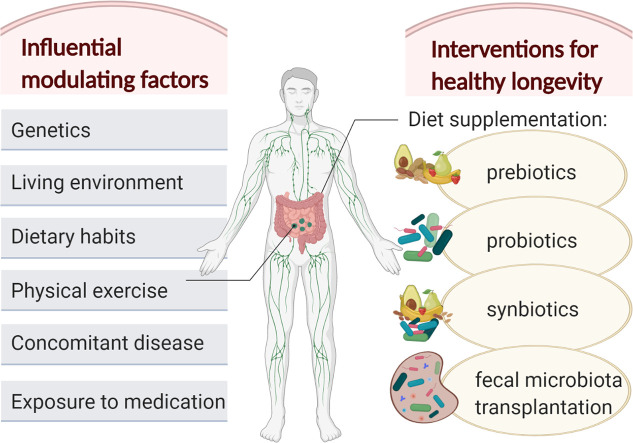
Abstract
Our gut microbiota is a complex and dynamic ecosystem with a paramount role in shaping our metabolic and immunological functions. Recent research suggests that aging may negatively affect the composition, diversity, and function of our microbiota mainly due to alterations in diet and immunologic reactivity (i.e. immunosenescence), and increased incidence of certain diseases and, therefore, increased exposure to certain medication (e.g. antibiotics, proton pump inhibitors). In turn, this aging-related gut dysbiosis may contribute to the initiation and/or progress of other metabolic diseases, and consequently, to a decrease in healthy longevity. On the positive side, promising therapeutic interventions, such as diet supplementation with prebiotics, probiotics and synbiotics, or fecal microbiota transplantation, aimed to counteract these aging-related deleterious consequences, could improve our health, and extend our healthy lifespan. In this context, the current review aims to assess the latest progress in identifying the key elements affecting the gut microbiota of the older adults and their mechanism of action, and the effectiveness of the therapeutic interventions aimed at restoring the diversity and healthy functions of the gut microbiota in older individuals.
Keywords: Aging, Human gut microbiota, Dietary alterations, Concomitant disease, Therapeutic interventions, Healthy longevity
Graphical abstract
1. Introduction
Over the past two centuries, improvements in living conditions, education, nutrition, and medical services have roughly doubled human life expectancy (Oeppen and Vaupel, 2002). Current estimates show that life expectancy surpasses 60, 70 and 80 years (yr) of age in 87.7% (200/228), 73.7% (168/228) and 20.6% (47/228) of the world countries, respectively (Geoba, 2020). In terms of forecasts, the population ≥ 60 yr is expected to reach ~2 billion by 2050 (WHO, 2017). The percentage of those ≥80 yr in the European Union will presumably have doubled between 2017 (5.5%) and 2080 (12.7%) (EUROSTAT, 2019). From a societal perspective, these figures present several challenges in terms of a constant increase in healthcare costs (~5-fold in the past 50 years) (Cutler et al., 2006; Harper, 2014; Jenkner and Leive, 2010) and special dietary requirements for the older population (especially for those affected by malnutrition) (Milne et al., 2009), but also may create opportunities for multidisciplinary approaches to improve the quality of life of the older adults (Jedrusek-Golinska et al., 2020). In terms of healthcare costs, some studies argue that although aging affects health spending, it is not the main driver; progress in medical technology and income growth were suggested to be the most important factors in increasing healthcare expenditure (Smith et al., 2009).
From a healthcare perspective, a longer life does not necessarily mean more health life-years (WHO, 2019). Unfortunately, aging often manifests itself negatively through frailty, defined as “a state of increased vulnerability to poor resolution of homoeostasis after a stressor event, which increases the risk of adverse outcomes, including falls, delirium, and disability” (Clegg et al., 2013). Mammalian aging is a complex, multifactorial process, with nine hallmarks being defined and described relatively recently (López-Otín et al., 2013; López-Otín et al., 2016): genomic instability, telomere attrition, epigenetic alterations, loss of proteostasis, deregulated nutrient-sensing, mitochondrial dysfunction, cellular senescence, stem cell exhaustion, and altered intercellular communication. All these hallmarks are linked to metabolic perturbations (López-Otín et al., 2016), which are associated in older adults with irreversible declines in physical and cognitive functions, leading to possible combinations and clustering of chronic diseases (Finkel and Holbrook, 2000; Kennedy et al., 2014), and to the onset of frailty (Ferrucci and Fabbri, 2018).
Recent research has suggested that aging may be also associated with a different gut microbiota composition (i.e. a rearrangement) and with the increased presence of certain pathobionts, as compared to younger adults (Biagi et al., 2010; Claesson et al., 2011; Odamaki et al., 2016). This may be related to the heightened disease progression (and exposure to medication) and the decrease in immunocompetence in the older population (Hopkins et al., 2001). According to WHO, most degenerative diseases (cardiovascular and cerebrovascular disease, diabetes, osteoporosis and cancer) are affected by our long-term dietary habits and lifestyle (WHO, 2019). Diet is also one of the most influential factors affecting our gut microbiota composition, diversity and function (Cotillard et al., 2013; David et al., 2014; Gibson et al., 2004; Sonnenburg and Bäckhed, 2016; Zmora et al., 2019). Therefore, age-related dietary alterations could negatively impact the gut microbiota health in the older adults, and, consequently, their healthy longevity (Clements and Carding, 2018). To counteract these negative aspects, dietary interventions targeting the gut microbiota of the older adults could be a suitable solution associated with health benefits (Cotillard et al., 2013; Flanagan et al., 2020; Ghosh et al., 2020; Lynch and Pedersen, 2016).
How aging affects the gut microbiota, the effects of the age-related changes on the health status of the host, and possible therapeutic approaches to counteract the negative aspects of dysbiosis have recently received considerable research interest (Biagi et al., 2011; Bischoff, 2016; Maffei et al., 2017; Marchesi et al., 2016; O'Toole and Jeffery, 2015, O'Toole and Jeffery, 2018; Saraswati and Sitaraman, 2014; Sarkar and Pitchumoni, 2017; Stavropoulou and Bezirtzoglou, 2019; Valle Gottlieb et al., 2018; Xu et al., 2019a). Although this research area has been expanding tremendously over the past years, there are still major gaps in our understanding of the functional interactions between the complex microbial community and the human host, especially at an advanced age. For example, which are the main factors which negatively influence our gut microbiota diversity and functions? Are they different in the older population as compared to younger adults? Are there specific microbiome signatures for longevity or are they rather reflective of health status? Are short-term effects (e.g. antibiotic exposure) as important as the long-term ones (e.g. food preferences), and which ones are more difficult to counterbalance? Are therapeutic interventions targeting the gut microbiota less effective or less safe in the older adults? Are these interventions suitable for entire populations, or should they be targeted individually (i.e. personalized modulation of the microbiota)? Which therapeutic intervention should be recommended against a certain condition (e.g. chronic constipation)? In this context, our narrative review aims to summarize the role and mechanisms of the major factors influencing a healthy microbiota and the most recent dietary and therapeutic advancements in counteracting these deleterious consequences in the older adults in comparison to younger adults.
2. The main factors affecting the gut microbiota of the older people
The human gut contains 1013–1014 microorganisms (the microbiota) with 100–150 times more genes (the microbiome) than our own genome (Gill et al., 2006; Huttenhower et al., 2012; Qin et al., 2010). This complex ecosystem provides a remarkable enzymatic capability, able to break down food components that are not digested directly by the host, to extract nutrients and energy from them, and to produce metabolites (such as short-chain fatty acids; SCFAs) with important roles in modulating endocrine and immunologic functions (Clemente et al., 2012; Hooper et al., 2012; Huttenhower et al., 2012; Tremaroli and Bäckhed, 2012). A healthy gut microbiota also plays an essential role in protecting the host against opportunistic strains and pathogens (Nagpal et al., 2018; Sullivan, 2001). On the other hand, gut dysbiosis, translated as imbalances in the gut microbiota in terms of composition, diversity and functions, can have a serios impact on the well-being of the host, with consequences ranging from gastrointestinal disorders to cardiometabolic and neurologic conditions (Hsiao et al., 2013; Lynch and Pedersen, 2016; Nicholson et al., 2012; Round and Mazmanian, 2009). Fortunately for us, research has shown that the gut microbiota is rather stable in individuals and can even be restored after perturbation (i.e. dysbiosis) (Costello et al., 2009; Lozupone et al., 2012; Seksik et al., 2003; Tannock et al., 2000; Wu et al., 2011).
Population- and age-specific differences in gut microbiomes show unique characteristics specific to different locations and lifestyles (Yatsunenko et al., 2012). Landmark studies have shown that the microbiota of older adults has more inter-individual variation and that the composition differs as compared to younger adults (Biagi et al., 2010; Claesson et al., 2011; Odamaki et al., 2016). Additionally, factors such as infections and antibiotics, can transiently alter the stability of the gut microbiota and thereby have negative effects on the well-being of the host (Forsythe et al., 2010). An analysis of gut microbial communities proposed three predominant enterotypes, dominated by Bacteroides, Prevotella, and Ruminococcus, respectively (Arumugam et al., 2011), which were found to be independent of nationality, sex, age, or body mass index. Details on the taxonomic gut microbiota composition are given in Supplementary Table 1. Besides, 12 genes that significantly correlate with age were identified (Arumugam et al., 2011). Starch degradation enzymes were found to increase with age (as a possible response to a decline in the host breakdown of dietary carbohydrates), while gut stress response was shown to decrease (possibly related to immunosenescence) (Franceschi et al., 2000).
Two studies from 2010 to 2011, investigating the age-related changes in the gut microbiota, found a lower Firmicutes to Bacteroidetes (F:B) ratio in the older adults as compared to younger adults, and a reduction in species producing SCFAs, and in particular butyrate (Clostridium clusters) (Biagi et al., 2010; Claesson et al., 2011), reflecting a maladaptive remodeling of the gut microbiota with age (Santoro et al., 2018). The microbiota remodeling was confirmed in 2015, by the results of a study depicting the evolution of the gut microbiota composition from birth to extreme aging (i.e. over 100 yr) in a large Japanese cohort (N = 367) (Odamaki et al., 2016).
The consequences of microbiota remodeling (e.g. changes in F:B ratio) are not fully understood, but animal and human studies have indicated that a lower relative abundance of Bacteroidetes and higher relative abundance of Firmicutes can influence energy balance (Ley et al., 2005; Ley et al., 2006; Schwiertz et al., 2010; Turnbaugh et al., 2008). A mutualistic host–microbe relationship (both partners benefit) was shown to promote more efficient energy extraction from diet (e.g. higher monosaccharides and SCFA production from complex polysaccharides) and higher energy storage in adipocytes (Bäckhed et al., 2004; Bäckhed et al., 2005; Ley et al., 2005; Turnbaugh et al., 2006). Additionally, some human studies have also shown a correlation with diabetes (Larsen et al., 2010; Murri et al., 2013), carbohydrate consumption (Duncan et al., 2007), and SCFA production (Fernandes et al., 2014; Schwiertz et al., 2010). SCFAs have key roles in gut homeostasis (promoting the expansion and differentiation of regulatory T cells) (Lathrop et al., 2011), acting as energy sources, and inflammation and signaling modulators (Rios-Covian et al., 2016; Skelly et al., 2019; Smith et al., 2013).
Alterations in the microbiota composition were also associated with frailty in a small cohort of older adults (N = 23; range: 73–100 yr) from a single residence location in The Netherlands (Van Tongeren et al., 2005). The impact of microbiota composition on immunosenescence and frailty across the community was later confirmed in 2 large Irish cohorts of older adults by Claesson et al. (N = 178; age range 64–102 yr) and Jeffery et al. (N = 371; age range 64–102 yr) (Claesson et al., 2012; Jeffery et al., 2016).
In the study from 2012, Claesson et al. showed a significant difference in gut microbiota structure between long-stay care and community-dwelling older adults in terms of bacterial diversity, correlating with long-stay-associated frailty and poorer health (Claesson et al., 2012). The community dwellers had a higher similarity with healthy young adults, showing the important effect of diet and lifestyle on the gut microbiota composition (Claesson et al., 2012). Dietary analysis revealed significant differences between the two groups: participants in the community had a ‘low–moderate fat/high fiber’ diet, while a majority of participants (83%) from long-stay care homes had a ‘moderate–high fat/low fiber’ diet; these differences correlated with their structural changes in gut microbiota ecology (Claesson et al., 2012).
In the later analysis from 2016, Jeffery et al. showed that fraily-related changes in bacterial populations were gradual (for at least 18 months) in older adults from long term residential care and that individuals with low initial microbiota diversity were prone to larger changes (Jeffery et al., 2016). The authors suggest that this timescale should be taken into consideration in intervention studies to accurately assess the effects of the intervention versus normal background changes.
In general, the diversity and stability of the adult gut microbiota declines with age, in connection to frequent comorbidities and decreased immunocompetence (Lynch and Pedersen, 2016). However, there are exceptions to this, namely in the healthy and active centenarians (Biagi et al., 2017; Cătoi et al., 2020; O'Toole and Jeffery, 2018; Santoro et al., 2018), underlying the importance of stratifying populations by health status in studies related to microbiome composition and aging. Several studies published in 2015–2017 enrolled exceptionally old people from Guangxi, China (8 participants; 100–108 yr) (Wang et al., 2015), Sichuan, China (67 participants; 90–102 yr) (Kong et al., 2016), and Emilia Romagna, Italy (15 participants; 99–104 yr) (Biagi et al., 2016). These studies showed that healthy centenarians are a special population showing high diversity in terms of species composition and an increased occurrence of butyrate-producing bacteria (e.g. Roseburia of Clostridium cluster XIVa) (Kong et al., 2016; Wang et al., 2015). Butyrate was found to play important roles in human health, such as establishing immunologic homeostasis in the gut (Furusawa et al., 2013). Additionally, butyrate producers such as Faecalibacterium and Coprococcus have been recently associated with higher quality of life indicators (Valles-Colomer et al., 2019).
Another recent study from Jiangsu, China, including 198 healthy, active centenarians (94 yr and above), showed that age-related fecal microbiota dysbiosis is mitigated in this group; the overall composition of their gut microbiota showed no significant difference from that of middle-age adults (30–50 yr), suggesting an active involvement of the gut microbiota in healthy aging (Bian et al., 2017). Of note, all these centenarian populations were recruited from small, homogeneous geographical areas (i.e. from the same village, city, or province). They also lived with families in the same place all their life, had a high quality of life, and enjoyed daily activities.
The critical importance of changing the geographical environment and diet on the health of our gut microbiota has been once again demonstrated by two recent studies (Jha et al., 2018; Vangay et al., 2018). The first one, conducted in four rural villages in Nepal, investigated how the gut community changes in response to shifts in traditional human lifestyles, i.e. living on a gradient from foraging to farming, and showed that the shifts in microbiota composition are more pronounced in the population that switched to farming two-three centuries ago and that the higher abundance of taxa was maintained in the population that kept foraging lifestyle aspects (Jha et al., 2018). In the second study, Vangay et al. (2018) showed that immigrating from a Thai refugee camp to the USA was associated with profound changes in the gut microbiota composition and diversity (loss of native strains, loss of fiber degradation capabilities), including shifts from Prevotella to Bacteroides dominance (Vangay et al., 2018).
Having surveyed the relevant literature in the field, we can assume that some the most influential factors which can modulate the composition and overall health of the human gut microbiota (and especially of the older adults) are: genetics and geographical environment, diet and exercise, and concomitant disease and medication. These factors are treated comprehensively in terms of impact and mechanism of action in the following subsections.
2.1. Genetic makeup versus environmental influence on the gut microbiota
Based on the fact that family members were shown to have more similar microbiotas than unrelated individuals (Turnbaugh et al., 2009a; Yatsunenko et al., 2012), and that direct relationships were established between metabolic disease and both the gut microbiome (Ley et al., 2005; Turnbaugh et al., 2006) and variation in host genetics (Barroso and McCarthy, 2019; Frazer et al., 2009), previous studies in twins and first-degree relatives have identified a substantial role of genetics in modulating the gut microbiota composition, with several heritable taxa (e.g., the family Christensenellaceae and the methanogen Methanobrevibacter smithii) (Goodrich et al., 2016; Goodrich et al., 2014; Turpin et al., 2016). However, a more recent study from 2018 argues that host genetics have only a minor role in determining our microbiome composition and that environmental factors such as diet, lifestyle, and drug interactions are the main influencers (Rothschild et al., 2018). This study was conducted in 1046 healthy Israeli individuals of several different ancestral origins and found that diet and lifestyle may be responsible for over 20% of the variance in our microbiome β-diversity (Rothschild et al., 2018). The same researchers also estimated the genetic influence on the gut microbiome taxa of 2252 twins from a previously studied UK cohort (Goodrich et al., 2016), and found that the average heritability can be as low as 2% (Rothschild et al., 2018). These findings suggest that therapeutic interventions targeting microbiome alterations could be carried out across various populations, irrespective of their genetic background.
Our living environment can have profound influences on our life quality, and our gut microbiota composition (Mueller et al., 2006). A very recent study has even shown correlations between yard vegetation in the living area and the abundance of some beneficial bacterial taxa (e.g. increased F:B ratio) (Parajuli et al., 2020). Hospitalization is another important factor in altering the gut microbiota of the older adults. A marked reduction in the Bacteroides-Prevotella group and an increase in Enterobacteria were observed following hospitalization as compared to healthy older volunteers living in the local community (Bartosch et al., 2004). Claesson et al. (2012) have also shown that people from long-stay care homes have a significantly less diverse microbiota than those living in a community, and this translates into increased frailty, reduced muscle mass, and poorer mental activity (Claesson et al., 2012).
Future large-scale studies should continue to elucidate the exact role of genetics in our gut microbiome composition and consequences for human health. Although environment appears to be the main contributor in shaping our gut microbiota, host genotype could also influence microbiome composition indirectly via gene-diet interactions (food preference), as shown in the case of regulation of Bifidobacterium abundance in individuals with lactose intolerance (Bonder et al., 2016). Additionally, epigenetic mechanisms (e.g. DNA methylation, histone modification, small RNAs) may play a role in transmitting to children behaviors acquired by parents via adverse exposure to external factors (e.g. stress, toxins, over- or under-nutrition, obesity, diabetes) (Barrès and Zierath, 2016; Sales et al., 2017).
2.2. Dietary habits, physical activity, and the gut microbiota
Based on current scientific evidence, diet is one of the most important determining factors of gut microbiota community composition, diversity and function (Cotillard et al., 2013; Gibson et al., 2004; Sonnenburg and Bäckhed, 2016; Zmora et al., 2019). Therefore, variations among individuals in terms of relative abundance of gut microorganisms and enterotype partitioning can derive from different long-term dietary habits (Cordain et al., 2005; Wu et al., 2011). Therefore, if there is a direct link between an enterotype and disease, long-term dietary interventions may be essential to modulate a certain enterotype to improve health. Short-term changes can be significant and rapid, but a healthy microbiome can rapidly adapt and restore perturbations (David et al., 2014; Wu et al., 2011). However, this ability decreases with increased age, as some gastrointestinal disorders become more prevalent in the older population (Nagpal et al., 2018; Saffrey, 2014).
The influence of transgenerational dietary habits on the gut microbiota composition was clearly demonstrated 10 years ago by a study comparing the fecal microbiota of western European and African children from a village in Burkina Faso (De Filippo et al., 2010). The diet of the 14 healthy children from Africa was rich in fiber (10.0 g/d [2.26%] in 1–2 yr and 14.2 g/d [3.19%] in 2–6 yr), similar to that of ancient populations living mainly from agriculture (Cordain et al., 2005), while the 15 Italian children had a typical western diet, high in animal protein, sugar, starch, and fat and low in fiber (5.6 g/d [0.67%] in 1–2 yr and 8.4 g/d [0.9%] in 2–6 yr). Using 16S rDNA sequencing and biochemical analyses, the study found significant differences between the two study groups in terms of gut microbiota composition: African children had more Bacteroidetes and less Firmicutes (p < 0.001), many bacterial genes for cellulose and xylan hydrolysis from the genera Xylanibacter, Prevotella, Butyrivibrio, and Treponema (absent in the European children), and more SCFAs (p < 0.001); European children had significantly more Enterobacteriaceae (Shigella and Escherichia). These findings suggested that the gut microbiota of the African children evolved to maximize energy intake from their polysaccharide diet and to protect them from inflammations and non-infectious colonic diseases. The study results have suggested that diet has a major role in shaping the gut microbiota, surpassing in importance variables such as ethnicity, sanitation, hygiene, geography, and climate. Additionally, the authors hypothesized that a diet rich in plant polysaccharides and low in sugar and fat could increase the proportion of SCFA-producing bacteria (De Filippo et al., 2010). In 2018, Zhao et al. used successfully a high-fiber diet (84 days) for a targeted restoration of SCFA producers to improve hemoglobin A1c levels in type 2 diabetes patients (Zhao et al., 2018a).
A later study from 2012, has shown similar differences between healthy Amerindians from the Amazonas of Venezuela, residents of rural Malawian communities, and inhabitants of USA metropolitan areas (Yatsunenko et al., 2012). The included individuals covered a broad age range (0–70 yr). The first finding of this study was that in all three populations, it took 3 years from birth for the phylogenetic composition of the microbiota to evolve towards an adult-like configuration. In comparison, an earlier study has suggested that the microbial ecosystem may have the characteristics of the adult gastrointestinal tract within a year after birth (Palmer et al., 2007). Second, the study found more pronounced differences between children from the different populations than between adults (Yatsunenko et al., 2012). Third, there was a clear separation across ages in the phylogenetic composition of fecal microbiota between USA compared to Malawian and Amerindian gut communities. Finally, the bacterial diversity increased with age in all populations, but the fecal microbiota of the USA adults was the least diverse when compared to the other populations (Yatsunenko et al., 2012).
Further insights have been given by studies of the gut microbiota of the Hadza hunter–gatherers from Tanzania (Fragiadakis et al., 2019; Rampelli et al., 2015; Schnorr et al., 2014; Smits et al., 2017). These studies showed a high degree of bacterial diversity as compared to industrialized populations, and a unique enrichment in metabolic pathways and seasonal cycling of species. The seasonal cycling was adapted to seasonal diet: complex polysaccharides (fiber-rich tubers and baobab) during the wet season and meat from hunting during the dry season (Fragiadakis et al., 2019; Smits et al., 2017). Having considered the data from the aforementioned studies and several more examples reviewed by (Moeller, 2017), we have summarized the main differences between the traditional microbiota (i.e. hunter gatherers, remote rural populations) and the westernized/industrialized microbiota in Table 1 .
Table 1.
Comparison between traditional and modern (industrialized) microbiotas.
| Parameter | Traditional microbiota | Industrialized microbiota | References |
|---|---|---|---|
| Diet | Low fat, low sugar, high fiber Low added salt (low Na—K ratio) Low ratio of n-6 to n-3 polyunsaturated fatty acids (3:1) |
High fat, high sugar, low fiber High added salt (high Na—K ratio) High ratio of n-6 to n-3 polyunsaturated fatty acids (10:1) |
(Cordain et al., 2005; De Filippo et al., 2010; Sonnenburg and Sonnenburg, 2019a; Sonnenburg and Sonnenburg, 2019b; Vangay et al., 2018; Yatsunenko et al., 2012) |
| Diversity | High microbial richness and biodiversity | Low microbial richness and biodiversity | (De Filippo et al., 2010; Moeller, 2017; Sonnenburg and Sonnenburg, 2019a; Sonnenburg and Sonnenburg, 2019b; Vangay et al., 2018; Yatsunenko et al., 2012) |
| Functions | Increased fiber degraders and SCFA producers | Increased mucus degraders and antibiotic-resistant strains | (De Filippo et al., 2010; Sonnenburg and Sonnenburg, 2019a; Vangay et al., 2018) |
| Composition | Higher relative abundance of Prevotella, reduction of Bacteroides | Higher relative abundance of A. muciniphila, increased Bacteroides | (De Filippo et al., 2010; Fragiadakis et al., 2019; Schnorr et al., 2014; Vangay et al., 2018) |
| Significant presence (30% of taxa) of Prevotellaceae, Spirochaetaceae and Succinivibrionaceae | Low presence (3% of taxa) of Prevotellaceae, Spirochaetaceae and Succinivibrionaceae | (Fragiadakis et al., 2019; Schnorr et al., 2014) | |
| Flexibility | Seasonal dynamics | No seasonal dynamics | (Fragiadakis et al., 2019; Smits et al., 2017) |
| Disease incidence | Low incidence of non-communicable chronic diseases | High incidence of non-communicable chronic diseases | (Cordain et al., 2005; Moeller, 2017; Schnorr et al., 2014; Sonnenburg and Sonnenburg, 2019a) |
Data from these studies suggest that the significant changes in diet and living conditions promoted especially by industrial advances (e.g. processed foods, antibiotics, sanitized environments) have negatively altered our gut microbiota in terms of diversity and functions, with profound implications for the current prevalence of chronic diseases in the Western civilization (Cordain et al., 2005; Sonnenburg and Sonnenburg, 2019a; Sonnenburg and Sonnenburg, 2019b).
The seasonal dynamics seem to be lost within our ‘industrialized’ microbiota, together with a significant reduction in dietary fiber digestibility, which can occur in a relatively short time post significant changes in diet or geographical location (Vangay et al., 2018). However, an enhanced human dietary flexibility was shown in young adults (21–33 yr) in a study conducted in the USA showing that a short-term (i.e. five days) consumption of diets composed entirely of animal or plant products can alter significantly the microbial community structure (David et al., 2014).
In the older adults, optimal dietary requirements may be hindered by several additional factors on top of gastrointestinal disorders (Nagpal et al., 2018; Saffrey, 2014). For example, deficient oral health could negatively influence nutritional status via a lower desire and ability to eat solid foods, for example (Razak et al., 2014). Missing and/or painful teeth can lead to chewing and swallowing deficiencies, which in turn may also contribute to malnutrition (Mann et al., 2013; Salazar et al., 2017). The prevalence of oral bacterial transition to the gut may be higher in older people than in younger adults (Iwauchi et al., 2019); this could be influenced by long-term exposure to proton pump inhibitors (Imhann et al., 2016; Jackson et al., 2016). Aging is also frequently associated with chronic constipation, possibly due to suboptimal intake of dietary fiber, poor fluid intake and prolonged exposure to medication (Gallegos-Orozco et al., 2012).
Regular exercise and physical activity have been associated with a better quality of life and better health for decades (Penedo and Dahn, 2005). Physical exercise was also shown to positively impact the gut microbiota, by restoring the F:B ratio, improving SCFA production, and boosting intestinal mucosal immunity (Clarke et al., 2014; Mach and Fuster-Botella, 2017; Mailing et al., 2019; Monda et al., 2017). Animal studies have shown that exercise can induce alterations in the gut microbiota at the genus level, which are different from the dietary effects (Evans et al., 2014; Matsumoto et al., 2008; Petriz et al., 2014; Queipo-Ortuno et al., 2013). Older adults (human) data on the possible effects of exercise on the gut microbiota are scarce, but some examples have been published recently (Taniguchi et al., 2018; Zhu et al., 2020).
The first study (Taniguchi et al., 2018), a randomized crossover trial conducted in Japan with 33 older men (62–76 yr), evaluated the short-term (5 weeks) endurance exercise effects, and found very little impact on diversity and composition on the gut microbiota (e.g. reduced relative abundance of Clostridium difficile, positively correlated with changes in the visceral fat area). In comparison, data collected in a study involving younger athletes (31.3 ± 6.1 yr) has revealed more evident metabolic changes (i.e., increased species richness, increased organic acids, decreased nucleic acids) in the gut environment after running a half-marathon (Zhao et al., 2018b).
The second recent study, evaluated the impact of exercise frequency on gut microbiota health of 897 older adults (>61 yr) as compared to younger adults (18–60 yr) (Zhu et al., 2020). The analysis revealed significant age-related differences both at the phylum and family levels. At the phylum level, Proteobacteria was higher, while Actinobacteria was lower in the older group; the F:B ratio was not significantly different between the two groups. At the family level, Desulfovibrionaceae and Enterobacteriaceae were significantly increased, while Bifidobacteriaceae was significantly decreased in the older adults. Some of these differences were alleviated in the daily exercise older group: Actinobacteria increased while Cyanobacteria gradually decreased, reaching comparable values to the younger adults. Additionally, differences in gut composition were found between normal weight and overweight older adults (increased F:B ratio and decreased variety in the overweight individuals). Regular (daily) exercise had a positive role in maintaining the stability of the gut microbiota especially in the overweight group (Zhu et al., 2020).
2.3. Concomitant disease and medication
Aging generally increases the prevalence of chronic disease and disability (Vos et al., 2017). The management of these conditions often requires multiple medications, and their long-term use has been frequently associated with an increased risk for negative health outcomes (Maher et al., 2014). In the gut, the interaction between different medications and the microbiota impacts both the therapeutic effect of the drug and the composition and metabolic functionality of the microbial ecosystem. Our gut microbiota was shown to contribute to the metabolism of many medical drugs, which may partially explain the interindividual variability in responses (efficacy and safety) (Zimmermann et al., 2019). For example, gut microbiota composition was shown to affect the response of melanoma patients to immunotherapy (Gopalakrishnan et al., 2018; Matson et al., 2018; Routy et al., 2018). Recently, exposure to pharmaceuticals targeting human cells and not bacteria (antibiotics) (Blaser, 2016), such proton pump inhibitors (Imhann et al., 2016; Jackson et al., 2016), antidiabetics (i.e. metformin) (Forslund et al., 2015), laxatives (Vich Vila et al., 2020), nonsteroidal anti-inflammatory drugs (Rogers and Aronoff, 2016), and atypical antipsychotics (Flowers et al., 2017), was shown to affect the gut microbiota composition. Many non-antibiotics were shown to have antibiotic-like effects by inhibiting the growth of certain bacterial strains, increasing the risk of antibiotic resistance (Maier et al., 2018).
2.3.1. Medication exposure causing dysbiosis (antibiotics and proton pump inhibitors)
2.3.1.1. Antibiotics
The discovery, development and use of effective antibiotics over the past 80 years have revolutionized medicine, saving countless human lives. However, the widespread use (and often overuse) of broad-spectrum antibiotics (WHO, 2018), not only in medical settings but also in agriculture (Chang et al., 2015), has generated two main problems in terms of human microbial ecology: antibiotic resistance (Davies and Davies, 2010; Laxminarayan et al., 2013), and reduced diversity of microbial species within our ‘industrialized’ microbiota (Blaser and Falkow, 2009; Sonnenburg and Sonnenburg, 2019b) – as compared to more traditional rural populations (Martínez et al., 2015; O'Keefe et al., 2015; Pasolli et al., 2019).
In our gut, the number and variety of antibiotic resistance genes were shown to vary depending on the country where we live in (mainly driven by antibiotic overuse) (Forslund et al., 2013; Hu et al., 2013) and the antibiotic administration route (oral vs intravenous injection) (Zhang et al., 2013). Given its enormous density of bacterial cells and diversity of species, the human gut can be a significant reservoir of antibiotic resistant genes (Francino, 2015; Sommer et al., 2009), which can also promote lateral (horizontal) gene transfer events (Ochman et al., 2000) and thus spread antibiotic resistance traits to non-resistant bacteria.
While a healthy microbiota possesses colonization resistance (protection against opportunistic strains and pathogens) (Nagpal et al., 2018; Sullivan, 2001), a microbiota affected by antibiotics in terms of reduced numbers and species can promote the proliferation of opportunistic species such as Enterococcus faecalis (Bartosch et al., 2004), and even increase the risk of infection with C. difficile (Kwok et al., 2012). In the older adults, antibiotic exposure was also shown to lower the production SCFAs (Woodmansey et al., 2004), the main metabolites of bacterial fermentation, which are thought to be key biological mediators that modulate immunologic and inflammatory responses (Chambers et al., 2018; Rios-Covian et al., 2016; Sanna et al., 2019), and even influence psychological responses. However, the pathways and mechanisms by which SCFAs may influence affective and cognitive process in humans are not fully elucidated (Dalile et al., 2019).
Since antibiotic treatments are crucial in many situations, better management of the antibiotic use and the prescription of narrow-spectrum antibiotics would possibly have less severe dysbiotic effects on our gut microbiota. More research focus is also needed in the direction of correcting the negative effects of antibiotics post-treatment, by using suitable combinations of pre- and probiotics, especially in the older population (Vemuri et al., 2017).
2.3.1.2. Proton-pump inhibitors
Proton-pump inhibitors are used frequently to treat gastrointestinal disorders such as gastroesophageal reflux disease and peptic ulcers (Olbe et al., 2003). Being considered low-risk, they are among the most commonly used medications. However, recent studies have found an association with an increased risk of enteric infections, and especially C. difficile (Kwok et al., 2012; McDonald et al., 2015). Larger studies have shown that long-term use of proton-pump inhibitors can alter the gut microbiome (Imhann et al., 2016; Jackson et al., 2016), with more prominent impact than that of antibiotics or other commonly used medications (Imhann et al., 2016). These studies also found an increased presence in the gut of oral and pharyngeal commensals (e.g. Streptococcaceae) and other possible pathogens. Regarding older adults, a recent review paper evaluated the potential adverse effects of long-term use of proton-pump inhibitors and found associations with osteoporotic-related fractures, C. difficile infection, community-acquired pneumonia, vitamin B12 deficiency, kidney disease, and dementia (Maes et al., 2017).
2.3.2. Diabetes and therapeutic role of metformin
Metformin is a first line agent for the treatment of type 2 diabetes (T2D). Metformin lowers blood-glucose mainly by exerting an antigluconeogenic effect in the liver (Pernicova and Korbonits, 2014). Two previous metagenomic analyses of the gut microbial content in T2D patients with microbial dysbiosis yielded divergent conclusions, namely that the discriminant metagenomic markers for T2D differed between populations (European vs Chinese) (Karlsson et al., 2013; Qin et al., 2012). However, these two studies were unstratified for treatment, and a later analysis showed that a unified signature of gut microbiome shifts in T2D with depletion of butyrate-producing taxa can be revealed when controlling for the microbial mediated effects of metformin (Forslund et al., 2015). Metformin has strong effects on the gut microbiome of the T2D patients by increasing the abundance of Akkermansia muciniphila and other SCFA producers (de la Cuesta-Zuluaga et al., 2017; Wu et al., 2017). Thus, metformin has a beneficial health effect not only in the T2D patients but it has also been shown to decrease all-cause mortality and incidence of age-related diseases (Campbell et al., 2017; Pascale et al., 2019).
2.3.3. Chronic constipation, laxatives and dysbiosis
Chronic constipation is a frequent gastrointestinal disorder in the general population, and especially in the older adults, with a prevalence of about 20% in adults over 65 and 30% of adults over 84 (significantly higher percentages in women) (Gallegos-Orozco et al., 2012). Stool consistency itself which affects colon transit times, was shown to correlate strongly with major microbiome markers (species diversity, F:B ratio, and abundance of Akkermansia and Methanobrevibacter) (Vandeputte et al., 2016).
Laxatives are usually used to treat the symptoms of constipation (Ford and Suares, 2011). However, a short course of some laxatives can have long-lasting effects on the gut microbiome, as recently shown in a study on mice (Tropini et al., 2018). The researchers treated mice for six days with polyethylene glycol (a common component of many laxatives) and observed a reduction in the diversity of their gut microbiome even at two weeks after the treatment was stopped (Tropini et al., 2018). Although the long-term effect in humans has not yet been described, laxatives were found to exhibit one of the strongest associations with the microbiome in a recent Dutch cohort study investigating the effect of multiple medications on the composition and metabolic function of the gut microbiota (Vich Vila et al., 2020).
Increased intake of fibers or fiber supplementation was also shown to have a moderate positive effect in chronic constipation, increasing stool frequency and softening stool consistency (Christodoulides et al., 2016; Rao et al., 2015). Some adverse events such as bloating, distension, flatulence, and cramping may occur, especially if the increases in fiber intake are sudden, and therefore, they should be introduced gradually (Ford et al., 2014a).
2.3.4. Obesity and dysbiosis
The prevalence of obesity and its associated comorbidities has been continually increasing around the world over the past decades (WHO, 2020), including within the older age groups (Han et al., 2011; Mathus-Vliegen and Obesity Management Task Force of the European Association for the Study of, 2012). Although the key driver of obesity is the excess energy intake versus expenditure (San-Cristobal et al., 2020), the additional effects of a host-bacterial mutualistic relationship promoting better energy harvest from diet and storage were shown in murine models (Bäckhed et al., 2005; Ley et al., 2005; Turnbaugh et al., 2006). This mutualism contributes significantly to the pathophysiology of obesity using a complex bidirectional communication system, namely the gut-brain axis (Bliss and Whiteside, 2018). At the gut level, obesity translates into reduced bacterial diversity and genes, altered metabolism and functions, and phylum-level changes in the relative abundance of the two dominant bacterial divisions, i.e. Bacteroidetes and Firmicutes; and these characteristics can be transmitted via microbiota transplantation in mice (Ridaura et al., 2013a; Turnbaugh et al., 2009a). In turn, changes in gut microbiota can increase intestinal permeability while affecting metabolic endotoxemia, inflammation, and insulin sensitivity (Cani et al., 2007; Cani et al., 2008). Metabolic endotoxemia refers to the presence in obese individuals of high concentrations of systemic lipopolysaccharides, the major components of the outer cell membrane of gram-negative bacteria (Cani et al., 2007). Lipopolysaccharides have been thought to initiate the occurrence of chronic low-grade inflammation (Cani et al., 2007; Cani et al., 2008), which is the primary hallmark of obesity (Gregor and Hotamisligil, 2011). This low grade inflammation is also a key triggering factor in diabetes, cardiovascular disease and some cancers (Cani and Jordan, 2018).
Overall, these results have indicated that the human gut microbiome may be used as a biomarker and a new therapeutic target for obesity (Villanueva-Millán et al., 2015). However, the solution may not be straightforward, as more recent meta analyses have shown that previously reported signatures of obesity (e.g. reduced diversity and F:B ratio) may not be consistent between multiple studies and could not be generalized to large human populations (Finucane et al., 2014; Sze and Schloss, 2016; Walters et al., 2014). A possible explanation may be that many associations found in individual studies may not be disease-specific, but rather part of a more complex response to disease in general (Duvallet et al., 2017). Nevertheless, the gut microbiota has become an exciting new research area into metabolic health over the past decade (Boulange et al., 2016; Sonnenburg and Bäckhed, 2016).
One area of high interest has been the promising use of probiotics and prebiotics in modulating the gut microbiota and, therefore, act against obesity and related metabolic diseases (Abenavoli et al., 2019; Delzenne et al., 2011; Kobyliak et al., 2016). Additionally, numerous preclinical and clinical studies have shown positive effects of various plant polyphenols (e.g. hydroxycinnamic acids) in reducing obesity-related dysbiosis (Coman and Vodnar, 2020). A more recent research direction has been the investigation of fecal microbiota transplantation (FMT), which has been successfully used to treat recurrent or refractory C. difficile infection (Cammarota et al., 2015; Kelly et al., 2016; Lee et al., 2016; Van Nood et al., 2013; Youngster et al., 2014), as a possible future therapeutic option for obesity and diabetes (Aron-Wisnewsky et al., 2019; Lee et al., 2019; Zhang et al., 2019).
3. Therapeutic interventions targeting the gut microbiota in the older adults
Since gut microbiota deviations from a healthy state (i.e. dysbiosis) have been associated with numerous human diseases (from metabolic to neurological disorders) (Cryan and Dinan, 2012; Round and Mazmanian, 2009; Sarkar et al., 2020; Zmora et al., 2019), a tremendous research effort has been channeled in investigating approaches to restore a healthy microbiota with hopes to improve certain health outcomes. In terms of links with disease, human-microbiota associated (HMA) (Arrieta et al., 2016) rodents have been used extensively in microbiome research, and the results of these studies have been extrapolated to establish causal relationships in humans (Britton et al., 2019; Ridaura et al., 2013b; Turnbaugh et al., 2009b). However, some concern has arisen very recently related to this extensive extrapolation from animal studies to humans due to the limitations of this approach (e.g., metabolic differences, replication of ecological factors driving the dysbiosis event in humans) (Walter et al., 2020). The authors of this leading-edge perspective paper launch a warning about the possibility of overstating the role of the gut microbiota in human disease and “advocate for a more rigorous and critical approach for inferring causality to avoid false concepts and prevent unrealistic expectations that may undermine the credibility of microbiome science and delay its translation” (Walter et al., 2020).
3.1. Pre-, pro- and synbiotic interventions
Over the past two decades, numerous randomized clinical trials have been conducted in various target populations, including the older adults, to investigate the effectiveness of several therapeutic approaches impacting out gut microbiota and improving our health. Among these, supplementation of the human diet with beneficial microorganisms (probiotics), substrates to promote the proliferation of these beneficial microbes (prebiotics), or a combination of both (synbiotics) represent the most investigated health interventions.
To provide a more comprehensive picture of the evaluated outcomes in randomized clinical trials (RCTs), we analyzed several recent studies in older populations with prebiotics (Chung et al., 2007; Scheid et al., 2014; Vulevic et al., 2015), probiotics (Lefevre et al., 2015; Moro-Garcia et al., 2013; Ouwehand et al., 2008; Valentini et al., 2015), and synbiotics (Bartosch et al., 2005; Bjorklund et al., 2012; Cicero et al., 2020; Costabile et al., 2017; Macfarlane et al., 2013; Maneerat et al., 2013; Neto et al., 2013; Ouwehand et al., 2009), and their main findings are given in Table 2 . Most of the studies were conducted in a single center, having a small number of included individuals, and mostly evaluated changes in microbiota composition, but also in inflammatory and immunologic responses, and in blood and biochemical parameters. Of note, direct comparisons between individual studies are exceedingly difficult, as study settings and parameters differ significantly. Moreover, several technical and methodological issues (e.g. sample collection and storage, DNA extraction and amplification, different methods for detection and quantification of microbiota from stool samples, quality control, sequence alignment) could introduce errors and biases in the results (Fiedorova et al., 2019; Panek et al., 2018).
Table 2.
Randomized clinical trials with prebiotics, probiotics and synbiotics in older adults target groups.
| Study | Time | Intervention | Cohort (T/P) | Age (Y) | Assessed effects | Summary of results | Ref. |
|---|---|---|---|---|---|---|---|
| Randomized, placebo-controlled; Taiwan | 3 wk | Prebiotic (XOS; 4 g/d) | 22 (13/9) | 79.8 ± 6.6 (T) 77.5 ± 6.7 (P) |
Anthropometric and nutrient parameters, fecal moisture content, pH, Bifidobacterium and Clostridium counts. | Increases in bifidobacteria and fecal moisture; decreased fecal pH. No differences for nutrient intakes, GI function, and blood parameters. | (Chung et al., 2007) |
| Double-blind, placebo-controlled, crossover; UK | 28 wka | Prebiotic (GOS mixture; 5.5 g/d) | 40 | 70.4 ± 3.8 (all) | Fecal metabolites, cytokine production, NK cell activity, microbiota composition. | Increases in bifidobacteria and bacteroides, and lactic acid. Higher IL-10, IL-8, NK cell activity and C-reactive protein and lower IL-1b. | (Vulevic et al., 2015) |
| Double-blind placebo controlled; Brazil | 9 wk | Prebiotic (FOS as freeze-dried yacon powder; 7.4 g/d) | 74 (37/37) | 67.11 ± 6.12 (T) 67.11 ± 5.53 (P) |
Anthropometric measurements, intestinal transit, blood parameters (glucose, insulin, lipids, C-reactive protein). | Decrease in serum glucose (p = 0.013); no change in serum lipids. No adverse effect on intestinal transit; no bloating, flatulence, or intestinal discomfort. | (Scheid et al., 2014) |
| Double-blind placebo controlled; Finland | 6 M | Probiotic (oat-based drink with 109 CFU/d B. longum 2C and 46) | 209 (56/67/86b) | 84.3 ± 0.98 (all) | Serum levels of the cytokines IL-10, tumor necrosis factor (TNF)-a and transforming growth factor (TGF)-b1. | Modest increases in Bifidobacteria. Serum cytokines levels correlated with the presence and concentration of specific strains. | (Ouwehand et al., 2008) |
| Double-blind, placebo-controlled; Spain | 6 M | Probiotic (3 × 107L. delbrueckii subsp. Bulgaricus 8481) | 47 (26/21) | 71.6 ± 5.46 (T) 69.5 ± 9.19 (P) |
Cell subpopulations, cytokines, T cell receptor excision circle (TREC), human β-defensin-2 (hBD-2) and CMV titers. | Increased percentage of NK cells and immature T cells numbers; decreased concentrations of IL-8 but increased hBD-2. No differences in CMV titers. | (Moro-Garcia et al., 2013) |
| Double-blind, placebo-controlled; France | 4 Mc | Probiotic (2 × 109B. subtilis CU1 spores daily) | 100 (50/50) | 63.3 ± 2.8 (T) 63.0 ± 2.4 (P) |
Immune stimulation and resistance to common infectious disease episodes. | Significant increase in fecal and salivary secretory IgA concentrations; decreased frequency of respiratory infections. | (Lefevre et al., 2015) |
| Double-blind, placebo-controlled; Europe | 8 wk | Probiotic VSL#3® (2 capsules/d).d | 62 (31/31) | 70.1 ± 3.9 (all) | C-reactive protein; oxidative stress, inflammatory status, and gut microbiota composition. | Reduction in homocysteine concentration and increase in folate, vitamin B12 and bifidobacteria. No other effect on inflammation. | (Valentini et al., 2015) |
| Double-blind, placebo-controlled; UK | 4 wk | Synbiotic (7 × 1010 CFU B. bifidum BB-02 and B. lactis BL-01 and 6 g Synergy1e) | 18 (9/9) | 73 (68–90) (T) 71 (63–85) (P) |
Fecal bifidobacterial and lactobacilli levels. | Significantly higher total numbers of fecal bifidobacteria, and lactobacilli. Bifidobacteria was found in the synbiotic group 3 weeks post-intervention. | (Bartosch et al., 2005) |
| Double-blind placebo controlled; Finland | 2 wk | Synbiotic (10 g lactitol and 2 × 1010 L. acidophilus NCFM) | 47 (23/24) | 71.7 ± 6.2 (T) 70.3 ± 7.2 (P) |
Relative changes in total microbiota composition. | Increased numbers of endogenous lactobacilli and bifidobacteria. Other commensals were not impacted by the treatment. | (Bjorklund et al., 2012; Ouwehand et al., 2009) |
| Double-blind, placebo-controlled, crossover; UK | 4 × 3 wk | Prebiotic (GOS; 8 g/d); probiotic B. lactis Bi-07 (109 CFU/d); synbiotic (Bi-07 + GOS); and maltodextrin (8 g/d) | 36f | 67.2 (all) | Phagocytic activity of monocytes and granulocytes, whole-blood response to LPS, chemokines and salivary IgA levels; microbiota and SCFAs. | Improved phagocytic activity of monocytes (P < 0.001) and granulocytes (P = 0.02). Other parameters were unchanged. | (Maneerat et al., 2013) |
| Double-blind, placebo-controlled; Brazil | 3 M | Synbiotic (6 g FOS; 108–109 CFU L. paracasei, L. rhamnosus, L. acidophilus; B. lactis) | 17 (9/8) | 67.9 ± 4.5 (all) | Anthropometric measurements, bioelectric impedance with vectorial analysis, IL-6 and TNF-α. | No significant changes in inflammatory cytokines or body composition; a subtle trend of improved hydration status. | (Neto et al., 2013) |
| Double-blind, placebo-controlled, crossover; UK | 4 wk | Synbiotic (2 × 1011 freeze-dried viable B. longum and 6 g prebiotic Synergy I™, twice daily. | 43 (23/20) | 71.9 ± 5.4 (all) | Microbiota composition, SCFAs concentration, cytokine production, bowel habits, clinical parameters | Increased Bifidobacteria, Actinobacteria and Firmicutes; decreased Proteobacteria. Increased butyrate production. Decreased pro-inflammatory cytokine TNF-a. | (Macfarlane et al., 2013) |
| Double-blind, placebo-controlled, crossover; UK | 3 wkg | Synbiotic (1010 CF g L. rhamnosus GG and 6 g Promitor™ Soluble Corn Fiber) | 37 | 60–80 (all) | Blood lipids, glucose, cytokines, NK cell activity, phenotype, and microbiota composition. | Reduced total cholesterol and LDL-cholesterol. Increased NK cell activity; decreased pro-inflammatory cytokine IL-6. | (Costabile et al., 2017) |
| Double-blind, placebo-controlled; Italy | 2 M | Synbiotic (2 × 109 CFU L. plantarum, L. acidophilus, L. reuteri and inulin and FOS) | 60 (30/30) | 72 ± 3 (T) 71 ± 3 (P) |
Anthropometric measurements, blood lipids, blood pressure, fasting plasma glucose and insulin. | Reduced blood lipids, arterial pressure, and fasting plasma glucose. Reduced waist circumference and visceral adiposity. | (Cicero et al., 2020) |
T – treatment; P – placebo; wk. – week; M -month; XOS - xylo-oligosaccharides; GOS - galacto-oligosaccharides; FOS - fructo-oligosaccharides; NK – natural killer (cells); LPS – lipopolysaccharide; SCFA – short chain fatty acids.
10-wk intervention, 4-wk washout, 10-wk placebo, 4-wk follow-up.
Commercial control: fermented oat drink (B. lactis Bb-12 at ~109 CFU/day).
4 interventions for 10 days intermittently, alternating with 18-day breaks.
VSL#3 contains 112 billion lyophilized bacteria consisting of the following strains: Bifidobacterium infantis DSM 24737, B. longum DSM 24736, B. breve DSM 24732, Lactobacillus acidophilus DSM 24735, L. delbrückii ssp. bulgaricus DSM 24734, L. paracasei DSM 24733, L. plantarum DSM 24730, and Streptococcus thermophilus DSM 24731, in defined ratios.
Synergy1 is a combination of chicory inulin enriched by a fraction of chicory oligofructose produced by partial enzymic hydrolysis of chicory inulin.
Volunteers received prebiotic, probiotic, their combination, and maltodextrin control in 4 × 3-wk periods separated by 4-wk wash-out periods.
3 wk. for each treatment, with 3-wk washout periods.
3.1.1. Prebiotics
For interventions with prebiotics, defined as “a selectively fermented ingredient that allows specific changes, both in the composition and/or activity in the gastrointestinal microflora that confers benefits upon host wellbeing and health” (Gibson et al., 2004), we analyzed comparatively three studies with different nondigestible oligosaccharides: xylo-oligosaccharides (XOS) (Chung et al., 2007), galacto-oligosaccharides (GOS) (Vulevic et al., 2015), and fructo-oligosaccharides (FOS) (Scheid et al., 2014). XOS are made-up of xylose subunits and have important food-related applications (Vázquez et al., 2000), and significant prebiotic potential (Aachary and Prapulla, 2011). GOS consist of β-linked galactose subunits (with galactose or glucose at the reducing end), and are important prebiotic components of functional foods (Sangwan et al., 2011; Torres et al., 2010). FOS consist of linear chains of fructose linked by β (2–1) bonds, and are used consistently in foods due to prebiotic effects (Sabater-Molina et al., 2009; Sangeetha et al., 2005).
In the first analyzed study (Chung et al., 2007), conducted in Taiwan mostly in middle-old healthy individuals, the treatment group (N = 13) received a supplement of XOS at 4 g/d for 3 weeks, while the control group (N = 9) received sucrose (4 g/d). As the main outcome, XOS consumption was shown to significantly increase the fecal bifidobacteria counts within the 3-week supplementation (+2.2 log10 CFU/g feces vs baseline), increase the fecal moisture and decrease fecal pH; however, these proved to be short-term benefits – as the parameters were found to decrease to baseline levels after a 3-week washout period post-intervention. Bifidobacteria are known to be able to degrade a large variety of oligo- and polysaccharides within the human gut (Pokusaeva et al., 2011), and to have a beneficial impact on human health (O'Callaghan and van Sinderen, 2016). No adverse effects on gastrointestinal (GI) health, blood parameters or nutrient intake were observed during the study (Chung et al., 2007). The duration of the intervention is an important factor for sustained effects, as also suggested by a recent meta-analysis (23 RCTs) evaluating the role of symbiotic consumption on lipid profiles (Hadi et al., 2020). This analysis has shown that the expected effects are more pronounced in case of interventions longer than 8 weeks.
In the second study (randomized, cross-over study with 40 older adults, conducted in the UK) (Vulevic et al., 2015), the intervention consisted of GOS at 5.5 g/d for 10 weeks (vs maltodextrin), while evaluating the effects on the fecal microbiota and immunosenescence (i.e. the deterioration of immune responses in the older adults (Shaw et al., 2010)). Here, a bifidogenic effect was also observed, as well as some immunomodulatory effects reflected in the increased production of IL-10 (an anti-inflammatory cytokine (Couper et al., 2008)) and a significant increase in the activity of NK cells (lymphocytes with a major role against viral infections and some malignancies). The bifidogenic effect seems to be very common in prebiotic interventions in adults, as shown by a recent meta-analysis of 64 RCTs (So et al., 2018). The authors of this study also highlight the importance of the prebiotic source and of the enzymes used in its production, aspects that can crucially impact its selectivity towards bifidobacteria (Vulevic et al., 2015).
Finally, the third study conducted in Brazil, evaluated the effects of FOS supplementation (7.4 g/d for 9 weeks) vs placebo (maltodextrin) on the blood parameters (glucose, lipids) and intestinal transit of healthy and relatively young older adults (60–75 years old) (Scheid et al., 2014). The intervention resulted in a significant decrease in serum glucose as compared to placebo, but no effect of FOS was seen on serum lipids or intestinal transit. No adverse events such as bloating, flatulence or intestinal discomfort were observed. The effect on the glycemic response is also supported by the findings of a recent meta-analysis of 33 RCTs, which shows that supplementation with fructans reduces significantly four glycemic indicators (i.e. fasting blood glucose, glycosylated hemoglobin, fasting insulin, and insulin resistance), especially in prediabetic and type 2 diabetic populations (Wang et al., 2019).
3.1.2. Probiotics
Probiotics are “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” (Hill et al., 2014). Recent meta-analyses of RCTs in adult populations have shown that probiotics could have beneficial effects in treatment and prevention of gastrointestinal diseases in general (74 RCTs) (Ritchie and Romanuk, 2012), irritable bowel syndrome (IBS) (43 RCTs) (Ford et al., 2014b), blood pressure (9 RCTs) (Khalesi et al., 2014), and depressive symptoms (10 RCTs) (Wallace and Milev, 2017). Pre- and probiotics have been also associated with a positive role in modulating immune responses to respiratory viruses, resulting, for example, in an increase in seroconversion and seroprotection rates in adults (and especially in healthy older adults) vaccinated against influenza (Lei et al., 2017). Based on such results, several authors have recently hypothesized that diet supplementation with pre- and probiotics may be beneficial in the current context of the severe acute respiratory syndrome coronavirus 2 (Sars-CoV-2) pandemic (Baud et al., 2020; Dhar and Mohanty, 2020; Infusino et al., 2020), especially for older adults, who are at a higher risk for developing more serious complications from COVID-19 illness (Bialek et al., 2020; Wu et al., 2020a). However, rigorous RCTs need to be carried out to confirm any positive results in the current pandemic, especially since there are also many conflicting results published, which did not find any positive effects of probiotic supplementation. This may be due to many parameters needing to be controlled both from the probiotic and host sides. For the probiotic, the choice of strain(s), dosage (adequate amount) and duration of intervention can strongly influence the beneficial outcome. Host factors that could influence the success of the intervention are diet and lifestyle, age, concomitant disease, antibiotic exposure, and baseline microbiota composition and function (Suez et al., 2020).
Studies in the older population are a fraction of the total numbers, but sufficient findings have been made public to allow for the successful publication of several systematic reviews and meta-analyses (Gui et al., 2020; Jafarnejad et al., 2016; Martinez-Martinez et al., 2017; Miller et al., 2017, Miller et al., 2019; Qu et al., 2019). In terms of finding, one meta-analysis found that probiotics did not reduce the risk of antibiotic-associated diarrhea in the older adults (>65 years) (Jafarnejad et al., 2016), while another found a positive effect of probiotics on constipation (10–40% improvement compared with controls) (Martinez-Martinez et al., 2017). An improvement of cellular immune function in terms of polymorphonuclear cell phagocytic capacity or natural killer (NK) cell tumoricidal activity was found in an analysis of 17 studies with short-term administration of probiotics (Miller et al., 2019); this is consistent with a previous meta-analysis of a smaller number of clinical trials (N = 4) investigating the effect of B. lactis HN019 supplementation on cellular immune activity in healthy older participants (Miller et al., 2017). Another very recent meta-analysis of 6 RCTs found that probiotics (mainly Lactobacillus interventions) significantly increased (P < 0.05) NK cell activity in healthy older individuals (Gui et al., 2020). Finally, one recent meta-analysis analyzing 10 RCTs found no significant benefit of microbiota-driven therapy (pre-, pro-, and synbiotics) on inflammatory responses in older individuals (Qu et al., 2019).
However, most of these meta-analyses have been usually limited by the reduced number of included studies and the heterogeneity of the original studies in terms of study design, population, and risk of bias, and their authors acknowledge that high-quality, large scale, RCTs with long-term follow-up are further needed to provide convincing evidence for clear associations between interventions and health outcomes. Although systematic reviews and meta-analyses are known to reduce methodological and analytical limitations of individual studies, and to reveal general trends, the inclusion of outlier studies or grouping studies evaluating non-related probiotics for the same outcome, may lead to conflicting findings between meta-analyses evaluating the same outcome (Suez et al., 2019).
Several recent individual studies with probiotic interventions were conducted in connection with inflamm-aging in the older adults (Lefevre et al., 2015; Moro-Garcia et al., 2013; Ouwehand et al., 2008; Valentini et al., 2015), evaluating the inflammatory status and cytokine production as compared to a placebo. An earlier study from 2008 was conducted for 6 months in two nursing homes in Finland and involved 209 older adults (84.3 ± 0.98 yr) randomized to an intervention group (n = 56) – an oat-based drink supplemented with 109 CFU/day of both B. longum; a placebo group (n = 67) – no probiotics; and a control group (n = 86) – a commercial product with 109 CFU/day B. animalis ssp. lactis Bb-12 (Ouwehand et al., 2008). The study found that probiotic supplementation can modestly influence the microbiota composition even at this advanced age, correlated with changes in the levels of certain cytokines (e.g. lower IL-10 levels in the intervention group vs placebo).
Another study conducted in Spain showed positive effects on the immunologic response of older adults given 3 × 107 Lactobacillus delbrueckii subsp. bulgaricus 8481 daily for 6 months (Moro-Garcia et al., 2013). However, these effects disappeared 6 months after the treatment ended, suggesting that the probiotic did not colonize the intestinal tract. A stimulation of the immune response during the winter months was also shown by (Lefevre et al., 2015) in a French cohort receiving intermittently 2 × 109 B. subtilis CU1 spores daily for 4 months. The immunologic stimulation translated into a reduction of respiratory infections in the probiotic group. Another multicentric study conducted in Europe showed a reduction in homocysteine concentration and an increase in folate and vitamin B12 following a 8 week supplementation with VSL#3 (a mixture of 8 bacterial strains) (Valentini et al., 2015).
3.1.3. Synbiotics
A synbiotic contains both a probiotic and a prebiotic that work synergistically, i.e. the beneficial effect is higher than that of the probiotic alone (De Vrese and Schrezenmeir, 2008; Pandey et al., 2015; Schrezenmeir and De Vrese, 2001). Although numerous studies have been conducted to assess this synergism, the results have been modest so far, with no clear benefits for some conditions such as IBS (Ford et al., 2018), renal function (Firouzi and Haghighatdoost, 2018), or nonalcoholic fatty liver disease (Hadi et al., 2019); but with beneficial outcomes for others, such as obesity (lipid profiles) (Hadi et al., 2020), high fasting blood glucose (Nikbakht et al., 2018), postoperative infections (Yang et al., 2017), and some inflammatory markers (Kazemi et al., 2020).
As with pre- and probiotics, only a limited number of studies were conducted specifically in older populations. Several examples are detailed in Table 2. Synbiotics have been shown to alter the composition of the gut microbiota in older adults, especially by inducing significant increases in the fecal amount of bifidobacteria and lactobacilli, which persisted in some cases even weeks after the treatment ended (Bartosch et al., 2005; Bjorklund et al., 2012; Ouwehand et al., 2009). Bartosch et al. (2005) investigated the effects of a synbiotic containing Bifidobacterium bifidum BB-02 and B. lactis BL-01 administered together with an inulin-based prebiotic in 18 healthy older volunteers (mean 72 yr) for 4 weeks vs placebo (maltooligosaccharides). Ouwehand et al. (2009) and Bjorklund et al. (2012) used L. acidophilus NCFM (Danisco) and lactitol in 47 healthy older volunteers (mean 71 yr) with regular use of non-steroidal anti-inflammatory drugs, for 2 weeks vs placebo (sucrose).
The impact on the immune function of the older adults has been frequently investigated in the context of immunosenescence. Maneerat et al. (2013) showed an improvement in phagocytic activity of monocytes and granulocytes (major mechanism in the clearance of pathogens) with Bifidobacterium lactis Bi-07 (109 CFU/d) for 4 × 3-week periods separated by 4-week wash-out periods in 36 healthy older adults (mean 67.2 yr). However, the results were not improved with the synbiotic treatment (using GOS as prebiotic) vs probiotic alone. Neto et al. (2013) did not see any influence on the inflammatory response or body composition in 9 older individuals with a risk of frailty treated with a synbiotic (6 g FOS and 108–109 CFU for each of: L. paracasei, L. rhamnosus, L. acidophilus and B. lactis) for 3 months. Macfarlane et al. (2013) showed a reduced pro-inflammatory cytokine TNF-α in peripheral blood after 2 and 4 weeks of synbiotics (2 × 1011 B. longum and 6 g inulin-based prebiotic) vs placebo (maltodextrose) in 43 older adults. Additionally, there were changes in microbiota composition (increased Bifidobacteria, Firmicutes and Actinobacteria vs decreased Proteobacteria) and an increased butyrate production.
Finally, the effect of synbiotics on cardiovascular risk factors in the older adults has been investigated in a few recent studies (Cicero et al., 2020; Costabile et al., 2017). Costabile et al. (2017) showed a reduction of hypercholesteremia with a synbiotic formulation (L. rhamnosus GG and soluble corn fiber) for 3 weeks in 40 healthy older participants (60–80 yr). Similarly, Cicero et al. (2020) showed marked improvements in several cardiovascular risk factors and markers of insulin resistance in 30 older adults treated for 2 months with a synbiotic made of three lactobacilli, inulin and FOS.
Recent scientific and medical advances have shown that probiotic interventions may not be successful in a an entire population, the response being driven both by host and microbiota characteristics, suggesting shifting the future research avenue towards personalized nutrition strategies for better health, taking into consideration the specific host and microbiome features and their mutualistic relationship (Bashiardes et al., 2018; Kolodziejczyk et al., 2019; Lam et al., 2019; Suez et al., 2020). While host factors (e.g. age and concomitant disease) may not be easy to modulate, microbiota factors (diversity and composition, interspecies competition) (Patnode et al., 2019) may be easier to control with specific dietary interventions, tailored to the individual (Bashiardes et al., 2018).
One relevant example for a personally tailored diet intervention is the work of Zeevi et al. (2015), who used artificial intelligence to predict postprandial glucose responses by integrating microbiome composition, blood tests and anthropometrics of 800 people. Another recent research direction aimed to take advantage of exclusive metabolic niches (i.e. a food source to which a certain probiotic has exclusive access) to promote a controlled probiotic colonization of the gut (Kearney et al., 2018; Shepherd et al., 2018). Shepherd et al. (2018) used a marine polysaccharide, porphyran, and an exogenous Bacteroides strain harboring a rare gene cluster for porphyran utilization, and showed that the abundance of the strain in the gut can be fine-tuned over several orders of magnitude. Kearney et al. (2018) showed that co-introducing a bacterial commensal (i.e. Bacteroides plebeius DSM 17135) along with a specific resource, seaweed, enables colonization of the mouse gut microbiota at a median abundance between 1 and 10%. Alternatively, another recent report has shown the possibility to chemically inhibit the expansion of a specific bacterial family (i.e. Enterobacteriaceae) and to reduce intestinal inflammation in murine models of colitis (Zhu et al., 2018).
Colonization resistance is an important feature of our microbiota by which it protects us from pathologic infections. Simultaneously, the same mechanism may preclude probiotic colonization, and this colonization resistance may be person-specific (Zmora et al., 2018). Pre-treatments with antibiotics may improve probiotic colonization (Zmora et al., 2018), but post-antibiotic probiotic benefits may be counteracted by a compromised gut mucosal recovery (Suez et al., 2018). Adhesive properties of probiotics (and prebiotics) are also important to be considered (Monteagudo-Mera et al., 2019), and future research may benefit from recent advancements in synthetic biology, opening the way towards engineered live bacterial therapeutics (Charbonneau et al., 2020).
3.2. Fecal microbiota transplantation
Based on the results of several RCTs showing fecal microbiota transplantation (FMT) as a viable alternative treatment approach against C. difficile infection (Cammarota et al., 2015; Kelly et al., 2016; Lee et al., 2016; Van Nood et al., 2013; Youngster et al., 2014), current clinical guidelines from the USA recommend FMT for “patients with multiple recurrences of C. difficile infection who have failed appropriate antibiotic treatments (strong recommendation, moderate quality of evidence)” (McDonald et al., 2018). C. difficile infection is known to affect the older population disproportionately (Keller and Surawicz, 2014; Smits et al., 2016), mainly due to immunosenescence, increased exposure to healthcare settings, and frequent use of antibiotics and proton-pump inhibitors (Loo et al., 2011).
Although FMT interventions against C. difficile infection are performed mostly in older adults, only a limited number of large cohort studies have been conducted specifically in this age group (Agrawal et al., 2016; Luo et al., 2020). In a multicenter, long-term follow-up study conducted in the USA, Canada and Australia, Agrawal et al. (2016) investigated the long-term efficacy and safety of FMT in 146 older patients (mean 78.6 yr) with either recurrent, severe, or complicated C. difficile infection. The primary and secondary cure rates were 82.9% (121 of 146) and 95.9% (140 of 146), respectively. Thus, FMT proved to be a safe and effective treatment option for C. difficile infection in the older adults (Agrawal et al., 2016). Another recent long-term follow-up study investigated the efficacy and safety of FMT in 75 older patients (mean 76.4 yr), out of which 34 (45.3%) were “higher-risk”, namely immunocompromised, with a history of inflammatory bowel disease, or presenting with severe or fulminant colitis (Luo et al., 2020). In this study, the adjusted primary cure rate was only 67.2% and post-FMT C. difficile infection recurrence rate was 29.9%. These results suggest that FMT should be used early in the treatment of C. difficile infection to avoid disease progression and recurrence, especially in older patients (Agrawal, 2020). Additionally, multiple FMT interventions may be needed for a good result.
Following the success in C. difficile infection treatment, the potential of FMT has also been investigated against Crohn's disease (Sokol et al., 2020), irritable bowel syndrome (Xu et al., 2019b), cirrhosis (Woodhouse et al., 2019), and even neurological and behavioral conditions (Vendrik et al., 2020). The results are promising, but still modest. One important aspect in the success of FMT is the diversity and the composition of the stool donor, which plays an essential role in restoring metabolic deficits in recipients (Wilson et al., 2019).
Although a rapidly emerging new therapy (Ooijevaar et al., 2019), there is a need for further larger studies and controlled clinical trials to assess the long-term benefits of FMT outside C. difficile infection treatment and any related safety issues associated with it. Sometimes, unintended consequences may arise from the transfer of antibiotic-resistant bacteria or other unwanted phenotypes from the donor (Alang and Kelly, 2015; DeFilipp et al., 2019), or pathogenic viruses (Chehoud et al., 2016). Donor screening and sample testing are especially important to be considered in the current pandemic situation with SARS-CoV-2, where a potential risk of transmission by FMT may occur due to the documented prolonged presence of the virus in stool samples of infected individuals (Wang et al., 2020; Wu et al., 2020b). To address this issue, the FDA issued a safety alert, highlighting the additional precautions needed for FMT clinical use, including screening, and testing of donor and donor samples for SARS-CoV-2 (FDA, 2020).
4. Concluding remarks
Our gut microbiota is a dynamic ecosystem, which adapts continuously to changes in lifestyle, nutrition, hygiene, and exposure to medication. Establishing and maintaining positive interactions between us and our gut microbiota are essential for our health. The longer the exposure to certain stressors, the more significant the changes, which may explain why recent research has found that older populations have a less diverse microbiota than younger individuals, and more pathobionts. With advanced age, the prevalence of certain diseases increases as well, which can also contribute to an increased risk of frailty leading to microbiota dysfunctionalities, and therefore, to progression of other metabolic diseases.
Our diet is the main shaping factor of our gut microbiota. Although the gut microbiota responds rapidly to significant short-term changes in diet, long-term dietary habits (e.g. transgenerational patterns) are the main factor in shaping the microbiota composition and diversity in individuals. Comparisons with populations with traditional lifestyles, such as remote rural populations or hunter gatherer tribes, have shown that our microbiota is less diverse and has lower fiber degrading capabilities (and thus lower production of relevant metabolites such as SCFAs). These changes have been attributed mainly to modern advancements in the food industry (highly processed foods, high in fat and carbohydrates and low in fiber), and in the medical sector (broad range antibiotics and other medication affecting our microbiota composition and function).
The responses to similar dietary changes may vary individually due to particularities of individual microbiotas, underlying the future importance of personalized nutritional interventions. Clinical trials involving dietary interventions and supplementation with pre-, pro- and synbiotics have revealed that there is no single intervention to benefit complex populations, mainly due to these interindividual differences. Future studies should consider the baseline microbiota characteristics of included individuals and tailor the interventions based on their needs. The results of such studies should provide the scientific clues for the rational design of food products specific to certain groups of older adults (e.g. those affected by chronic constipation, obese individuals).
Regarding the older adults, and especially the residents of long-term care facilities, microbiota-targeted interventions should be made early and often, to attenuate the occurrence of critical conditions such as frailty. Any long-term medication exposure, and especially antibiotic treatments, should be followed by a microbiota restauration therapy, to prevent the occurrence of dangerous infections such as C. difficile and the proliferation of other opportunistic strains.
The following is the supplementary data related to this article.
The 6 major phyla of the human gut microbiota and their predominant families (with examples of species).
CRediT authorship contribution statement
Vasile Coman: Conceptualization; Methodology; Writing - original draft.
Dan Cristian Vodnar: Funding acquisition; Supervision; Writing - review & editing.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported by the National Research, Development and Innovations Programme for 2015-2020-PNII, developed with the support of UEFISCDI (Project No. PN-III-P1-1.2-PCCDI-2017-0056; 2PCCDI).
Section Editor: Cheryl Conover
References
- Aachary A.A., Prapulla S.G. Xylooligosaccharides (XOS) as an emerging prebiotic: microbial synthesis, utilization, structural characterization, bioactive properties, and applications. Compr. Rev. Food Sci. Food Saf. 2011;10:2–16. [Google Scholar]
- Abenavoli L., Scarpellini E., Colica C., Boccuto L., Salehi B., Sharifi-Rad J., Aiello V., Romano B., De Lorenzo A., Izzo A.A., Capasso R. Gut microbiota and obesity: a role for probiotics. Nutrients. 2019;11 doi: 10.3390/nu11112690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal G. Fecal microbiota transplantation for Clostridioides difficile in high-risk older adults: treat early, treat often. Dig. Dis. Sci. 2020 doi: 10.1007/s10620-020-06291-6. [DOI] [PubMed] [Google Scholar]
- Agrawal M., Aroniadis O.C., Brandt L.J., Kelly C., Freeman S., Surawicz C., Broussard E., Stollman N., Giovanelli A., Smith B., Yen E., Trivedi A., Hubble L., Kao D., Borody T., Finlayson S., Ray A., Smith R. The long-term efficacy and safety of fecal microbiota transplant for recurrent, severe, and complicated clostridium difficile infection in 146 elderly individuals. J. Clin. Gastroenterol. 2016;50:403–407. doi: 10.1097/MCG.0000000000000410. [DOI] [PubMed] [Google Scholar]
- Alang N., Kelly C.R. Weight gain after fecal microbiota transplantation. Open Forum Infect Dis. 2015;2:ofv004. doi: 10.1093/ofid/ofv004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron-Wisnewsky J., Clement K., Nieuwdorp M. Fecal microbiota transplantation: a future therapeutic option for obesity/diabetes? Curr. Diab. Rep. 2019;19:51. doi: 10.1007/s11892-019-1180-z. [DOI] [PubMed] [Google Scholar]
- Arrieta M.C., Walter J., Finlay B.B. Human microbiota-associated mice: a model with challenges. Cell Host Microbe. 2016;19:575–578. doi: 10.1016/j.chom.2016.04.014. [DOI] [PubMed] [Google Scholar]
- Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D.R., Fernandes G.R., Tap J., Bruls T., Batto J.M., Bertalan M., Borruel N., Casellas F., Fernandez L., Gautier L., Hansen T., Hattori M., Hayashi T., Kleerebezem M., Kurokawa K., Leclerc M., Levenez F., Manichanh C., Nielsen H.B., Nielsen T., Pons N., Poulain J., Qin J., Sicheritz-Ponten T., Tims S., Torrents D., Ugarte E., Zoetendal E.G., Wang J., Guarner F., Pedersen O., De Vos W.M., Brunak S., Doré J., Weissenbach J., Ehrlich S.D., Bork P. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F., Ding H., Wang T., Hooper L.V., Gou Y.K., Nagy A., Semenkovich C.F., Gordon J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F., Ley R.E., Sonnenburg J.L., Peterson D.A., Gordon J.I. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Barrès R., Zierath J.R. The role of diet and exercise in the transgenerational epigenetic landscape of T2DM. Nat. Rev. Endocrinol. 2016;12:441–451. doi: 10.1038/nrendo.2016.87. [DOI] [PubMed] [Google Scholar]
- Barroso I., McCarthy M.I. The genetic basis of metabolic disease. Cell. 2019;177:146–161. doi: 10.1016/j.cell.2019.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosch S., Fite A., Macfarlane G.T., McMurdo M.E.T. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl. Environ. Microbiol. 2004;70:3575–3581. doi: 10.1128/AEM.70.6.3575-3581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosch S., Woodmansey E.J., Paterson J.C.M., McMurdo M.E.T., Macfarlane G.T. Microbiological effects of consuming a synbiotic containing Bifidobacterium bifidum, Bifidobacterium lactis, and oligofructose in elderly persons, determined by real-time polymerase chain reaction and counting of viable bacteria. Clin. Infect. Dis. 2005;40:28–37. doi: 10.1086/426027. [DOI] [PubMed] [Google Scholar]
- Bashiardes S., Godneva A., Elinav E., Segal E. Towards utilization of the human genome and microbiome for personalized nutrition. Curr. Opin. Biotechnol. 2018;51:57–63. doi: 10.1016/j.copbio.2017.11.013. [DOI] [PubMed] [Google Scholar]
- Baud D., Dimopoulou Agri V., Gibson G.R., Reid G., Giannoni E. Using probiotics to flatten the curve of coronavirus disease COVID-2019 pandemic. Front. Public Health. 2020;8 doi: 10.3389/fpubh.2020.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biagi E., Nylund L., Candela M., Ostan R., Bucci L., Pini E., Nikkila J., Monti D., Satokari R., Franceschi C., Brigidi P., De Vos W. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One. 2010;5:e10667. doi: 10.1371/journal.pone.0010667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biagi E., Candela M., Franceschi C., Brigidi P. The aging gut microbiota: new perspectives. Ageing Res. Rev. 2011;10:428–429. doi: 10.1016/j.arr.2011.03.004. [DOI] [PubMed] [Google Scholar]
- Biagi E., Franceschi C., Rampelli S., Severgnini M., Ostan R., Turroni S., Consolandi C., Quercia S., Scurti M., Monti D., Capri M., Brigidi P., Candela M. Gut microbiota and extreme longevity. Curr. Biol. 2016;26:1480–1485. doi: 10.1016/j.cub.2016.04.016. [DOI] [PubMed] [Google Scholar]
- Biagi E., Rampelli S., Turroni S., Quercia S., Candela M., Brigidi P. The gut microbiota of centenarians: signatures of longevity in the gut microbiota profile. Mech. Ageing Dev. 2017;165:180–184. doi: 10.1016/j.mad.2016.12.013. [DOI] [PubMed] [Google Scholar]
- Bialek S., Boundy E., Bowen V., Chow N., Cohn A., Dowling N., Ellington S., Gierke R., Hall A., MacNeil J., Patel P., Peacock G., Pilishvili T., Razzaghi H., Reed N., Ritchey M., Sauber-Schatz E. Severe outcomes among patients with coronavirus disease 2019 (COVID-19) - United States, February 12-march 16, 2020. Morb. Mortal. Weekly Rep. 2020;69:343–346. doi: 10.15585/mmwr.mm6912e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian G., Gloor G.B., Gong A., Jia C., Zhang W., Hu J., Zhang H., Zhang Y., Zhou Z., Zhang J., Burton J.P., Reid G., Xiao Y., Zeng Q., Yang K., Li J. The gut microbiota of healthy aged Chinese is similar to that of the healthy young. mSphere. 2017;2 doi: 10.1128/mSphere.00327-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff S.C. Microbiota and aging. Curr. Opin. Clin. Nutr. Metab. Care. 2016;19:26–30. doi: 10.1097/MCO.0000000000000242. [DOI] [PubMed] [Google Scholar]
- Bjorklund M., Ouwehand A.C., Forssten S.D., Nikkila J., Tiihonen K., Rautonen N., Lahtinen S.J. Gut microbiota of healthy elderly NSAID users is selectively modified with the administration of Lactobacillus acidophilus NCFM and lactitol. Age. 2012;34:987–999. doi: 10.1007/s11357-011-9294-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser M.J. Antibiotic use and its consequences for the normal microbiome. Science. 2016;352:544–545. doi: 10.1126/science.aad9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser M.J., Falkow S. What are the consequences of the disappearing human microbiota? Nat. Rev. Microbiol. 2009;7:887–894. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss E.S., Whiteside E. The gut-brain Axis, the human gut microbiota and their integration in the development of obesity. Front. Physiol. 2018;9:900. doi: 10.3389/fphys.2018.00900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonder M.J., Kurilshikov A., Tigchelaar E.F., Mujagic Z., Imhann F., Vila A.V., Deelen P., Vatanen T., Schirmer M., Smeekens S.P., Zhernakova D.V., Jankipersadsing S.A., Jaeger M., Oosting M., Cenit M.C., Masclee A.A.M., Swertz M.A., Li Y., Kumar V., Joosten L., Harmsen H., Weersma R.K., Franke L., Hofker M.H., Xavier R.J., Jonkers D., Netea M.G., Wijmenga C., Fu J., Zhernakova A. The effect of host genetics on the gut microbiome. Nat. Genet. 2016;48:1407–1412. doi: 10.1038/ng.3663. [DOI] [PubMed] [Google Scholar]
- Boulange C.L., Neves A.L., Chilloux J., Nicholson J.K., Dumas M.E. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. 2016;8:42. doi: 10.1186/s13073-016-0303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton G.J., Contijoch E.J., Mogno I., Vennaro O.H., Llewellyn S.R., Ng R., Li Z., Mortha A., Merad M., Das A., Gevers D., McGovern D.P.B., Singh N., Braun J., Jacobs J.P., Clemente J.C., Grinspan A., Sands B.E., Colombel J.F., Dubinsky M.C., Faith J.J. Microbiotas from humans with inflammatory bowel disease Alter the balance of Gut Th17 and RORγt(+) regulatory T cells and exacerbate colitis in mice. Immunity. 2019;e214(50):212–224. doi: 10.1016/j.immuni.2018.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammarota G., Masucci L., Ianiro G., Bibbò S., Dinoi G., Costamagna G., Sanguinetti M., Gasbarrini A. Randomised clinical trial: Faecal microbiota transplantation by colonoscopy vs. vancomycin for the treatment of recurrent Clostridium difficile infection. Aliment. Pharmacol. Ther. 2015;41:835–843. doi: 10.1111/apt.13144. [DOI] [PubMed] [Google Scholar]
- Campbell J.M., Bellman S.M., Stephenson M.D., Lisy K. Metformin reduces all-cause mortality and diseases of ageing independent of its effect on diabetes control: a systematic review and meta-analysis. Ageing Res. Rev. 2017;40:31–44. doi: 10.1016/j.arr.2017.08.003. [DOI] [PubMed] [Google Scholar]
- Cani P.D., Jordan B.F. Gut microbiota-mediated inflammation in obesity: a link with gastrointestinal cancer. Nature Reviews Gastroenterology and Hepatology. 2018;15:671–682. doi: 10.1038/s41575-018-0025-6. [DOI] [PubMed] [Google Scholar]
- Cani P.D., Amar J., Iglesias M.A., Poggi M., Knauf C., Bastelica D., Neyrinck A.M., Fava F., Tuohy K.M., Chabo C., Waget A., Delmée E., Cousin B., Sulpice T., Chamontin B., Ferrières J., Tanti J.F., Gibson G.R., Casteilla L., Delzenne N.M., Alessi M.C., Burcelin R. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- Cani P.D., Bibiloni R., Knauf C., Waget A., Neyrinck A.M., Delzenne N.M., Burcelin R. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- Cătoi A.F., Corina A., Katsiki N., Vodnar D.C., Andreicut A.D., Stoian A.P., Rizzo M., Perez-Martinez P. Gut microbiota and aging-a focus on centenarians. Biochim. Biophys. Acta Mol. basis Dis. 2020;1866:165765. doi: 10.1016/j.bbadis.2020.165765. [DOI] [PubMed] [Google Scholar]
- Chambers E.S., Preston T., Frost G., Morrison D.J. Role of gut microbiota-generated short-chain fatty acids in metabolic and cardiovascular health. Curr Nutr Rep. 2018;7:198–206. doi: 10.1007/s13668-018-0248-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Q., Wang W., Regev-Yochay G., Lipsitch M., Hanage W.P. Antibiotics in agriculture and the risk to human health: how worried should we be? Evol. Appl. 2015;8:240–247. doi: 10.1111/eva.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonneau M.R., Isabella V.M., Li N., Kurtz C.B. Developing a new class of engineered live bacterial therapeutics to treat human diseases. Nat. Commun. 2020;11:1738. doi: 10.1038/s41467-020-15508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehoud C., Dryga A., Hwang Y., Nagy-Szakal D., Hollister E.B., Luna R.A., Versalovic J., Kellermayer R., Bushman F.D. Transfer of viral communities between human individuals during fecal microbiota transplantation. mBio. 2016;7 doi: 10.1128/mBio.00322-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulides S., Dimidi E., Fragkos K.C., Farmer A.D., Whelan K., Scott S.M. Systematic review with meta-analysis: effect of fibre supplementation on chronic idiopathic constipation in adults. Aliment. Pharmacol. Ther. 2016;44:103–116. doi: 10.1111/apt.13662. [DOI] [PubMed] [Google Scholar]
- Chung Y.C., Hsu C.K., Ko C.Y., Chan Y.C. Dietary intake of xylooligosaccharides improves the intestinal microbiota, fecal moisture, and pH value in the elderly. Nutr. Res. 2007;27:756–761. [Google Scholar]
- Cicero A.F.G., Fogacci F., Bove M., Giovannini M., Borghi C. Impact of a short-term synbiotic supplementation on metabolic syndrome and systemic inflammation in elderly patients: a randomized placebo-controlled clinical trial. Eur. J. Nutr. 2020 doi: 10.1007/s00394-020-02271-8. [DOI] [PubMed] [Google Scholar]
- Claesson M.J., Cusack S., O'Sullivan O., Greene-Diniz R., De Weerd H., Flannery E., Marchesi J.R., Falush D., Dinan T., Fitzgerald G., Stanton C., Van Sinderen D., O'Connor M., Harnedy N., O'Connor K., Henry C., O'Mahony D., Fitzgerald A.P., Shanahan F., Twomey C., Hill C., Ross R.P., O'Toole P.W. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc. Natl. Acad. Sci. U. S. A. 2011;108:4586–4591. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson M.J., Jeffery I.B., Conde S., Power S.E., O'Connor E.M., Cusack S., Harris H.M.B., Coakley M., Lakshminarayanan B., O'Sullivan O., Fitzgerald G.F., Deane J., O'Connor M., Harnedy N., O'Connor K., O'Mahony D., Van Sinderen D., Wallace M., Brennan L., Stanton C., Marchesi J.R., Fitzgerald A.P., Shanahan F., Hill C., Paul Ross R., O'Toole P.W. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- Clarke S.F., Murphy E.F., O'Sullivan O., Lucey A.J., Humphreys M., Hogan A., Hayes P., O'Reilly M., Jeffery I.B., Wood-Martin R., Kerins D.M., Quigley E., Ross R.P., O'Toole P.W., Molloy M.G., Falvey E., Shanahan F., Cotter P.D. Exercise and associated dietary extremes impact on gut microbial diversity. Gut. 2014;63:1913–1920. doi: 10.1136/gutjnl-2013-306541. [DOI] [PubMed] [Google Scholar]
- Clegg A., Young J., Iliffe S., Rikkert M.O., Rockwood K. Frailty in elderly people. Lancet. 2013;381:752–762. doi: 10.1016/S0140-6736(12)62167-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente J.C., Ursell L.K., Parfrey L.W., Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements S.J., Carding S.R. Diet, the intestinal microbiota, and immune health in aging. Crit. Rev. Food Sci. Nutr. 2018;58:651–661. doi: 10.1080/10408398.2016.1211086. [DOI] [PubMed] [Google Scholar]
- Coman V., Vodnar D.C. Hydroxycinnamic acids and human health. Recent advances. J. Sci. Food Agric. 2020;100:483–499. doi: 10.1002/jsfa.10010. [DOI] [PubMed] [Google Scholar]
- Cordain L., Eaton S.B., Sebastian A., Mann N., Lindeberg S., Watkins B.A., O’Keefe J.H., Brand-Miller J. Origins and evolution of the Western diet: health implications for the 21st century. Am. J. Clin. Nutr. 2005;81:341–354. doi: 10.1093/ajcn.81.2.341. [DOI] [PubMed] [Google Scholar]
- Costabile A., Bergillos-Meca T., Rasinkangas P., Korpela K., de Vos W.M., Gibson G.R. Effects of soluble corn Fiber alone or in synbiotic combination with Lactobacillus rhamnosus GG and the pilus-deficient derivative GG-PB12 on fecal microbiota, metabolism, and markers of immune function: a randomized, double-blind, placebo-controlled, crossover study in healthy elderly (Saimes study) Front. Immunol. 2017;8:1443. doi: 10.3389/fimmu.2017.01443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello E.K., Lauber C.L., Hamady M., Fierer N., Gordon J.I., Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotillard A., Kennedy S.P., Kong L.C., Prifti E., Pons N., Le Chatelier E., Almeida M., Quinquis B., Levenez F., Galleron N., Gougis S., Rizkalla S., Batto J.M., Renault P., Doré J., Zucker J.D., Clément K., Ehrlich S.D., Blottière H., Leclerc M., Juste C., De Wouters T., Lepage P., Fouqueray C., Basdevant A., Henegar C., Godard C., Fondacci M., Rohia A., Hajduch F., Weissenbach J., Pelletier E., Le Paslier D., Gauchi J.P., Gibrat J.F., Loux V., Carré W., Maguin E., Van De Guchte M., Jamet A., Boumezbeur F., Layec S. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- Couper K.N., Blount D.G., Riley E.M. IL-10: the master regulator of immunity to infection. J. Immunol. 2008;180:5771–5777. doi: 10.4049/jimmunol.180.9.5771. [DOI] [PubMed] [Google Scholar]
- Cryan J.F., Dinan T.G. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 2012;13:701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- de la Cuesta-Zuluaga J., Mueller N.T., Corrales-Agudelo V., Velasquez-Mejia E.P., Carmona J.A., Abad J.M., Escobar J.S. Metformin is associated with higher relative abundance of mucin-degrading Akkermansia muciniphila and several short-Chain fatty acid-producing microbiota in the gut. Diabetes Care. 2017;40:54–62. doi: 10.2337/dc16-1324. [DOI] [PubMed] [Google Scholar]
- Cutler D.M., Rosen A.B., Vijan S. The value of medical spending in the United States, 1960-2000. New Engl. J. Med. 2006;355:920–927. doi: 10.1056/NEJMsa054744. [DOI] [PubMed] [Google Scholar]
- Dalile B., Van Oudenhove L., Vervliet B., Verbeke K. The role of short-chain fatty acids in microbiota-gut-brain communication. Nat. Rev. Gastroenterol. Hepatol. 2019;16:461–478. doi: 10.1038/s41575-019-0157-3. [DOI] [PubMed] [Google Scholar]
- David L.A., Maurice C.F., Carmody R.N., Gootenberg D.B., Button J.E., Wolfe B.E., Ling A.V., Devlin A.S., Varma Y., Fischbach M.A., Biddinger S.B., Dutton R.J., Turnbaugh P.J. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J., Davies D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo C., Cavalieri D., Di Paola M., Ramazzotti M., Poullet J.B., Massart S., Collini S., Pieraccini G., Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. U. S. A. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vrese M., Schrezenmeir J. In: Adv. Biochem. Eng. Biotechnol. Stahl U., Donalies U.E.B., Nevoigt E., editors. 2008. Probiotics, prebiotics, and synbiotics; pp. 1–66. [DOI] [PubMed] [Google Scholar]
- DeFilipp Z., Bloom P.P., Soto M.T., Mansour M.K., Sater M.R.A., Huntley M.H., Turbett S., Chung R.T., Chen Y.B., Hohmann E.L. Drug-Resistant E. coli bacteremia transmitted by fecal microbiota transplant. New Engl. J. Med. 2019;381:2043–2050. doi: 10.1056/NEJMoa1910437. [DOI] [PubMed] [Google Scholar]
- Delzenne N.M., Neyrinck A.M., Bäckhed F., Cani P.D. Targeting gut microbiota in obesity: effects of prebiotics and probiotics. Nat. Rev. Endocrionol. 2011;7:639–646. doi: 10.1038/nrendo.2011.126. [DOI] [PubMed] [Google Scholar]
- Dhar D., Mohanty A. Gut microbiota and Covid-19- possible link and implications. Virus Res. 2020;285:198018. doi: 10.1016/j.virusres.2020.198018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan S.H., Belenguer A., Holtrop G., Johnstone A.M., Flint H.J., Lobley G.E. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl. Environ. Microbiol. 2007;73:1073–1078. doi: 10.1128/AEM.02340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvallet C., Gibbons S.M., Gurry T., Irizarry R.A., Alm E.J. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat. Commun. 2017;8:1784. doi: 10.1038/s41467-017-01973-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EUROSTAT, 2019. Increase in the share of the population aged 65 years or over between 2007 and 2017. Available at https://ec.europa.eu/eurostat/statistics-explained/index.php/Population_structure_and_ageing#The_share_of_elderly_people_continues_to_increase. Accessed January 05, 2020.
- Evans C.C., LePard K.J., Kwak J.W., Stancukas M.C., Laskowski S., Dougherty J., Moulton L., Glawe A., Wang Y., Leone V., Antonopoulos D.A., Smith D., Chang E.B., Ciancio M.J. Exercise prevents weight gain and alters the gut microbiota in a mouse model of high fat diet-induced obesity. PLoS One. 2014;9:e92193. doi: 10.1371/journal.pone.0092193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA, 2020. Fecal microbiota for transplantation: new safety information - regarding additional protections for screening donors for COVID-19 and exposure to SARS-CoV-2 and testing for SARS-CoV-2. Availble at: https://www.fda.gov/safety. Accesed on June 17, 2020.
- Fernandes J., Su W., Rahat-Rozenbloom S., Wolever T.M., Comelli E.M. Adiposity, gut microbiota and faecal short chain fatty acids are linked in adult humans. Nutr. Diabetes. 2014;4:e121. doi: 10.1038/nutd.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrucci L., Fabbri E. Inflammageing: chronic inflammation in ageing, cardiovascular disease, and frailty. Nat. Rev. Cardiol. 2018;15:505–522. doi: 10.1038/s41569-018-0064-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedorova K., Radvansky M., Nemcova E., Grombirikova H., Bosak J., Cernochova M., Lexa M., Smajs D., Freiberger T. The impact of DNA extraction methods on stool bacterial and fungal microbiota community recovery. Front. Microbiol. 2019;10:821. doi: 10.3389/fmicb.2019.00821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T., Holbrook N.J. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- Finucane M.M., Sharpton T.J., Laurent T.J., Pollard K.S. A taxonomic signature of obesity in the microbiome? Getting to the guts of the matter. PLoS One. 2014;9 doi: 10.1371/journal.pone.0084689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firouzi S., Haghighatdoost F. The effects of prebiotic, probiotic, and synbiotic supplementation on blood parameters of renal function: a systematic review and meta-analysis of clinical trials. Nutrition. 2018;51-52:104–113. doi: 10.1016/j.nut.2018.01.007. [DOI] [PubMed] [Google Scholar]
- Flanagan E., Lamport D., Brennan L., Burnet P., Calabrese V., Cunnane S.C., de Wilde M.C., Dye L., Farrimond J.A., Emerson Lombardo N., Hartmann T., Hartung T., Kalliomaki M., Kuhnle G.G., La Fata G., Sala-Vila A., Samieri C., Smith A.D., Spencer J.P.E., Thuret S., Tuohy K., Turroni S., Vanden Berghe W., Verkuijl M., Verzijden K., Yannakoulia M., Geurts L., Vauzour D. Nutrition and the ageing brain: moving towards clinical applications. Ageing Res. Rev. 2020;62 doi: 10.1016/j.arr.2020.101079. [DOI] [PubMed] [Google Scholar]
- Flowers S.A., Evans S.J., Ward K.M., McInnis M.G., Ellingrod V.L. Interaction between atypical antipsychotics and the gut microbiome in a bipolar disease cohort. Pharmacotherapy. 2017;37:261–267. doi: 10.1002/phar.1890. [DOI] [PubMed] [Google Scholar]
- Ford A.C., Suares N.C. Effect of laxatives and pharmacological therapies in chronic idiopathic constipation: systematic review and meta-analysis. Gut. 2011;60:209–218. doi: 10.1136/gut.2010.227132. [DOI] [PubMed] [Google Scholar]
- Ford A.C., Moayyedi P., Lacy B.E., Lembo A.J., Saito Y.A., Schiller L.R., Soffer E.E., Spiegel B.M., Quigley E.M. American College of Gastroenterology monograph on the management of irritable bowel syndrome and chronic idiopathic constipation. Am. J. Gastroenterol. 2014;109(Suppl. 1):S2–26. doi: 10.1038/ajg.2014.187. (quiz S27) [DOI] [PubMed] [Google Scholar]
- Ford A.C., Quigley E.M., Lacy B.E., Lembo A.J., Saito Y.A., Schiller L.R., Soffer E.E., Spiegel B.M., Moayyedi P. Efficacy of prebiotics, probiotics, and synbiotics in irritable bowel syndrome and chronic idiopathic constipation: systematic review and meta-analysis. Am. J. Gastroenterol. 2014;109(1562):1547–1561. doi: 10.1038/ajg.2014.202. quiz 1546. [DOI] [PubMed] [Google Scholar]
- Ford A.C., Harris L.A., Lacy B.E., Quigley E.M.M., Moayyedi P. Systematic review with meta-analysis: the efficacy of prebiotics, probiotics, synbiotics and antibiotics in irritable bowel syndrome. Aliment. Pharmacol. Ther. 2018;48:1044–1060. doi: 10.1111/apt.15001. [DOI] [PubMed] [Google Scholar]
- Forslund K., Sunagawa S., Kultima J.R., Mende D.R., Arumugam M., Typas A., Bork P. Country-specific antibiotic use practices impact the human gut resistome. Genome Res. 2013;23:1163–1169. doi: 10.1101/gr.155465.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund K., Hildebrand F., Nielsen T., Falony G., Le Chatelier E., Sunagawa S., Prifti E., Vieira-Silva S., Gudmundsdottir V., Krogh Pedersen H., Arumugam M., Kristiansen K., Yvonne Voigt A., Vestergaard H., Hercog R., Igor Costea P., Roat Kultima J., Li J., Jørgensen T., Levenez F., Dore J., Bjørn Nielsen H., Brunak S., Raes J., Hansen T., Wang J., Dusko Ehrlich S., Bork P., Pedersen O. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528:262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsythe P., Sudo N., Dinan T., Taylor V.H., Bienenstock J. Mood and gut feelings. Brain Behav. Immun. 2010;24:9–16. doi: 10.1016/j.bbi.2009.05.058. [DOI] [PubMed] [Google Scholar]
- Fragiadakis G.K., Smits S.A., Sonnenburg E.D., Van Treuren W., Reid G., Knight R., Manjurano A., Changalucha J., Dominguez-Bello M.G., Leach J., Sonnenburg J.L. Links between environment, diet, and the hunter-gatherer microbiome. Gut Microbes. 2019;10:216–227. doi: 10.1080/19490976.2018.1494103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi C., Bonafè M., Valensin S., Olivieri F., De Luca M., Ottaviani E., De Benedictis G. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann. N. Y. Acad. Sci. 2000;908:244–254. doi: 10.1111/j.1749-6632.2000.tb06651.x. [DOI] [PubMed] [Google Scholar]
- Francino M.P. Antibiotics and the human gut microbiome: Dysbioses and accumulation of resistances. Front. Microbiol. 2015;6:1543. doi: 10.3389/fmicb.2015.01543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer K.A., Murray S.S., Schork N.J., Topol E.J. Human genetic variation and its contribution to complex traits. Nat. Rev. Gen. 2009;10:241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- Furusawa Y., Obata Y., Fukuda S., Endo T.A., Nakato G., Takahashi D., Nakanishi Y., Uetake C., Kato K., Kato T., Takahashi M., Fukuda N.N., Murakami S., Miyauchi E., Hino S., Atarashi K., Onawa S., Fujimura Y., Lockett T., Clarke J.M., Topping D.L., Tomita M., Hori S., Ohara O., Morita T., Koseki H., Kikuchi J., Honda K., Hase K., Ohno H. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- Gallegos-Orozco J.F., Foxx-Orenstein A.E., Sterler S.M., Stoa J.M. Chronic constipation in the elderly. Am. J. Gastroenterol. 2012;107:18–25. doi: 10.1038/ajg.2011.349. (quiz 26) [DOI] [PubMed] [Google Scholar]
- Geoba, 2020. The world: life expectancy (2019). Available at http://www.geoba.se. Accesed on May 05, 2020.
- Ghosh T.S., Rampelli S., Jeffery I.B., Santoro A., Neto M., Capri M., Giampieri E., Jennings A., Candela M., Turroni S., Zoetendal E.G., Hermes G.D.A., Elodie C., Meunier N., Brugere C.M., Pujos-Guillot E., Berendsen A.M., De Groot L., Feskins E.J.M., Kaluza J., Pietruszka B., Bielak M.J., Comte B., Maijo-Ferre M., Nicoletti C., De Vos W.M., Fairweather-Tait S., Cassidy A., Brigidi P., Franceschi C., O'Toole P.W. Mediterranean diet intervention alters the gut microbiome in older people reducing frailty and improving health status: the NU-AGE 1-year dietary intervention across five European countries. Gut. 2020;69:1218–1228. doi: 10.1136/gutjnl-2019-319654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G.R., Probert H.M., Van Loo J., Rastall R.A., Roberfroid M.B. Dietary modulation of the human colonic microbiota: updating the concept of prebiotics. Nut. Res. Rev. 2004;17:259–275. doi: 10.1079/NRR200479. [DOI] [PubMed] [Google Scholar]
- Gill S.R., Pop M., DeBoy R.T., Eckburg P.B., Turnbaugh P.J., Samuel B.S., Gordon J.I., Relman D.A., Fraser-Liggett C.M., Nelson K.E. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J.K., Waters J.L., Poole A.C., Sutter J.L., Koren O., Blekhman R., Beaumont M., Van Treuren W., Knight R., Bell J.T., Spector T.D., Clark A.G., Ley R.E. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J.K., Davenport E.R., Beaumont M., Jackson M.A., Knight R., Ober C., Spector T.D., Bell J.T., Clark A.G., Ley R.E. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe. 2016;19:731–743. doi: 10.1016/j.chom.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan V., Spencer C.N., Nezi L., Reuben A., Andrews M.C., Karpinets T.V., Prieto P.A., Vicente D., Hoffman K., Wei S.C., Cogdill A.P., Zhao L., Hudgens C.W., Hutchinson D.S., Manzo T., Petaccia De Macedo M., Cotechini T., Kumar T., Chen W.S., Reddy S.M., Szczepaniak Sloane R., Galloway-Pena J., Jiang H., Chen P.L., Shpall E.J., Rezvani K., Alousi A.M., Chemaly R.F., Shelburne S., Vence L.M., Okhuysen P.C., Jensen V.B., Swennes A.G., McAllister F., Marcelo Riquelme Sanchez E., Zhang Y., Le Chatelier E., Zitvogel L., Pons N., Austin-Breneman J.L., Haydu L.E., Burton E.M., Gardner J.M., Sirmans E., Hu J., Lazar A.J., Tsujikawa T., Diab A., Tawbi H., Glitza I.C., Hwu W.J., Patel S.P., Woodman S.E., Amaria R.N., Davies M.A., Gershenwald J.E., Hwu P., Lee J.E., Zhang J., Coussens L.M., Cooper Z.A., Futreal P.A., Daniel C.R., Ajami N.J., Petrosino J.F., Tetzlaff M.T., Sharma P., Allison J.P., Jenq R.R., Wargo J.A. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor M.F., Hotamisligil G.S. Inflammatory mechanisms in obesity. Annu. Rev. Immunol. 2011:415–445. doi: 10.1146/annurev-immunol-031210-101322. [DOI] [PubMed] [Google Scholar]
- Gui Q., Wang A., Zhao X., Huang S., Tan Z., Xiao C., Yang Y. Effects of probiotic supplementation on natural killer cell function in healthy elderly individuals: a meta-analysis of randomized controlled trials. Eur. J. Clin. Nutr. 2020 doi: 10.1038/s41430-020-0670-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadi A., Mohammadi H., Miraghajani M., Ghaedi E. Efficacy of synbiotic supplementation in patients with nonalcoholic fatty liver disease: a systematic review and meta-analysis of clinical trials: Synbiotic supplementation and NAFLD. Crit. Rev. Food Sci. Nutr. 2019;59:2494–2505. doi: 10.1080/10408398.2018.1458021. [DOI] [PubMed] [Google Scholar]
- Hadi A., Ghaedi E., Khalesi S., et al. Effects of synbiotic consumption on lipid profile: a systematic review and meta-analysis of randomized controlled clinical trials. Eur. J. Nutr. 2020;59:2857–2874. doi: 10.1007/s00394-020-02248-7. [DOI] [PubMed] [Google Scholar]
- Han T.S., Tajar A., Lean M.E.J. Obesity and weight management in the elderly. Br. Med. Bull. 2011;97:169–196. doi: 10.1093/bmb/ldr002. [DOI] [PubMed] [Google Scholar]
- Harper S. Economic and social implications of aging societies. Science. 2014;346:587–591. doi: 10.1126/science.1254405. [DOI] [PubMed] [Google Scholar]
- Hill C., Guarner F., Reid G., Gibson G.R., Merenstein D.J., Pot B., Morelli L., Canani R.B., Flint H.J., Salminen S., Calder P.C., Sanders M.E. Expert consensus document. The international scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014;11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- Hooper L.V., Littman D.R., Macpherson A.J. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins M.J., Sharp R., Macfarlane G.T. Age and disease related changes in intestinal bacterial populations assessed by cell culture, 16S rRNA abundance, and community cellular fatty acid profiles. Gut. 2001;48:198–205. doi: 10.1136/gut.48.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao E.Y., McBride S.W., Hsien S., Sharon G., Hyde E.R., McCue T., Codelli J.A., Chow J., Reisman S.E., Petrosino J.F., Patterson P.H., Mazmanian S.K. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Yang X., Qin J., Lu N., Cheng G., Wu N., Pan Y., Li J., Zhu L., Wang X., Meng Z., Zhao F., Liu D., Ma J., Qin N., Xiang C., Xiao Y., Li L., Yang H., Wang J., Yang R., Gao G.F., Wang J., Zhu B. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat. Commun. 2013;4:2151. doi: 10.1038/ncomms3151. [DOI] [PubMed] [Google Scholar]
- Huttenhower, C., Gevers, D., Knight, R., Abubucker, S., Badger, J.H., Chinwalla, A.T., Creasy, H.H., Earl, A.M., Fitzgerald, M.G., Fulton, R.S., Giglio, M.G., Hallsworth-Pepin, K., Lobos, E.A., Madupu, R., Magrini, V., Martin, J.C., Mitreva, M., Muzny, D.M., Sodergren, E.J., Versalovic, J., Wollam, A.M., Worley, K.C., Wortman, J.R., Young, S.K., Zeng, Q., Aagaard, K.M., Abolude, O.O., Allen-Vercoe, E., Alm, E.J., Alvarado, L., Andersen, G.L., Anderson, S., Appelbaum, E., Arachchi, H.M., Armitage, G., Arze, C.A., Ayvaz, T., Baker, C.C., Begg, L., Belachew, T., Bhonagiri, V., Bihan, M., Blaser, M.J., Bloom, T., Bonazzi, V., Paul Brooks, J., Buck, G.A., Buhay, C.J., Busam, D.A., Campbell, J.L., Canon, S.R., Cantarel, B.L., Chain, P.S.G., Chen, I.M.A., Chen, L., Chhibba, S., Chu, K., Ciulla, D.M., Clemente, J.C., Clifton, S.W., Conlan, S., Crabtree, J., Cutting, M.A., Davidovics, N.J., Davis, C.C., Desantis, T.Z., Deal, C., Delehaunty, K.D., Dewhirst, F.E., Deych, E., Ding, Y., Dooling, D.J., Dugan, S.P., Michael Dunne, W., Scott Durkin, A., Edgar, R.C., Erlich, R.L., Farmer, C.N., Farrell, R.M., Faust, K., Feldgarden, M., Felix, V.M., Fisher, S., Fodor, A.A., Forney, L.J., Foster, L., Di Francesco, V., Friedman, J., Friedrich, D.C., Fronick, C.C., Fulton, L.L., Gao, H., Garcia, N., Giannoukos, G., Giblin, C., Giovanni, M.Y., Goldberg, J.M., Goll, J., Gonzalez, A., Griggs, A., Gujja, S., Kinder Haake, S., Haas, B.J., Hamilton, H.A., Harris, E.L., Hepburn, T.A., Herter, B., Hoffmann, D.E., Holder, M.E., Howarth, C., Huang, K.H., Huse, S.M., Izard, J., Jansson, J.K., Jiang, H., Jordan, C., Joshi, V., Katancik, J.A., Keitel, W.A., Kelley, S.T., Kells, C., King, N.B., Knights, D., Kong, H.H., Koren, O., Koren, S., Kota, K.C., Kovar, C.L., Kyrpides, N.C., La Rosa, P.S., Lee, S.L., Lemon, K.P., Lennon, N., Lewis, C.M., Lewis, L., Ley, R.E., Li, K., Liolios, K., Liu, B., Liu, Y., Lo, C.C., Lozupone, C.A., Dwayne Lunsford, R., Madden, T., Mahurkar, A.A., Mannon, P.J., Mardis, E.R., Markowitz, V.M., Mavromatis, K., McCorrison, J.M., McDonald, D., McEwen, J., McGuire, A.L., McInnes, P., Mehta, T., Mihindukulasuriya, K.A., Miller, J.R., Minx, P.J., Newsham, I., Nusbaum, C., Oglaughlin, M., Orvis, J., Pagani, I., Palaniappan, K., Patel, S.M., Pearson, M., Peterson, J., Podar, M., Pohl, C., Pollard, K.S., Pop, M., Priest, M.E., Proctor, L.M., Qin, X., Raes, J., Ravel, J., Reid, J.G., Rho, M., Rhodes, R., Riehle, K.P., Rivera, M.C., Rodriguez-Mueller, B., Rogers, Y.H., Ross, M.C., Russ, C., Sanka, R.K., Sankar, P., Fah Sathirapongsasuti, J., Schloss, J.A., Schloss, P.D., Schmidt, T.M., Scholz, M., Schriml, L., Schubert, A.M., Segata, N., Segre, J.A., Shannon, W.D., Sharp, R.R., Sharpton, T.J., Shenoy, N., Sheth, N.U., Simone, G.A., Singh, I., Smillie, C.S., Sobel, J.D., Sommer, D.D., Spicer, P., Sutton, G.G., Sykes, S.M., Tabbaa, D.G., Thiagarajan, M., Tomlinson, C.M., Torralba, M., Treangen, T.J., Truty, R.M., Vishnivetskaya, T.A., Walker, J., Wang, L., Wang, Z., Ward, D.V., Warren, W., Watson, M.A., Wellington, C., Wetterstrand, K.A., White, J.R., Wilczek-Boney, K., Wu, Y., Wylie, K.M., Wylie, T., Yandava, C., Ye, L., Ye, Y., Yooseph, S., Youmans, B.P., Zhang, L., Zhou, Y., Zhu, Y., Zoloth, L., Zucker, J.D., Birren, B.W., Gibbs, R.A., Highlander, S.K., Methé, B.A., Nelson, K.E., Petrosino, J.F., Weinstock, G.M., Wilson, R.K., White, O., 2012. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214.
- Imhann F., Bonder M.J., Vila A.V., Fu J.Y., Mujagic Z., Vork L., Tigchelaar E.F., Jankipersadsing S.A., Cenit M.C., Harmsen H.J.M., Dijkstra G., Franke L., Xavier R.J., Jonkers D., Wijmenga C., Weersma R.K., Zhernakova A. Proton pump inhibitors affect the gut microbiome. Gut. 2016;65:740–748. doi: 10.1136/gutjnl-2015-310376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infusino F., Marazzato M., Mancone M., Fedele F., Mastroianni C.M., Severino P., Ceccarelli G., Santinelli L., Cavarretta E., Marullo A.G.M., Miraldi F., Carnevale R., Nocella C., Biondi-Zoccai G., Pagnini C., Schiavon S., Pugliese F., Frati G., d’Ettorre G. Diet supplementation, probiotics, and nutraceuticals in SARS-CoV-2 infection: a scoping review. Nutrients. 2020;12 doi: 10.3390/nu12061718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwauchi M., Horigome A., Ishikawa K., Mikuni A., Nakano M., Xiao J.Z., Odamaki T., Hironaka S. Relationship between oral and gut microbiota in elderly people. Immun. Inflamm. Dis. 2019;7:229–236. doi: 10.1002/iid3.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M.A., Goodrich J.K., Maxan M.E., Freedberg D.E., Abrams J.A., Poole A.C., Sutter J.L., Welter D., Ley R.E., Bell J.T., Spector T.D., Steves C.J. Proton pump inhibitors alter the composition of the gut microbiota. Gut. 2016;65:749–756. doi: 10.1136/gutjnl-2015-310861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafarnejad S., Shab-Bidar S., Speakman J.R., Parastui K., Daneshi-Maskooni M., Djafarian K. Probiotics reduce the risk of antibiotic-associated diarrhea in adults (18-64 years) but not the elderly (>65 years): a meta-analysis. Nutr. Clin. Pract. 2016;31:502–513. doi: 10.1177/0884533616639399. [DOI] [PubMed] [Google Scholar]
- Jedrusek-Golinska A., Gorecka D., Buchowski M., Wieczorowska-Tobis K., Gramza-Michalowska A., Szymandera-Buszka K. Recent progress in the use of functional foods for older adults: a narrative review. Compr Rev Food Sci F. 2020;19:835–856. doi: 10.1111/1541-4337.12530. [DOI] [PubMed] [Google Scholar]
- Jeffery I.B., Lynch D.B., O’Toole P.W. Composition and temporal stability of the gut microbiota in older persons. ISME J. 2016;10:170–182. doi: 10.1038/ismej.2015.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkner E., Leive A. IMF. Fiscal Affairs Department; 2010. Health Care Spending Issues in Advanced Economies. [Google Scholar]
- Jha A.R., Davenport E.R., Gautam Y., Bhandari D., Tandukar S., Ng K.M., Fragiadakis G.K., Holmes S., Gautam G.P., Leach J., Sherchand J.B., Bustamante C.D., Sonnenburg J.L. Gut microbiome transition across a lifestyle gradient in Himalaya. PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.2005396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson F.H., Tremaroli V., Nookaew I., Bergström G., Behre C.J., Fagerberg B., Nielsen J., Bäckhed F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- Kazemi A., Soltani S., Ghorabi S., Keshtkar A., Daneshzad E., Nasri F., Mazloomi S.M. Effect of probiotic and synbiotic supplementation on inflammatory markers in health and disease status: a systematic review and meta-analysis of clinical trials. Clin. Nutr. 2020;39:789–819. doi: 10.1016/j.clnu.2019.04.004. [DOI] [PubMed] [Google Scholar]
- Kearney S.M., Gibbons S.M., Erdman S.E., Alm E.J. Orthogonal dietary niche enables reversible engraftment of a gut bacterial commensal. Cell Rep. 2018;24:1842–1851. doi: 10.1016/j.celrep.2018.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller J.M., Surawicz C.M. Clostridium difficile infection in the elderly. Clin. Geriatr. Med. 2014;30:79–93. doi: 10.1016/j.cger.2013.10.008. [DOI] [PubMed] [Google Scholar]
- Kelly C.R., Khoruts A., Staley C., Sadowsky M.J., Abd M., Alani M., Bakow B., Curran P., McKenney J., Tisch A., Reinert S.E., MacHan J.T., Brandt L.J. Effect of fecal microbiota transplantation on recurrence in multiply recurrent clostridium difficile infection a randomized trial. Ann. Intern. Med. 2016;165:609–616. doi: 10.7326/M16-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy B.K., Berger S.L., Brunet A., Campisi J., Cuervo A.M., Epel E.S., Franceschi C., Lithgow G.J., Morimoto R.I., Pessin J.E., Rando T.A., Richardson A., Schadt E.E., Wyss-Coray T., Sierra F. Geroscience: linking aging to chronic disease. Cell. 2014;159:709–713. doi: 10.1016/j.cell.2014.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalesi S., Sun J., Buys N., Jayasinghe R. Effect of probiotics on blood pressure: a systematic review and meta-analysis of randomized, controlled trials. Hypertension. 2014;64:897–903. doi: 10.1161/HYPERTENSIONAHA.114.03469. [DOI] [PubMed] [Google Scholar]
- Kobyliak N., Conte C., Cammarota G., Haley A.P., Styriak I., Gaspar L., Fusek J., Rodrigo L., Kruzliak P. Probiotics in prevention and treatment of obesity: a critical view. Nutr. Metab. (Lond.) 2016;13:14. doi: 10.1186/s12986-016-0067-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziejczyk A.A., Zheng D., Elinav E. Diet–microbiota interactions and personalized nutrition. Nat. Rev. Microbiol. 2019;17:742–753. doi: 10.1038/s41579-019-0256-8. [DOI] [PubMed] [Google Scholar]
- Kong F., Hua Y., Zeng B., Ning R., Li Y., Zhao J. Gut microbiota signatures of longevity. Curr. Biol. 2016;26:R832–R833. doi: 10.1016/j.cub.2016.08.015. [DOI] [PubMed] [Google Scholar]
- Kwok C.S., Arthur A.K., Anibueze C.I., Singh S., Cavallazzi R., Loke Y.K. Risk of clostridium difficile infection with acid suppressing drugs and antibiotics: Meta-analysis. Am. J. Gastroenterol. 2012;107:1011–1019. doi: 10.1038/ajg.2012.108. [DOI] [PubMed] [Google Scholar]
- Lam K.N., Alexander M., Turnbaugh P.J. Precision medicine Goes microscopic: engineering the microbiome to improve drug outcomes. Cell Host Microbe. 2019;26:22–34. doi: 10.1016/j.chom.2019.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen N., Vogensen F.K., van den Berg F.W., Nielsen D.S., Andreasen A.S., Pedersen B.K., Al-Soud W.A., Sorensen S.J., Hansen L.H., Jakobsen M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One. 2010;5 doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop S.K., Bloom S.M., Rao S.M., Nutsch K., Lio C.W., Santacruz N., Peterson D.A., Stappenbeck T.S., Hsieh C.S. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxminarayan R., Duse A., Wattal C., Zaidi A.K., Wertheim H.F., Sumpradit N., Vlieghe E., Hara G.L., Gould I.M., Goossens H., Greko C., So A.D., Bigdeli M., Tomson G., Woodhouse W., Ombaka E., Peralta A.Q., Qamar F.N., Mir F., Kariuki S., Bhutta Z.A., Coates A., Bergstrom R., Wright G.D., Brown E.D., Cars O. Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 2013;13:1057–1098. doi: 10.1016/S1473-3099(13)70318-9. [DOI] [PubMed] [Google Scholar]
- Lee C.H., Steiner T., Petrof E.O., Smieja M., Roscoe D., Nematallah A., Weese J.S., Collins S., Moayyedi P., Crowther M., Ropeleski M.J., Jayaratne P., Higgins D., Li Y., Rau N.V., Kim P.T. Frozen vs fresh fecal microbiota transplantation and clinical resolution of diarrhea in patients with recurrent Clostridium difficile infection: a randomized clinical trial. JAMA. 2016;315:142–149. doi: 10.1001/jama.2015.18098. [DOI] [PubMed] [Google Scholar]
- Lee P., Yacyshyn B.R., Yacyshyn M.B. Gut microbiota and obesity: an opportunity to alter obesity through faecal microbiota transplant (FMT) Diabetes Obes. Metab. 2019;21:479–490. doi: 10.1111/dom.13561. [DOI] [PubMed] [Google Scholar]
- Lefevre M., Racedo S.M., Ripert G., Housez B., Cazaubiel M., Maudet C., Justen P., Marteau P., Urdaci M.C. Probiotic strain Bacillus subtilis CU1 stimulates immune system of elderly during common infectious disease period: a randomized, double-blind placebo-controlled study. Immun. Ageing. 2015;12:24. doi: 10.1186/s12979-015-0051-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei W.T., Shih P.C., Liu S.J., Lin C.Y., Yeh T.L. Effect of probiotics and prebiotics on immune response to influenza vaccination in adults: a systematic review and Meta-analysis of randomized controlled trials. Nutrients. 2017;9 doi: 10.3390/nu9111175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R.E., Bäckhed F., Turnbaugh P., Lozupone C.A., Knight R.D., Gordon J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R.E., Turnbaugh P.J., Klein S., Gordon J.I. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Loo V.G., Bourgault A.M., Poirier L., Lamothe F., Michaud S., Turgeon N., Toye B., Beaudoin A., Frost E.H., Gilca R., Brassard P., Dendukuri N., Béliveau C., Oughton M., Brukner I., Dascal A. Host and pathogen factors for Clostridium difficile infection and colonization. New Engl. J. Med. 2011;365:1693–1703. doi: 10.1056/NEJMoa1012413. [DOI] [PubMed] [Google Scholar]
- López-Otín C., Blasco M.A., Partridge L., Serrano M., Kroemer G. The hallmarks of aging. Cell. 2013;153:1194. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C., Galluzzi L., Freije J.M.P., Madeo F., Kroemer G. Metabolic control of longevity. Cell. 2016;166:802–821. doi: 10.1016/j.cell.2016.07.031. [DOI] [PubMed] [Google Scholar]
- Lozupone C.A., Stombaugh J.I., Gordon J.I., Jansson J.K., Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Tixier E.N., Grinspan A.M. Fecal microbiota transplantation for Clostridioides difficile in high-risk older adults is associated with early recurrence. Dig. Dis. Sci. 2020 doi: 10.1007/s10620-020-06147-z. [DOI] [PubMed] [Google Scholar]
- Lynch S.V., Pedersen O. The human intestinal microbiome in health and disease. New Engl. J. Med. 2016;375:2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- Macfarlane S., Cleary S., Bahrami B., Reynolds N., Macfarlane G.T. Synbiotic consumption changes the metabolism and composition of the gut microbiota in older people and modifies inflammatory processes: a randomised, double-blind, placebo-controlled crossover study. Aliment. Pharmacol. Ther. 2013;38:804–816. doi: 10.1111/apt.12453. [DOI] [PubMed] [Google Scholar]
- Mach N., Fuster-Botella D. Endurance exercise and gut microbiota: a review. J. Sport Health Sci. 2017;6:179–197. doi: 10.1016/j.jshs.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes M.L., Fixen D.R., Linnebur S.A. Adverse effects of proton-pump inhibitor use in older adults: a review of the evidence. Therapeutic advances in drug safety. 2017;8:273–297. doi: 10.1177/2042098617715381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffei V.J., Kim S., Blanchard E.t., Luo M., Jazwinski S.M., Taylor C.M., Welsh D.A. Biological aging and the human gut microbiota. J. Gerontol. A Biol. Sci. Med. Sci. 2017;72:1474–1482. doi: 10.1093/gerona/glx042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher R.L., Hanlon J., Hajjar E.R. Clinical consequences of polypharmacy in elderly. Expert Opin. Drug Saf. 2014;13:57–65. doi: 10.1517/14740338.2013.827660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier L., Pruteanu M., Kuhn M., Zeller G., Telzerow A., Anderson E.E., Brochado A.R., Fernandez K.C., Dose H., Mori H., Patil K.R., Bork P., Typas A. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018;555:623–628. doi: 10.1038/nature25979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailing L.J., Allen J.M., Buford T.W., Fields C.J., Woods J.A. Exercise and the gut microbiome: a review of the evidence, potential mechanisms, and implications for human health. Exerc. Sport Sci. Rev. 2019;47:75–85. doi: 10.1249/JES.0000000000000183. [DOI] [PubMed] [Google Scholar]
- Maneerat S., Lehtinen M.J., Childs C.E., Forssten S.D., Alhoniemi E., Tiphaine M., Yaqoob P., Ouwehand A.C., Rastall R.A. Consumption of Bifidobacterium lactis Bi-07 by healthy elderly adults enhances phagocytic activity of monocytes and granulocytes. J. Nutr. Sci. 2013;2:e44. doi: 10.1017/jns.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann T., Heuberger R., Wong H. The association between chewing and swallowing difficulties and nutritional status in older adults. Aust. Dent. J. 2013;58:200–206. doi: 10.1111/adj.12064. [DOI] [PubMed] [Google Scholar]
- Marchesi J.R., Adams D.H., Fava F., Hermes G.D.A., Hirschfield G.M., Hold G., Quraishi M.N., Kinross J., Smidt H., Tuohy K.M., Thomas L.V., Zoetendal E.G., Hart A. The gut microbiota and host health: a new clinical frontier. Gut. 2016;65:330–339. doi: 10.1136/gutjnl-2015-309990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez I., Stegen J.C., Maldonado-Gómez M.X., Eren M.A., Siba P.M., Greenhill A.R., Walter J. The gut microbiota of rural Papua New Guineans: composition, diversity patterns, and ecological processes. Cell Rep. 2015;11:527–538. doi: 10.1016/j.celrep.2015.03.049. [DOI] [PubMed] [Google Scholar]
- Martinez-Martinez M.I., Calabuig-Tolsa R., Cauli O. The effect of probiotics as a treatment for constipation in elderly people: a systematic review. Arch. Gerontol. Geriatr. 2017;71:142–149. doi: 10.1016/j.archger.2017.04.004. [DOI] [PubMed] [Google Scholar]
- Mathus-Vliegen E.M., Obesity Management Task Force of the European Association for the Study of, O Prevalence, pathophysiology, health consequences and treatment options of obesity in the elderly: a guideline. Obes Facts. 2012;5:460–483. doi: 10.1159/000341193. [DOI] [PubMed] [Google Scholar]
- Matson V., Fessler J., Bao R., Chongsuwat T., Zha Y., Alegre M.L., Luke J.J., Gajewski T.F. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M., Inoue R., Tsukahara T., Ushida K., Chiji H., Matsubara N., Hara H. Voluntary running exercise alters microbiota composition and increases n-butyrate concentration in the rat cecum. Biosci. Biotechnol. Biochem. 2008;72:572–576. doi: 10.1271/bbb.70474. [DOI] [PubMed] [Google Scholar]
- McDonald E.G., Milligan J., Frenette C., Lee T.C. Continuous proton pump inhibitor therapy and the associated risk of recurrent Clostridium difficile infection. JAMA Intern. Med. 2015;175:784–791. doi: 10.1001/jamainternmed.2015.42. [DOI] [PubMed] [Google Scholar]
- McDonald L.C., Gerding D.N., Johnson S., Bakken J.S., Carroll K.C., Coffin S.E., Dubberke E.R., Garey K.W., Gould C.V., Kelly C., Loo V., Shaklee Sammons J., Sandora T.J., Wilcox M.H. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA) Clin. Infect. Dis. 2018;66:e1–e48. doi: 10.1093/cid/cix1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L.E., Lehtoranta L., Lehtinen M.J. The effect of Bifidobacterium animalis ssp lactis HN019 on cellular immune function in healthy elderly subjects: systematic review and Meta-analysis. Nutrients. 2017;9 doi: 10.3390/nu9030191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L.E., Lehtoranta L., Lehtinen M.J. Short-term probiotic supplementation enhances cellular immune function in healthy elderly: systematic review and meta-analysis of controlled studies. Nutr. Res. 2019;64:1–8. doi: 10.1016/j.nutres.2018.12.011. [DOI] [PubMed] [Google Scholar]
- Milne A.C., Potter J., Vivanti A., Avenell A. Protein and energy supplementation in elderly people at risk from malnutrition. Cochrane Database Syst. Rev. 2009;2 doi: 10.1002/14651858.CD003288.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller A.H. The shrinking human gut microbiome. Curr. Opin. Microbiol. 2017;38:30–35. doi: 10.1016/j.mib.2017.04.002. [DOI] [PubMed] [Google Scholar]
- Monda V., Villano I., Messina A., Valenzano A., Esposito T., Moscatelli F., Viggiano A., Cibelli G., Chieffi S., Monda M., Messina G. Exercise modifies the gut microbiota with positive health effects. Oxidative Med. Cell. Longev. 2017;2017 doi: 10.1155/2017/3831972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteagudo-Mera A., Rastall R.A., Gibson G.R., Charalampopoulos D., Chatzifragkou A. Adhesion mechanisms mediated by probiotics and prebiotics and their potential impact on human health. Appl. Microbiol. Biotechnol. 2019;103:6463–6472. doi: 10.1007/s00253-019-09978-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro-Garcia M.A., Alonso-Arias R., Baltadjieva M., Benitez C.F., Barrial M.A.F., Ruisánchez E.D., Santos R.A., Sánchez M.A., Miján J.S., López-Larrea C. Oral supplementation with Lactobacillus delbrueckii subsp. bulgaricus 8481 enhances systemic immunity in elderly subjects. Age. 2013;35:1311–1326. doi: 10.1007/s11357-012-9434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S., Saunier K., Hanisch C., Norin E., Alm L., Midtvedt T., Cresci A., Silvi S., Orpianesi C., Verdenelli M.C., Clavel T., Koebnick C., Zunft H.J.F., Doré J., Blaut M. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl. Environ. Microbiol. 2006;72:1027–1033. doi: 10.1128/AEM.72.2.1027-1033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murri M., Leiva I., Gomez-Zumaquero J.M., Tinahones F.J., Cardona F., Soriguer F., Queipo-Ortuno M.I. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC Med. 2013;11:46. doi: 10.1186/1741-7015-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal R., Mainali R., Ahmadi S., Wang S., Singh R., Kavanagh K., Kitzman D.W., Kushugulova A., Marotta F., Yadav H. Gut microbiome and aging: physiological and mechanistic insights. Nutr. Heal. Aging. 2018;4:267–285. doi: 10.3233/NHA-170030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto J.V., de Melo C.M., Ribeiro S.M.L. Effects of three-month intake of synbiotic on inflammation and body composition in the elderly: a pilot study. Nutrients. 2013;5:1276–1286. doi: 10.3390/nu5041276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson J.K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- Nikbakht E., Khalesi S., Singh I., Williams L.T., West N.P., Colson N. Effect of probiotics and synbiotics on blood glucose: a systematic review and meta-analysis of controlled trials. Eur. J. Nutr. 2018;57:95–106. doi: 10.1007/s00394-016-1300-3. [DOI] [PubMed] [Google Scholar]
- O’Callaghan A., van Sinderen D. Bifidobacteria and their role as members of the human gut microbiota. Front. Microbiol. 2016;7:925. doi: 10.3389/fmicb.2016.00925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H., Lawrence J.G., Grolsman E.A. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- Odamaki T., Kato K., Sugahara H., Hashikura N., Takahashi S., Xiao J.Z., Abe F., Osawa R. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol. 2016;16:90. doi: 10.1186/s12866-016-0708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeppen J., Vaupel J.W. Demography: broken limits to life expectancy. Science. 2002;296:1029–1031. doi: 10.1126/science.1069675. [DOI] [PubMed] [Google Scholar]
- O’Keefe S.J., Li J.V., Lahti L., Ou J., Carbonero F., Mohammed K., Posma J.M., Kinross J., Wahl E., Ruder E., Vipperla K., Naidoo V., Mtshali L., Tims S., Puylaert P.G., DeLany J., Krasinskas A., Benefiel A.C., Kaseb H.O., Newton K., Nicholson J.K., de Vos W.M., Gaskins H.R., Zoetendal E.G. Fat, fibre and cancer risk in African Americans and rural Africans. Nat. Commun. 2015;6:6342. doi: 10.1038/ncomms7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olbe L., Carlsson E., Lindberg P. A proton-pump inhibitor expedition: the case histories of omeprazole and esomeprazole. Nat. Rev. Drug Discov. 2003;2:132–139. doi: 10.1038/nrd1010. [DOI] [PubMed] [Google Scholar]
- Ooijevaar R.E., Terveer E.M., Verspaget H.W., Kuijper E.J., Keller J.J. Clinical application and potential of fecal microbiota transplantation. Annu. Rev. Med. 2019:335–351. doi: 10.1146/annurev-med-111717-122956. [DOI] [PubMed] [Google Scholar]
- O'Toole P.W., Jeffery I.B. Gut microbiota and aging. Science. 2015;350:1214–1215. doi: 10.1126/science.aac8469. [DOI] [PubMed] [Google Scholar]
- O'Toole P.W., Jeffery I.B. Microbiome-health interactions in older people. Cell. Mol. Life Sci. 2018;75:119–128. doi: 10.1007/s00018-017-2673-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouwehand A.C., Bergsma N., Parhiala R., Lahtinen S., Gueimonde M., Finne-Soveri H., Strandberg T., Pitkälä K., Salminen S. Bifidobacterium microbiota and parameters of immune function in elderly subjects. FEMS Immunol. Med. Microbiol. 2008;53:18–25. doi: 10.1111/j.1574-695X.2008.00392.x. [DOI] [PubMed] [Google Scholar]
- Ouwehand A.C., Tiihonen K., Saarinen M., Putaala H., Rautonen N. Influence of a combination of Lactobacillus acidophilus NCFM and lactitol on healthy elderly: intestinal and immune parameters. Br. J. Nutr. 2009;101:367–375. doi: 10.1017/S0007114508003097. [DOI] [PubMed] [Google Scholar]
- Palmer C., Bik E.M., DiGiulio D.B., Relman D.A., Brown P.O. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:1556–1573. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey K.R., Naik S.R., Vakil B.V. Probiotics, prebiotics and synbiotics- a review. J. Food Sci. Technol. 2015;52:7577–7587. doi: 10.1007/s13197-015-1921-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panek M., Cipcic Paljetak H., Baresic A., Peric M., Matijasic M., Lojkic I., Vranesic Bender D., Krznaric Z., Verbanac D. Methodology challenges in studying human gut microbiota - effects of collection, storage, DNA extraction and next generation sequencing technologies. Sci. Rep. 2018;8:5143. doi: 10.1038/s41598-018-23296-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parajuli A., Hui N., Puhakka R., Oikarinen S., Grönroos M., Selonen V.A.O., Siter N., Kramna L., Roslund M.I., Vari H.K., Nurminen N., Honkanen H., Hintikka J., Sarkkinen H., Romantschuk M., Kauppi M., Valve R., Cinek O., Laitinen O.H., Rajaniemi J., Hyöty H., Sinkkonen A., group, A.s Yard vegetation is associated with gut microbiota composition. Sci. Total Environ. 2020;713 doi: 10.1016/j.scitotenv.2020.136707. [DOI] [PubMed] [Google Scholar]
- Pascale A., Marchesi N., Govoni S., Coppola A., Gazzaruso C. The role of gut microbiota in obesity, diabetes mellitus, and effect of metformin: new insights into old diseases. Curr. Opin. Pharm. 2019;49:1–5. doi: 10.1016/j.coph.2019.03.011. [DOI] [PubMed] [Google Scholar]
- Pasolli E., Asnicar F., Manara S., Zolfo M., Karcher N., Armanini F., Beghini F., Manghi P., Tett A., Ghensi P., Collado M.C., Rice B.L., DuLong C., Morgan X.C., Golden C.D., Quince C., Huttenhower C., Segata N. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell 176. 2019;e620:649–662. doi: 10.1016/j.cell.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnode M.L., Beller Z.W., Han N.D., Cheng J., Peters S.L., Terrapon N., Henrissat B., Le Gall S., Saulnier L., Hayashi D.K., Meynier A., Vinoy S., Giannone R.J., Hettich R.L., Gordon J.I. Interspecies competition impacts targeted manipulation of human gut bacteria by fiber-derived glycans. Cell. 2019;179:59–73.e13. doi: 10.1016/j.cell.2019.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penedo F.J., Dahn J.R. Exercise and well-being: a review of mental and physical health benefits associated with physical activity. Curr Opin Psychiatry. 2005;18:189–193. doi: 10.1097/00001504-200503000-00013. [DOI] [PubMed] [Google Scholar]
- Pernicova I., Korbonits M. Metformin-mode of action and clinical implications for diabetes and cancer. Nat. Rev. Endocrionol. 2014;10:143–156. doi: 10.1038/nrendo.2013.256. [DOI] [PubMed] [Google Scholar]
- Petriz B.A., Castro A.P., Almeida J.A., Gomes C.P., Fernandes G.R., Kruger R.H., Pereira R.W., Franco O.L. Exercise induction of gut microbiota modifications in obese, non-obese and hypertensive rats. BMC Genomics. 2014;15:511. doi: 10.1186/1471-2164-15-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokusaeva K., Fitzgerald G.F., van Sinderen D. Carbohydrate metabolism in Bifidobacteria. Genes Nutr. 2011;6:285–306. doi: 10.1007/s12263-010-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T., Mende D.R., Li J., Xu J., Li S., Li D., Cao J., Wang B., Liang H., Zheng H., Xie Y., Tap J., Lepage P., Bertalan M., Batto J.M., Hansen T., Le Paslier D., Linneberg A., Nielsen H.B., Pelletier E., Renault P., Sicheritz-Ponten T., Turner K., Zhu H., Yu C., Li S., Jian M., Zhou Y., Li Y., Zhang X., Li S., Qin N., Yang H., Wang J., Brunak S., Doré J., Guarner F., Kristiansen K., Pedersen O., Parkhill J., Weissenbach J., Bork P., Ehrlich S.D., Wang J., Antolin M., Artiguenave F., Blottiere H., Borruel N., Bruls T., Casellas F., Chervaux C., Cultrone A., Delorme C., Denariaz G., Dervyn R., Forte M., Friss C., Van De Guchte M., Guedon E., Haimet F., Jamet A., Juste C., Kaci G., Kleerebezem M., Knol J., Kristensen M., Layec S., Le Roux K., Leclerc M., Maguin E., Melo Minardi R., Oozeer R., Rescigno M., Sanchez N., Tims S., Torrejon T., Varela E., De Vos W., Winogradsky Y., Zoetendal E. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Li Y., Cai Z., Li S., Zhu J., Zhang F., Liang S., Zhang W., Guan Y., Shen D., Peng Y., Zhang D., Jie Z., Wu W., Qin Y., Xue W., Li J., Han L., Lu D., Wu P., Dai Y., Sun X., Li Z., Tang A., Zhong S., Li X., Chen W., Xu R., Wang M., Feng Q., Gong M., Yu J., Zhang Y., Zhang M., Hansen T., Sanchez G., Raes J., Falony G., Okuda S., Almeida M., Lechatelier E., Renault P., Pons N., Batto J.M., Zhang Z., Chen H., Yang R., Zheng W., Li S., Yang H., Ehrlich S.D., Nielsen R., Pedersen O., Kristiansen K., Wang J. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- Qu H., Zhang Y., Chai H., Gao Z.Y., Shi D.Z. Effects of microbiota-driven therapy on inflammatory responses in elderly individuals: a systematic review and meta-analysis. PLoS One. 2019;14 doi: 10.1371/journal.pone.0211233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queipo-Ortuno M.I., Seoane L.M., Murri M., Pardo M., Gomez-Zumaquero J.M., Cardona F., Casanueva F., Tinahones F.J. Gut microbiota composition in male rat models under different nutritional status and physical activity and its association with serum leptin and ghrelin levels. PLoS One. 2013;8 doi: 10.1371/journal.pone.0065465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampelli S., Schnorr S.L., Consolandi C., Turroni S., Severgnini M., Peano C., Brigidi P., Crittenden A.N., Henry A.G., Candela M. Metagenome sequencing of the Hadza hunter-gatherer gut microbiota. Curr. Biol. 2015;25:1682–1693. doi: 10.1016/j.cub.2015.04.055. [DOI] [PubMed] [Google Scholar]
- Rao S.S.C., Yu S., Fedewa A. Systematic review: dietary fibre and FODMAP-restricted diet in the management of constipation and irritable bowel syndrome. Aliment. Pharmacol. Ther. 2015;41:1256–1270. doi: 10.1111/apt.13167. [DOI] [PubMed] [Google Scholar]
- Razak P.A., Richard K.M., Thankachan R.P., Hafiz K.A., Kumar K.N., Sameer K.M. Geriatric oral health: a review article. Journal of international oral health : JIOH. 2014;6:110–116. [PMC free article] [PubMed] [Google Scholar]
- Ridaura V.K., Faith J.J., Rey F.E., Cheng J., Duncan A.E., Kau A.L., Griffin N.W., Lombard V., Henrissat B., Bain J.R., Muehlbauer M.J., Ilkayeva O., Semenkovich C.F., Funai K., Hayashi D.K., Lyle B.J., Martini M.C., Ursell L.K., Clemente J.C., Van Treuren W., Walters W.A., Knight R., Newgard C.B., Heath A.C., Gordon J.I. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341 doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura V.K., Faith J.J., Rey F.E., Cheng J., Duncan A.E., Kau A.L., Griffin N.W., Lombard V., Henrissat B., Bain J.R., Muehlbauer M.J., Ilkayeva O., Semenkovich C.F., Funai K., Hayashi D.K., Lyle B.J., Martini M.C., Ursell L.K., Clemente J.C., Van Treuren W., Walters W.A., Knight R., Newgard C.B., Heath A.C., Gordon J.I. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341 doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios-Covian D., Ruas-Madiedo P., Margolles A., Gueimonde M., de Los Reyes-Gavilan C.G., Salazar N. Intestinal short chain fatty acids and their link with diet and human health. Front. Microbiol. 2016;7:185. doi: 10.3389/fmicb.2016.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.L., Romanuk T.N. A meta-analysis of probiotic efficacy for gastrointestinal diseases. PLoS One. 2012;7 doi: 10.1371/journal.pone.0034938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M.A.M., Aronoff D.M. The influence of non-steroidal anti-inflammatory drugs on the gut microbiome. Clin. Microbiol. Infect. 2016;22:178.e171–178.e179. doi: 10.1016/j.cmi.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild D., Weissbrod O., Barkan E., Kurilshikov A., Korem T., Zeevi D., Costea P.I., Godneva A., Kalka I.N., Bar N., Shilo S., Lador D., Vila A.V., Zmora N., Pevsner-Fischer M., Israeli D., Kosower N., Malka G., Wolf B.C., Avnit-Sagi T., Lotan-Pompan M., Weinberger A., Halpern Z., Carmi S., Fu J., Wijmenga C., Zhernakova A., Elinav E., Segal E. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–215. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- Round J.L., Mazmanian S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routy B., Le Chatelier E., Derosa L., Duong C.P.M., Alou M.T., Daillère R., Fluckiger A., Messaoudene M., Rauber C., Roberti M.P., Fidelle M., Flament C., Poirier-Colame V., Opolon P., Klein C., Iribarren K., Mondragón L., Jacquelot N., Qu B., Ferrere G., Clémenson C., Mezquita L., Masip J.R., Naltet C., Brosseau S., Kaderbhai C., Richard C., Rizvi H., Levenez F., Galleron N., Quinquis B., Pons N., Ryffel B., Minard-Colin V., Gonin P., Soria J.C., Deutsch E., Loriot Y., Ghiringhelli F., Zalcman G., Goldwasser F., Escudier B., Hellmann M.D., Eggermont A., Raoult D., Albiges L., Kroemer G., Zitvogel L. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- Sabater-Molina M., Larqué E., Torrella F., Zamora S. Dietary fructooligosaccharides and potential benefits on health. J. Physiol. Biochem. 2009;65:315–328. doi: 10.1007/BF03180584. [DOI] [PubMed] [Google Scholar]
- Saffrey M.J. Aging of the mammalian gastrointestinal tract: a complex organ system. Age. 2014;36:1019–1032. doi: 10.1007/s11357-013-9603-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar N., Valdes-Varela L., Gonzalez S., Gueimonde M., de Los Reyes-Gavilan C.G. Nutrition and the gut microbiome in the elderly. Gut Microbes. 2017;8:82–97. doi: 10.1080/19490976.2016.1256525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sales V.M., Ferguson-Smith A.C., Patti M.E. Epigenetic mechanisms of transmission of metabolic disease across generations. Cell Metab. 2017;25:559–571. doi: 10.1016/j.cmet.2017.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San-Cristobal R., Navas-Carretero S., Martínez-González M., Ordovas J.M., Martínez J.A. Contribution of macronutrients to obesity: implications for precision nutrition. Nat. Rev. Endocrinol. 2020;16:305–320. doi: 10.1038/s41574-020-0346-8. [DOI] [PubMed] [Google Scholar]
- Sangeetha P.T., Ramesh M.N., Prapulla S.G. Recent trends in the microbial production, analysis and application of Fructooligosaccharides. Trends Food Sci. Technol. 2005;16:442–457. [Google Scholar]
- Sangwan V., Tomar S.K., Singh R.R.B., Singh A.K., Ali B. Galactooligosaccharides: novel components of designer foods. J. Food Sci. 2011;76:R103–R111. doi: 10.1111/j.1750-3841.2011.02131.x. [DOI] [PubMed] [Google Scholar]
- Sanna S., van Zuydam N.R., Mahajan A., Kurilshikov A., Vich Vila A., Võsa U., Mujagic Z., Masclee A.A.M., Jonkers D.M.A.E., Oosting M., Joosten L.A.B., Netea M.G., Franke L., Zhernakova A., Fu J., Wijmenga C., McCarthy M.I. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat. Genet. 2019;51:600–605. doi: 10.1038/s41588-019-0350-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro A., Ostan R., Candela M., Biagi E., Brigidi P., Capri M., Franceschi C. Gut microbiota changes in the extreme decades of human life: a focus on centenarians. Cell. Mol. Life Sci. 2018;75:129–148. doi: 10.1007/s00018-017-2674-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraswati S., Sitaraman R. Aging and the human gut microbiota-from correlation to causality. Front. Microbiol. 2014;5:764. doi: 10.3389/fmicb.2014.00764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A., Pitchumoni C.S. The Microbiota in Gastrointestinal Pathophysiology: Implications for Human Health, Prebiotics, Probiotics, and Dysbiosis. Elsevier Inc; 2017. Identification of the microbiota in the aging process; pp. 37–56. [Google Scholar]
- Sarkar A., Harty S., Johnson K.V., Moeller A.H., Carmody R.N., Lehto S.M., Erdman S.E., Dunbar R.I.M., Burnet P.W.J. The role of the microbiome in the neurobiology of social behaviour. Biol. Rev. Camb. Philos. Soc. 2020;95:1131–1166. doi: 10.1111/brv.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid M.M., Genaro P.S., Moreno Y.M., Pastore G.M. Freeze-dried powdered yacon: effects of FOS on serum glucose, lipids and intestinal transit in the elderly. Eur. J. Nutr. 2014;53:1457–1464. doi: 10.1007/s00394-013-0648-x. [DOI] [PubMed] [Google Scholar]
- Schnorr S.L., Candela M., Rampelli S., Centanni M., Consolandi C., Basaglia G., Turroni S., Biagi E., Peano C., Severgnini M., Fiori J., Gotti R., De Bellis G., Luiselli D., Brigidi P., Mabulla A., Marlowe F., Henry A.G., Crittenden A.N. Gut microbiome of the Hadza hunter-gatherers. Nat. Commun. 2014;5:3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrezenmeir J., De Vrese M. Probiotics, prebiotics, and synbiotics - approaching a definition. Am. J. Clin. Nutr. 2001;73:361S–364S. doi: 10.1093/ajcn/73.2.361s. [DOI] [PubMed] [Google Scholar]
- Schwiertz A., Taras D., Schäfer K., Beijer S., Bos N.A., Donus C., Hardt P.D. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring, Md.) 2010;18:190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- Seksik P., Rigottier-Gois L., Gramet G., Sutren M., Pochart P., Marteau P., Jian R., Doré J. Alterations of the dominant faecal bacterial groups in patients with Crohn's disease of the colon. Gut. 2003;52:237–242. doi: 10.1136/gut.52.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw A.C., Joshi S., Greenwood H., Panda A., Lord J.M. Aging of the innate immune system. Curr. Opin. Immunol. 2010;22:507–513. doi: 10.1016/j.coi.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd E.S., Deloache W.C., Pruss K.M., Whitaker W.R., Sonnenburg J.L. An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature. 2018;557:434–438. doi: 10.1038/s41586-018-0092-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelly A.N., Sato Y., Kearney S., Honda K. Mining the microbiota for microbial and metabolite-based immunotherapies. Nat. Rev. Immunol. 2019;19:305–323. doi: 10.1038/s41577-019-0144-5. [DOI] [PubMed] [Google Scholar]
- Smith S., Newhouse J.P., Freeland M.S. Income, insurance, and technology: why does health spending outpace economic growth? Health Aff. 2009;28:1276–1284. doi: 10.1377/hlthaff.28.5.1276. [DOI] [PubMed] [Google Scholar]
- Smith P.M., Howitt M.R., Panikov N., Michaud M., Gallini C.A., Bohlooly-Y M., Glickman J.N., Garrett W.S. The microbial metabolites, short-chain fatty acids, regulate colonic T reg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits W.K., Lyras D., Lacy D.B., Wilcox M.H., Kuijper E.J. Clostridium difficile infection. Nat Rev Dis Primers. 2016;2 doi: 10.1038/nrdp.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits S.A., Leach J., Sonnenburg E.D., Gonzalez C.G., Lichtman J.S., Reid G., Knight R., Manjurano A., Changalucha J., Elias J.E., Dominguez-Bello M.G., Sonnenburg J.L. Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science. 2017;357:802–805. doi: 10.1126/science.aan4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So D., Whelan K., Rossi M., Morrison M., Holtmann G., Kelly J.T., Shanahan E.R., Staudacher H.M., Campbell K.L. Dietary fiber intervention on gut microbiota composition in healthy adults: a systematic review and meta-analysis. Am. J. Clin. Nutr. 2018;107:965–983. doi: 10.1093/ajcn/nqy041. [DOI] [PubMed] [Google Scholar]
- Sokol H., Landman C., Seksik P., Berard L., Montil M., Nion-Larmurier I., Bourrier A., Le Gall G., Lalande V., De Rougemont A., Kirchgesner J., Daguenel A., Cachanado M., Rousseau A., Drouet E., Rosenzwajg M., Hagege H., Dray X., Klatzman D., Marteau P., Saint-Antoine I.B.D.N., Beaugerie L., Simon T. Fecal microbiota transplantation to maintain remission in Crohn's disease: a pilot randomized controlled study. Microbiome. 2020;8:12. doi: 10.1186/s40168-020-0792-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer M.O.A., Dantas G., Church G.M. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science. 2009;325:1128–1131. doi: 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg J.L., Bäckhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535:56–64. doi: 10.1038/nature18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg E.D., Sonnenburg J.L. The ancestral and industrialized gut microbiota and implications for human health. Nat. Rev. Microbiol. 2019;17:383–390. doi: 10.1038/s41579-019-0191-8. [DOI] [PubMed] [Google Scholar]
- Sonnenburg J.L., Sonnenburg E.D. Vulnerability of the industrialized microbiota. Science. 2019;366 doi: 10.1126/science.aaw9255. [DOI] [PubMed] [Google Scholar]
- Stavropoulou E., Bezirtzoglou E. Human microbiota in aging and infection: a review. Crit. Rev. Food Sci. Nutr. 2019;59:537–545. doi: 10.1080/10408398.2017.1379469. [DOI] [PubMed] [Google Scholar]
- Suez J., Zmora N., Zilberman-Schapira G., Mor U., Dori-Bachash M., Bashiardes S., Zur M., Regev-Lehavi D., Ben-Zeev Brik R., Federici S., Horn M., Cohen Y., Moor A.E., Zeevi D., Korem T., Kotler E., Harmelin A., Itzkovitz S., Maharshak N., Shibolet O., Pevsner-Fischer M., Shapiro H., Sharon I., Halpern Z., Segal E., Elinav E. Post-antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell. 2018;174:1406–1423. doi: 10.1016/j.cell.2018.08.047. (e1416) [DOI] [PubMed] [Google Scholar]
- Suez J., Zmora N., Segal E., Elinav E. The pros, cons, and many unknowns of probiotics. Nat. Med. 2019;25:716–729. doi: 10.1038/s41591-019-0439-x. [DOI] [PubMed] [Google Scholar]
- Suez J., Zmora N., Elinav E. Probiotics in the next-generation sequencing era. Gut Microbes. 2020;11:77–93. doi: 10.1080/19490976.2019.1586039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan A. Effect of antimicrobial agents on the ecological balance of human microflora. Lancet Infect. Dis. 2001;1:101–114. doi: 10.1016/S1473-3099(01)00066-4. [DOI] [PubMed] [Google Scholar]
- Sze M.A., Schloss P.D. Looking for a signal in the noise: revisiting obesity and the microbiome. mBio. 2016:7. doi: 10.1128/mBio.01018-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H., Tanisawa K., Sun X.M., Kubo T., Hoshino Y., Hosokawa M., Takeyama H., Higuchi M. Effects of short-term endurance exercise on gut microbiota in elderly men. Physiol. Rep. 2018;6:16. doi: 10.14814/phy2.13935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock G.W., Munro K., Harmsen H.J., Welling G.W., Smart J., Gopal P.K. Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl. Environ. Microbiol. 2000;66:2578–2588. doi: 10.1128/aem.66.6.2578-2588.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres D.P., Gonçalves M., Teixeira J.A., Rodrigues L.R. Galacto-oligosaccharides: production, properties, applications, and significance as prebiotics. Compr. Rev. Food Sci. Food Saf. 2010;9:438–454. doi: 10.1111/j.1541-4337.2010.00119.x. [DOI] [PubMed] [Google Scholar]
- Tremaroli V., Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- Tropini C., Moss E.L., Merrill B.D., Ng K.M., Higginbottom S.K., Casavant E.P., Gonzalez C.G., Fremin B., Bouley D.M., Elias J.E., Bhatt A.S., Huang K.C., Sonnenburg J.L. Transient osmotic perturbation causes long-term alteration to the gut microbiota. Cell. 2018;173:1742–1754.e1717. doi: 10.1016/j.cell.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Turnbaugh P.J., Bäckhed F., Fulton L., Gordon J.I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host and Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P.J., Hamady M., Yatsunenko T., Cantarel B.L., Duncan A., Ley R.E., Sogin M.L., Jones W.J., Roe B.A., Affourtit J.P., Egholm M., Henrissat B., Heath A.C., Knight R., Gordon J.I. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P.J., Ridaura V.K., Faith J.J., Rey F.E., Knight R., Gordon J.I. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turpin W., Espin-Garcia O., Xu W., Silverberg M.S., Kevans D., Smith M.I., Guttman D.S., Griffiths A., Panaccione R., Otley A., Xu L., Shestopaloff K., Moreno-Hagelsieb G., Paterson A.D., Croitoru K. Association of host genome with intestinal microbial composition in a large healthy cohort. Nat. Genet. 2016;48:1413–1417. doi: 10.1038/ng.3693. [DOI] [PubMed] [Google Scholar]
- Valentini L., Pinto A., Bourdel-Marchasson I., Ostan R., Brigidi P., Turroni S., Hrelia S., Hrelia P., Bereswill S., Fischer A., Leoncini E., Malaguti M., Blanc-Bisson C., Durrieu J., Spazzafumo L., Buccolini F., Pryen F., Donini L.M., Franceschi C., Lochs H. Impact of personalized diet and probiotic supplementation on inflammation, nutritional parameters and intestinal microbiota - the “RISTOMED project”: randomized controlled trial in healthy older people. Clin. Nutr. 2015;34:593–602. doi: 10.1016/j.clnu.2014.09.023. [DOI] [PubMed] [Google Scholar]
- Valle Gottlieb M.G., Closs V.E., Junges V.M., Schwanke C.H.A. Impact of human aging and modern lifestyle on gut microbiota. Crit. Rev. Food Sci. Nutr. 2018;58:1557–1564. doi: 10.1080/10408398.2016.1269054. [DOI] [PubMed] [Google Scholar]
- Valles-Colomer M., Falony G., Darzi Y., Tigchelaar E.F., Wang J., Tito R.Y., Schiweck C., Kurilshikov A., Joossens M., Wijmenga C., Claes S., Van Oudenhove L., Zhernakova A., Vieira-Silva S., Raes J. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat. Microbiol. 2019;4:623–632. doi: 10.1038/s41564-018-0337-x. [DOI] [PubMed] [Google Scholar]
- Van Nood E., Vrieze A., Nieuwdorp M., Fuentes S., Zoetendal E.G., De Vos W.M., Visser C.E., Kuijper E.J., Bartelsman J.F.W.M., Tijssen J.G.P., Speelman P., Dijkgraaf M.G.W., Keller J.J. Duodenal infusion of donor feces for recurrent clostridium difficile. New Engl. J. Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- Van Tongeren S.P., Slaets J.P.J., Harmsen H.J.M., Welling G.W. Fecal microbiota composition and frailty. Appl. Environ. Microbiol. 2005;71:6438–6442. doi: 10.1128/AEM.71.10.6438-6442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeputte D., Falony G., Vieira-Silva S., Tito R.Y., Joossens M., Raes J. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut. 2016;65:57–62. doi: 10.1136/gutjnl-2015-309618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangay P., Johnson A.J., Ward T.L., Al-Ghalith G.A., Shields-Cutler R.R., Hillmann B.M., Lucas S.K., Beura L.K., Thompson E.A., Till L.M., Batres R., Paw B., Pergament S.L., Saenyakul P., Xiong M., Kim A.D., Kim G., Masopust D., Martens E.C., Angkurawaranon C., McGready R., Kashyap P.C., Culhane-Pera K.A., Knights D. US immigration westernizes the human gut microbiome. Cell. 2018;175:962–972. doi: 10.1016/j.cell.2018.10.029. (e910) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez M.J., Alonso J.L., Domínguez H., Parajó J.C. Xylooligosaccharides: manufacture and applications. Trends Food Sci. Technol. 2000;11:387–393. [Google Scholar]
- Vemuri R.C., Gundamaraju R., Shinde T., Eri R. Therapeutic interventions for gut dysbiosis and related disorders in the elderly: antibiotics, probiotics or faecal microbiota transplantation? Benefic. Microbes. 2017;8:179–192. doi: 10.3920/BM2016.0115. [DOI] [PubMed] [Google Scholar]
- Vendrik K.E.W., Ooijevaar R.E., de Jong P.R.C., Laman J.D., van Oosten B.W., van Hilten J.J., Ducarmon Q.R., Keller J.J., Kuijper E.J., Contarino M.F. Fecal microbiota transplantation in neurological disorders. Front. Cell. Infect. Microbiol. 2020;10:98. doi: 10.3389/fcimb.2020.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vich Vila A., Collij V., Sanna S., Sinha T., Imhann F., Bourgonje A.R., Mujagic Z., Jonkers D.M.A.E., Masclee A.A.M., Fu J., Kurilshikov A., Wijmenga C., Zhernakova A., Weersma R.K. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat. Commun. 2020;11 doi: 10.1038/s41467-019-14177-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva-Millán M.J., Pérez-Matute P., Oteo J.A. Gut microbiota: a key player in health and disease. A review focused on obesity. J. Physiol. Biochem. 2015;71:509–525. doi: 10.1007/s13105-015-0390-3. [DOI] [PubMed] [Google Scholar]
- Vos, T., Abajobir, A.A., Abbafati, C., Abbas, K.M., Abate, K.H., Abd-Allah, F., Abdulle, A.M., Abebo, T.A., Abera, S.F., Aboyans, V., Abu-Raddad, L.J., Ackerman, I.N., Adamu, A.A., Adetokunboh, O., Afarideh, M., Afshin, A., Agarwal, S.K., Aggarwal, R., Agrawal, A., Agrawal, S., Ahmad Kiadaliri, A., Ahmadieh, H., Ahmed, M.B., Aichour, A.N., Aichour, I., Aichour, M.T.E., Aiyar, S., Akinyemi, R.O., Akseer, N., Al Lami, F.H., Alahdab, F., Al-Aly, Z., Alam, K., Alam, N., Alam, T., Alasfoor, D., Alene, K.A., Ali, R., Alizadeh-Navaei, R., Alkerwi, A., Alla, F., Allebeck, P., Allen, C., Al-Maskari, F., Al-Raddadi, R., Alsharif, U., Alsowaidi, S., Altirkawi, K.A., Amare, A.T., Amini, E., Ammar, W., Amoako, Y.A., Andersen, H.H., Antonio, C.A.T., Anwari, P., Ärnlöv, J., Artaman, A., Aryal, K.K., Asayesh, H., Asgedom, S.W., Assadi, R., Atey, T.M., Atnafu, N.T., Atre, S.R., Avila-Burgos, L., Avokpaho, E.F.G.A., Awasthi, A., Ayala Quintanilla, B.P., Ba Saleem, H.O., Bacha, U., Badawi, A., Balakrishnan, K., Banerjee, A., Bannick, M.S., Barac, A., Barber, R.M., Barker-Collo, S.L., Bärnighausen, T., Barquera, S., Barregard, L., Barrero, L.H., Basu, S., Battista, B., Battle, K.E., Baune, B.T., Bazargan-Hejazi, S., Beardsley, J., Bedi, N., Beghi, E., Béjot, Y., Bekele, B.B., Bell, M.L., Bennett, D.A., Bensenor, I.M., Benson, J., Berhane, A., Berhe, D.F., Bernabé, E., Betsu, B.D., Beuran, M., Beyene, A.S., Bhala, N., Bhansali, A., Bhatt, S., Bhutta, Z.A., Biadgilign, S., Bienhoff, K., Bikbov, B., Birungi, C., Biryukov, S., Bisanzio, D., Bizuayehu, H.M., Boneya, D.J., Boufous, S., Bourne, R.R.A., Brazinova, A., Brugha, T.S., Buchbinder, R., Bulto, L.N.B., Bumgarner, B.R., Butt, Z.A., Cahuana-Hurtado, L., Cameron, E., Car, M., Carabin, H., Carapetis, J.R., Cárdenas, R., Carpenter, D.O., Carrero, J.J., Carter, A., Carvalho, F., Casey, D.C., Caso, V., Castañeda-Orjuela, C.A., Castle, C.D., Catalá-López, F., Chang, H.Y., Chang, J.C., Charlson, F.J., Chen, H., Chibalabala, M., Chibueze, C.E., Chisumpa, V.H., Chitheer, A.A., Christopher, D.J., Ciobanu, L.G., Cirillo, M., Colombara, D., Cooper, C., Cortesi, P.A., Criqui, M.H., Crump, J.A., Dadi, A.F., Dalal, K., Dandona, L., Dandona, R., Das Neves, J., Davitoiu, D.V., De Courten, B., De Leo, D., Degenhardt, L., Deiparine, S., Dellavalle, R.P., Deribe, K., Des Jarlais, D.C., Dey, S., Dharmaratne, S.D., Dhillon, P.K., Dicker, D., Ding, E.L., Djalalinia, S., Do, H.P., Dorsey, E.R., Dos Santos, K.P.B., Douwes-Schultz, D., Doyle, K.E., Driscoll, T.R., Dubey, M., Duncan, B.B., El-Khatib, Z.Z., Ellerstrand, J., Enayati, A., Endries, A.Y., Ermakov, S.P., Erskine, H.E., Eshrati, B., Eskandarieh, S., Esteghamati, A., Estep, K., Fanuel, F.B.B., Farinha, C.S.E.S., Faro, A., Farzadfar, F., Fazeli, M.S., Feigin, V.L., Fereshtehnejad, S.M., Fernandes, J.C., Ferrari, A.J., Feyissa, T.R., Filip, I., Fischer, F., Fitzmaurice, C., Flaxman, A.D., Flor, L.S., Foigt, N., Foreman, K.J., Franklin, R.C., Fullman, N., Fürst, T., Furtado, J.M., Futran, N.D., Gakidou, E., Ganji, M., Garcia-Basteiro, A.L., Gebre, T., Gebrehiwot, T.T., Geleto, A., Gemechu, B.L., Gesesew, H.A., Gething, P.W., Ghajar, A., Gibney, K.B., Gill, P.S., Gillum, R.F., Ginawi, I.A.M., Giref, A.Z., Gishu, M.D., Giussani, G., Godwin, W.W., Gold, A.L., Goldberg, E.M., Gona, P.N., Goodridge, A., Gopalani, S.V., Goto, A., Goulart, A.C., Griswold, M., Gugnani, H.C., Gupta, R., Gupta, R., Gupta, T., Gupta, V., Hafezi-Nejad, N., Hailu, A.D., Hailu, G.B., Hamadeh, R.R., Hamidi, S., Handal, A.J., Hankey, G.J., Hao, Y., Harb, H.L., Hareri, H.A., Haro, J.M., Harvey, J., Hassanvand, M.S., Havmoeller, R., Hawley, C., Hay, R.J., Hay, S.I., Henry, N.J., Heredia-Pi, I.B., Heydarpour, P., Hoek, H.W., Hoffman, H.J., Horita, N., Hosgood, H.D., Hostiuc, S., Hotez, P.J., Hoy, D.G., Htet, A.S., Hu, G., Huang, H., Huynh, C., Iburg, K.M., Igumbor, E.U., Ikeda, C., Irvine, C.M.S., Jacobsen, K.H., Jahanmehr, N., Jakovljevic, M.B., Jassal, S.K., Javanbakht, M., Jayaraman, S.P., Jeemon, P., Jensen, P.N., Jha, V., Jiang, G., John, D., Johnson, C.O., Johnson, S.C., Jonas, J.B., Jürisson, M., Kabir, Z., Kadel, R., Kahsay, A., Kamal, R., Kan, H., Karam, N.E., Karch, A., Karema, C.K., Kasaeian, A., Kassa, G.M., Kassaw, N.A., Kassebaum, N.J., Kastor, A., Katikireddi, S.V., Kaul, A., Kawakami, N., Keiyoro, P.N., Kengne, A.P., Keren, A., Khader, Y.S., Khalil, I.A., Khan, E.A., Khang, Y.H., Khosravi, A., Khubchandani, J., Kieling, C., Kim, D., Kim, P., Kim, Y.J., Kimokoti, R.W., Kinfu, Y., Kisa, A., Kissimova-Skarbek, K.A., Kivimaki, M., Knudsen, A.K., Kokubo, Y., Kolte, D., Kopec, J.A., Kosen, S., Koul, P.A., Koyanagi, A., Kravchenko, M., Krishnaswami, S., Krohn, K.J., Kuate Defo, B., Kucuk Bicer, B., Kumar, G.A., Kumar, P., Kumar, S., Kyu, H.H., Lal, D.K., Lalloo, R., Lambert, N., Lan, Q., Larsson, A., Lavados, P.M., Leasher, J.L., Lee, J.T., Lee, P.H., Leigh, J., Leshargie, C.T., Leung, J., Leung, R., Levi, M., Li, Y., Li, Y., Li Kappe, D., Liang, X., Liben, M.L., Lim, S.S., Linn, S., Liu, A., Liu, P.Y., Liu, S., Liu, Y., Lodha, R., Logroscino, G., London, S.J., Looker, K.J., Lopez, A.D., Lorkowski, S., Lotufo, P.A., Low, N., Lozano, R., Lucas, T.C.D., Macarayan, E.R.K., Magdy Abd El Razek, H., Magdy Abd El Razek, M., Mahdavi, M., Majdan, M., Majdzadeh, R., Majeed, A., Malekzadeh, R., Malhotra, R., Malta, D.C., Mamun, A.A., Manguerra, H., Manhertz, T., Mantilla, A., Mantovani, L.G., Mapoma, C.C., Marczak, L.B., Martinez-Raga, J., Martins-Melo, F.R., Martopullo, I., März, W., Mathur, M.R., Mazidi, M., McAlinden, C., McGaughey, M., McGrath, J.J., McKee, M., McNellan, C., Mehata, S., Mehndiratta, M.M., Mekonnen, T.C., Memiah, P., Memish, Z.A., Mendoza, W., Mengistie, M.A., Mengistu, D.T., Mensah, G.A., Meretoja, A., Meretoja, T.J., Mezgebe, H.B., Micha, R., Millear, A., Miller, T.R., Mills, E.J., Mirarefin, M., Mirrakhimov, E.M., Misganaw, A., Mishra, S.R., Mitchell, P.B., Mohammad, K.A., Mohammadi, A., Mohammed, K.E., Mohammed, S., Mohanty, S.K., Mokdad, A.H., Mollenkopf, S.K., Monasta, L., Hernandez, J.M., Montico, M., Moradi-Lakeh, M., Moraga, P., Mori, R., Morozoff, C., Morrison, S.D., Moses, M., Mountjoy-Venning, C., Mruts, K.B., Mueller, U.O., Muller, K., Murdoch, M.E., Murthy, G.V.S., Musa, K.I., Nachega, J.B., Nagel, G., Naghavi, M., Naheed, A., Naidoo, K.S., Naldi, L., Nangia, V., Natarajan, G., Negasa, D.E., Negoi, I., Negoi, R.I., Newton, C.R., Ngunjiri, J.W., Nguyen, C.T., Nguyen, G., Nguyen, M., Nguyen, Q.L., Nguyen, T.H., Nichols, E., Ningrum, D.N.A., Nolte, S., Nong, V.M., Norrving, B., Noubiap, J.J.N., O'Donnell, M.J., Ogbo, F.A., Oh, I.H., Okoro, A., Oladimeji, O., Olagunju, A.T., Olagunju, T.O., Olsen, H.E., Olusanya, B.O., Olusanya, J.O., Ong, K., Opio, J.N., Oren, E., Ortiz, A., Osgood-Zimmerman, A., Osman, M., Owolabi, M.O., Pa, M., Pacella, R.E., Pana, A., Panda, B.K., Papachristou, C., Park, E.K., Parry, C.D., Parsaeian, M., Patten, S.B., Patton, G.C., Paulson, K., Pearce, N., Pereira, D.M., Perico, N., Pesudovs, K., Peterson, C.B., Petzold, M., Phillips, M.R., Pigott, D.M., Pillay, J.D., Pinho, C., Plass, D., Pletcher, M.A., Popova, S., Poulton, R.G., Pourmalek, F., Prabhakaran, D., Prasad, N., Prasad, N.M., Purcell, C., Qorbani, M., Quansah, R., Rabiee, R.H.S., Radfar, A., Rafay, A., Rahimi, K., Rahimi-Movaghar, A., Rahimi-Movaghar, V., Rahman, M., Rahman, M.H.U., Rai, R.K., Rajsic, S., Ram, U., Ranabhat, C.L., Rankin, Z., Rao, P.V., Rao, P.C., Rawaf, S., Ray, S.E., Reiner, R.C., Reinig, N., Reitsma, M.B., Remuzzi, G., Renzaho, A.M.N., Resnikoff, S., Rezaei, S., Ribeiro, A.L., Ronfani, L., Roshandel, G., Roth, G.A., Roy, A., Rubagotti, E., Ruhago, G.M., Saadat, S., Sadat, N., Safdarian, M., Safi, S., Safiri, S., Sagar, R., Sahathevan, R., Salama, J., Salomon, J.A., Salvi, S.S., Samy, A.M., Sanabria, J.R., Santomauro, D., Santos, I.S., Santos, J.V., Santric Milicevic, M.M., Sartorius, B., Satpathy, M., Sawhney, M., Saxena, S., Schmidt, M.I., Schneider, I.J.C., Schöttker, B., Schwebel, D.C., Schwendicke, F., Seedat, S., Sepanlou, S.G., Servan-Mori, E.E., Setegn, T., Shackelford, K.A., Shaheen, A., Shaikh, M.A., Shamsipour, M., Shariful Islam, S.M., Sharma, J., Sharma, R., She, J., Shi, P., Shields, C., Shigematsu, M., Shinohara, Y., Shiri, R., Shirkoohi, R., Shirude, S., Shishani, K., Shrime, M.G., Sibai, A.M., Sigfusdottir, I.D., Silva, D.A.S., Silva, J.P., Silveira, D.G.A., Singh, J.A., Singh, N.P., Sinha, D.N., Skiadaresi, E., Skirbekk, V., Slepak, E.L., Sligar, A., Smith, D.L., Smith, M., Sobaih, B.H.A., Sobngwi, E., Sorensen, R.J.D., Sousa, T.C.M., Sposato, L.A., Sreeramareddy, C.T., Srinivasan, V., Stanaway, J.D., Stathopoulou, V., Steel, N., Stein, D.J., Stein, M.B., Steiner, C., Steiner, T.J., Steinke, S., Stokes, M.A., Stovner, L.J., Strub, B., Subart, M., Sufiyan, M.B., Suliankatchi Abdulkader, R., Sunguya, B.F., Sur, P.J., Swaminathan, S., Sykes, B.L., Sylte, D.O., Tabarés-Seisdedos, R., Taffere, G.R., Takala, J.S., Tandon, N., Tavakkoli, M., Taveira, N., Taylor, H.R., Tehrani-Banihashemi, A., Tekelab, T., Temam Shifa, G., Terkawi, A.S., Tesfaye, D.J., Tesssema, B., Thamsuwan, O., Thomas, K.E., Thrift, A.G., Tiruye, T.Y., Tobe-Gai, R., Tollanes, M.C., Tonelli, M., Topor-Madry, R., Tortajada, M., Touvier, M., Tran, B.X., Tripathi, S., Troeger, C., Truelsen, T., Tsoi, D., Tuem, K.B., Tuzcu, E.M., Tyrovolas, S., Ukwaja, K.N., Undurraga, E.A., Uneke, C.J., Updike, R., Uthman, O.A., Uzochukwu, B.S.C., Van Boven, J.F.M., Varughese, S., Vasankari, T., Venkatesh, S., Venketasubramanian, N., Vidavalur, R., Violante, F.S., Vladimirov, S.K., Vlassov, V.V., Vollset, S.E., Wadilo, F., Wakayo, T., Wang, Y.P., Weaver, M., Weichenthal, S., Weiderpass, E., Weintraub, R.G., Werdecker, A., Westerman, R., Whiteford, H.A., Wijeratne, T., Wiysonge, C.S., Wolfe, C.D.A., Woodbrook, R., Woolf, A.D., Workicho, A., Wulf Hanson, S., Xavier, D., Xu, G., Yadgir, S., Yaghoubi, M., Yakob, B., Yan, L.L., Yano, Y., Ye, P., Yimam, H.H., Yip, P., Yonemoto, N., Yoon, S.J., Yotebieng, M., Younis, M.Z., Zaidi, Z., Zaki, M.E.S., Zegeye, E.A., Zenebe, Z.M., Zhang, X., Zhou, M., Zipkin, B., Zodpey, S., Zuhlke, L.J., Murray, C.J.L., 2017. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990-2016: a systematic analysis for the global burden of disease study 2016. Lancet 390, 1211–1259. [DOI] [PMC free article] [PubMed]
- Vulevic J., Juric A., Walton G.E., Claus S.P., Tzortzis G., Toward R.E., Gibson G.R. Influence of galacto-oligosaccharide mixture (B-GOS) on gut microbiota, immune parameters and metabonomics in elderly persons. Br. J. Nutr. 2015;114:586–595. doi: 10.1017/S0007114515001889. [DOI] [PubMed] [Google Scholar]
- Wallace C.J.K., Milev R. The effects of probiotics on depressive symptoms in humans: a systematic review. Ann. General Psychiatry. 2017;16:14. doi: 10.1186/s12991-017-0138-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J., Armet A.M., Finlay B.B., Shanahan F. Establishing or exaggerating causality for the gut microbiome: lessons from human microbiota-associated rodents. Cell. 2020;180:221–232. doi: 10.1016/j.cell.2019.12.025. [DOI] [PubMed] [Google Scholar]
- Walters W.A., Xu Z., Knight R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 2014;588:4223–4233. doi: 10.1016/j.febslet.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Yu T., Huang G., Cai D., Liang X., Su H., Zhu Z., Li D., Yang Y., Shen P., Mao R., Yu L., Zhao M., Li Q. Gut microbiota community and its assembly associated with age and diet in Chinese centenarians. J. Microbiol. Biotechnol. 2015;25:1195–1204. doi: 10.4014/jmb.1410.10014. [DOI] [PubMed] [Google Scholar]
- Wang L., Yang H., Huang H., Zhang C., Zuo H.X., Xu P., Niu Y.M., Wu S.S. Inulin-type fructans supplementation improves glycemic control for the prediabetes and type 2 diabetes populations: results from a GRADE-assessed systematic review and dose-response meta-analysis of 33 randomized controlled trials. J. Transl. Med. 2019;17:410. doi: 10.1186/s12967-019-02159-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO, 2017. Ageing and health fact sheet. Available at http://www.who.int/features/factfiles/ageing/en/. Accessed January 05, 2020.
- WHO, 2018. Report on surveillance of antibiotic consumption. 2016 - 2018 early implementation. Available at: https://www.who.int/medicines/areas/rational_use/oms-amr-amc-report-2016-2018/en/. Accessed January 05, 2020.
- WHO, 2019. Nutrition for older persons. Available at https://www.who.int/nutrition/topics/ageing/en/. Accessed January 05, 2020.
- WHO, 2020. Obesity and overweight. Available at: https://www.who.int/en/news-room/fact-sheets/detail/obesity-and-overweight. Accessed on April 01, 2020.
- Wilson B.C., Vatanen T., Cutfield W.S., O’Sullivan J.M. The super-donor phenomenon in fecal microbiota transplantation. Front. Cell. Infect. Microbiol. 2019;9:2. doi: 10.3389/fcimb.2019.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse C., Kronsten V., Zamalloa A., Hatton G., Patel V.C., Goldenberg S., Shawcross D.L. PROFIT: PROspective, randomised placebo-controlled feasibility trial of Faecal microbiota transplantation in cirrhosis interim analysis. Am. J. Gastroenterol. 2019;114:S5–S7. doi: 10.1136/bmjopen-2018-023518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodmansey E.J., McMurdo M.E.T., Macfarlane G.T., Macfarlane S. Comparison of compositions and metabolic activities of fecal microbiotas in young adults and in antibiotic-treated and non-antibiotic-treated elderly subjects. Appl. Environ. Microbiol. 2004;70:6113–6122. doi: 10.1128/AEM.70.10.6113-6122.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G.D., Chen J., Hoffmann C., Bittinger K., Chen Y.Y., Keilbaugh S.A., Bewtra M., Knights D., Walters W.A., Knight R., Sinha R., Gilroy E., Gupta K., Baldassano R., Nessel L., Li H., Bushman F.D., Lewis J.D. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Esteve E., Tremaroli V., Khan M.T., Caesar R., Mannerås-Holm L., Ståhlman M., Olsson L.M., Serino M., Planas-Fèlix M., Xifra G., Mercader J.M., Torrents D., Burcelin R., Ricart W., Perkins R., Fernàndez-Real J.M., Bäckhed F. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med. 2017;23:850–858. doi: 10.1038/nm.4345. [DOI] [PubMed] [Google Scholar]
- Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S., Huang H., Zhang L., Zhou X., Du C., Zhang Y., Song J., Wang S., Chao Y., Yang Z., Xu J., Zhou X., Chen D., Xiong W., Xu L., Zhou F., Jiang J., Bai C., Zheng J., Song Y. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., Yin H., Xiao Q., Tang Y., Qu X., Kuang L., Fang X., Mishra N., Lu J., Shan H., Jiang G., Huang X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Zhu H., Qiu P. Aging progression of human gut microbiota. BMC Microbiol. 2019;19:236. doi: 10.1186/s12866-019-1616-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Chen V.L., Steiner C.A., Berinstein J.A., Eswaran S., Waljee A.K., Higgins P.D.R., Owyang C. Efficacy of fecal microbiota transplantation in irritable bowel syndrome: a systematic review and Meta-analysis. Am. J. Gastroenterol. 2019;114:1043–1050. doi: 10.14309/ajg.0000000000000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Wu Q., Liu Y., Fan D. Effect of perioperative probiotics and synbiotics on postoperative infections after gastrointestinal surgery: a systematic review with meta-analysis. J. Parenter. Enter. Nutr. 2017;41:1051–1062. doi: 10.1177/0148607116629670. [DOI] [PubMed] [Google Scholar]
- Yatsunenko T., Rey F.E., Manary M.J., Trehan I., Dominguez-Bello M.G., Contreras M., Magris M., Hidalgo G., Baldassano R.N., Anokhin A.P., Heath A.C., Warner B., Reeder J., Kuczynski J., Caporaso J.G., Lozupone C.A., Lauber C., Clemente J.C., Knights D., Knight R., Gordon J.I. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngster I., Russell G.H., Pindar C., Ziv-Baran T., Sauk J., Hohmann E.L. Oral, capsulized, frozen fecal microbiota transplantation for relapsing Clostridium difficile infection. JAMA. 2014;312:1772–1778. doi: 10.1001/jama.2014.13875. [DOI] [PubMed] [Google Scholar]
- Zeevi D., Korem T., Zmora N., Israeli D., Rothschild D., Weinberger A., Ben-Yacov O., Lador D., Avnit-Sagi T., Lotan-Pompan M., Suez J., Mahdi J.A., Matot E., Malka G., Kosower N., Rein M., Zilberman-Schapira G., Dohnalová L., Pevsner-Fischer M., Bikovsky R., Halpern Z., Elinav E., Segal E. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163:1079–1094. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Zhang L., Huang Y., Zhou Y., Buckley T., Wang H.H. Antibiotic administration routes significantly influence the levels of antibiotic resistance in gut microbiota. Antimicrob. Agents Chemother. 2013;57:3659–3666. doi: 10.1128/AAC.00670-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Mocanu V., Cai C., Dang J., Slater L., Deehan E.C., Walter J., Madsen K.L. Impact of fecal microbiota transplantation on obesity and metabolic syndrome-a systematic review. Nutrients. 2019;11 doi: 10.3390/nu11102291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Zhang F., Ding X., Wu G., Lam Y.Y., Wang X., Fu H., Xue X., Lu C., Ma J., Yu L., Xu C., Ren Z., Xu Y., Xu S., Shen H., Zhu X., Shi Y., Shen Q., Dong W., Liu R., Ling Y., Zeng Y., Wang X., Zhang Q., Wang J., Wang L., Wu Y., Zeng B., Wei H., Zhang M., Peng Y., Zhang C. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science. 2018;359:1151–1156. doi: 10.1126/science.aao5774. [DOI] [PubMed] [Google Scholar]
- Zhao X., Zhang Z., Hu B., Huang W., Yuan C., Zou L. Response of gut microbiota to metabolite changes induced by endurance exercise. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Winter M.G., Byndloss M.X., Spiga L., Duerkop B.A., Hughes E.R., Büttner L., De Lima Romão E., Behrendt C.L., Lopez C.A., Sifuentes-Dominguez L., Huff-Hardy K., Wilson R.P., Gillis C.C., Tükel Ç., Koh A.Y., Burstein E., Hooper L.V., Bäumler A.J., Winter S.E. Precision editing of the gut microbiota ameliorates colitis. Nature. 2018;553:208–211. doi: 10.1038/nature25172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Jiang S., Du G. Effects of exercise frequency on the gut microbiota in elderly individuals. MicrobiologyOpen. 2020;9 doi: 10.1002/mbo3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M., Zimmermann-Kogadeeva M., Wegmann R., Goodman A.L. Separating host and microbiome contributions to drug pharmacokinetics and toxicity. Science. 2019;363 doi: 10.1126/science.aat9931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmora N., Zilberman-Schapira G., Suez J., Mor U., Dori-Bachash M., Bashiardes S., Kotler E., Zur M., Regev-Lehavi D., Brik R.B.Z., Federici S., Cohen Y., Linevsky R., Rothschild D., Moor A.E., Ben-Moshe S., Harmelin A., Itzkovitz S., Maharshak N., Shibolet O., Shapiro H., Pevsner-Fischer M., Sharon I., Halpern Z., Segal E., Elinav E. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell. 2018;174:1388–1405. doi: 10.1016/j.cell.2018.08.041. (e1321) [DOI] [PubMed] [Google Scholar]
- Zmora N., Suez J., Elinav E. You are what you eat: diet, health and the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 2019;16:35–56. doi: 10.1038/s41575-018-0061-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The 6 major phyla of the human gut microbiota and their predominant families (with examples of species).