Highlights
-
•
The sensitivity and specificity of seven serological assays for SARS-CoV-2 were compared.
-
•
No single assay offered a combination of very high sensitivity and very high specificity.
-
•
To maximize sensitivity and specificity two assays should be combined.
Keywords: COVID-19, ELISA, Serology, SARS-CoV-2
Abstract
There is an ongoing need for highly reliable serological assays to detect individuals with past SARS-CoV-2 infection. Using 75 sera from patients tested positive or negative by SARS-CoV-2 PCR, we investigated the sensitivity and specificity of the Liaison SARS-CoV-2 S1/S2 IgG assay (DiaSorin), the Elecsys Anti-SARS-CoV-2 assay (Roche), and the ID Screen SARS-CoV-2-N IgG indirect kit (IDVet). We determined a sensitivity of 95.5 %, 95.5 %, and 100 % and a specificity of 90.5 %, 96.2 %, and 92.5 % for the DiaSorin assay, the Roche assay, and the IDVet assay, respectively. We conclude that serologic assays combining very high sensitivity and specificity are still not commercially available for SARS-CoV-2. For maximizing sensitivity and specificity of SARS-CoV-2 serological diagnostics, the combination of two assays may be helpful.
1. Introduction
The new coronavirus SARS-CoV-2 emerged in 2019 (Zhou et al., 2020; Zhu et al., 2020) causing an ongoing pandemic. Diagnostic laboratories are facing the challenge of correctly identifying acute and past infections with SARS-CoV-2. Whereas nucleic acid amplification assays are the gold standard for detection of acute infections (Eis-Hübinger et al., 2020), sensitive and specific serologic assays are needed for detection of past infections.
Developing accurate serological assays for coronaviruses is considered a technical challenge (Meyer et al., 2014). We and others have published results of the assessment of the first commercially available serological assays, such as the Anti SARS-CoV-2 ELISA (IgG) from Euroimmun (Krüttgen et al., 2020; Okba et al., 2020).
Most recently, highly anticipated chemoluminescence immunoassays (CLIA) for widely-implemented automated high-throughput platforms (Cobas, LIAISON) and further ELISAs became available. To allow the comparison of performance indicators, these assays have to be compared using identical collections of serum samples. We therefore compared these three new assays with respect to their sensitivity and specificity to detect SARS-CoV-2 specific antibodies using a collection of serum samples employed previously for the analysis of four other assays. These values were compared to four assays previously evaluated in our laboratory (Krüttgen et al., 2020): the anti SARS-CoV-2 ELISA (IgG) (Euroimmun, Germany), the EDI Novel Coronavirus COVID-19 IgG ELISA, (Epitope diagnostics (EDI), USA), the recomWell SARS-CoV-2 IgG ELISA (Mikrogen, Germany), and the SARS-CoV-2 Virachip IgG (Viramed, Germany).
Thus we aimed at directly comparing the performance of seven commercially available serological assays (two CLIAs, four ELISAs and one immunoblot) in terms of sensitivity and specificity.
2. Materials and methods
We used 75 previously characterized sera of 56 different patients hospitalized in the University Hospital RWTH Aachen, Germany; approval of Medical Ethics Committee EK093/2020). In brief, 25 sera were collected from 25 patients with a negative SARS-CoV-2 PCR result in respiratory specimens and 50 sera were collected from 31 patients with a positive SARS-CoV-2 PCR result in respiratory specimens. The sera were stored for about four weeks before use with the assays described in this manuscript. One freeze thaw cycle was applied.
22 of the 75 sera were defined SARS-CoV-2 IgG positive and 53 sera were defined SARS-CoV-2 IgG negative. In analogy to a previous study (de Ory et al., 2018) the SARS-CoV-2 IgG status of the sera was defined as follows: A serum was regarded as SARS-CoV-2 IgG negative if at least three of the four assays compared here had a negative test result applying the manufacturer’s interpretation criteria. On the other hand, a serum was regarded as SARS-CoV-2 IgG positive if at least two of the four assays had a positive test result.
Borderline samples were handled as not positive for determination of the sensitivity and not negative for determination of the specificity.
Three semiquantitative enzyme immune assays, the Liaison SARS-CoV-2 S1/S2 IgG assay (DiaSorin, Italy), the Elecsys Anti-SARS-CoV-2 assay (Roche, USA), and the ID Screen SARS-CoV-2-N IgG indirect kit (IDVet, France) were compared for their ability to detect SARS-CoV-2-specific antibodies. To allow comparison of semiquantitative values between assays, the values were divided by the assay-specific cut off value for normalization. Normalized values of > = 1 thereby represented a positive test result.
3. Results
We included 75 previously characterized sera in this study (Krüttgen et al., 2020), 25 of which were collected from patients with a negative SARS-CoV-2 PCR result and 50 from patients with a positive SARS-CoV-2 PCR result in respiratory specimens. The sera of patients with negative SARS-CoV-2 PCR result were obtained at the same day as the PCR analysis of the respiratory specimen. The sera of the 31 patients with positive SARS-CoV-2 PCR were collected 11.9 days (± 5.0 days) post onset of symptoms. Each serum was tested in parallel for the presence of SARS-CoV-2 IgG antibodies as recommended by the manufacturers.
Using the Diasorin assay, 26 sera were classified as SARS-CoV-2 antibody positive, 49 were classified as negative. Using the Roche assay 23 sera had a positive result and 52 a negative result. The IDvet assay yielded 26 positive test results as well as 49 negative results.
The rate of correct positive and the rate of correct negative test results of the assays is displayed in Table 1 a. They resulted in a sensitivity of 95.5 %, 95.5 %, and 100 % for the Diasorin assay, the Roche assay, and the IDvet assay, respectively. The corresponding results for the specificity were 90.5 %, 96.2 %, and 92.5%.
Table 1.
Sensitivity and specificity of SARS-CoV-2 antibody assays determined by us in this study (a) and in a recent study (b) (6) using the identical 75 serum samples.
| a) | ||||
|---|---|---|---|---|
| Manufacturer | Test results | Sensitivity | Specificity | |
| Rate of correct positive test results | Rate of correct negative test results | |||
| Diasorin | 21/22 | 48/53 | 95.5 % | 90.5 % |
| RocheI | 21/22 | 51/53 | 95.5 % | 96.2 % |
| IDvet | 22/22 | 49/53 | 100 % | 92.5 % |
| b) | ||||
|---|---|---|---|---|
| Manufacturer | Test results |
Sensitivity | Specificity | |
| Rate of correct positive test results | Rate of correct negative test results | |||
| Euroimmun | 19/22 | 51/53 | 86.4 % | 96.2 % |
| EDI | 22/22 | 47/53 | 100 % | 88.7 % |
| Mikrogen | 19/22 | 53/53 | 86.4 % | 100 % |
| Viramed | 17/22 | 53/53 | 77.3 % | 100 % |
These values were comparable to four assays previously evaluated in our laboratory (Krüttgen et al., 2020; from Euroimmun, Epitope diagnostics (EDI), Mikrogen, and Viramed) which exhibited a sensitivity between 86.4 % and 100 % and a specificity between 88.7 % and 100 % (Table 1b).
Thus, none of the seven assays combined a very high sensitivity with a very high specificity. Two assays reached a sensitivity of 100 % (IDvet and EDI) whereas two other assay had a specificity of 100 % (Mikrogen and Viramed). The Roche assay offered the best balance between high sensitivity and high specificity (95.5 % and 96.2 %).
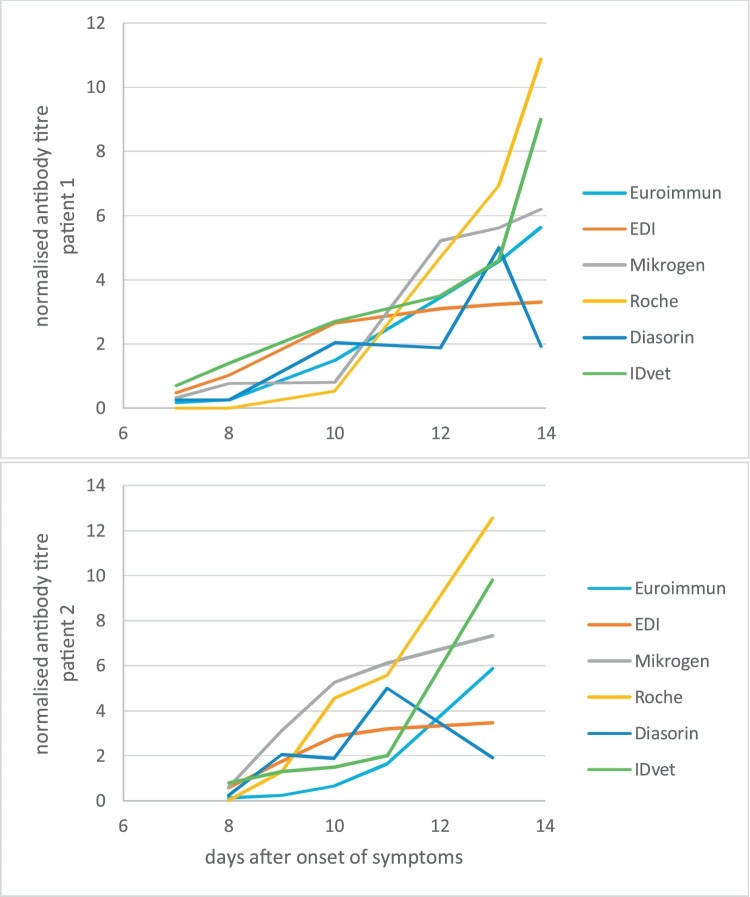
Using the assays of Roche, DiaSorin and IDvet we also compared the kinetic of antibody titers for two patients of whom consecutive sera were available. We found that all three assay delivered positive results 9–10 days after onset of symptoms for both patients (Fig. 1 ). In comparison with the three previously characterized semiquantitative assays (Euroimmun, Mikrogen and EDI), the two new assays confirmed the previously defined time span of 8–13 days after onset of clinical symptoms to obtain a positive serological test result. The assays target two different antigens: the nucleocapsid protein and the spike protein. However, neither an individual assays nor a group of assays using the same target protein is positive first or shows the strongest quantitative increase for both patients. This is also true for other patients with consecutive sera available (data not shown).
Fig. 1.
Time course of serological responses in two patients after onset of symptoms using different assay.
4. Discussion
Reliable serological assays are urgently needed to supplement the diagnostic repertoire and identify patients with past SARS-CoV-2 infection. This would allow the detection of patients presenting during later stage of the disease when direct pathogen detection has turned negative due to viral clearance (Vogel, 2020). During the initial stage of the outbreak, the ELISA of Euroimmun was a commonly used commercial ELISA and first evaluated by Okba and coworkers (Okba et al., 2020). In the meantime, several additional assay using different methodologies (CLIA, ELISA, immunoblot) targeting different virus antigens (such as the S- and N- antigen) have become available, increasing the commercially available diagnostic tools of clinical laboratories.
We found that the highly anticipated CLIAs for the widely-used automatons from Diasorin and Roche both offer a high sensitivity of >95 %. Using our collection of samples, the Roche assay offers a higher specificity (96.5 % Roche vs 90.5 % Diasorin). This difference might be related to the different antigens targeted by the assay (the nucleocapsid protein by Roche, IDVet, EDI and Microgen; the spike protein by DiaSorin and Euroimmun; the Viramed assay targets both the spike protein and the nucleocapsid protein), However, based on our data there is no general trend indicating a higher sensitivity of assays targeting the nucleocapsid protein. The higher sensitivity of the Roche assay may be based on the determination of total antibody (IgG and IgM). The Diasorin assay is targeting IgG antibodies only. Although somewhat less specific, the DiaSorin assay offers the attractive possibility of measuring neutralizing antibodies targeting the viral spike protein (although no serological assay for SARS-CoV-2 has so far been demonstrated to measure neutralizing antibodies). Compared to the assays of Roche and Diasorin, the IDvet ELISA had the highest sensitivity and an intermediary specificity.
Our comparative approach to test in total seven different SARS-CoV-2 antibody assays with an identical collection of serum samples allowed for the first time the direct comparison of performance indicators of such a large number of automated assays. A perfect serological test would offer >99 % sensitivity and >99 % specificity (Mallapaty, 2020). We found that two assay reached a sensitivity of 100 % (IDvet and EDI) whereas two other assay offered a specificity of 100 % (Microgen and Viramed). The requested very high sensitivity in combination with very high specificity was not observed in any of the seven assays. Thus, the combination of two different assays (one test with 100 % sensitivity and another test with 100 % specificity) might provide a maximum diagnostic value. However, if a laboratory decided to implement a single assay approach, the Roche assay offers the best balance of high sensitivity and high specificity (95.5 and 96.2 %). A general advantage in terms of sensitivity or specificity by the use of either the nucleocapsid protein or the spike protein as a target could not be observed.
The interpretation of this study is limited by a lack of neutralization information for the samples included. Further, the use of different targets from two different viral proteins affects the assay’s results in an unknown way.
Taken together, our study will assist diagnostic laboratories to select the appropriate assays or assay combinations according to their sample load, equipment, and diagnostic question.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sector.
CRediT authorship contribution statement
Alexander Krüttgen: conceptualization, data curation, formal analysis, investigation, methodology, Project administration, Supervision, Validation, writing -original draft and final review+editing. Christian G. Cornelissen: Resources, Writing - review & editing. Michael Dreher: Resources, Writing - review & editing. Mathias W. Hornef: Writing - review & editing. Matthias Imöhl: Resources, Writing - review & editing. Michael Kleines: conceptualization, data curation, formal analysis, investigation, methodology, Project administration, Supervision, Validation, writing -original draft and final review+editing.
Declaration of Competing Interest
We declare no conflict of interest
References
- de Ory F., Guisasola M.E., Balfagon P., Sanz J.C. Comparison of commercial methods of immunoblot, ELISA, and chemiluminescent immunoassay for detecting type-specific herpes simplex viruses-1 and -2 IgG. J. Clin. Lab. Anal. 2018:32. doi: 10.1002/jcla.22203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eis-Hübinger A.M., Hönemann M., Wenzel J.J., Berger A., Widera M., Schmidt B., Aldabbagh S., Marx B., Streeck H., Ciesek S., Liebert U.G., Huzly D., Hengel H., Panning M. Ad hoc laboratory-based surveillance of SARS-CoV-2 by real-time RT-PCR using minipools of RNA prepared from routine respiratory samples. J. Clin. Virol. 2020;127 doi: 10.1016/j.jcv.2020.104381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüttgen A., Cornelissen C.G., Dreher M., Hornef M., Imöhl M., Kleines M. Comparison of four new commercial serologic assays for determination of SARS-CoV-2 IgG. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallapaty S. Will antibody tests for the coronavirus really change everything? Nature. 2020;580:571–572. doi: 10.1038/d41586-020-01115-z. [DOI] [PubMed] [Google Scholar]
- Meyer B., Drosten C., Müller M.A. Serological assays for emerging coronaviruses: challenges and pitfalls. Virus Res. 2014;194:175–183. doi: 10.1016/j.virusres.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okba N.M.A., Muller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., Lamers M.M., Sikkema R.S., de Bruin E., Chandler F.D., Yazdanpanah Y., Le Hingrat Q., Descamps D., Houhou-Fidouh N., Reusken C., Bosch B.J., Drosten C., Koopmans M.P.G., Haagmans B.L. Severe acute respiratory syndrome coronavirus 2-Specific antibody responses in coronavirus disease 2019 patients. Emerging Infect. Dis. 2020:26. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel G. First antibody surveys draw fire for quality, bias. Science. 2020;368:350–351. doi: 10.1126/science.368.6489.350. [DOI] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]