Abstract
Riccia fluitans L. is the most common species in Riccia genus. To investigate intraspecific variations on mitochondrial genomes of R. fluitans, we completed mitochondrial genome of R. fluitans. Its length is 185,640 bp, longer than that of NC_043906 by 19 bp and it contains 74 genes (42 protein-coding genes, 3 rRNAs, 28 tRNAs, and 1 pseudogene). 18 single nucleotide polymorphisms (SNPs) and 19 insertions and deletions are identified, higher than that of Marchantia polymorpha subsp. ruderalis. One non-synonymous SNP is found in ccmFN. Phylogenetic trees show that R. fluitans is clustered with Dumortiera hirsuta, requiring additional mitogenome to clarify the phylogenetic relationship.
Keywords: Riccia fluitans, mitochondrial genome, Ricciaceae, intraspecific variation, ccmFN
Riccia fluitans L., the most common species in Riccia genus, is distributed all over the world (Manju et al. 2012). In order to investigate its organellar genome sequences, its chloroplast genome was sequenced previously (Kwon, Min, Xi, et al. 2019). In addition, the mitochondrial genome of R. fluitans was also uncovered to understand RNA editing events on mitochondrial genomes of early land plants (Myszczyński et al. 2019). To understand intraspecific variations of mitochondrial genome of R. fluitans, we completed its mitochondrial genome sequences.
The thallus of R. fluitans collected in Namyangju city, Korea (Voucher in InfoBoss Cyber Herbarium (IN); W. Kwon, IB-50004; 37.583486 N, 127.238337E) was used for extracting DNA with DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using HiSeqX at Macrogen Inc., Korea. Mitochondrial genome was completed by Velvet 1.2.10 (Zerbino and Birney 2008), SOAPGapCloser 1.12 (Zhao et al. 2011), BWA 0.7.17 (Li 2013), and SAMtools 1.9 (Li et al. 2009). Geneious R11 11.0.5 (Biomatters Ltd, Auckland, New Zealand) was used for annotation based on R. fluitans mitochondrial genome (NC_043906; Myszczyński et al. 2019).
The mitochondrial genome of R. fluitans (GenBank accession is MN927134) is 185,640 bp, which is longer than NC_043906 by 19 bp. It contains 74 genes (42 protein-coding genes, 3 rRNAs, 28 tRNAs, and 1 pseudogene) and overall GC content is 42.4%. nad7 gene was revealed as pseudogene in both R. fluitans mitochondrial genomes.
From alignment of two mitochondrial genomes, 18 single nucleotide polymorphisms (SNPs; 0.0096%) and 19 insertions and deletions (INDELs; 0.010%) were identified. Number and ratio of sequence variations are higher than those of Marchantia polymorpha subsp. ruderalis (7 SNPs; 0.0038%; Kwon et al. 2019c); however, it is still less than those of vascular plants, such as Liriodendron tulifipera (365 SNPs; 0.066%; 2,117 INDELs; 0.38%; Park et al. 2019) and Arabidopsis thaliana (64 SNPs; 0.017% and 1,089 INDELs; 0.30%; Park et al., in preparation), addressing hypothesis that the number of variations on bryophytes mitochondrial genomes is less than those of vascular plants. Only one non-synonymous SNP is found in ccmFN and the remaining variations are in intergenic space.
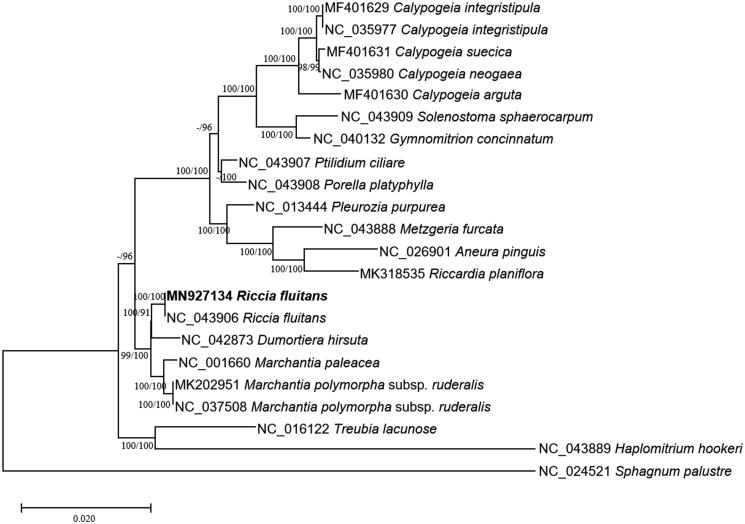
Twenty-two complete mitochondrial genomes including R. fluitans mitochondrial genome were used for drawing neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) trees based on alignments of 26 conserved protein-coding genes based on annotation of all used mitochondrial genomes by MAFFT 7.450 (Katoh and Standley 2013) and MEGA X (Kumar et al. 2018). Phylogenetic trees present that R. fluitans is clustered with Dumortiera hirsuta (Figure 1), which is not congruent with the tree that D. hirsuta is clustered with Marchantia genus (Kwon et al. 2019a) but agrees with three of four trees which display that R. fluitans is clustered with D. hirsuta (Kwon, Min, Xi, et al. 2019). Because of incongruence in the topology of R. fluitans, D. hirsuta, and Marchantia genus, additional mitochondrial genomes such as Reboulia hemisphaerica clustered with D. hirsuta (Kwon et al. 2019b; Kwon, Min, Kim, et al. 2019) will be required to clearify their phylogenetic relationship.
Figure 1.
Neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees of 22 complete mitochondrial genomes: Riccia fluitans (MN927134 in this study and NC_043906), Marchantia polymorpha subsp. ruderalis (NC_037508 and MK202951), Marchantia paleacea (NC_001660), Pleurozia purpurea (NC_013444), Aneura pinguis (NC_026901), Dumortiera hirsuta (NC_042873), Calypogeia neogaea (NC_035980), Calypogeia integristipula (NC_035977), Calypogeia suecica (MF401631), Calypogeia arguta (MF401630), Calypogeia integristipula (MF401629), Riccardia planiflora (MK318535), Solenostoma sphaerocarpum (NC_043909), Porella platyphylla (NC_043908), Ptilidium ciliare (NC_043907), Metzgeria furcata (NC_043888), Haplomitrium hookeri (NC_043889), Tritomaria quinquedentata (NC_037041), Gymnomitrion concinnatum (NC_040132), Calypogeia integristipula (NC_035977), Treubia lacunose (NC_016122), and Sphagnum palustre (NC_024521) as an outgroup. The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor-joining phylogenetic trees, respectively.
Funding Statement
This work was supported by InfoBoss Research Grant [IBG-0027] and ‘The Consortium of Korea Biodiversity and Sustainable Use’ [TFDV2019051609821691] at the Ministry of Environment, Korea.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon W, Kim Y, Park J. 2019a. The complete chloroplast genome sequence of Dumortiera hirsuta (Sw.) Nees (Marchantiophyta, Dumortieraceae). Mitochondrial DNA Part B. 4(1):318–319. [Google Scholar]
- Kwon W, Kim Y, Park J. 2019b. The complete mitochondrial genome of Dumortiera hirsuta (Sw.) Nees (Dumortieraceae, Marchantiophyta). Mitochondrial DNA Part B. 4(1):1586–1587. [Google Scholar]
- Kwon W, Kim Y, Park J. 2019c. The complete mitochondrial genome of Korean Marchantia polymorpha subsp. ruderalis Bischl. & Boisselier: inverted repeats on mitochondrial genome between Korean and Japanese isolates. Mitochondrial DNA Part B. 4(1):769–770. [Google Scholar]
- Kwon W, Min J, Kim Y, Park J. 2019. The complete chloroplast genome of Reboulia hemisphaerica (L.) Raddi (Aytoniaceae, Marchantiophyta). Mitochondrial DNA Part B. 4(1):1459–1460. [Google Scholar]
- Kwon W, Min J, Xi H, Park J. 2019. The complete chloroplast genome of Riccia fluitans L.(Ricciaceae, Marchantiophyta). Mitochondrial DNA Part B. 4(1):1895–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997. [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manju C, Rajesh K, Prakashkumar R. 2012. On the identity of Riccia fluitans (Ricciaceae: Marchantiophyta) in India. Acta Biol Plantarum Agriensis. 2:115–124. [Google Scholar]
- Myszczyński K, Ślipiko M, Sawicki J. 2019. Potential of transcript editing across mitogenomes of early land plants shows novel and familiar trends. IJMS. 20(12):2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Kim Y, Kwon M. 2019. The complete mitochondrial genome of tulip tree, Liriodendron tulipifera L. (Magnoliaceae): intra-species variations on mitochondrial genome. Mitochondrial DNA Part B. 4(1):1308–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinf. 12(Suppl 14):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]