Abstract
Disanthus cercidifolius subsp. longipes is a rare and endangered plant species. In our study, the complete chloroplast genome was assembled by using high-throughput DNA sequencing data. The whole CP genome is 158,076 bp in length, comprising of a large single-copy region of 87,148 bp, a small single-copy region of 18,300 bp, and two inverted repeat regions of 26,314 bp each. There are 136 genes in the genome, including 86 protein-coding genes, 40 transfer RNA genes, eight ribosomal RNA genes, and two pseudogenes (ndhK and ycf1). Phylogenetic results demonstrated that D. cercidifolius subsp. longipes grouped with other Hamamelidaceae species, with a support rate of 100%.
Keywords: Disanthus cercidifolius subsp. longipes, chloroplast genome, phylogenetic analysis, rare plant species
The genus Disanthus belongs to Hamamelidaceae family, and it is a monotypic genus with a plant species of Disanthus cercidifolius (Wu et al. 2003; Gao et al. 2009). Disanthus cercidifolius subsp. longipes is a subspecies of D. cercidifolius in China–Japan flora region (Yu et al. 2014). This plant is a shrub growing to about four meters, with brown branchlets, broadly ovate-rounded leaves, and red petals. D. cercidifolius subsp. longipes is listed as an endangered plant species by the IUCN, and in China, it distributes only in provinces of Hunan, Jiangxi, and Zhejiang, with a small number of individuals. In Zhejiang province, the species is now listed as a key protected wild plant. In this study, we assembled and annotated the complete chloroplast (CP) genome of D. cercidifolius subsp. longipes to understand its features as well as its phylogenetic relationship with other plant species.
Fresh leaves were sampled in Longquanshan (27°53′42′′N, 119°10′11′′E), Longquan, Zhejiang Province, and they were kept in plastic bags before taking to the laboratory. A voucher specimen (CHS2018012) was deposited in the Molecular Biology Laboratory of Taizhou University. Total genomic DNA was isolated according to the CTAB protocol described by Doyle and Doyle (1987). A DNA library was prepared following the protocol supplied by Illumina Inc. (San Diego, CA) and was then sequenced using an Illumina Hiseq X Ten system. Approximately 6.3 GB of 150 bp paired-end reads were generated, and they were filtered by NGS QC Toolkit v2.3.3 to trim off adapters and remove low quality reads (Patel and Jain 2012). The CP genome was then assembled by running the Perl program in NOVOPlasty package (Dierckxsens et al. 2017), and it was annotated using DOGMA (Wyman et al. 2004). The complete plastome (GenBank accession: MN527332) is 158,076 bp in length with a typical quadripartite structure. The sizes of large single-copy region (LSC), small single-copy region (SSC), and inverted repeat regions (IRs) are 87,148, 18,300, and 26,314 bp, respectively. Totally, 136 genes are annotated in the CP genome, these include 86 protein-coding genes, 40 transfer RNA genes, eight ribosomal RNA genes, and two pseudogenes. Two protein-coding genes, ndhK and ycf1, were identified as pseudogenes. The overall GC content in the CP genome is 37.9%, while in LSC, SSC, and IR, the GC contents are 36.0, 32.9, and 43.1%, respectively.
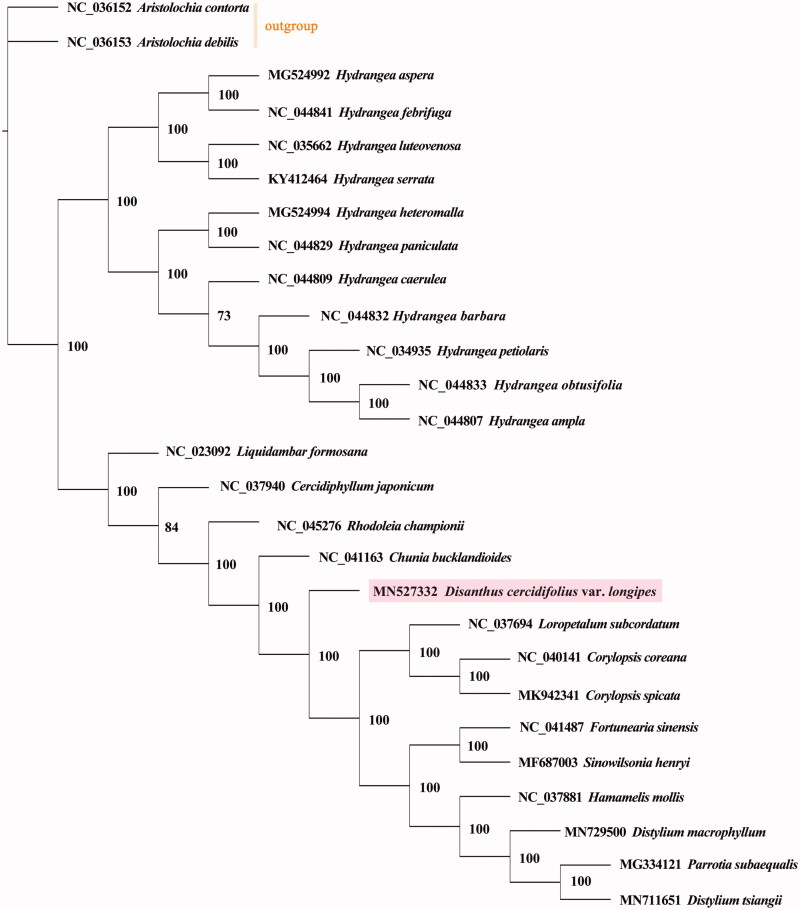
To understand the evolutionary relationship between D. cercidifolius subsp. longipes and related plant species whose CP genomes were assembled, the complete CP genomes of Chunia bucklandioides (NC_041163), Corylopsis coreana (NC_040141), Corylopsis spicata (MK942341), Sinowilsonia henryi (MF687003), Liquidambar formosana (NC_023092), Cercidiphyllum japonicum (NC_037940), Fortunearia sinensis (NC_041487), Rhodoleia championii (NC_045276), Parrotia subaequalis (MG334121), Hamamelis mollis (NC_037881), and 11 Hydrangea plants were downloaded from NCBI. A maximum-likelihood tree was constructed based on GTR + G+I model with both Aristolochia contorta and Aristolochia debilis as the outgroup by using PhyML 3.1 (Guindon et al. 2010). The phylogenetic analysis results indicated D. cercidifolius subsp. longipes and other nine Hamamelidaceae species grouped in the same clade, with a support rate of 100% (Figure 1).
Figure 1.
A maximum-likelihood tree based on the complete chloroplast genome sequences of Disanthus cercidifolius subsp. longipes (Hamamelidaceae) and related species, with both Aristolochia contorta and A. debilis (Aristolochiaceae) as the outgroup. The numbers next to nodes indicate bootstrap support values.
Funding Statement
This work was supported by 211 Talent Training Fund of Taizhou.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Gao P, Yang A, Yao X, Huang H. 2009. Isolation and characterization of nine polymorphic microsatellite loci in the endangered shrub Disanthus cercidifolius var. longipes (Hamamelidaceae). Mol Ecol Resour. 9(3):1047–1049. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. [DOI] [PubMed] [Google Scholar]
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7(2):e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZY, Raven P, Hong D. 2003. Flora of China (Vol. 9). Beijing (China): Science Press; St. Louis (MO): Missouri Botanical Garden Press. [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255. [DOI] [PubMed] [Google Scholar]
- Yu Y, Fan Q, Shen R, Guo W, Jin J, Cui D, Liao W. 2014. Genetic variability and population structure of Disanthus cercidifolius subsp. longipes (Hamamelidaceae) based on AFLP analysis. PLoS One. 9(9):e107769. [DOI] [PMC free article] [PubMed] [Google Scholar]