Highlights
-
•
Inflammatory innate immunity can be described as prognostic factors.
-
•
The balancing between Th1 and Th2 response can be associated with mortality in patients with moderate to severe COVID-19 infection.
-
•
IFN-γ was an independent risk factor associated with mortality in patients with SARS-Cov2.
Keywords: COVID-19, Immune response, Interleukin, Cytokines
Abstract
Background
Innate and adaptive immune responses have been evaluated in infected patients with COVID-19. The severity of the disease has been supposed to be associated with some profile not reported with other bacterial and viral pneumonia. We proposed a study in patients with moderate to severe COVID-19 infection to evaluate the interleukin patterns and its role as prognosis factors.
Methods
A prospective cohort with moderate and severe cases of COVID-19 infection from June to July 2020. Blood samples from patients were collected regularly to evaluate IFN-γ, TNF-α, IL-4, IL-6, and IL-10. Clinical, laboratory, radiological data, and outcomes were recorded. The outcome variable was in-hospital death, survival, mechanical ventilation, and admission at the intensive care unit. Data are presented in median and interquartile range [IQR].
Results
We evaluated the Th1 and Th2 responses according to evolution, distinguishing possible predictive markers. The IFN-γ median of 323 pg/mL [IQR 166−570] was found in patients who died and 208 pg/mL [IQR 155−392] in the survival group (p = 0.017). IFN-γ was also higher in the early stages of the disease (394 pg/mL [IQR 229–575] against 162 pg/mL [IQR 117–259], p < 0.001). IL-4 that was increased in late-stage (182 pg/mL [IQR 162–199] against 131 pg/mL [IQR 124–152], p < 0.001) but not associated with mortality. Also, death was also related to male gender (relative risk = 1.5 [95 % confidence interval = 1.1−2.0]).
Conclusion
Our results suggest that the activation of the host immune response between Th1 or Th2 in COVID-19 infection may be related to the final result between discharge or death. This implies an attempt to control cytokines, such as IFN-γ, with combined therapies for clinical treatment.
1. Introduction
In early 2020, the World Health Organization declared pandemic due to Coronavirus Disease 2019 (COVID-19). Since then, a large number of researches have been published, from epidemiological to experimental studies, trying better to understand immunological pathways of COVID-19 and possible treatments. CD3+, CD4+, and CD8+ lymphocytes count are usually decreased according to disease stages (Zhang et al., 2020a). Besides it, cytokine storm is also present in severe patients due to elevation of interleukins such as TNF-α, IL-6, IL-8, and IL-10 (Huang et al., 2020; Han et al., 2020). Thus, the differences among host immune responses play a major role in COVID-19 severity.
Older age, elevated C-reactive protein, serum lactate dehydrogenase, bilirubin, blood urea nitrogen, and decreased albumin are described as prognostic factors of severe COVID-19 (Gong et al., 2020). Comorbidities are also described as prognosis factors and the higher the number of them, the poorer the prognosis (i.e., hypertension, diabetes, chronic pulmonary disease, coronary artery disease, malignancies) (Liang et al., 2020). Interestingly, in an Italian and Spanish cohort, genetic factors related to ABO blood-group system were reported as susceptibility to COVID-19 respiratory failure (Ellinghaus et al., 2020).
Different interleukins are described as prognostic factors. A reasonable hypothesis is that (i) pro-inflammatory innate immunity and (ii) anti-inflammatory system are related to disease severity or death once IL-6, IL-8, and IL10 are closely described as prognostic factors in patients diagnosed with COVID-19 (Zhang et al., 2020a; Han et al., 2020; Luo et al., 2020). Nevertheless, few studies have focused on the adaptive immune system to evaluate the balancing between Th1 and Th2 response. Consequently, IL-4 is usually not studied concomitant with IFN-γ and IL-6 to better characterize Th2, Th1, and innate immune response. IFN-γ is the type II IFN produced by NK cells and T lymphocytes, although cells of different phases of the immune response (innate and adaptative, respectively), this cytokine is important in all phases of the immune response (Robinson et al., 2010). IFN- γ system is essential for antiviral defense. IFN-γ downregulate virus replication and it activates cytokine production by T cells, augmenting the cytotoxic T lymphocyte killing activity (Levy and Garcia-Sastre, 2001). However, persistent high levels of IFN-γ worsens the systemic inflammation, and increasing tissue injury and organ failure (Yin et al., 2005). Considering this ambiguous role of IFN-γ in the outcome, it is important to understand the serum pattern of this and other cytokines in patients with COVID-19.
The aim of this study was to investigate the interleukins patterns in patients admitted due to COVID-19 and its role as prognosis factors.
2. Methods
2.1. Study design
This was a prospective cohort of hospitalized patients diagnosed with COVID-19 conducted in the South of Brazil, Curitiba, Paraná, from June to July 2020. Patients were admitted only after approval of the research ethics committee and an approved consent form.
Blood samples from patients were collected to evaluate IFN-γ, TNF-α, IL-4, IL-6, and IL-10. Clinical, laboratory, radiological data, and outcomes were recorded.
2.2. Inclusion criteria
COVID-19 infection was defined by clinical-radiological presentation plus a nasopharyngeal swab polymerase chain reaction (PCR) positive to COVID-19. Inclusion criteria were hospitalized patients with moderate or severe confirmed COVID-19 infection.
The moderate disease was defined as an adult with clinical signs of pneumonia (fever, cough, dyspnea, fast breathing) but no signs of severe pneumonia, including SpO2 ≥ 90 % on room air; and severe disease as an adult with clinical signs of pneumonia (fever, cough, dyspnea, fast breathing) plus one of the following: respiratory rate > 30 breaths/min; severe respiratory distress; or SpO2 < 90 % on room air (WHO, 2020).
2.3. Exclusion criteria
Patients diagnosed with other viral infections, such as HIV, HCV, HBV, or another common respiratory virus, were excluded, as well as solid organ or hematological transplantation patients. Patients who used tocilizumab were also excluded.
2.4. Cytokine evaluation
Patients were classified according to days of symptoms: 0–10 days and >10 days. Blood samples were collected using a standard coagulation tube (SST II Advance, BD Biosciences) to obtain the serum, which was aliquoted and stored at -80 °C until analysis. The cytokines were measured using commercially available ELISA kits for TNF-α, IFN-γ, IL-6, IL-4, and IL-10 (ImmunoTools, Friesoythe, Germany), according to the manufacture instructions.
2.5. Clinical data
Several clinical and laboratory data were evaluated. The outcome variable was in-hospital death, survival, mechanical ventilation, and admission to the intensive care unit (ICU).
2.6. Statistical analysis
Continuous variables were expressed as median values and interquartile range (IQR) and analyzed by Mann Whitney test. Categorical variables were expressed as absolute frequencies with proportions and analyzed by chi-square or Fisher exact test. A p-value < 0.05 was considered significant. All variables in the univariate model meeting a cut-off of p <0.1 were included in the multivariable model. SPSS v23.0 (IBM, Chicago, IL) and GraphPad Prism v7 (GraphPad, San Diego, CA) were used for statistical analysis. The variable days of symptoms was split in >10 days or ≤ 10 days using the optimal binning procedure on SPSS for all cytokines included in the analysis.
3. Results
3.1. General characteristics of the study population
Fifty-six patients were included in all samples. Median age was 61 [47−73] years, and 69.6 % (n = 39) were male. Most common comorbidities were systemic arterial hypertension (50 %, n = 28), diabetes (25 %, n = 14) and chronic obstructive pulmonary disease (16.1 %, n = 9). At hospital admission, moderate and severe patients were composed of 85.7 % (n = 48) and 14.3 % (n = 8) from the sample, respectively. General clinical characteristics are in Table 1 . Steroids were used in 58.9 % (n = 33) and hydroxychloroquine in 12.5 % (n = 7). The median length of hospitalization was 13 [8–21] days. Specific characteristics from each patient group (0–10d vs. >10d) are demonstrated in Table 2 .
Table 1.
Clinical and laboratory data from patients with moderate and severe COVID-19 infection according to the outcome.
| Total (n = 56) |
Death (n = 18) |
Survival (n = 38) |
RR [CI95 %] | P value | ||||
|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | |||
| Male | 39 | 69.6 | 16 | 41% | 23 | 59% | 1.5[1.1−2.0] | 0.029 |
| Female | 17 | 30.4 | 2 | 12% | 15 | 88% | ||
| Intensive Care Unit | 33 | 58.9 | 18 | 55% | 15 | 45% | 2.2[1.5−3.1] | <0.001 |
| Intubation | 18 | 30.9 | 11 | 61% | 7 | 39% | 2.2[1.1−4.4] | 0.002 |
| Death | 18 | 32.1 | ||||||
| Arterial hypertension | 28 | 50.0 | 18 | 64% | 10 | 36% | 0.506 | |
| Diabetes mellitus | 14 | 25.0 | 5 | 36% | 9 | 64% | 0.562 | |
| Chronic heart failure | 6 | 10.7 | 2 | 33% | 4 | 67% | 0.674 | |
| Chronic coronary disease | 7 | 12.5 | 4 | 57% | 3 | 43% | 0.190 | |
| Previous myocardial infarct | 1 | 1.8 | 1 | 100 % | 0 | 0% | 0.340 | |
| Previous stroke | 1 | 1.8 | 1 | 100 % | 0 | 0% | 0.340 | |
| Arrhytmia | 5 | 8.9 | 3 | 60% | 3 | 60% | 0.326 | |
| Asthma | 5 | 8.9 | 2 | 40% | 3 | 60% | 0.560 | |
| Chronic pulmonary disease | 9 | 16.1 | 5 | 56% | 4 | 44% | 0.132 | |
| Chronic renal failure | 2 | 3.6 | 1 | 50 % | 1 | 50 % | 0.569 | |
| Neoplasm | 5 | 8.9 | 2 | 40% | 3 | 60% | 0.560 | |
| Dislipidemia | 4 | 7.1 | 1 | 25 % | 3 | 75% | 0.580 | |
| Symptoms | ||||||||
| Dyspnea | 30 | 53.6 | 11 | 37% | 19 | 63% | 0.430 | |
| Cough | 38 | 67.9 | 11 | 29% | 27 | 71% | 0.160 | |
| Fever | 25 | 44.6 | 5 | 20% | 20 | 80% | 0.6[0.4−0.9] | 0.036 |
| Diarrhea | 10 | 17.9 | 3 | 30% | 7 | 70% | 0.539 | |
| Throat pain | 8 | 14.3 | 2 | 25 % | 6 | 75% | 0.423 | |
| Coryza | 3 | 5.4 | 0 | 0% | 3 | 100 % | 0.260 | |
| Continuous variables (Median with IQR) on admission | ||||||||
| Age | 61 [47−73] | 66 [56−77] | 56 [43−72] | 0.568 | ||||
| Lenght of hospitalization | 13 [8–21] | 17 [12.5−26.5] | 10.5 [7−17.7] | 0.025 | ||||
| Temperature | 36.5 [36.1−37.1] | 36 [35.5−36.4] | 36.6 [35.5−37.3] | 0.046 | ||||
| Systolic blood pressure | 134 [121−147] | 1340 [125−152] | 130 [120−140] | 0.432 | ||||
| Diastolic blood preassure | 80 [72−90] | 80 [50−90] | 80 [80−90] | 0.355 | ||||
| Heart rate | 91 [77−100] | 95 [83−100] | 90 [76−108] | 1.00 | ||||
| Respiratory rate | 19 [16–20] | 20 [16–22] | 18 [16–20] | 0.675 | ||||
| Oxygen saturation | 94 [91−96] | 95 [88−96] | 94 [91−96] | 0.748 | ||||
| Hematocrit | 37.8 [33.2−40.5] | 36.9 [30.6−38.9] | 37.8 [34−40.6] | 0.845 | ||||
| Leucocytes | 7.6 [5.1−10.1] | 10.7 [5.7−18.5] | 7.4 [4.6−9.0] | 0.278 | ||||
| Lymph cells | 1.048 [655−1.212] | 1148 [759−1651] | 864 [550−1177] | 0.514 | ||||
| Monocytes | 393 [226−537] | 436 [227−557] | 352 [148−587] | 0.514 | ||||
| Platelets | 199 [146−259] | 219 [152−296] | 195 [136−244] | 0.845 | ||||
| Creatinine | 0.9 [0.6−1.5] | 1.3 [0.7−2] | 0.7 [0.5−1.0] | 0.065 | ||||
| C-reactive protein | 6.55 [3.95−16.85] | 11.7 [3.5−23.6] | 6.2 [3−16.6] | 0.821 | ||||
| Citokines levels | ||||||||
| IL4 (pg/mL) | 159 [128−184] | 150 [124−188] | 174 [130−182] | 0.775 | ||||
| TNF-alpha (pg/mL) | 670 [475−726] | 616 [454−705] | 678 [507−738] | 0.287 | ||||
| IL-10 (pg/mL) | 309 [217−447] | 293 [226−456] | 318 [217−446] | 0.775 | ||||
| IFN-gamma (pg/mL) | 239 [159−475] | 323 [166−570] | 208 [155−392] | 0.017 | ||||
| IL-6 (pg/mL) | 889 [653−1091] | 895 [679−1107] | 883 [645−1090] | 0.775 | ||||
Table 2.
Clinical and outcome variables in patients with more or less than ten days of symptoms of moderate and severe COVID-19 infection.
| <10 days of symptoms (n = 28) |
>10 days of symptoms (n = 28) |
||||
|---|---|---|---|---|---|
| n | % | n | % | P value | |
| Male | 19 | 68 | 21 | 75 | 0.777 |
| Intensive Care Unit | 14 | 50 | 14 | 50 | 0.182 |
| Intubation | 10 | 36 | 8 | 28 | 0.569 |
| Death | 11 | 39 | 8 | 28 | 0.408 |
| Continuous variables (Median with IQR) | |||||
| Age | 59 [45−73] | 63 [49−74] | 1.0 | ||
| Lenght of hospitalization | 10 [7–15] | 17 [11–22] | 0.75 | ||
| Temperature | 36.3 [36−36.6] | 36.9 [36.4−37.6] | 0.049 | ||
| SAP | 140 [127−151] | 130 [120−140] | 0.858 | ||
| DAP | 80 [70−85] | 82 [80−90] | 0.117 | ||
| HR | 86 [71−100] | 96 [84−103] | 0.343 | ||
| sO2 | 95 [90−96] | 93 [91−95] | 0.417 | ||
3.2. Laboratory results
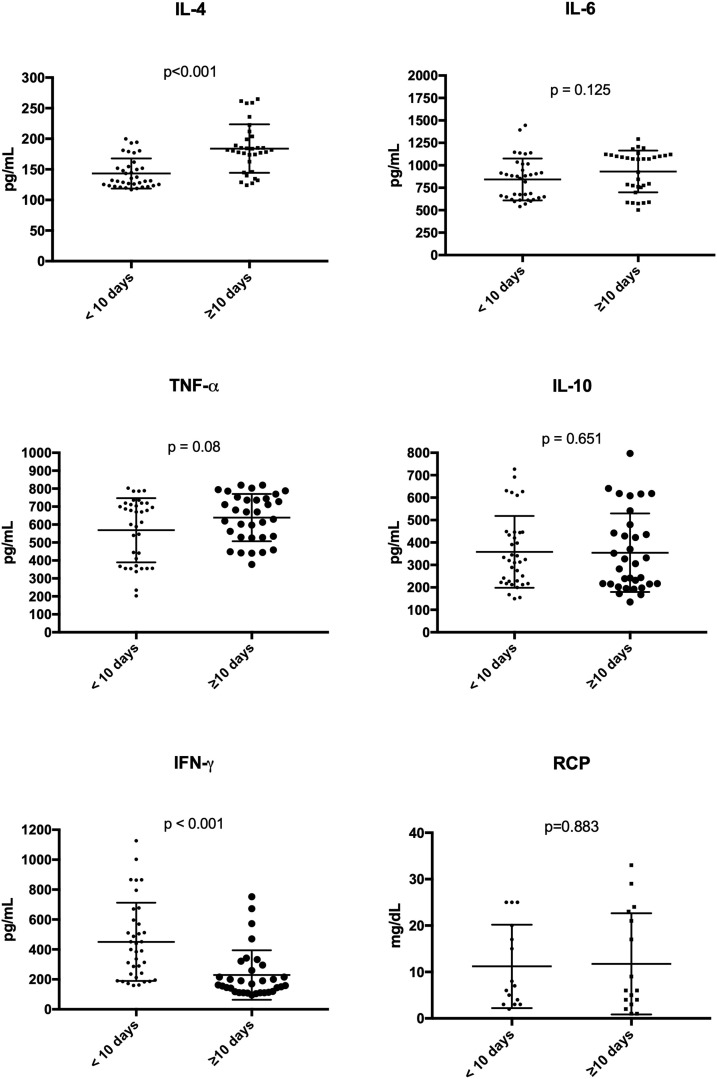
At admission, median leukocytes, lymph cells and platelets counts were 7600 [5100–10,100] cells/mm3, 1048 [655−1,212] cells/mm3, and 199 [146−259] platelets/mm3, respectively. Median creatinine was 0.9 [0.6−1.5] mg/dL. General laboratorial results are in Table 1. Median IL-6 and TNF-α were 889 [653−1091] pg/mL and 670 [475−726] pg/mL, respectively. Median IFN-γ and IL-10 were 239 [159−475] pg/mL and 309 [217−447] pg/mL, respectively. Median IL-4 was 159 [128−184] pg/mL. Differences between the groups (0–10d vs. >10d) are shown in Fig. 1 . IFN-γ levels were higher in patients from group 0–10d of symptoms (p < 0.001); while IL-4 levels were higher in patients from group >10d of symptoms (p < 0.001). Thus, IFN-γ and IL-4 was inversely proportional related (p = 0.03) (Table 3 ). There was no relation between use of steroids during treatment of COVID-19 and citokines levels (p > 0.05).
Fig. 1.
Levels of cytokines from patients with moderate and severe COVID-19 infection according to days of symptoms (data in the median with IQR).
Table 3.
Correlation of cytokines from patients with moderate and severe COVID-19 infection.
| IFN-γ vs. IL-4 | IFN-γ vs. TNF-α | IFN-γ vs. IL-10 | IFN-γ vs IL-6 | |
|---|---|---|---|---|
| Pearson r | −0.2895 | −0.07336 | 0.113 | −0.06725 |
| 95 % confidence interval | −0.5134 to -0.02882 | −0.3299 to 0.1933 | −0.1545 to 0.365 | −0.3244 to 0.1992 |
| R squared | 0.08382 | 0.005381 | 0.01276 | 0.004523 |
| P (two-tailed) | 0.0304 | 0.5911 | 0.4072 | 0.6224 |
3.3. Outcomes: ICU admission, oral intubation, and death
ICU admission and oral tube intubation (OTI) occurred in 58.9 % (n = 33) and 30.9 % (n = 18) of patients during the treatment. Global death rate was 32 % (n = 18). Outcomes (death or survival) according to each group of patients are demonstrated in Table 2.
3.4. Prognosis factors
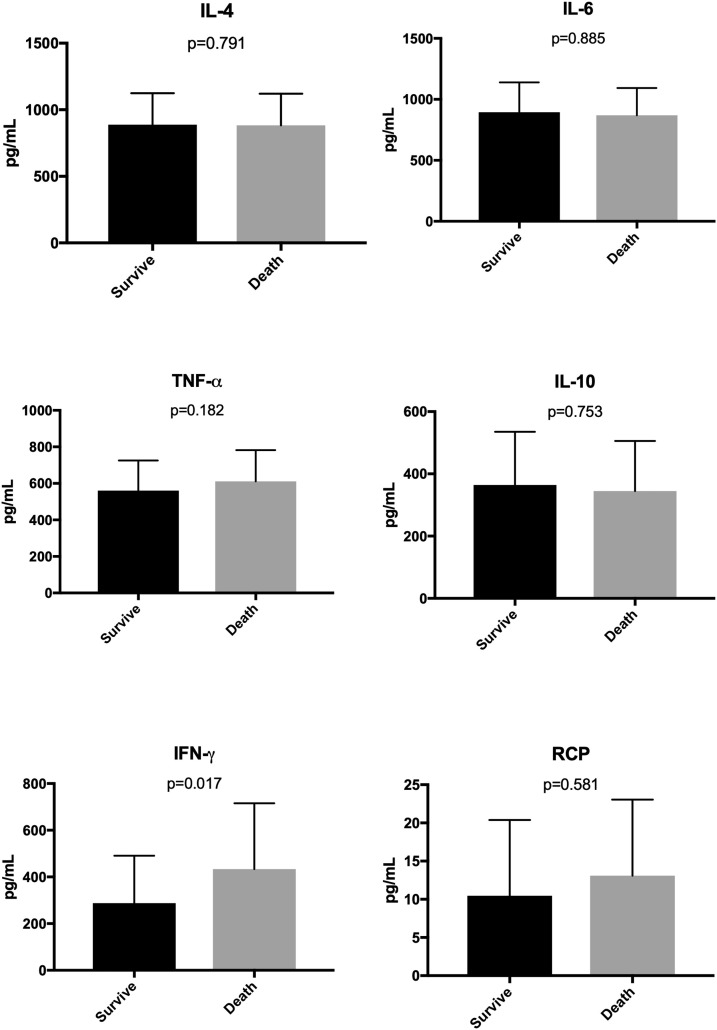
In univariate analysis, death was related to gender (p = 0.029), ICU (p < 0.001), OTI (p = 0.002), IFN-γ (p = 0.017) and length of hospitalization (p = 0.025); while survival was related to fever (p = 0.036). Cytokines analysis is shown in Fig. 2 . Nevertheless, in multivariate analysis, death remained related to age, gender, and IFN-γ (p < 0.05).
Fig. 2.
Levels of cytokines from patients with moderate and severe COVID-19 infection according to the outcome (data in the median with IQR).
4. Discussion
In this prospective cohort of patients, higher levels of IFN-γ were related to a poorer prognosis. These results demonstrate that an effective Th1 response is not sustained and probable it is followed by the development of Th2 immune adaptive response. IL-6 and IL-10 did not differ when compared to days of symptoms, evolution, nor outcomes. Previous studies elucidated the capacity of NK and NK T cells to produce IFN-γ before the specific Th1 adaptive immune response (Stetson et al., 2003, Vidal et al., 2011). Our results converge with this hypothesis once higher IFN-γ levels were detected in early COVID-19 infection than healthy populations (Forti et al., 1985). Nevertheless, these levels were not sustained after ten days of symptoms. In those with sustained IFN-γ levels, the mortality increased.
Th2 profile cytokine IL-4 presents an upward trend in disease progression in critically ill patients (Lucas et al., 2020). IL-4 levels may (i) inhibit naive CD4+ cells from proceeding to Th1 maturation or (ii) block IFN-γ gene transcription (Wurtz et al., 2004). On the other hand, the differentiation of T cells to the Th1 profile is driven by IFN-γ, IL-12, and IFNs, while IFN-γ exclusively acts as an inhibitor of the Th2 pathway, preventing the proliferation of Th2 cells (Murphy et al., 2000). Thus, our results might emphasize the dominant Th2 response in COVID-19 patients.
In our cohort, gender, and IFN-γ levels were related to poorer prognosis. Besides it, since the COVID-19 beginning, it was demonstrated that hospital admission was more likely in the male gender with comorbidities (Chen et al., 2020a; Yang et al., 2020). Hypertension, cardiovascular diseases, and diabetes were the most frequent comorbidities in our patients. These factors may explain the high ICU admission rate presented in this study. Furthermore, the death rate found was 32 %, which is higher than other general cohorts (Wu et al., 2020). Nevertheless, given the high ICU admission rate, it is expected that outcomes results become closer to cohorts of critically-ill patients (Abate et al., 2020).
Previous studies have not reported the association between IFN-γ and death, even evaluating the COVID-19-reactive CD69+ expressing IFN-γ producing CD8 + T in 25 patients with severe and moderate disease (Gimenez et al., 2020). Most studies have described the evident immunological dysfunction in the moderate and severe disease, with reduced expression of IFN-γ by CD4 + T, CD8 + T, and NK cells (Chen et al., 2020b). The same author found that IFN-γ by CD4 + T cells tended to be lower in severe cases than moderate cases. However, none of these studies evaluated the levels of IFN-γ with death. Some subsets of T lymphocytes correlated with in-hospital death and severity of illness, but only the innate immune cytokines were evaluated (Xu et al., 2020a). It was recently demonstrated that levels of the IFN-γ secreted by CD4+ and CD8 + T cells from patients with moderate disease were compatible with those in critically ill patients. In this case, IFN-γ increased over time in critically ill patients, but with decreased levels in moderate patients, contrasting with the results presented here. However, IFN-γ was highly correlated with viral load, suggesting that the virus can boost the secretion of these cytokines (Lucas et al., 2020).
In a previous report, the proportion of memory and naïve helper T cells decreased in severe cases (Qin et al., 2020). Patients with COVID-19 also have a lower level of regulatory T cells, suggesting that the adaptive immune response is prejudiced. However, another study showed a higher number of CD4 T cells producing IFN-γ in comparison with a control group (without disease) (De Biasi et al., 2020). In children with COVID-19, IFN-γ is increased, but with inferior levels than we found, suggesting a less severe disease in the pediatric population (Xiong et al., 2020). In cardiomyocytes, there is an up-regulation of genes responsible for IFN-γ suppression, also suggesting an effect IFN-γ in the cardiac function, in a sepsis-like pattern (Xu et al., 2020b). Furthermore, the relationship of IFN-γ disruption is not only associated with COVID-19 and the immune system, but there is a relationship of microbiome modification, altering the cell transcriptome with gene overexpression, which can be associated with the cytokine storm (Zhang et al., 2020b; Arnaldez et al., 2020).
Interleukin-4 plays a critical role in the Th2 pathway, being predominantly associated with fibrogenic inflammatory remodeling, while Th1 cells exert anti-fibrotic activity by secreting gamma interferon (IFN-γ) and interleukin 2. IL-4 also induce alternative activation of M2 macrophages, promoting the release of TGF-β and platelet-derived factor. This phase is characterized by the expansion of resident fibroblasts and the matrix remodelling (Wu et al., 2020). Since severe COVID-19 can lead to diffuse alveolar damage, which has a potential of developing septal fibrosis; recovering patients with high levels of IL-4 may might progress to pulmonary fibrosis aggravating impairment of the lung functions.
Our study has some limitations and strengths that merit consideration. As limitations, our study evaluated the first patients admitted during the outbreak in our city. Therefore, changes in clinical management during the evolving epidemics might have a differential impact on our studied outcomes. Our limited sample size might have decreased our power; however, also because of the pandemic, the findings of this study offer new, potentially useful information for this patient population. On the other hand, our grouping by symptom days may have standardized the disease's different stages when patients were admitted. Another concern is memory bias for onset of symptoms, because some patients could be in the wrong group (more or less 10 days).
We evaluated the Th1 and Th2 responses according to the time of evolution, so we were able to identify possible predictive markers such as (i) IFN-γ in early-stage and (ii) IL-4 in late-stage for the outcomes of discharge or death. Our results suggest that the moderate to the severe progression of COVID-19 may have been one of the causes of the immune responses developed by the host at the beginning of the COVID-19 infection, influencing the need for combination therapy to block these inflammatory mediators. The activation of the host immune response between Th1 or Th2 in COVID-19 infection may be related to the final result between discharge or death. Any source to control cytokines, such as IFN-γ, with combined therapies for clinical treatment will be important in the future for COVID-19 infection.
Transparency declaration
The authors have no interest conflicts. Felipe Tuon and Lucia de Noronha are CNPQ researchers,
Funding
This study has no funding.
CRediT authorship contribution statement
Ana Carolina Gadotti: Investigation. Marina de Castro Deus: Investigation. Joao Paulo Telles: Formal analysis, Writing - original draft. Rafael Wind: Investigation. Marina Goes: Investigation. Roberta Garcia Charello Ossoski: Supervision. Alessandra Michalski de Padua: Project administration. Lucia de Noronha: Project administration, Writing - original draft. Andrea Moreno-Amaral: Methodology, Writing - original draft. Cristina Pellegrino Baena: Data curation, Formal analysis, Methodology, Writing - original draft. Felipe Francisco Tuon: Formal analysis, Conceptualization, Writing - review & editing.
Acknowledgments
None.
References
- Zhang X., Tan Y., Ling Y. Viral and host factors related to the clinical outcome of COVID-19. Nature. 2020;583(July 7816):437–440. doi: 10.1038/s41586-020-2355-0. [DOI] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. February 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H., Ma Q., Li C. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg. Microbes Infect. 2020;9(December1):1123–1130. doi: 10.1080/22221751.2020.1770129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J., Ou J., Qiu X. A tool for early prediction of severe coronavirus disease 2019 (COVID-19): A multicenter study using the risk nomogram in Wuhan and Guangdong, China. Clin. Infect. Dis. 2020;71(15):833–840. doi: 10.1093/cid/ciaa443. July 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W., Liang H., Ou L. Development and validation of a clinical risk score to predict the occurrence of critical illness in hospitalized patients with COVID-19. JAMA Intern. Med. 2020;(May 12) doi: 10.1001/jamainternmed.2020.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D., Degenhardt F., Bujanda L. Genomewide association study of severe Covid-19 with respiratory failure. N. Engl. J. Med. 2020;(Jun 17) doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Mao L., Yuan X. Prediction model based on the combination of cytokines and lymphocyte subsets for prognosis of SARS-CoV-2 infection. J. Clin. Immunol. 2020;(Jul 13) doi: 10.1007/s10875-020-00821-00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson C.M., O’Dee D., Hamilton T., Nau G.J. Cytokines involved in interferon-gamma production by human macrophages. J. Innate Immun. 2010;2(1):56–65. doi: 10.1159/000247156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D.E., Garcia-Sastre A. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 2001;12(2–3):143–156. doi: 10.1016/s1359-6101(00)00027-7. Jun-Sep. [DOI] [PubMed] [Google Scholar]
- Yin K., Gribbin E., Wang H. Interferon-gamma inhibition attenuates lethality after cecal ligation and puncture in rats: implication of high mobility group box-1. Shock. 2005;24(October 4):396–401. doi: 10.1097/01.shk.0000175556.03300.c6. [DOI] [PubMed] [Google Scholar]
- WHO . World Health Organization; 2020. Clinical Management of Severe Acute Respiratory Infection When Novel Coronavirus (2019-nCoV) Infection is Suspected: Interim Guidance, 28 January 2020. [Google Scholar]
- Stetson D.B., Mohrs M., Reinhardt R.L. Constitutive cytokine mRNAs mark natural killer (NK) and NK T cells poised for rapid effector function. J. Exp. Med. 2003;198(7):1069–1076. doi: 10.1084/jem.20030630. Oct 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal S.M., Khakoo S.I., Biron C.A. Natural killer cell responses during viral infections: flexibility and conditioning of innate immunity by experience. Curr. Opin. Virol. 2011;1(December 6):497–512. doi: 10.1016/j.coviro.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forti R.L., Moldovan R.A., Mitchell W.M. Application of an objective biological assay of human interferons to clinical specimens and a survey of a normal population. J. Clin. Microbiol. 1985;21(May 5):689–693. doi: 10.1128/JCM.21.5.689-693.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas C., Wong P., Klein J. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;(Jul 27) doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtz O., Bajenoff M., Guerder S. IL-4-mediated inhibition of IFN-gamma production by CD4+ T cells proceeds by several developmentally regulated mechanisms. Int. Immunol. 2004;16(March 3):501–508. doi: 10.1093/intimm/dxh050. [DOI] [PubMed] [Google Scholar]
- Murphy K.M., Ouyang W., Farrar J.D. Signaling and transcription in T helper development. Annu. Rev. Immunol. 2000;18:451–494. doi: 10.1146/annurev.immunol.18.1.451. [DOI] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. Feb 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Yu Y., Xu J. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8(May 5):475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Chen X., Cai Y. Risk factors associated with acute respiratory distress syndrome and death in patients with Coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020;(Mar 13) doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abate S.M., Ahmed Ali S., Mantfardo B., Basu B. Rate of Intensive Care Unit admission and outcomes among patients with coronavirus: a systematic review and Meta-analysis. PLoS One. 2020;15(7) doi: 10.1371/journal.pone.0235653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez E., Albert E., Torres I. SARS-CoV-2-reactive interferon-gamma-producing CD8+ T cells in patients hospitalized with coronavirus disease 2019. J. Med. Virol. 2020;(Jun 24) doi: 10.1002/jmv.26213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Wu D., Guo W. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130(5):2620–2629. doi: 10.1172/JCI137244. May 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Fan C.Y., Wang A.L. Suppressed T cell-mediated immunity in patients with COVID-19: a clinical retrospective study in Wuhan, China. J. Infect. 2020;81(July 1):e51–e60. doi: 10.1016/j.jinf.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C., Zhou L., Hu Z. Dysregulation of Immune Response in Patients With Coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2020;71(15):762–768. doi: 10.1093/cid/ciaa248. Jul 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Biasi S., Meschiari M., Gibellini L. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat. Commun. 2020;11(1):3434. doi: 10.1038/s41467-020-17292-4. Jul 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X., Chua G.T., Chi S. Haematological and immunological data of Chinese children infected with coronavirus disease 2019. Data Brief. 2020;31(August) doi: 10.1016/j.dib.2020.105953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Ma M., Xu Y. Single-cell transcriptome analysis indicates new potential regulation mechanism of ACE2 and NPs signaling among heart failure patients infected with SARS-CoV-2. medRxiv. 2020;(May 15) doi: 10.1101/2020.04.30.20081257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Ai J.W., Yang W. Metatranscriptomic characterization of COVID-19 identified a host transcriptional classifier associated with immune signaling. Clin. Infect. Dis. 2020;(May 28) doi: 10.1093/cid/ciaa663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaldez F.I., O’Day S.J., Drake C.G. The Society for Immunotherapy of Cancer perspective on regulation of interleukin-6 signaling in COVID-19-related systemic inflammatory response. J. Immuno Ther. Cancer. 2020;8(May 1) doi: 10.1136/jitc-2020-000930. [DOI] [PMC free article] [PubMed] [Google Scholar]