, E. , Shiba, H. , Kamada, H. , Hasegawa, P. M. , & Murata, Y. (2013). SIZ1 deficiency causes reduced stomatal aperture and enhanced drought tolerance via controlling salicylic acid‐induced ROS accumulation in Arabidopsis . The Plant Journal, 73, 91–104. [DOI] [PubMed] [Google Scholar]
- Mutka, A. M. , Fawley, S. , Tsao, T. , & Kunkel, B. N. (2013). Auxin promotes susceptibility to Pseudomonas syringae via a mechanism independent of suppression of salicylic acid‐mediated defenses. The Plant Journal, 74, 746–754. [DOI] [PubMed] [Google Scholar]
- Nagpal, P. , Ellis, C. M. , Weber, H. , Ploense, S. E. , Barkawi, L. S. , Guilfoyle, T. J. , Hagen, G. , Alonso, J. M. , Cohen, J. D. , Farmer, E. E. , & Ecker, J. R. (2005). Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development, 132, 4107–4118. 10.1242/dev.01955 [DOI] [PubMed] [Google Scholar]
- Navarro, L. , Dunoyer, P. , Jay, F. , Arnold, B. , Dharmasiri, N. , Estelle, M. , Voinnet, O. , & Jones, J. D. G. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science, 312, 436–439. 10.1126/science.1126088 [DOI] [PubMed] [Google Scholar]
- Nimchuk, Z. , Eulgem, T. , Holt, B. F. 3rd , & Dangl, J. L. (2003). Recognition and response in the plant immune system. Annual Review of Genetics, 37, 579–609. 10.1146/annurev.genet.37.110801.142628 [DOI] [PubMed] [Google Scholar]
- Nomura, K. , Melotto, M. , & He, S. Y. (2005). Suppression of host defense in compatible plant‐Pseudomonas syringae interactions. Current Opinion in Plant Biology, 8, 361–368. 10.1016/j.pbi.2005.05.005 [DOI] [PubMed] [Google Scholar]
- Padmanabhan, C. , Zhang, X. , & Jin, H. (2009). Host small RNAs are big contributors to plant innate immunity. Current Opinion in Plant Biology, 12, 465–472. 10.1016/j.pbi.2009.06.005 [DOI] [PubMed] [Google Scholar]
- Park, J.‐E. , Park, J. Y. , Kim, J.‐S. , Staswick, P. E. , Jeon, J. , Yun, J. , Kim, S. Y. , Kim, J. , Lee, Y. H. , & Park, C. M. (2007). GH3‐mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis . Journal of Biological Chemistry, 282, 10036–10046. [DOI] [PubMed] [Google Scholar]
- Park, Y. J. , Lee, H. J. , Kwak, K. J. , Lee, K. , Hong, S. W. , & Kang, H. (2014). MicroRNA400‐guided cleavage of Pentatricopeptide repeat protein mRNAs renders Arabidopsis thaliana more susceptible to pathogenic Bacteria and Fungi. Plant and Cell Physiology, 55(9), 1660–1668. 10.1093/pcp/pcu096 [DOI] [PubMed] [Google Scholar]
- Reeves, P. H. , Ellis, C. M. , Ploense, S. E. , Wu, M. F. , Yadav, V. , Tholl, D. , Chételat, A. , Haupt, I. , Kennerley, B. J. , Hodgens, C. , & Farmer, E. E. (2012). A regulatory network for coordinated plant maturation. PLoS Genetics, 8, e1002506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert‐Seilaniantz, A. , MacLean, D. , Jikumaru, Y. , Hill, L. , Yamaguchi, S. , Kamiya, Y. , & Jones, J. D. (2011). The microRNA miR393 re‐directs secondary metabolite biosynthesis away from camalexin and towards glucosinolates. The Plant Journal, 67, 218–231. 10.1111/j.1365-313X.2011.04591.x [DOI] [PubMed] [Google Scholar]
- Rose, L. E. , Overdijk, E. , & van Damme, M. (2019). Small RNA molecules and their role in plant disease. European Journal of Plant Pathology, 153(1), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz‐Ferrer, V. , & Voinnet, O. (2009). Roles of plant small RNAs in biotic stress responses. Annual Review of Plant Biology, 60, 485–510. 10.1146/annurev.arplant.043008.092111 [DOI] [PubMed] [Google Scholar]
- Seo, J. K. , Wu, J. , Lii, Y. , Li, Y. , & Jin, H. (2013). Contribution of small RNA pathway components in plant immunity. Molecular Plant‐Microbe Interactions, 26, 617–625. 10.1094/MPMI-10-12-0255-IA [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger, D. , Korneli, C. , Lummer, M. , & Navarro, L. (2013). Emerging role for RNA‐based regulation in plant immunity. New Phytologist, 197, 394–404. 10.1111/nph.12022 [DOI] [PubMed] [Google Scholar]
- Tabata, R. , Ikezaki, M. , Fujibe, T. , Aida, M. , Tian, C.‐E. , Ueno, Y. , Yamamoto, K. T. , Machida, Y. , Nakamura, K. , & Ishiguro, S. (2010). Arabidopsis auxin response factor6 and 8 regulate jasmonic acid biosynthesis and floral organ development via repression of class 1 KNOX genes. Plant and Cell Physiology, 51, 164–175. 10.1093/pcp/pcp176 [DOI] [PubMed] [Google Scholar]
- Tian, C. E. , Muto, H. , Higuchi, K. , Matamura, T. , Tatematsu, K. , Koshiba, T. , & Yamamoto, K. T. (2004). Disruption and overexpression of auxin response factor 8 gene of Arabidopsis affect hypocotyl elongation and root growth habit, indicating its possible involvement in auxin homeostasis in light condition. The Plant Journal, 40, 333–343. [DOI] [PubMed] [Google Scholar]
- Tornero, P. , & Dangl, J. L. (2001). A high‐throughput method for quantifying growth of phytopathogenic bacteria in Arabidopsis thaliana . The Plant Journal, 28, 475–481. 10.1046/j.1365-313X.2001.01136.x [DOI] [PubMed] [Google Scholar]
- Truman, W. , Bennett, M. H. , Kubigsteltig, I. , Turnbull, C. , & Grant, M. (2007). Arabidopsis systemic immunity uses conserved defense signaling pathways and is mediated by jasmonates. Proceedings of the National Academy of Sciences of the United States of America, 104, 1075–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman, W. M. , Bennett, M. H. , Turnbull, C. G. , & Grant, M. R. (2010). Arabidopsis auxin mutants are compromised in systemic acquired resistance and exhibit aberrant accumulation of various indolic compounds. Plant Physiology, 152, 1562–1573. 10.1104/pp.109.152173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T. , Murfett, J. , Hagen, G. , & Guilfoyle, T. J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. The Plant Cell, 9, 1963–1971. 10.1105/tpc.9.11.1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D. , Pajerowska‐Mukhtar, K. , Culler, A. H. , & Dong, X. (2007). Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Current Biology, 17, 1784–1790. 10.1016/j.cub.2007.09.025 [DOI] [PubMed] [Google Scholar]
- Wildermuth, M. C. , Dewdney, J. , Wu, G. , & Ausubel, F. M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature, 414, 562–565. 10.1038/35107108 [DOI] [PubMed] [Google Scholar]
- Wu, M. F. , Tian, Q. , & Reed, J. W. (2006). Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development, 133, 4211–4218. 10.1242/dev.02602 [DOI] [PubMed] [Google Scholar]
- Yang, L. , & Huang, H. (2014). Roles of small RNAs in plant disease resistance. Journal of Integrative Plant Biology, 56, 962–970. 10.1111/jipb.12200 [DOI] [PubMed] [Google Scholar]
- Zeng, W. , Brutus, A. , Kremer, J. M. , Withers, J. C. , Gao, X. , Jones, A. D. , & He, S. Y. (2011). A genetic screen reveals arabidopsis stomatal and/or apoplastic defenses against pseudomonas syringae pv. tomato DC3000. PLoS Path, 7, e1002291 10.1371/journal.ppat.1002291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, W. , & He, S. Y. (2010). A prominent role of the flagellin receptor FLAGELLIN‐SENSING2 in mediating stomatal response to Pseudomonas syringae pv tomato DC3000 in Arabidopsis . Plant Physiology, 153, 1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W. , Gao, S. , Zhou, X. , Chellappan, P. , Chen, Z. , Zhou, X. , Zhang, X. , Fromuth, N. , Coutino, G. , Coffey, M. , & Jin, H. (2011). Bacteria‐responsive microRNAs regulate plant innate immunity by modulating plant hormone networks. Plant Molecular Biology, 75, 93–105. 10.1007/s11103-010-9710-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Li, Q. , Li, Z. , Staswick, P. E. , Wang, M. , Zhu, Y. , & He, Z. (2007). Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis‐Pseudomonas syringae interaction. Plant Physiology, 145, 450–464. 10.1104/pp.107.106021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Wei, L. , Zou, X. , Tao, Y. , Liu, Z. , & Zheng, Y. (2008). Submergence‐responsive MicroRNAs are potentially involved in the regulation of morphological and metabolic adaptations in maize root cells. Annals of Botany, 102, 509–519. 10.1093/aob/mcn129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, X. Y. , Spivey, N. W. , Zeng, W. , Liu, P. P. , Fu, Z. Q. , Klessig, D. F. , He, S. Y. , & Dong, X. (2012). Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host & Microbe, 11, 587–596. 10.1016/j.chom.2012.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐S4‐Table S1


