Abstract
The complete mitochondrial genome of the black citrus aphid from Sichuan Province of China, Aphis aurantii, was sequenced and analyzed. The mitochondrial genome was a double strand, circular molecule with 15,296 bp and an A + T content of 83.5%, comprising 13 PCGs, 22 tRNA genes, and two rRNA genes. Gene arrangement was conserved in the mitogenome of A. aurantii. A 631-bp long control region was found, with a high A + T content of 82.6%. All PCGs used standard ATN start codons and most PCGs ended with complete TAA stop codons. The phylogenetic analysis supported that A. aurantii was closely related to other five congeners of the genus Aphis.
Keywords: Mitochondrial genome, Aphis aurantii, phylogeny
Aphis aurantii (Boyer de Fonscolombe) (Hemiptera: Aphididae), also known as the black citrus aphid, is a worldwide transmission of citrus tristeza virus (CTV) and major pest of citrus (Wang and Tsai 2001). Genetic biodiversity of this pest in China is still unclear, which can be resolved by sequencing more genetic data of different geographical populations. To provide more genetic data for A. aurantii, we sequenced the complete mitogenome of Sichuan’s population of A. aurantii using Illumina Hiseq 4000 (Shanghai BIOZERON Co., Ltd). The specimens of A. aurantii were collected from Chenjiaba Town, Beichuan County, Sichuan Province, China (31°55′53.99″N, 104°35′16.13″E) in April 2019. All specimens and isolated DNA samples were stored in the Insect Collection of Sichuan Academy of Agricultural Sciences (ICSAAS, No. ICSAAS-HEM-APHI1), Chengdu, China. The mitogenome sequence was deposited in GenBank with the accession number MN871977.
The complete mitogenome of A. aurantii has a length of 15,296 bp and contains an A + T content of 83.5% (A: 44.8%, T: 38.7%, C: 10.6%, G: 5.9%). A total of 37 genes (13 PCGs, 22 tRNA genes, and two rRNA genes), and a non-coding control region were annotated. The control region was located between rrnS and trnIle, 631 bp in length with an A + T content of 82.6%. In the mitogenome of A. aurantii, no gene rearrangement was detected.
All the PCGs started with the standard start codon ATN (ATA, ATT, and ATG); most PCGs terminated with the complete stop codon TAA, whereas nad4 ended with an incomplete codon T—. The 22 tRNA genes varied in length from 62 bp to 80 bp, mostly showing clover-leaf structures except for trnSer1 (AGN), the DHU arm of which was completely lost. Two rRNA genes, rrnL and rrnS, were found in the conserved locations between trnSer1 (AGN) and the control region. The rrnL gene was 1275 bp in length with an A + T content of 84.6%. The rrnS gene was 769 bp in length with an A + T content of 84.3%. There were 73 overlapped nucleotides between 14 gene pairs with the longest overlap between atp6 and atp8. A total of 272 intergenic nucleotides were found between 13 gene pairs, with the longest 179-bp intergenic sequence between trnPhe (F) and trnGlu (E).
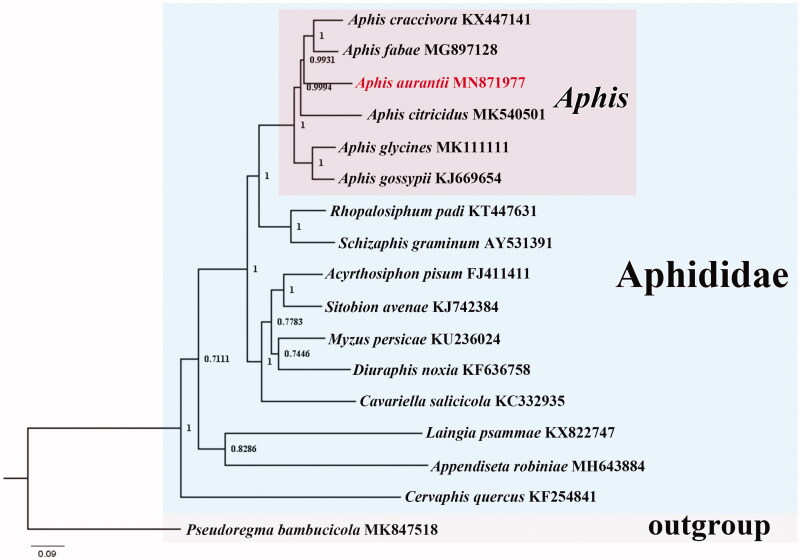
Phylogenetic relationship within Aphididae was reconstructed based on the nucleotide sequences of 13 PCGs. The Bayesian inference (BI) method generated a tree topology identical to that of Wang et al. (2019) (Figure 1). Aphis aurantii sequenced in this study was grouped with other five congeners of Aphis, which supported the efficiency of mitogenome data in phylogenetic analysis and confirmed the morphological identification of the sequenced specimens. Cervaphis quercus was recovered as a relatively basal species to other aphids used in this study. The phylogeny of Aphididae is still unresolved, requiring more molecular data including mitogenome data to solve this problem.
Figure 1.
Phylogenetic tree of 16 species of Aphididae. Numbers at the nodes are posterior probabilities. The GenBank accession numbers are indicated after the scientific names. The tree is rooted with Pseudoregma bambucicola (MK847518).
Funding Statement
This work was provided by the Frontier discipline Fund of Sichuan Academy of Agricultural Sciences [2019QYXK032], National Key R & D Program of China [2016YFD0200900] and Sichuan Tea Innovation Team of National Modern Agricultural Industry Technology System.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Wang J, Tsai J.. 2001. Development, survival and reproduction of black citrus aphid, Toxoptera aurantii (Hemiptera: Aphididae), as a function of temperature. B Entomol Res. 91(6):477–487. [PubMed] [Google Scholar]
- Wang Y, Ding M, Du YM, Huang AJ.. 2019. Phylogenetic relationship and characterization of the complete mitochondrial genome of the black citrus aphid, Aphis aurantii (Hemiptera: Aphididae). Mitochondrial DNA B. 4(2):3567–3568. [DOI] [PMC free article] [PubMed] [Google Scholar]