Abstract
Objective
To conduct a multireader validation study to evaluate the interobserver variability and the diagnostic accuracy for the lung involvement by COVID-19 of COVID-19 Reporting and Data System (CO-RADS) score.
Methods
This retrospective study included consecutive symptomatic patients who underwent chest CT and reverse transcriptase-polymerase chain reaction (RT-PCR) from March 2020 to May 2020 for suspected COVID-19. Twelve readers with different levels of expertise independently scored each CT using the CO-RADS scheme for detecting pulmonary involvement by COVID-19. Receiver operating characteristic (ROC) curves were computed to investigate diagnostic yield. Fleiss’ kappa statistics was used to evaluate interreader agreement.
Results
A total of 572 patients (mean age, 63 ± 20 [standard deviation]; 329 men; 142 patients with COVID-19 and 430 patients without COVID-19) were evaluated. There was a moderate agreement for CO-RADS rating among all readers (Fleiss’ K = 0.43 [95% CI 0.42–0.44]) with a substantial agreement for CO-RADS 1 category (Fleiss’ K = 0.61 [95% CI 0.60–0.62]) and moderate agreement for CO-RADS 5 category (Fleiss’ K = 0.60 [95% CI 0.58–0.61]). ROC analysis showed the CO-RADS score ≥ 4 as the optimal threshold, with a cumulative area under the curve of 0.72 (95% CI 66–78%), sensitivity 61% (95% CI 52–69%), and specificity 81% (95% CI 77–84%).
Conclusion
CO-RADS showed high diagnostic accuracy and moderate interrater agreement across readers with different levels of expertise. Specificity is higher than previously thought and that could lead to reconsider the role of CT in this clinical setting.
Key Points
• COVID-19 Reporting and Data System (CO-RADS) demonstrated a good diagnostic accuracy for lung involvement by COVID-19 with an average AUC of 0.72 (95% CI 67–75%).
• When a threshold of ≥ 4 was used, sensitivity and specificity were 61% (95% CI 52–69%) and 81% (95% CI 76–84%), respectively.
• There was an overall moderate agreement for CO-RADS rating across readers with different levels of expertise (Fleiss’ K = 0.43 [95% CI 0.42–0.44]).
Electronic supplementary material
The online version of this article (10.1007/s00330-020-07273-y) contains supplementary material, which is available to authorized users.
Keywords: COVID-19; Severe acute respiratory syndrome coronavirus 2; Tomography, X-ray computed; ROC curve; Sensitivity and specificity
Introduction
The coronavirus disease-2019 (COVID-19) outbreak is currently straining numerous health systems throughout the world. Timely identification of SARS-CoV-2 positive patients is proving to be the key to contain the spread of the virus effectively. Reverse transcriptase-polymerase chain reaction (RT-PCR) is considered the reference standard for the diagnosis of COVID-19. However, a not negligible number of initial false-negative results have been reported [1–3], and many areas throughout the world are facing the challenge of relative shortage and unavailability of RT-PCR tests [4].
On the other hand, the role of CT in the workup of patients with suspected COVID-19 is still under debate. Whereas the early evidence from China reported a high sensitivity [5], suggesting that CT could play a pivotal role in the rapid detection of infected patients, many scientific societies have recommended not to use CT for disease screening, especially in asymptomatic patients, due to its low specificity in differentiating COVID-19 from other viral infections that could present with overlapping imaging features [6, 7]. Moreover, as underlined in two recently published meta-analyses, the high variability of CT findings considered to define a CT scan consistent with SARS-CoV-2 pneumonia introduced a relevant bias hampering the applicability of results and leading to an underestimation of the real specificity of CT [1, 8].
In this scenario, the introduction of standardized reporting systems for patients with suspected COVID-19 has been extensively advocated with the aim to improve communication with referring physicians and make CT diagnostic accuracy data comparable across different institutions to gather scientific evidence [9, 10]. Trying to accomplish this task, the Dutch Radiological Society has recently developed the COVID-19 Reporting and Data System (CO-RADS) and tested it on a sample of symptomatic patients, demonstrating a satisfying diagnostic yield for predicting COVID-19 with substantial interreader agreement [11].
However, to promote its introduction into clinical practice, a more extensive validation of the CO-RADS should be provided, especially considering a larger patient sample and radiologists with different levels of experience as readers.
Therefore, the purpose of our study was to investigate, in a large sample of consecutive patients and through the analysis of multiple readers with different levels of expertise, whether the CO-RADS score is a valid tool in order to identify lung involvement in patients with suspected COVID-19.
Materials and methods
This retrospective, single-center study was approved by our institutional review board and written informed consent was waived. Reporting was done in accordance with the Standards for Reporting Diagnostic Accuracy Initiative (STARD) recommendations [12]
Study participants
We retrospectively reviewed medical records from our university hospital archives to search for consecutive patients who underwent chest CT and RT-PCR testing for suspected COVID-19, from March 9, 2020, to May 3, 2020. Chest CT and RT-PCR testing were performed for clinical suspicion of COVID-19 based on the presence of at least one of the following respiratory tract infection symptoms: (I) fever higher > 37.5 °C, (II) cough, and (III) clinically relevant dyspnea, with or without a history suggestive of exposure to SARS-CoV-2 including (a) close relationship with a confirmed positive individual, (b) travel or residential history in areas with high prevalence of disease, or (c) contact with individuals with fever or respiratory symptoms from those areas within 14 days prior to CT scan.
Exclusion criteria were as follows: (I) lack of RT-PCR testing results, (II) time interval between CT scan and RT-PCR longer than 7 days, and (III) uninterpretable CT scans due to motion artifacts or incomplete scanning.
CT Technique and image analysis
All CT scans were performed using a single 64-slice CT scanner (LightSpeed VCT, GE Healthcare). All patients were scanned on supine position during single deep-inspiration breath-hold. No contrast medium was administered. Scanning parameters were as follows: tube voltage of 100 or 120 kV according to the patient’s body size, variable tube current with automatic mAs modulation (Smart mA, GE Healthcare), 0.6-mm section thickness and a pitch of 1.388, and iterative reconstruction techniques (ASIR) at 40%.
All CT scans were retrieved from the Picture Archiving and Communication Systems, anonymized and uploaded onto a dedicated workstation (SuiteEstensa 2.0, EBIT - Esaote Group Company) for image analysis.
Each CT scan was independently analyzed by twelve readers, stratified into four different groups according to their experience as follows: high-experience group (R1, R2, and R3 [D.B., U.D., and E.D.], board-certified radiologists with more than 10 years of experience in thoracic imaging and more than 100 COVID-19 positive CTs reported); intermediate-experience group (R4, R5, and R6 [S.M., M.G.C., and M.I.], board-certified radiologists with more than 50 and less than 100 COVID-19-positive CTs reported); low-experience group (R7, R8, and R9 [M.D.I., F.G., and E.O.], radiologists in-training with less than 50 COVID-19-positive CTs reported); and group of radiographers (R10, R11, and R12 [S.P., V.C., and C.G.], all with a background of more than 50 CTs performed on COVID-19-positive patients). A training set of 30 CTs, in which findings corresponding to each CO-RADS category were equally distributed, was provided to each reader. Furthermore, all readers had a general familiarity with CO-RADS, having adopted it at our institution since its introduction, approximately a month before the start of our study.
All readers scored each CT scan assigning a CO-RADS category reflecting their overall suspicion of COVID-19 lung involvement as follows: CO-RADS 1, very low probability; CO-RADS 2, low probability; CO-RADS 3, equivocal/unsure probability; CO-RADS 4, high probability; and CO-RADS 5, very high probability. For a detailed description of all the CT findings associated with each CO-RADS category, please refer to the original paper by Prokop et al [11]. All readers were blinded to the RT-PCR results, to the clinical information and radiological reports of individual patients, and to the disease prevalence in the study sample.
Reference standard
RT-PCR testing performed on respiratory specimens obtained by nasopharyngeal and throat swabs served as a reference standard for the diagnosis of COVID-19. Clinical information and index test results were not available to the assessors of the reference standard. As per our institution guidelines, patients with initial negative RT-PCR, but CT findings suggestive of COVID-19, underwent repeated RT-PCR testing up to a maximum of three times within 7 days after CT scan. Patients who showed at least one positive RT-PCR were considered to be positive for COVID-19; otherwise, they were considered negative. Nonetheless, patients with initial negative RT-PCR and negative CT findings underwent a 14-day follow-up and were considered to be negative if no symptoms’ worsening or laboratory findings consistent with COVID-19 occurred.
Statistical analysis
Categorical variables were expressed as frequencies or percentages. Continuous variables were expressed as means ± standard deviations (SD). The Χ2 test was used to calculate differences in sex, symptoms, and number of RT-PCR testing between COVID+ and COVID− participants. The Mann-Whitney U test was performed to assess differences in age between the two groups.
Fleiss’ kappa statistics were used to evaluate interreader agreement for CO-RADS rating both among all readers and among each group of reader. The following coefficients were applied: κ ≤ 0.20, slight agreement; κ = 0.21–0.40, fair agreement; κ = 0.41–0.60, moderate agreement; κ = 0.61–0.80, substantial agreement; and κ = 0.81–1.0, almost perfect agreement [13].
For each reader, the receiver operating characteristics curve (ROC) and the corresponding area under the curve (AUC) were calculated by using the DeLong et al method [14], to assess the CO-RADS diagnostic performance. Mean AUC across observers from the four different readers’ groups and their corresponding 95% confidence intervals (95% CI) were computed and a pairwise comparison of AUCs from all readers was performed by means of the DeLong et al method [14].
For each reader, the highest Youden index (J = sensitivity + specificity− 1) was calculated to select the optimal threshold to discriminate between COVID+ and COVID− participants, and the corresponding sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were computed. Inconclusive results (i.e., CO-RADS 3) were included in the analysis of the diagnostic performance and whether to treat them as positive or negative results depended on the ROC curve and Youden’s index analysis results.
In addition, the number and the percentage of readings assigned to each CO-RADS category were determined for both COVID+ and COVID− participants. False-positive CO-RADS 4 and 5 patients and false-negative CO-RADS 1 and 2 patients were subsequently investigated to clarify the reason for erroneous classification.
In all cases, p < 0.05 was considered the threshold for assessing statistical significance. All statistical analyses were performed with commercially available software (MedCalc Statistical Software version 19.2.5, MedCalc Software Ltd).
Results
Study participants
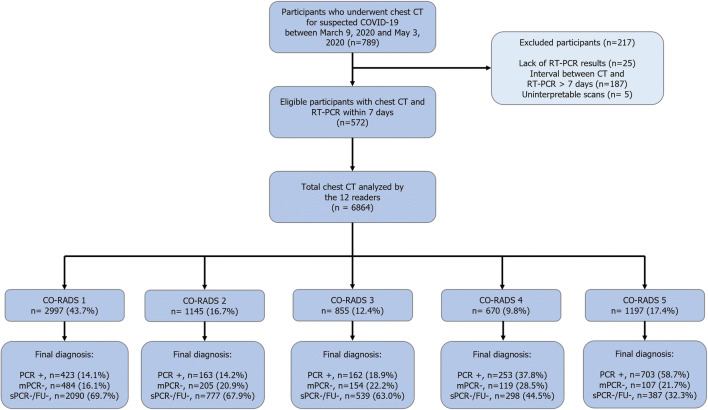
Figure 1 portrays the study participants’ STARD flowchart. From 789 consecutive patients initially screened from our database, we included in the final study sample 572 patients (mean age, 63 years ± 20standard deviation [SD]; range, 8–96 years; including 332 men [mean age, 62 years ± 19 SD; range, 8–96 years] and 240 women [mean age, 64 years ± 21 SD; range, 12–96 years]). Two hundred and seventeen patients were excluded due to the lack of reference standard (n = 25), interval between CT scan and reference standard longer than 1 week (n = 187), or uninterpretable CT scans (n = 5). Detailed baseline characteristics of the patient study groups are reported in Table 1.
Fig. 1.
Study flowchart for the inclusion and exclusion criteria of the patient sample and CO-RADS ratings. Note: CO-RADS, COVID-19 Reporting and Data System; COVID-19, coronavirus disease-2019; RT-PCR, reverse transcriptase-polymerase chain reaction; PCR+, single or multiple RT-PCR testing positive for SARS-CoV-2 infection; mPCR−, multiple negative RT-PCR testing; sPCR-/FU-, single negative RT-PCR testing and negative follow-up during the 14 days following the CT scan
Table 1.
Patient characteristics
| Characteristic | All participants (n = 572) | COVID-19+ (n = 142) | COVID-19– (n = 430) | p value |
|---|---|---|---|---|
| Sex | ||||
| Male | 329 (58%) | 77 (54%) | 252 (59%) | 0.36 |
| Female | 243 (42%) | 65 (46%) | 178 (41%) | |
| Age, years * | 63 ± 20 (8–96) | 63 ± 20 (8–96) | 61 ± 19 (12–96) | 0.11 |
| Symptoms | ||||
| Fever | 267 (47%) | 86 (61%) | 181 (53%) | 0.14 |
| Cough | 317 (55%) | 88 (62%) | 229 (67%) | 0.26 |
| Dyspnea | 233 (41%) | 77 (54%) | 156 (46%) | 0.09 |
| RT-PCR testing | ||||
| 1 | 355 (62%) | 16 (12%) | 339 (79%) | < 0.001 |
| 2 | 85 (15%) | 19 (13%) | 66 (15%) | |
| 3 | 132 (23%) | 107 (75%) | 25 (6%) | |
Unless otherwise indicated, data are numbers with percentages in parentheses. COVID-19, coronavirus disease-2019; RT-PCR, reverse transcription polymerase chain reaction
*Data are means ± standard deviations, with ranges in parentheses
Reference standard
Three hundred and fifty-five participants underwent a single RT-PCR testing, whereas 217 participants underwent multiple RT-PCR testing, including 85 patients who were tested twice, and 132 participants who underwent three consecutive RT-PCR testing. The median time interval between CT scan and reference standard was 2 days (range 0–7).
One hundred and forty-two patients had at least one RT-PCR testing positive for SARS-CoV-2 infection, resulting in a disease prevalence in our study sample of 24.8%. Four hundred and thirty patients were tested negative for SARS-CoV-2 infection, including 91 patients with multiple negative RT-PCR testing, and 339 (78.8%) patients with initial negative RT-PCR and negative follow-up during the 14 days following the CT scan.
Interrater agreement
There was a moderate agreement for overall CO-RADS rating among all observers (Fleiss’ K = 0.43 [95% CI 0.42–0.44]). Regarding each individual CO-RADS category, there was a substantial agreement for CO-RADS 1 category (Fleiss’ K = 0.61 [95% CI 0.60–0.62], moderate agreement for CO-RADS 5 category (Fleiss’ K = 0.60 [95% CI 0.58–0.61]), and fair agreement for CO-RADS categories 2 and 4 (Fleiss’ K = 0.28 [95% CI 0.27–0.29] and 0.23 [95% CI 0.22–0.24], respectively), and poor agreement for CO-RADS 3 category (Fleiss’ K = 0.16 [95% CI 0.15–0.17]). Fleiss’ K values among each group of readers were as follows: high-experience group: 0.54 (95% CI 0.52–0.57), intermediate-experience group: 0.38 (95% CI 0.36–0.41), low-experience group: 0.66 (95% CI 0.64–0.69), and group of radiographers: 0.30 (95% CI 0.27–0.32).
CO-RADS diagnostic performance
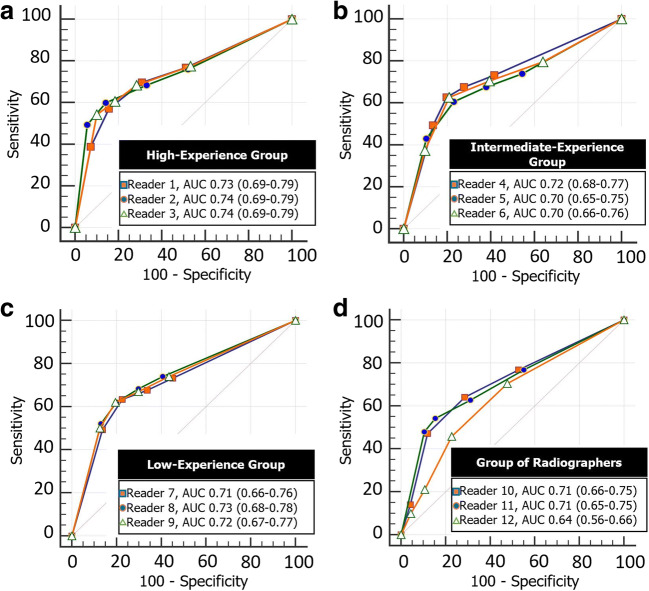
Receiver operating characteristic analysis showed the CO-RADS score ≥ 4 as the optimal threshold to discern between patients with PCR+ from those with PCR−. Considering the high-experience reader group and CO-RADS score ≥ 4 as a threshold, the average value of area under the curve was 0.74 (95% CI 0.67–0.79) (Fig. 2), with a sensitivity of 59.1% (95% CI 0.52–0.67) and a specificity of 84.1% (95% CI 0.81–0.87).
Fig. 2.
Receiver operating characteristic curve of high-experience group (a), intermediate-experience group (b), low-experience group (c), and group of radiographers (d) for predicting lung involvement by coronavirus disease-2019 using the COVID-19 Reporting and Data System (CO-RADS). Note: AUC, area under the curve
The average value of area under the curve for intermediate-experience readers was 0.70 (95% CI 0.66–0.76) (Fig. 2), with a sensitivity of 62% (95% CI 54.2–69.9) and a specificity of 78.7% (95% CI 74.5–82.5). The average value of area under the curve for low-experience readers was 0.73 (95% CI 0.67-0.79) (Fig. 2), with a sensitivity of 62.7% (95% CI 53.2–70.1) and a specificity of 79.3% (95% CI 75.1–83). Averaging data from radiographers, we obtained an area under the curve of 0.68 (95% CI 0.62–0.75) (Fig. 2), a sensitivity of 40.8% (95% CI 33–49), and a specificity of 87.3% (95% CI 83.8–90.3).
The diagnostic accuracy values for all readers are supplied in Table 2. When all datasets from each radiologist were blended, the optimal diagnostic threshold of ≥ 4 yielded an area under the curve of 0.72 (95% CI 66–78%) with a sensitivity of 61% (95% CI 52–69%) and specificity of 81% (95% CI 77–84%). Detailed results from pairwise comparison of the different readers’ AUC are displayed in Table E1 of the supplemental material; briefly, there was no difference in diagnostic accuracy between readers with the highest AUC from the three different radiologists’ groups (p > 0.05 for all comparisons).
Table 2.
Readers diagnostic performance for lung involvement by COVID-19 when CO-RADS ≥ 4 was used as a positive threshold
| Reader | TP | TN | FP | FN | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | AUC |
|---|---|---|---|---|---|---|---|---|---|
| R1 | 81 | 363 | 67 | 61 |
57.0 (81/142) [48.5–65.3] |
84.4 (363/430) [80.6–87.7] |
54.7 (81/148) [48.2–61.1] |
85.6 (363/424) [83.1–87.8] |
0.73 [0.69–0.79] |
| R2 | 85 | 369 | 61 | 57 |
59.9 (85/142) [51.3–68.0] |
85.8 (369/430) [82.2–89.0] |
58.2 (85/146) [51.6–64.6] |
86.6 (369/426) [84.1–88.8] |
0.74 [0.69–0.79] |
| R3 | 86 | 351 | 79 | 56 |
60.6 (86/142) [52.0–68.7] |
81.6 (351/430) [77.6–85.2] |
52.1 (86/165) [46.1–58.0] |
86.2 (351/407) [83.6–88.5] |
0.74 [0.69–0.79] |
| R4 | 89 | 345 | 85 | 53 |
62.7 (89/142) [54.2–70.6] |
80.2 (345/430) [76.1–83.9] |
51.1 (89/174) [45.4–56.8] |
86.7 (345/398) [84.0–89.0] |
0.72 [0.68–0.77] |
| R5 | 86 | 331 | 99 | 56 |
60.6 (86/142) [52.0–68.7] |
77.0 (331/430) [72.7–80.9] |
46.5 (86/185) [41.1–51.9] |
85.5 (331/387) [82.7–87.9] |
0.70 [0.65–0.75] |
| R6 | 89 | 340 | 90 | 53 |
62.7 (89/142) [54.2–70.6] |
79.1 (340/430) [74.9–82.8] |
49.7 (89/179) [44.2–55.3] |
86.5 (340/393) [83.8–88.9] |
0.70 [0.66–0.76] |
| R7 | 90 | 333 | 97 | 52 |
63.4 (90/142) [54.9–71.3] |
77.4 (333/430) [73.2–81.3] |
48.1 (90/187) [42.8–53.5] |
86.5 (333/385) [83.7–88.9] |
0.71 [0.66–0.76] |
| R8 | 88 | 344 | 86 | 54 |
62.0 (88/142) [53.5–70.0] |
80.0 (344/430) [75.9–83.7] |
50.6 (88/174) [44.9–56.3] |
86.4 (344/398) [83.7–88.8] |
0.73 [0.68–0.78] |
| R9 | 88 | 346 | 84 | 54 |
62.0 (88/142) [53.5–70.0] |
80.5 (346/430) [76.4–84.1] |
51.2 (88/172) [45.4–56.9] |
86.5 (346/400) [83.8–8.88] |
0.72 [0.67–0.77] |
| R10 | 67 | 379 | 51 | 75 |
47.2 (67/142) [38.8–55.7] |
88.1 (379/430) [84.7–91.0] |
56.8 (67/118) [49.0–64.2] |
83.5 (379/454) [81.2–85.6] |
0.71 [0.66–0.75] |
| R11 | 77 | 364 | 66 | 65 |
54.2 (77/142) [45.7–62.6] |
84.6 (364/430) [80.9–87.9] |
53.8 (77/143) (47.1–60.4) |
84.8 (364/429) [82.3–87.1] |
0.71 [0.65–0.75] |
| R12 | 30 | 384 | 46 | 112 |
21.1 (30/142) [14.7–28.8] |
89.3 (384/430) [86.0–92.1] |
39.5 (30/76) [30.0–49.8] |
77.4 (384/496) [75.8–79.0] |
0.64 [0.56–0.66] |
Data in parentheses are numerators and denominators, with 95% confidence intervals in brackets
COVID-19, coronavirus disease-2019; CO-RADS, COVID-19 Reporting and Data System; TP, true positive; TN, true negative; FP, false positive; FN, false negative; PPV, positive predictive value; NPV, negative predictive value; AUC, area under the ROC curve
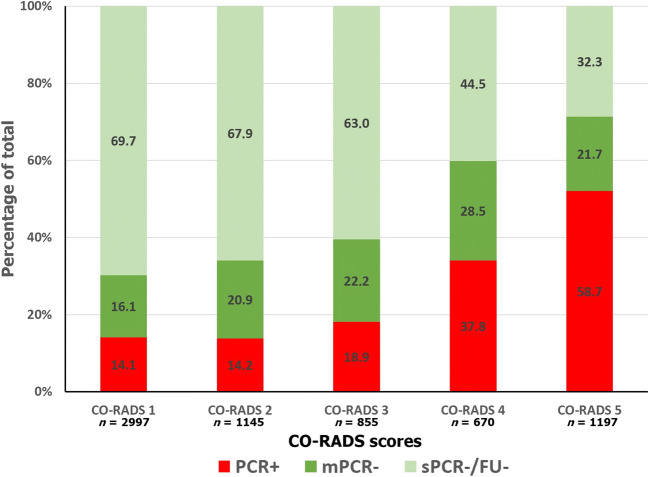
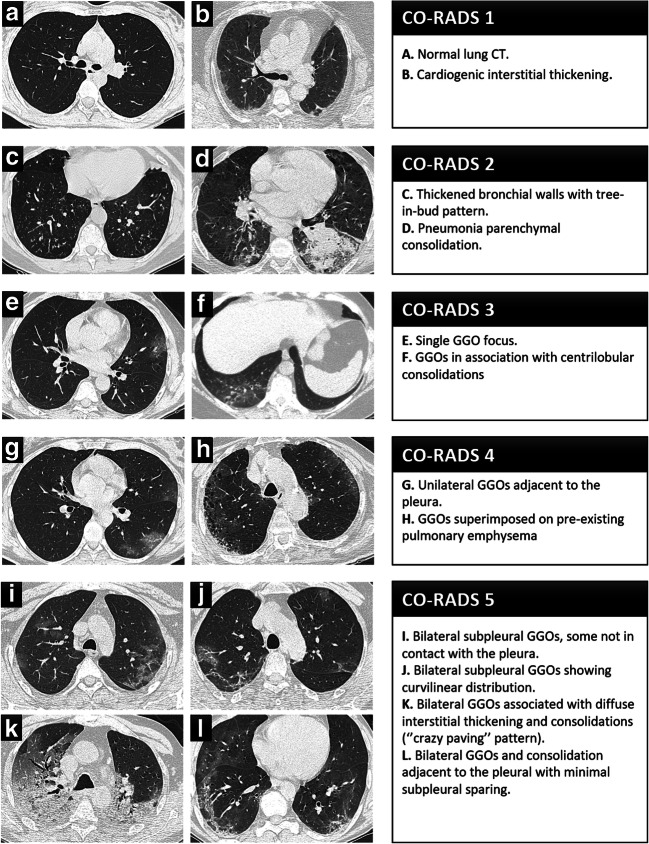
We reported a total number of 6864 CO-RADS scorings from twelve readers (Fig. 3). Among them, 911 (8.9%) in 185 negative patients for SARS-CoV-2 infection were scored as 4 and 5 and resulted in false-positive chest CT. The post hoc analysis of those cases showed that 37.8 % (70/185) were effectively a 4 and 5 CO-RADS pattern and 62.2% (115/185) were erroneous evaluations from readers. False-negative CO-RADS 1 and CO-RADS 2 were found in 586 (8.5%) readings (96 patients). Among them, 44% (42/96) were correctly identified as CO-RADS 1 and 2 categories, while 56% (54/96) were erroneous evaluations. Representative CT images for each CO-RADS category are illustrated in Fig. 4.
Fig. 3.
Distribution of final diagnosis among each cumulative CO-RADS score category. Red columns show the percentage of patients with positive RT-PCR (PCR+), dark green columns show the percentage of patients with multiple negative RT-PCR (mPCR−), and light green columns show the percentage of patients with initial negative RT-PCR and negative clinical follow-up the 14 days after CT scan (sPCR-/FU-). Note: CO-RADS, COVID-19 Reporting and Data System
Fig. 4.
Pictorial overview portraying axial chest CT images from our study population illustrating imaging findings characteristics of the CO-RADS 1 (a, b), CO-RADS 2 (c, d), CO-RADS 3 (e, f), CO-RADS 4 (g, h), and CO-RADS 5 (i, j, k, l) scores and their corresponding descriptions. Note: CO-RADS, COVID-19 Reporting and Data System
Discussion
Since the early stage of the coronavirus disease-2019 (COVID-19) outbreak, the value of CT in the workup of patients with suspected SARS-CoV-2 infection has been controversial [15, 16]. The adoption of standardized reporting systems for lung involvement in COVID-19 has been encouraged in an effort to enhance communication with referring physicians and multicentric gathering of CT diagnostic accuracy data [9–11]. In our retrospective study, we aimed to validate the proposed COVID-19 Reporting and Data System (CO-RADS) in a retrospective multireader diagnostic accuracy study. Our results show that, when a threshold of CO-RADS ≥ 4 was used, readers with different levels of expertise were able to discriminate in approximately 30 s between patients with positive and negative reverse transcriptase-polymerase chain reaction (RT-PCR) testing, with a sensitivity of 61% (95% CI 52–69%), a specificity of 81% (95% CI 76–84%), and an AUC of 0.72 (95% CI 67–75%).
Our findings are in contrast with those reported by the earlier literature on CT diagnostic performance for COVID-19. In detail, a recent meta-analysis estimated a pooled sensitivity and specificity for chest CT of 94% and 37%, respectively [8]. However, it has been underlined that most of the earlier studies investigating the diagnostic performance of CT for COVID-19 were flawed by interpretation bias [15, 17, 18]. Indeed, most of the studies published to date did not provide pre-specified criteria to differentiate between positive and negative CT scans. Therefore, it could be hypothesized that applying unclear CT signs to define a positive case may have resulted in an optimistic overestimation of CT sensitivity at the expense of a reduced specificity. On the other hand, our results suggest that the use of standardized threshold for CT findings evaluation provided by reporting systems such as CO-RADS could more realistically reflect the diagnostic value of CT in the daily clinical practice, characterized by a lower sensitivity and an improved specificity for COVID-19.
CO-RADS yielded an overall moderate interreader agreement for the assessment of lung involvement in COVID-19 (Fleiss’ K = 0.43 [95% CI 0.42–0.44]). This is comparable to what is reported by Prokop et al, who found an overall moderate interrater agreement among the 8 observers included (Fleiss’ K = 0.47 [95% CI 0.45–0.49]) [11].
In the current study, CO-RADS yielded a lower diagnostic accuracy compared with what was reported by Prokop et al, who found a mean AUC of 0.91 (95% CI 0.85–0.97) for positive RT-PCR testing. We believe that this discrepancy could be related to the higher degree of expertise and familiarity with the CO-RADS scoring of the readers involved in the study by Prokop et al. However, we observed that CO-RADS diagnostic accuracy was not significantly affected by readers’ experience, when CT scans were read by radiologists with different levels of expertise, including radiologists in-training. These results suggest that the adoption of the CO-RADS reporting scheme could represent an effective strategy to facilitate and speed up the learning curve of radiologists in-training in the evaluation of chest CT scans of patients with suspected lung involvement by COVID-19. This latter aspect is noteworthy as, at many institutions, radiologists in-training are the first to assess CT scans, and they should be able to rapidly recognize typical CT findings of COVID-19. Indeed, the finding of CT patterns consistent with COVID-19 even in asymptomatic patients is not uncommon, and their recognition by radiologists in-training at the moment of CT scanning might limit the risk of spreading the contamination, allowing suspicious cases to be directed promptly to dedicated care pathways.
This study has several limitations that should be addressed. First, this is a single-center, retrospective study; further multicenter studies with a prospective design are warranted to confirm our findings. Second, multiple RT-PCR testing to rule out SARS-CoV-2 infection was available only for 92 (21 %) of the participants who were tested negative at the initial RT-PCR; this could raise concerns about the reference standard reliability in this subset of participants, since a considerable rate of initial false-negative RT-PCR has been reported [1–3]. However, all patients with initial negative RT-PCR underwent clinical and laboratory monitoring for at least 14 days to confirm their negative status in accordance with guidelines from our institution. Third, our study sample is composed only of symptomatic patients admitted to the Emergency Department with suspected COVID-19, without including asymptomatic patients. We recognize that this may have biased patient selection toward participants with a more severe disease spectrum, thereby affecting the CO-RADS accuracy estimate. However, our study sample reflects the actual population for whom chest CT scan is recommended as a problem-solving tool [6, 7]. Moreover, the CO-RADS scheme has been developed specifically to be used in patients with moderate to severe symptoms [11]. Fourth, a final diagnosis was not available for patients with false-positive CT findings. Therefore, we were not able to assess the prevalence of co-morbidities or other respiratory tract infections, whose imaging findings may overlap with those typically observed in COVID-19. Additional studies addressing this issue are advisable to validate further the CO-RADS algorithm, since its performance may vary according to the different prevalence of conditions with overlapping CT characteristics.
In conclusion, CO-RADS score, tested on a large sample of symptomatic patients, should be considered a valid tool for the identification of lung involvement in patients with suspected COVID-19, showing high diagnostic accuracy across readers with different levels of expertise. Specificity, in particular, is higher than previously thought and that could lead to reconsider the role of CT in this clinical setting.
Electronic supplementary material
(DOCX 19 kb)
Abbreviations
- 95% CI
95% Confidence intervals
- CO-RADS
COVID-19 Reporting and Data System
- COVID-19
Coronavirus disease-2019
- RT-PCR
Reverse transcriptase-polymerase chain reaction
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- STARD
Standards for reporting diagnostic accuracy initiative
Funding
The authors state that this work has not received any funding.
Compliance with ethical standards
Guarantor
The scientific guarantor of this publication is Dr. Davide Bellini.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
One of the authors has significant statistical expertise.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• diagnostic or prognostic study
• performed at one institution
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Xu B, Xing Y, Peng J et al (2020) Chest CT for detecting COVID-19: a systematic review and meta-analysis of diagnostic accuracy. Eur Radiol. 10.1007/s00330-020-06934-2 [DOI] [PMC free article] [PubMed]
- 2.Fang Y, Zhang H, Xie J et al (2020) Sensitivity of Chest CT for COVID-19: Comparison to RT-PCR. Radiology. 10.1148/radiol.2020200432 [DOI] [PMC free article] [PubMed]
- 3.Xie X, Zhong Z, Zhao W, Zheng C, Wang F, Liu J (2020) Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 10.1148/radiol.2020200343 [DOI] [PMC free article] [PubMed]
- 4.Woloshin S, Patel N, Kesselheim AS (2020) False negative tests for SARS-CoV-2 infection — challenges and implications. N Engl J Med. 10.1056/NEJMp2015897 [DOI] [PubMed]
- 5.Ai T, Yang Z, Hou H et al (2020) Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 10.1148/radiol.2020200642 [DOI] [PMC free article] [PubMed]
- 6.Rubin GD, Ryerson CJ, Haramati LB et al (2020) The role of chest imaging in patient management during the COVID-19 pandemic: a multinational consensus statement from the Fleischner Society. Radiology. 10.1148/radiol.2020201365 [DOI] [PMC free article] [PubMed]
- 7.American College of Radiology (2020) ACR recommendations for the use of Chest radiography and computed tomography (CT) for suspected COVID-19 infection. Available via https://www.acr.org/Advocacy-and-Economics/ACR-Position-Statements/Recommendations-for-Chest-Radiography-and-CT-for-Suspected-COVID19-Infection. Accessed 21 May 2020
- 8.Kim H, Hong H, Yoon SH (2020) Diagnostic performance of CT and reverse transcriptase-polymerase chain reaction for coronavirus disease 2019: a meta-analysis. Radiology. 10.1148/radiol.2020201343 [DOI] [PMC free article] [PubMed]
- 9.Salehi S, Abedi A, Balakrishnan S, Gholamrezanezhad A (2020) Coronavirus disease 2019 (COVID-19) imaging reporting and data system (COVID-RADS) and common lexicon: a proposal based on the imaging data of 37 studies. Eur Radiol. 10.1007/s00330-020-06863-0 [DOI] [PMC free article] [PubMed]
- 10.Simpson S, Kay FU, Abbara S et al (2020) Radiological Society of North America Expert consensus statement on reporting chest CT findings related to COVID-19. Endorsed by the Society of Thoracic Radiology, the American College of Radiology, and RSNA. Radiology Cardiothoracic Imaging. 10.1148/ryct.2020200152 [DOI] [PMC free article] [PubMed]
- 11.Prokop M, van Everdingen W, van Rees Vellinga T et al (2020) CO-RADS – a categorical CT assessment scheme for patients with suspected COVID-19: definition and evaluation. Radiology. 10.1148/radiol.2020201473 [DOI] [PMC free article] [PubMed]
- 12.Bossuyt PM, Reitsma JB, Bruns DE, et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. Radiology. 2015;277:826–832. doi: 10.1148/radiol.2015151516. [DOI] [PubMed] [Google Scholar]
- 13.Landis JR, Koch GG. A one-way components of variance model for categorical data. Biometrics. 1977;33:671. doi: 10.2307/2529465. [DOI] [PubMed] [Google Scholar]
- 14.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
- 15.Raptis CA, Hammer MM, Short RG et al (2020) Chest CT and coronavirus disease (COVID-19): a critical review of the literature to date. AJR Am J Roentgenol. 10.2214/ajr.20.23202 [DOI] [PubMed]
- 16.Hope MD, Raptis CA, Henry TS (2020) Chest computed tomography for detection of coronavirus disease 2019 (COVID-19): don’t rush the science. Ann Intern Med. 10.7326/m20-1382 [DOI] [PMC free article] [PubMed]
- 17.Eng J, Bluemke DA (2020) Imaging publications in the COVID-19 pandemic: applying new research results to clinical practice. Radiology. 10.1148/radiol.2020201724 [DOI] [PMC free article] [PubMed]
- 18.Waller JV, Kaur P, Tucker A et al (2020) Diagnostic tools for coronavirus disease (COVID-19): comparing CT and RT-PCR viral nucleic acid testing. AJR Am J Roentgenol. 10.2214/ajr.20.23418 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 19 kb)