Abstract
Tanacetum coccineum, a perennial plant of the Tanacetum genus, cultivated as a natural pesticide or ornamental plant widely distributed in many countries. In this research, the complete chloroplast genome sequence of T. coccineum was determined to comprise a 150,143 bp double-stranded circular DNA, including a pair of 24,416 bp inverted repeat regions (IRs), small single copy (SSC) region of 18,389 bp and large single copy (LCS) region of 82,922 bp. An overall GC content was 37.49%, and the corresponding values in IRs, SSC, and LSC regions are 43.16%, 30.88%, and 35.61%, respectively. A total of 129 genes include 84 protein-coding genes, 37 tRNA, and eight rRNA. Four rRNA genes and seven tRNA genes were duplicated in IRs. A phylogenetic tree reconstructed by 38 Composite family chloroplast genomes sequence reveals that T. coccineum is mostly related to Ismelia carinata.
Keywords: Tanacetum coccineum, chloroplast genome, phylogeny
Tanacetum coccineum (also called Chrysanthemum coccineum or Pyrethrum coccineum) is a perennial plant of the Tanacetum genus which contains more than 160 species of flowing plants, but the extraction of few of them like T. coccineum can kill insects. Its product pyrethrin has been widely used as a green insecticide due to its quick knock-down effect on a wide range of insects and low harm to human (Casida 1973). So T.coccineum has been employed as the natural pesticide plant in ancient (Ghany 2012). The first cultivated red flower pyrethrum species T. coccineum, also called the Persia pyrethrum, is native to Caucasus (between the Black Sea and the Caspian Sea) (Contant 1976). ‘Persian Insect Powder’ made from the dried flowers of T. coccineum were traded in the first half of the 19th century, which were applied in fighting against ectoparasites such as lice and fleas for children in ancient Persia (Pavela 2016). Although it is common as ornamental plant due to its daisy-like or button-shaped flower heads in shades of white, pink, and yellow, however, its phylogenetic function is still unrecognized due to lack of genome of T. coccineum. In this research, we measure the complete chloroplast genome of T. coccineum using high throughput sequencing technology, which will provide useful information for the phylogeny of Tanacetum genus and further research on the evolution of Tanacetum.
The flesh leaves of T. coccineum were collected from Flower Nursery Stock Base of Huazhong Agricultural University in Wuhan, China (the specimen was deposited at the Museum of Huazhong Agricultural University accession number: ccau0011589). And the total genomic DNA was extracted from the fresh leaves of T.coccineum using Rapid Plant Genomic DNA lsolation Kit (Sangong, Shanghai, China). The whole genome sequencing was conducted by Shanghai Sangong biotechCo., Inc. (Shanghai, China) on the Illumina Hiseq. The filtered sequences were assembled using the NOVOPlasty 2.7.2 (Dierckxsens et al., 2017). Annotation was performed using GeSeq (Tillich et al., 2017). Then, submitted to Genbank (accession number: MT104463).
The cp genome of T. coccineum was determined to comprise a 150,143 bp double-stranded circular DNA, including a pair of 24,416 bp inverted repeat regions (IRs), small single copy(SSC) region of 18,389 bp and large single copy (LCS) region of 82,922 bp. An overall GC content was 37.49%, and the corresponding values in IRs, SSC, and LSC regions are 43.16%, 30.88%, and 35.61%, respectively. A total of 129 genes include 84 protein-coding genes, 37 tRNA, and eight rRNA. Four rRNA genes and seven tRNA genes were duplicated in IRs.
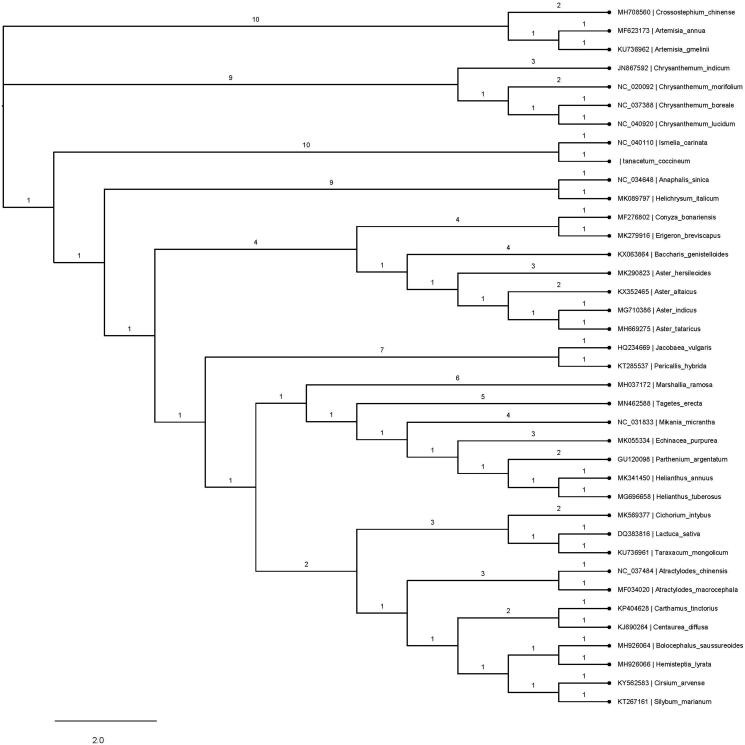
To investigate its taxonomic function, 38 Composite family chloroplast genome sequences were aligned by HomBlocks (Bi et al. 2018), and a maximum-likelihood (ML) tree was constructed by MEGAX (Kumar et al. 2018). The species T.coccineum was found to be relatively closely related to Ismelia carinata compared with other species in Composite (Figure 1). The complete chloroplast genome sequence of T.coccineum will provide a useful resource for the conservation family of Composite as well as for the phylogenetic studies of Tanacetum genus.
Figure 1.
Phylogenetic tree inferred by the maximum-likelihood (ML) method based on 38 representative species. A total of 1000 bootstrap replicates were computed, and the bootstrap support values are shown at the branches. GenBank accession numbers were also shown.
Funding Statement
This research was funded by Natural Science Research Project of Guizhou Education Department [KY[2018]129].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/nuccore/MT104463.1/, reference number [MT104463.1].
References
- Bi GQ, Mao YX, Xing QK, Cao M.. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110(1):18–22. [DOI] [PubMed] [Google Scholar]
- Casida JE. 1973. Pyrethrum: the natural insecticide. Saint Louis: Academic Press. [Google Scholar]
- Contant RB. 1976. Pyrethrum, Chrysanthemum spp. (Compositae). In: Simmonds NW, editor. Evolution of crop plants. London; New York: Longman. [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G.. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghany A. 2012. Plant extracts as effective compounds in pest control. Saarbrücken: LAPLAMBERT Academic Publishing. [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K.. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavela R. 2016. History, presence and perspective of using plant extracts as commercial botanical insecticides and farm products for protection against insects – a review. Plant Protect Sci. 52:229–241. [Google Scholar]
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S.. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/nuccore/MT104463.1/, reference number [MT104463.1].