Abstract
Salmonella is an important human pathogen and poultry products constitute an important source of human infections. This study investigated prevalence; identified serotypes based on whole genome sequence, described spatial distribution of Salmonella serotypes and predicted risk factors that could influence the prevalence of Salmonella infection in commercial poultry farms in Nigeria. A cross sectional approach was employed to collect 558 pooled shoe socks and dust samples from 165 commercial poultry farms in North West Nigeria. On-farm visitation questionnaires were administered to obtain information on farm management practices in order to assess risk factors for Salmonella prevalence. Salmonella was identified by culture, biotyping, serology and polymerase chain reaction (PCR). PCR confirmed isolates were paired-end Illumina- sequenced. Following de novo genome assembly, draft genomes were used to obtain serotypes by SeqSero2 and SISTR pipeline and sequence types by SISTR and Enterobase. Risk factor analysis was performed using the logit model. A farm prevalence of 47.9% (CI95 [40.3–55.5]) for Salmonella was observed, with a sample level prevalence of 15.9% (CI95 [12.9–18.9]). Twenty-three different serotypes were identified, with S. Kentucky and S. Isangi as the most prevalent (32.9% and 11%). Serotypes showed some geographic variation. Salmonella detection was strongly associated with disposal of poultry waste and with presence of other livestock on the farm. Salmonella was commonly detected on commercial poultry farms in North West Nigeria and S. Kentucky was found to be ubiquitous in the farms.
Introduction
Non-typhoidal Salmonella is one of the most common causes of food-borne diseases worldwide. It has been estimated to cause 93.8 million human infections and 155,000 deaths annually [1, 2]. Contaminated poultry products, especially undercooked meat and raw eggs are important sources of human salmonellosis [3, 4].
Serotyping is the first step to characterize Salmonella, because serovars often inform on possible pathogenic potential, host range and disease sequelae [5–7]. Serotyping therefore form the basis of national and international surveillance networks for Salmonella [8, 9]. Until recently, traditional serology based on reactions of rabbit antisera to the lipopolysaccharide and flagellar antigens and the surface antigen Vi was used to divide Salmonella into more than 2,600 serovars by the White Kauffmann Le Minor Scheme (WKL) [10]. However, whole genome sequencing (WGS) has now emerged as an alternative, rapid and more discriminatory method [7, 8]. By this method, prediction of serotypes can be done using freely available in silico pipelines, such as SeqSero, which utilizes surface antigen-encoding genes for predicting serotypes, and Salmonella In Silico Typing Resources (SISTR), which infers serovars from core genome MLST (cgMLST) and surface antigens [9, 11, 12]. Several studies have now used WGS in Salmonella surveillance and outbreak investigation [2, 13–15], and 91.9% concordance has been found between reported serovars by WKL scheme and predicted serovars using in silico resource [16], and 94.8% and 88.2% similarity was reported for SISTR and SeqSero, respectively [17].
Agricultural sector remains the largest contributor to the Nigerian economy, accounting for over 38% of the non-oil foreign exchange earnings, and employing about 70% of the active labour force of the population. The poultry sub-sector is the most commercialized of all the sub-sectors of the Nigeria’s agriculture [18] and has transformed the lives of the less privileged segment of the society with just a little investment and at low cost of technology. Annual production average 454 billion tonnes of meat and 3.8 million eggs, with a standing population of 180 million birds [19]. Poultry meat and eggs are the major sources of animal protein in Nigeria, as in many developing countries, because of their affordability and acceptability [20, 21]. Unfortunately, the sustainable growth of this important agricultural subsector is seriously threatened by several infectious diseases including, those caused by Salmonella species. So far, there are only a few published reports of circulating strains of Salmonella in poultry production in Nigeria [21–24], and very little has been done to understand the risk factors for the different types of Salmonella. The aim of the present study was to determine Salmonella prevalence, serotype distribution by WGS and risk factors for Salmonella obtained in commercial poultry farms in Nigeria.
Materials and methods
Ethical approval
Ethical approval for sampling and questionnaire investigation to obtain farm data was obtained from Sokoto State Ministry of Animal Health and Fisheries Developments, Kebbi State Ministry of Animal Health and Husbandry, and Zamfara State Directorate of Animal Health and Livestock Development with approval reference numbers MAH&FD/VET/166/11, MAHF/VET/VOL1, and DAHLD/SUB/VET/VOL.1 respectively.
Study area
The study was conducted in north-western Nigeria. The region occupies a total land mass of 226,662 km2, representing 24.5% of Nigeria's total land mass. There is an estimated human population of 48,942,307 (25.3% of Nigeria’s total population) majority of whom are involved in farming activities [25, 26]. The region also has an estimated exotic and backyard poultry population of 18,770,610 and 10,064,763 which respectively represent 16.2% and 46.2% of the total chicken populations in these categories in the country [18]. Sampling was conducted in Sokoto, Kebbi and Zamfara states due to significant poultry production in these areas.
Study design and sample collection
A cross sectional study design was employed to collect 558 pooled shoe socks and dust samples from 165 commercial poultry farms. On arrival to a farm, a pen was randomly selected from other pens as a sampling unit. From this representative unit (pen), sock samples were obtained by stepping on freshly dropped faeces while walking through the pen. Shoe covers were worn over fully covered leather shoes and were changed between farms using clean latex gloves. Shoe socks sample were immediately transferred into a sterile sampling bottle. Additionally, dust samples from that same pen were obtained from multiple spots by scooping up dust materials containing poultry litter materials, feeds and other composed materials into a sterile sample-bottle. The number of samples collected per farm depended on the categories of chicken raised in the farm. Two samples were collected per farm from 51 farms that reared either broilers or layers, while four samples were collected from 114 farms, two from each category of layers and broiler. Information about age of flock, chicken type and category of farm was recorded. A farm was consider positive when at least one of the samples collected was found to contain Salmonella species. All samples were adequately labelled and placed in cooling box containing ice-packs. Samples were transported to the laboratory in Central Veterinary Research Laboratory, Usmanu Danfodiyo Univesrity, Sokoto, Nigeria for immediate analysis.
Farm description
Poultry production system could be categorised in to five intermediate categories from the four operational classes of Food and Agricultural Organization (FAO), based on the number of chicken raised in a farm [18]. The size of farms ranged from backyard farms (less than 200 birds), semi- commercial farms (200–999 birds), small-scale farms (1,000–4,999 birds), and medium-scale farms (5,000–9,999 birds) to large-scale farms (more than 10,000 birds). The backyard farms represent the majority of the farms sampled in the study (S1 File). Grand-parent stocks are generally imported from Europe and breeding farms are concentrated outside the study area in the south of Nigeria. Day-old-chicks are likewise mostly produced in the south by big hatcheries and transported by road to different parts of the north-west Nigeria [18].
Isolation and characterization of Salmonella
Samples were investigated for presence of Salmonella according to ISO 6579 [27]. Briefly, one gram of sample was weighed (OHAUS, USA) before mixed with 9 ml of buffered peptone water (BPW, Oxoid UK) for non-selective pre-enrichment of samples at 37° C for 18 ± 2 hrs. Subsequently, an aliquot of 0.1 ml of the suspension was inoculated into 10 ml of Rappaport-Vassiliadis, (RV) broth (Oxoid, UK) for selective enrichment overnight at 41.5°C. Then selective plating was done in parallel on Xylose Lysine Deoxycholate, XLD (Oxoid, UK) and onto Brilliance Salmonella Agar, BSA (Oxoid, UK); plates were incubated at 37°C overnight. Plates were examined for the presence of Salmonella typical colonies, identified with a black centre or purple colour on XLD and BSA, respectively. One isolate was picked from a pure culture representing one sample unit. The reference strain Salmonella Typhimurium ATCC 14028 was spiked into selected samples for quality control purposes.
Presumptive Salmonella isolates were subjected to biochemical tests using commercially available media (Oxoid, UK). Briefly, a loopful of colonies was stabbed into citrate and sulphide, indole, motility (SIM) agar, and incubated at 37°C overnight. Isolates showing positive citrate, H2S production, and motility but a negative indole reaction were categorized as presumptive Salmonella and sub-cultured onto Nutrient agar (Oxoid, UK) and incubated at 37°C overnight. Colonies from this plate were subjected to serological confirmation by slide agglutination test using polyvalent Salmonella antisera (SSI, Denmark) and normal saline as a negative control and Salmonella Typhimurium ATCC 14028 as a positive control.
PCR-based Salmonella identification
As a final confirmation of Salmonella, isolates that were positive by serology were subjected to PCR identification using the invA-based method [28]. Briefly, one to two bacterial colonies were suspended into 100 μL of molecular grade water (Gibco, Life technologies, USA) and subjected to boiling at 100°C for 10 min. The mixture was centrifuged (Eppendorf, AG Germany) at 12,000 rpm for 2 min. PCR was performed using PuRe Taq Ready-To-Go PCR beads (illustra TM United Kingdom) containing buffers, dNTPs, enzyme, stabilizers and BSA in addition to 1 μL of sample DNA and 0.2 μL of the primers (inqaba biotec, Hartfield South Africa) (100 μM) invA forward (5'GTGAAATTATCGCCACGTTCGGGCA3') and invA reverse (5'TCATCGCACCGTCAAAGGAACC3') in 25 μl final volume reaction. Amplification was performed using Thermal cycler (Applied Biosystem, USA) with 95°C for 2 min, 95°C for 30 sec, 55°C for 30 sec and 72°C for 2 min for 35 cycles. A final cycle at 72°C for 5 min was used [29]. Amplicons were visualized in 1.5% agarose gels stained with SafeView nucleic acid stain using a UV trans-illuminator (UVP GelMax Imager, United Kingdom). Isolates that showed a band size of 284 bp was considered as Salmonella using 100 bp standard DNA ladder (New England BioLabs, United Kingdom). The reference strain Salmonella ATCC 14028 was used as positive control and water without DNA as negative control.
Serotype PCR of strains
Initial screening of isolates using serotype specific PCR was done at the Pharmaceutical Microbiology Laboratory University of Ibadan, Nigeria to identify S. Enteritidis and S. Typhimurium, which are some of the common non- typhoidal Salmonella in humans in the region [30].
The protocol developed by Tennant et al. (2010) was used to amplify specific genomic regions of strains to investigate whether they belonged to serotypes S. Enteritidis or S. Typhimurium; the SdfF and SdfR primers (inqaba biotec, Hartfield South Africa) were used to amplify SdfI, indicative of S. Enteritidis. Two sets of primers, FFLIB and RFLIA (inqaba biotec, Hartfield South Africa), which amplify the fliB-fliA intergenic region, and primers Sense-59 and Antisense-83, which amplify the Phase 2 (fljB) flagella gene, were used to detect S. Typhimurium including the monophasic variant [29, 31, 32]. PCR conditions and procedures were set as described above with primer concentrations of 1 μl each of 0.5 μL sdfF/ sdfR (5'CGTTCTTCTGGTACGATGAC3' forward, 5’TGTGTTTTATCTGATGCAAGAGG3’ reverse), FFLIB/RFLIB (5’GCGGTATACAGTGAATTCAC3’ forward, 5’CTGGCGACGATCTGTCGATG3’ reverse) sense-59/ Antisense-83 (5’GCCATATTTCAGCCTCTCGCCCG3’ forward, 5’CAACAACAACCTGCAGCGTGTGCG3’ reverse) for 100 μl reaction final volume respectively. Isolates that showed a band size of 333 bp and 1389/250 were considered as S. Enteritidis and S. Typhimurium respectively using 100 bp standard DNA ladder (New England BioLabs, United Kingdom).
DNA extraction and WGS analysis
Single colony of Salmonella on blood agar grown over night was suspended in 5 ml Luria broth (LB) (Difco, USA) for 16 hrs at 37°C in an incubator shaker (GFL, Germany). Genomic DNA was extracted using Promega Maxwell DNA automatic extraction robot and Maxwell RSC Cultured Cells DNA kit as described by the protocol of the manufacturer (Maxwell® RSC-16, USA). The concentration and quality of extracted DNA was evaluated using Nanodrop (Thermo Scientific, USA), with DNA concentration of greater 20 ng/μL and A260/A280 of 1.8–2.0 were sequenced. A sequencing library was prepared using Nextera XT kits as described by the manufacturer. Genomes were sequenced on an Illumina MiSeq platform using paired-end chemistry (2 x 250-bp) (Illumina, San Diego, California, USA). De novo genome assembly of sequence was done using SPAdes version 3.9 available on the Centre of Genomic Epidemiology server (cge.cbs.dtu.dk/services/SPAdes/). The quality of the assembled genome was evaluated using QUAST [33]. The draft genome sequences are available at the European Nucleotide Archive under study accession number PRJEB37477 (secondary accession ERP120792) and accession number for each genome is indicated in S2 File.
In Silico serotype and STs prediction
Because of high-throughput, and decreasing cost of next generation sequencing, WGS based serotyping is increasingly used as methods in Salmonella typing [34]. This method has been validated and found to be highly concordance with the results from conventional serotyping methods [17], with better efficiency. Assemblies with a genome size less than 4 Mb or greater than 6 Mb or with GC content of the genome less than 50% or greater than 54% were excluded (S1 Table). Also contaminated and genome assigned to different organism were excluded. Draft assembled genomes of Salmonella that satisfied the inclusion criteria were initially uploaded to the online version of SeqSero 2 v1.0.2 ( http://www.denglab.info/SeqSero2) [9, 35]. However, as some strains would not be assigned to serotypes by SeqSero2, draft assemblies were also uploaded to SISTR (https://lfz.corefacility.ca/sistr-app/) through the web application programming interface and the results of the predicted serovars were compared with that of SeqSero2 [11, 16]. Most of the strains were assigned multi-locus sequence types (STs) by SISTR pipeline using seven housekeeping genes (aroC, dnaN, hemD, hisD, purE, sucA, thrA) [36]. Some isolates could not be assigned ST type by SISTR; raw reads of these strains were submitted to Enterobase (http://enterobase.warwick.ac.uk/).
Risk factors analysis
A signed written consent was obtain from farmers prior to administration of questionnaire. A questionnaire (S3 File) and consent to collect information about risk factors for Salmonella at the poultry farms was designed and pre-tested with a small population of 10 farmers for validity and reliability before applied to 65 consented farmers. The questionnaire contained information about farm manager demography, farm size and management, farmer’s knowledge about Salmonella and salmonellosis, disease management, farm sanitation and biosecurity (S4 File). The interviews were done during the visits to the farm when the different samples for Salmonella analysis were collected. The questions were posed to the owner, farm manager, consulting veterinarian or animal health workers who were available at the time of the visit.
Data and statistical analysis
Serotype predictions by two pipelines were imported to SPSS version 26 (IBM, USA) to check for level of agreement between the two pipelines using Cohan's kappa statistics. Questionnaire responses were entered into Epi Info 7 (CDC, USA) and later exported to Microsoft Excel 2016 (Microsoft Corporation, Redmond, WA, USA) as a database. Risk factor analysis was done using Statistical software R using (Glm package) relevant installed packages [37]. Chi-square test of independence was used to test for association between Salmonella prevalence and categorical variables (farm category, type of chicken, sample and age of chicken). A two-step statistical procedure was used to evaluate relationship between variables and Salmonella farm status. In the first step, 11 potential risk factors (production system, report of previous outbreaks, frequency of Salmonellosis outbreaks, report of Salmonellosis outbreak in neighbouring farm, fencing of farm, poultry waste disposal, proximity with other poultry farms, provision of disinfection of boots, availability of toilets, presence of other livestock in the farm and frequency of farm cleaning) were selected for univariate regression analysis between specific variable and outcome of Salmonella status in a farm. In the second step, statistically significant predictors were selected for multiple logistic regression analysis to model between predictors and outcome. The significant level was p < 0.05 with results expressed as estimates and standard error.
Results
Prevalence of Salmonella
Among 165 commercial farms sampled, 47.9% (CI95 [40.3–55.5]) were positive for Salmonella, while 15.9% (CI95 [12.9–18.9]) of the individual samples were positive (Table 1).
Table 1. Prevalence of Salmonella in poultry farms in Nigeria.
| No of farms | No. of samples | aSalmonella-positive farms | bSalmonella-positive samples | |||
|---|---|---|---|---|---|---|
| State | Count | (%) | Count | (%) | ||
| Sokoto | 62 | 200 | 30 | 48.4 | 33 | 16.5 |
| Kebbi | 48 | 176 | 17 | 35.4 | 19 | 10.8 |
| Zamfara | 55 | 182 | 32 | 58.2 | 37 | 20.3 |
| Total | 165 | 558 | 79 | 47.9 | 89 | 15.9 |
aFarm Confidence Interval = CI95 (40.3–55.5)
bSample level Confidence Interval = CI95 (12.9–18.9)
Large-scale farms had significantly higher (p = 0.0001) Salmonella prevalence (33%; CI95 [29.1–36.9]) than other farm categories, while small-scale farms had the lowest prevalence. Layer chickens had significantly higher prevalence (20.6%; CI95 [17.2–24.0]) than broilers (10.9%; CI95 [8.3–13.5]) (p = 0.003). Sample type (shoe socks, dust) and age categories were not significantly associated with the prevalence of Salmonella in poultry farms (Table 2).
Table 2. Variation in prevalence of Salmonella based on selected parameters in commercial poultry farms in Nigeria.
| Parameters | Number sampled | Salmonella-positive | ||
|---|---|---|---|---|
| Farm categories | Count | % | p-value | |
| Backyard | 119 | 18 | 15.1 | p = 0.0001 |
| Semi-commercial | 81 | 18 | 22.2 | |
| Small-scale | 198 | 11 | 5.6 | |
| Medium-scale | 66 | 11 | 16.7 | |
| Large-scale | 94 | 31 | 33.0 | |
| Sample type | ||||
| Shoe socks | 279 | 43 | 15.4 | p = 0.82 |
| Dust | 279 | 46 | 16.5 | |
| Chicken type | ||||
| Layers | 292 | 60 | 20.6 | p = 0.003 |
| Broilers | 266 | 29 | 10.9 | |
| Age category | ||||
| Broiler Starter | 90 | 11 | 12.2 | p = 0.78 |
| Broiler Finisher | 176 | 18 | 10.2 | |
| Chicks | 28 | 6 | 21.4 | p = 0.19 |
| Growers | 50 | 5 | 10.0 | |
| Layers | 212 | 49 | 23.1 | |
| Spent layers | 2 | 0 | 0.0 |
Serotypes identified in poultry flocks
Characteristics of genomes submitted to in silico serotype prediction, and which failed the quality check, are shown in S1 Table. Seventy-four isolates were sequenced, and twenty-three serotypes, all belonging to S. enterica subspecies enterica were predicted from this analysis. Fourteen isolates could not be assigned serotypes by SeqSero2, but their serotype was predicted by SISTR. One isolate was assigned the same antigenic formula, but both pipelines did not predict the serotype. Seqsero2 uses the new antigenic numeric designation for O antigen, while SISTR use letters for O antigen nomenclature. Multiple serotype predictions were observed for four isolates, while two isolates had double prediction with 66 isolates having a unique serotype assigned. Cohen’s Kappa test was run to determine if there was an agreement between SeqSero2 and SISTR serotype predictions. There was a substantial agreement between the two pipelines (k = 0.76, p < 0.005). The serotypes and ST types obtained for individual isolates are shown in the S2 Table. Fifteen isolates, whose STs could not be predicted by SISTR, were assigned STs by Enterobase. All strains from same serotype belonged to a single ST. Among the 74 strains, S. Kentucky (ST-198) and S. Isangi (ST-216) appeared with the highest prevalence, 32.8% and 11% respectively, while S. Poona (ST-308), S. Virchow (ST-6166) and S. Waycross (ST-7745) were among the serotypes with lowest frequencies (1.4%) observed (Table 3).
Table 3. Frequency distribution of Salmonella serotypes identified at Nigerian poultry farms.
| S/N | Serotypes | Number of strains (n = 74) | Percentage (%) |
|---|---|---|---|
| 1 | S. Abadina | 2 | 2.7 |
| 2 | S. Aberdeen | 1 | 1.4 |
| 3 | S. Alachua | 1 | 1.4 |
| 4 | S. Birmingham | 1 | 1.4 |
| 5 | S. Bradford | 1 | 1.4 |
| 6 | S. Chester | 2 | 2.7 |
| 7 | S. Chomedey | 1 | 1.4 |
| 8 | S. Colindale | 1 | 1.4 |
| 9 | S. Corvalis | 2 | 2.7 |
| 10 | S. Esen | 1 | 1.4 |
| 11 | S. Give | 1 | 1.4 |
| 12 | S. Isangi | 8 | 10.8 |
| 13 | S. Ituri | 2 | 2.7 |
| 14 | S. Kentucky | 24 | 32.4 |
| 15 | S. Larochelle | 4 | 5.4 |
| 16 | S. Menston | 1 | 1.4 |
| 17 | S. Muenster | 4 | 5.4 |
| 18 | S. Poona | 1 | 1.5 |
| 19 | S. Schwarzengrund | 4 | 5.4 |
| 20 | S. Takoradi | 6 | 8.1 |
| 21 | S. Telelkebir | 3 | 4.1 |
| 22 | S. Virchow | 1 | 1.4 |
| 23 | S. Waycross | 1 | 1.4 |
| 24 | -:z13,z28:I,z13,z28 | 1 | 1.4 |
Serotyping remains the first step to characterize Salmonella isolates [5]. However, the traditional phenotypic method for serotyping is logistically challenging, as it requires the use of more than 150 specific antisera and well-trained personnel to interpret the results, and it may show low performance due to weak or non-specific agglutination, auto-agglutination or loss of antigen expression [38], which may lead to delay in rapid identification and false prediction of serovars involved in an outbreak. Alternative methods based on PCR amplification of specific genomic regions of O and H antigens were developed [39]. In view of this, we evaluated PCR based method [29] for serotyping. The result of serotype-specific PCR for S. Enteritidis showed that 13/73 isolates were S. Enteritidis, however, these were assigned different serotypes by WGS (four assigned to S. Kentucky, two to S. Chester and seven to other different serotypes). No strain was found positive in the S. Typhimurium-specific PCR (S2 Table).
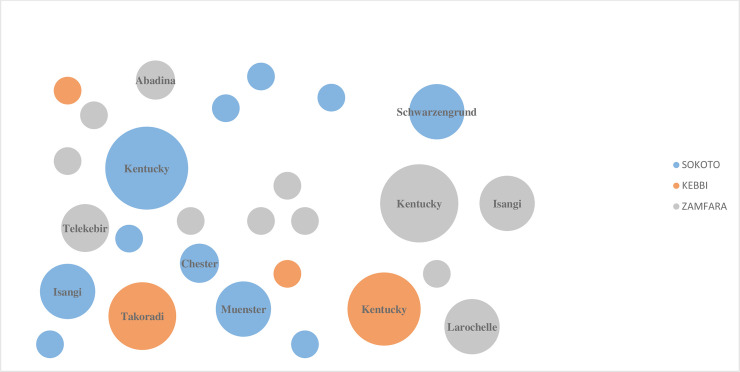
Spatial variation in the distribution of serotypes was evident. S. Larochelle (ST-22), S. Abadina and S. Telekebir (ST-2222) were exclusively identified in Zamfara state, while S. Schwarzengrund (ST-96) and S. Muenster (ST-321) were only identified in Sokoto state. Likewise, S. Takoradi (ST-531) and S. Poona (ST-308) were only identified in Kebbi state. However, S. Kentucky appeared with the highest prevalence in all three states and S. Isangi was common in Sokoto and Zamfara states (Fig 1).
Fig 1. Spatial bubble graph description of variation of Salmonella serotypes identified from poultry farms in different regions of Nigeria (colour marked).
The relative size of the bubble indicates the relative number of strains reported in that particular serovar.
Risk factors for presence of Salmonella in poultry farms
In the first univariate analysis of covariates from farm data, five factors were significantly associated with prevalence of Salmonella at the farm (p < 0.05); i.e. production system, report of salmonellosis outbreak in neighbouring farm, on-farm disposal of poultry waste, proximity to other poultry farms and presence of other livestock at the farm. In contrast, fencing of farm, provision of boot disinfection and staff lavatory were negatively associated with the prevalence of Salmonella (Table 4).
Table 4. Univariate analysis of variables associated with Salmonella infection in poultry farms in Nigeria.
| Variables | Responses (n = 65) | Positive for Salmonella (%) | Estimate ± SE | p-value |
|---|---|---|---|---|
| Production system | ||||
| Deep litter | 19 | 29.2 | 1.89±0.56 | 0.000713 |
| Battery cage | 8 | 12.3 | ||
| Previous outbreaks | ||||
| Yes | 27 | 41.5 | 20.46±2069.61 | 0.992 |
| No | 0 | 0 | ||
| Frequency of outbreak | ||||
| None | 0 | 0 | -20.95± 2109.0 | 0.9921 |
| Once | 3 | 4.6 | -1.90±0.92 | 0.0391 |
| Twice | 8 | 12.3 | -0.41±0.89 | 0.6442 |
| More | 16 | 24.6 | ||
| Outbreaks at neighbouring farms | ||||
| Yes | 17 | 26.2 | 2.99± 0.72 | 3.48e-05 |
| No | 10 | 22.2 | ||
| Farm fenced | ||||
| Yes | 6 | 9.2 | -3.40±0.70 | 1.37e-06 |
| No | 21 | 32.3 | ||
| Waste management | ||||
| On farm | 24 | 36.9 | 3.75±0.76 | 7.09e-07 |
| Off farm | 3 | 4.6 | ||
| Presence of other livestock | ||||
| Yes | 24 | 36.9 | 3.40±0.73 | 3.2e-06 |
| No | 3 | 4.6 | ||
| Proximity with farms (~1 km) | ||||
| Yes | 20 | 30.8 | 2.08±0.57 | 0.000286 |
| No | 7 | 10.8 | ||
| Disinfection of boots | ||||
| Yes | 3 | 4.6 | -3.97±0.78 | 3.43e-07 |
| No | 24 | 36.9 | ||
| Lavatory | ||||
| Yes | 2 | 3.1 | -3.85±0.84 | 4.14e-06 |
| No | 25 | 38.5 | ||
| Cleaning frequency | ||||
| Weekly | 1 | 1.5 | -3.72±1.08 | 0.000603 |
| Yearly | 7 | 10.8 | 18.11± 2465.3 | 0.994140 |
| Monthly | 19 | 29.2 |
In the second step logistic regression analysis, on-farm waste disposal and presence of other livestock in a farm showed statistically significant association with Salmonella infection. Using the logit model, the positive coefficient of the estimates indicated that disposing poultry waste on farm was associated with a three-fold higher chance that the farm was positive for Salmonella, while presence of other livestock increased the log odds by 2.6 units (Table 5).
Table 5. Logistic regression model of risk factors for presence of Salmonella in farms in Nigeria.
| Predictors | Estimate | ± SE | p value |
|---|---|---|---|
| Intercept | -5.0811 | 1.4741 | 0.000567 |
| Production system | |||
| Battery cage | 1.4472 | 1.1465 | 0.206834 |
| Neighbouring outbreak | |||
| Yes | 1.6299 | 1.2170 | 0.180491 |
| Waste management | |||
| On farm | 3.2436 | 1.1710 | 0.005605 |
| Presence of other livestock | |||
| Yes | 2.6157 | 1.1001 | 0.017425 |
| Proximity with farms (~1 km) | |||
| Yes | 0.7638 | 1.1249 | 0.497120 |
Discussion
In this study, a high farm prevalence (47.9%) of Salmonella infection was observed in commercial poultry farms in Nigeria. The results confirms observations from other parts of Nigeria by [21] who showed 43.6% farm prevalence in commercial layer farms. Relative high farm prevalence have also been reported in other sub-Saharan countries such as Ghana (44.0%), Uganda (20.7%), and Ethiopia (14.6%) [40–42] and likewise in developing Asian countries with report of 46.3% and 18% prevalence in central Vietnam and Bangladesh respectively [3, 43]. This is in contrast to many developed countries like Poland, where the total percentage of infected flocks was 1.57%, and where a decrease in prevalence of Salmonella spp. in broiler chickens was observed from 2.19% in 2014 to 1.22% in 2016. In Denmark, the prevalence for Salmonella infection poultry has been very low (0% to 1.8%) in the last decade, with the highest flock prevalence of 2.6% recorded in 2018 [44]. The reduction in European member countries can be attributed to implementation of specific control programmes [45], which are lacking in developing countries like Nigeria. Sample level prevalence (15.9%) of Salmonella from this study was similar to previously reported prevalence in other parts of Nigeria by Fagbamila et al., (2017) but higher than reported by Eguale (2018) (14.1% and 4.7% sample prevalence, respectively). Large scale farms were found to have higher Salmonella sample prevalence compared to other categories of farm levels, indication that once large farms were infected, the infection became more widespread in this farm type. Adesiyun et al. [46] observed a similar tendency for large farms compared to other farm categories from Caribbean countries. This might be attributed to large number of the flock making it difficult for the farmer to adhere to strict farm bio-securities and good farm management practices. The observations is not surprising, since there is conclusive evidence by European Food Safety Authority that larger poultry farms have higher chances of increased occurrence, persistence and spread of Salmonella [47, 48]. Furthermore, layer flocks, which spend longer time in the poultry house, had higher prevalence of Salmonella infection compared with broiler flocks. Wierup et al. [49] have also showed a substantially higher prevalence of Salmonella in layer flocks than in broilers among outdoor and indoor housing system.
A high number of Salmonella serotypes were observed in the farms investigated suggesting either a wide diversity of sources for introduction of Salmonella into the farms, or that common sources (such as contaminated feed) can contain different serotypes over time. Reports from other countries have also showed diversity in serotypes of non- typhoidal Salmonella in poultry farms [41, 42, 50–52]. Notably, S. Enteritidis was absent. This may be because the available vaccine used against Salmonella Gallinarum confers cross protection against other group D-strains [21, 53]. Also S. Typhimurium, which is commonly associated with poultry [54], was not observed in this study. This confirms observations by Fagbamila et al., (2017) and Useh et al., (2016) that these two serotypes play marginal role in the poultry industry in Nigeria.
S. Kentucky was the most commonly observed serotype. This serotype apparently has poultry as the main reservoir [55, 56], was also isolated from an health cattle [57]. And it has, over the years, emerged as a global zoonotic pathogen [58]. Human illnesses caused by this pathogen in North America and Europe are typically associated with a history of travel to Africa, Southeast Asia, and the Middle East, where this pathogen is established in poultry [59].
In general, the study observed predominantly Salmonella C group of the WKL scheme with few other members of B, E, G, O and S groups. Commonly isolated serotypes, besides S. Kentucky (ST-198), included S. Schwarzengrund (ST-96), S. Muenster, S. Poona, S. Isangi, S. Chester and S. Virchow. S. Schwarzengrund has been recorded from human in Denmark and the United States, where several isolates have shown multidrug resistance [54]. Recently it was isolated from diarrheal patients in a food poisoning event in China [60]. It was the fifth most common serovar isolated from retail meat in the United States in 2004, associated exclusively with poultry products, and other studies also suggest that poultry could be the most common reservoir [54]. S. Muenster has mainly been associated with salmonellosis in cattle [61]. In the current study, presence of other livestock in a farm was identified as a risk factor for Salmonella occurrence in poultry, and it may be that isolation in poultry is associated with horizontal transmission from other livestock. This serotype was associated with a nationwide outbreak of gastrointestinal illness in France, 2008 [62]. S. Poona has been reported from multistate outbreak in United States in 2015–2016 and was linked with the consumption of cucumber [55]. Reports from poultry are not common. Similarly, S. Isangi was isolated from a nosocomial infection outbreaks [63], while S. Chester accounted for 0.1% of all annual human salmonellosis cases notified in the EU/EEA [64]. S. Chester was also the second most common serotype in poultry, in 2010, in Burkina Faso [65]. S. Virchow is a serotype associated with poultry [66] and was reported to cause typhoid-like illness with fever and altered consciousness in human blood and stool culture [67]. This study also observed spatial variation in the distribution of serotypes, with some serotypes dominating a particular geographical area. This may reflect an ecologic niche established by those serotypes restricting them to a particular geographical region [68]. Similarly reported by Li et al. [69] and Pointon et al. [70] observed the dominance of one serovar over others in a particular geographical area.
Since Public Health England implemented whole genome sequencing (WGS) as a routine typing tool for public health surveillance of Salmonella [7], the use of WGS data for Salmonella serotyping has increased steadily. The method depends on publicly available databases. There was a substantial agreement between SeqSero2 and SISTR predictions (k = 0.76), and all the serotypes predicted by SeqSero2 were adequately predicted by SISTR. However, 14 isolates could not be predicted by SeqSero2 due to problems with adequate identification of O antigens. Six of these isolates were assigned multiple serovars in SISTR, while the serotype of the remaining isolates was resolved by this prediction too. Diep et al. (2019) likewise observed (1%) incomplete predictions of serotype when SeqSero2 was used, and explained this to be due to the same antigenic formula shared by strains from different subspecies, and that some serotypes in the WKL scheme require additional phenotypes for differentiation. Additionally, some serotypes in the WKL scheme differ only by minor epitopes of the same O antigen group. SISTR, in addition to using somatic (O) and flagella (H), utilizes the 330 genes in the SISTR cgMLST scheme, which provide an approximation of the genetic distance between serovars. This approximation is useful for disambiguating serovars with similar antigenic formula [16]. A recent study which assessed the performance of in silico serotyping of Salmonella spp. found the best performing prediction tool to be SISTR with 94% accuracy, followed by SeqSero2 (87%) [71]. However, SISTR could not assign ST types to some isolates, and these were assigned by Enterobase platform. This could be attributed to the fact that, Enterobase MLST database is synchronized and updated daily from pubMLST and other public databases [72], making the platform a more robust portal to get STs for large number of isolates.
There was discordant between the serotypes assigned by both in silico pipelines and the result from PCR serotyping. This may be due to the fact that, the primers (Salmonella difference fragment, Sdf I) we used to amplify our strains could also anneal to other genomic region in other serotypes. This could be explained by the report of Tennant et al. (2010) at the initial validation of the primers that, observed faint band amplicon products of the size of Sdf I in S. Meleagridis and S Livingstone. The Sdf genes was reported to be absent in only 34 serovars of Salmonella [32], so there is every possibility that some of these serotypes are among the remaining Salmonella serovars that possess the Sdf gene. The PCR-based method may be particularly unsuitable for assessing serotypes of livestock, wild-life and environmental isolates, as diverse serovars are often prevalent in these niches.
The finding from this study showed that Salmonella occurrence in poultry farms was influenced by several risk factors. In the final multivariate modeling, practicing deep liter system was observed as an important risk factor for the prevalence of Salmonella infection. The possible explanation could be that farmers seldom clean their deep litter poultry pen, which may lead to the persistent of Salmonella in poultry litter. Survival and persistence of Salmonella has been observed for 18 months in poultry litter [73, 74] which might result in higher chances of Salmonella infection than in battery cage system. A Study conducted by Mollenhorst et al. [75] showed that farms on deep litter system has a significant increased risk of Salmonella infection. Furthermore, farms located with close proximity with other farms and with report of neighboring farms having outbreaks of poultry salmonellosis was observed to have significantly increased risk of Salmonella infection. This could be due to personnel interaction and sharing of farm equipment, which could possibly introduce bacteria through contaminated tools or persons as previously described by Namata et al. (2009). Airborne transmission could also account for this risk factor, even though, based on available literature, aerosols do not appear to be important in the spread of salmonellosis. It has been earlier speculated that reported that Salmonella could become airborne, remain viable in the air and get transmitted among livestock over short distances [76, 77].
Improper waste disposal was observed to be at higher risk for infection with Salmonella in poultry farm as it allow for possible re-introduction of Salmonella through fomites into the poultry pen after cleaning. Furthermore, presence of other livestock in the farm was also identified as a risk factor to Salmonella prevalence. This could simply be explained by detection of serotypes that were associated with other farm animals like S. Muenster; a serotype which is associated with cattle [78]. This particular finding completely agrees with the study conducted by Djeffal et al. [79] who observed the presence of other livestock in a poultry farm as a risk factor to Salmonella infection.
Conclusion
Taken together, a high prevalence of Salmonella was observed in commercial poultry farms in Nigeria. Importantly, based on WGS data obtained in this study, we showed that a diverse non-typhoidal Salmonella serotypes circulate in commercial poultry farms in the study area with S. Kentucky (ST-198) having the highest prevalence and the widest geographical coverage. We also showed that WGS based serotyping with SISTR platform had higher chance of assigning serotypes than SeqSero2. Finally, presence of other livestock on farms and improper poultry waste-disposal have been identified as factors that increases the risk of having Salmonella infection in a farm.
Supporting information
(DOCX)
(DOCX)
(XLSX)
(XLSX)
(DOCX)
(XLSX)
Data Availability
The draft genome sequences are available at the European Nucleotide Archive under study accession number PRJEB37477 (secondary accession ERP120792) and accession number for each genome is indicated in S2 File.
Funding Statement
This work was part-supported by an African Research Leader Award to INO from the UK Medical Research Council (MRC) and the UK Department for International Development (DFID) under the MRC/DFID Concordat agreement that is also part of the EDCTP2 programme supported by the European Union.
References
- 1.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ, et al. The Global Burden of Nontyphoidal Salmonella Gastroenteritis. Clin Infect Dis. 2010. [DOI] [PubMed] [Google Scholar]
- 2.Leekitcharoenphon P, Raufu I, Nielsen MT, Rosenqvist Lund BS, Ameh JA, Ambali AG, et al. Investigating Salmonella Eko from various sources in Nigeria by whole genome sequencing to identify the source of human infections. PLoS One. 2016;11(5):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barua H, Biswas PK, Olsen KEP, Christensen JP. Prevalence and characterization of motile Salmonella in commercial layer poultry farms in Bangladesh. PLoS One. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foley SL, Nayak R, Hanning IB, Johnson TJ, Han J, Ricke SC. Population dynamics of Salmonella enterica serotypes in commercial egg and poultry production. Applied and Environmental Microbiology. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diep B, Barretto C, Portmann AC, Fournier C, Karczmarek A, Voets G, et al. Salmonella Serotyping; Comparison of the Traditional Method to a Microarray-Based Method and an in silico Platform Using Whole Genome Sequencing Data. Front Microbiol. 2019;10(November). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langridge GC, Fookesa M, Connora TR, Feltwella T, Feaseya N, Parsonsb BN, et al. Patterns of genome evolution that have accompanied host adaptation in Salmonella. Proc Natl Acad Sci U S A. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashton PM, Nair S, Peters TM, Bale JA, Powell DG, Painset A, et al. Identification of Salmonella for public health surveillance using whole genome sequencing. PeerJ. 2016;2016(4):1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ibrahim GM, Morin PM. Salmonella serotyping using whole genome sequencing. Front Microbiol. 2018;9(DEC):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang S, Yin Y, Jones MB, Zhang Z, Kaiser BLD, Dinsmore BA, et al. Salmonella serotype determination utilizing high-throughput genome sequencing data. J Clin Microbiol. 2015;53(5):1685–92. 10.1128/JCM.00323-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grimont PP, Weill FF-X. Antigenic Formulae of the Salmonella Serovars. 9th Edition, World Health Organization Collaborating Center for Reference and Research on Salmonella, Institut Pasteur, Paris. WHO Collaborating Centre for Reference and Research on Salmonella. 2007.
- 11.Yoshida CE, Kruczkiewicz P, Laing CR, Lingohr EJ, Gannon VPJ, Nash JHE, et al. The salmonella in silico typing resource (SISTR): An open web-accessible tool for rapidly typing and subtyping draft salmonella genome assemblies. PLoS One. 2016;11(1):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, Payne M, Lan R. In silico identification of serovar-specific genes for salmonella serotyping. Front Microbiol. 2019;10(APR):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gymoese P, Sørensen G, Litrup E, Olsen JE, Nielsen EM, Torpdahl M. Investigation of outbreaks of Salmonella enterica serovar typhimurium and its monophasic variants using whole-genome sequencing, Denmark. Emerg Infect Dis. 2017;23(10):1631–9. 10.3201/eid2310.161248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pornsukarom S, Van Vliet AHM, Thakur S. Whole genome sequencing analysis of multiple Salmonella serovars provides insights into phylogenetic relatedness, antimicrobial resistance, and virulence markers across humans, food animals and agriculture environmental sources. BMC Genomics. 2018;19(1):1–14. 10.1186/s12864-017-4368-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tay MYF, Pathirage S, Chandrasekaran L, Wickramasuriya U, Sadeepanie N, Waidyarathna KDK, et al. Whole-Genome Sequencing Analysis of Nontyphoidal Salmonella enterica of Chicken Meat and Human Origin Under Surveillance in Sri Lanka. Foodborne Pathog Dis. 2019;16(7):531–7. 10.1089/fpd.2018.2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robertson J, Yoshida C, Kruczkiewicz P, Nadon C, Nichani A, Taboada EN, et al. Comprehensive assessment of the quality of Salmonella whole genome sequence data available in public sequence databases using the Salmonella in silico Typing Resource (SISTR). Microb genomics. 2018;4(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yachison CA, Yoshida C, Robertson J, Nash JHE, Kruczkiewicz P, Taboada EN, et al. The validation and implications of using whole genome sequencing as a replacement for traditional serotyping for a national Salmonella reference laboratory. Front Microbiol. 2017;8(JUN):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adene D F and Oguntade A. Poultry sector review: Nigeria: FOOD Agric Organ UNITED NATIONS; 2008;1–95. [Google Scholar]
- 19.Food and Agriculture Organization. Livestock and livelihoods spotlight NIGERIA: Cattle and Poutry Sectors. 2018; Available from: http://www.fao.org/3/CA2149EN/ca2149en.pdf
- 20.Bettridge JM, Lynch SE, Brena MC, Melese K, Dessie T, Terfa ZG, et al. Infection-interactions in Ethiopian village chickens. Prev Vet Med. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fagbamila IO, Barco L, Mancin M, Kwaga J, Ngulukun SS, Zavagnin P, et al. Salmonella serovars and their distribution in Nigerian commercial chicken layer farms. PLoS One. 2017;12(3):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.M M., M L.U., A A.-G., M A.U., A S., B L. Prevalence of Salmonella associated with chick mortality at hatching and their susceptibility to antimicrobial agents. Vet Microbiol. 2010. [DOI] [PubMed] [Google Scholar]
- 23.Mshelbwala FM, Ibrahim NDG, Saidu SN, Azeez AA, Akinduti PA, Kwanashie CN, et al. Motile Salmonella serotypes causing high mortality in poultry farms in three South-Western States of Nigeria. Vet Rec Open. 2017;4(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raufu IA, Fashae K, Ameh JA, Ambali AG, Ogunsola FT, Coker AO, et al. Persistence of fluoroquinolone-resistant Salmonella enterica serovar Kentucky from poultry and poultry sources in Nigeria. J Infect Dev Ctries. 2014. [DOI] [PubMed] [Google Scholar]
- 25.Statistics NB of. Chapter 1 1.0 Population Projection. 2018;(May):7–8.
- 26.Mohammed S. Vegetation density and diversity in the dryland of northwestern nigeria. 2018;4(1):195–208. [Google Scholar]
- 27.INTERNATIONAL STANDARD Horizontal method for the detection, enumeration and serotyping of. 2017;2017.
- 28.Waghamare RN, Paturkar AM, Vaidya VM, Zende RJ, Dubal ZN, Dwivedi A, et al. Phenotypic and genotypic drug resistance profile of Salmonella serovars isolated from poultry farm and processing units located in and around Mumbai city, India. Vet World. 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tennant SM, Diallo S, Levy H, Livio S, Sow SO, Tapia M, et al. Identification by PCR of non-typhoidal Salmonella enterica serovars associated with invasive infections among febrile patients in Mali. PLoS Negl Trop Dis. 2010;4(3):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahmed AO, Raji MA, Mamman PH, Kwanashie CN, Raufu IA, Aremu A, et al. Salmonellosis: Serotypes, prevalence and multi-drug resistant profiles of Salmonella enterica in selected poultry farms, Kwara State, North Central Nigeria. Onderstepoort J Vet Res. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vanegas RA, Joys TM. Molecular analyses of the phase-2 antigen complex 1,2,.. of Salmonella spp. J Bacteriol. 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agron PG, Walker RL, Kinde H, Sawyer SJ, Hayes DC, Wollard J, et al. Identification by Subtractive Hybridization of Sequences Specific for Salmonella enterica Serovar Enteritidis. Appl Environ Microbiol. 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gurevich A, Saveliev V, Vyahhi N, Tesler G. F1000Prime recommendation of: QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng X, den Bakker HC, Hendriksen RS. Genomic Epidemiology: Whole-Genome-Sequencing–Powered Surveillance and Outbreak Investigation of Foodborne Bacterial Pathogens. Annu Rev Food Sci Technol. 2016. [DOI] [PubMed] [Google Scholar]
- 35.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. [BMC Bioinformatics. 2009]—PubMed—NCBI. BMC Bioinformatics. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Achtman M, Wain J, Weill FX, Nair S, Zhou Z, Sangal V, et al. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog. 2012;8(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Team RC. R: A Language and Environment for Statistical Computing. Vienna, Austria. 2019.
- 38.Wattiau P, Boland C, Bertrand S. Methodologies for Salmonella enterica subsp. Enterica Subtyping: Gold Standards and Alternatives. Appl Environ Microbiol. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herrera-León S, Ramiro R, Arroyo M, Díez R, Usera MA, Echeita MA. Blind comparison of traditional serotyping with three multiplex PCRs for the identification of Salmonella serotypes. Res Microbiol. 2007. [DOI] [PubMed] [Google Scholar]
- 40.Odoch T, Wasteson Y, L’Abée-Lund T, Muwonge A, Kankya C, Nyakarahuka L, et al. Prevalence, antimicrobial susceptibility and risk factors associated with non-typhoidal Salmonella on Ugandan layer hen farms. BMC Vet Res. 2017;13(1):1–10. 10.1186/s12917-016-0931-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eguale T. Non-typhoidal Salmonella serovars in poultry farms in central Ethiopia: Prevalence and antimicrobial resistance. BMC Vet Res. 2018;14(1):1–8. 10.1186/s12917-017-1323-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andoh LA, Dalsgaard A, Obiri-Danso K, Newman MJ, Barco L, Olsen JE. Prevalence and antimicrobial resistance of Salmonella serovars isolated from poultry in Ghana. Epidemiol Infect. 2016;144(15):3288–99. 10.1017/S0950268816001126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lettini AA, Vo Than T, Marafin E, Longo A, Antonello K, Zavagnin P, et al. Distribution of Salmonella Serovars and Antimicrobial Susceptibility from Poultry and Swine Farms in Central Vietnam. Zoonoses Public Health. 2016. [DOI] [PubMed] [Google Scholar]
- 44.DTU F. Annual Report on Zoonoses in Denmark 2018. 2018;1–64.
- 45.Food E, Authority S. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2014. EFSA J. 2015;13(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adesiyun A, Webb L, Musai L, Louison B, Joseph G, Stewart-Johnson A, et al. Survey of salmonella contamination in chicken layer farms in three caribbean countries. J Food Prot. 2014;77(9):1471–80. 10.4315/0362-028X.JFP-14-021 [DOI] [PubMed] [Google Scholar]
- 47.Koutsoumanis K, Allende A, Alvarez-Ordóñez A, Bolton D, Bover-Cid S, Chemaly M, et al. Salmonella control in poultry flocks and its public health impact. EFSA J. 2019;17(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Namata H, Welby S, Aerts M, Faes C, Abrahantes JC, Imberechts H, et al. Identification of risk factors for the prevalence and persistence of Salmonella in Belgian broiler chicken flocks. Prev Vet Med. 2009;90(3–4):211–22. 10.1016/j.prevetmed.2009.03.006 [DOI] [PubMed] [Google Scholar]
- 49.Wierup M, Wahlström H, Lahti E, Eriksson H, Jansson DS, Odelros Å, et al. Occurrence of Salmonella spp.: A comparison between indoor and outdoor housing of broilers and laying hens. Acta Vet Scand. 2017;59(1):1–8. 10.1186/s13028-016-0274-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Useh NM, Ngbede EO, Akange N, Thomas M, Foley A, Keena C, et al. Draft Genome Sequences of 37 Salmonella enterica Strains Isolated from Poultry Sources in Nigeria. 2016;4(3):15–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vinueza-Burgos C, Cevallos M, Ron-Garrido L, Bertrand S, De Zutter L. Prevalence and diversity of Salmonella serotypes in ecuadorian broilers at slaughter age. PLoS One. 2016;11(7):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raufu I, Bortolaia V, Svendsen CA, Ameh JA, Ambali AG, Aarestrup FM, et al. The first attempt of an active integrated laboratory-based Salmonella surveillance programme in the north-eastern region of Nigeria. J Appl Microbiol. 2013. [DOI] [PubMed] [Google Scholar]
- 53.Gast RK. Serotype-Specific and Serotype-Independent Strategies for Preharvest Control of Food-Borne Salmonella in Poultry. Avian Dis. 2007. [DOI] [PubMed] [Google Scholar]
- 54.Aarestrup FM, Hendriksen RS, Lockett J, Gay K, Teates K, McDermott PF, et al. International spread of multidrug-resistant Salmonella Schwarzengrund in food products. Emerg Infect Dis. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laughlin M, Bottichio L, Weiss J, Higa J, McDonald E, Sowadsky R, et al. Multistate outbreak of Salmonella Poona infections associated with imported cucumbers, 2015–2016. Epidemiol Infect. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vosik D, Tewari D, Dettinger L, M’Ikanatha NM, Shariat NW. CRISPR Typing and Antibiotic Resistance Correlates with Polyphyletic Distribution in Human Isolates of Salmonella Kentucky. Foodborne Pathog Dis. 2018. [DOI] [PubMed] [Google Scholar]
- 57.Rauch HE, Vosik D, Kariyawasam S, M’ikanatha N, Shariat NW. Prevalence of Group I Salmonella Kentucky in domestic food animals from Pennsylvania and overlap with human clinical CRISPR sequence types. Zoonoses Public Health. 2018. [DOI] [PubMed] [Google Scholar]
- 58.Bai J, Zhan Z, Wen J, Liao M, Zhang J. Ciprofloxacin-Resistant Salmonella enterica Serovar Kentucky ST198 in Broiler Chicken Supply Chain and. 2016;2010–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Le Hello S, Harrois D, Bouchrif B, Sontag L, Elhani D, Guibert V, et al. Highly drug-resistant Salmonella enterica serotype Kentucky ST198-X1: A microbiological study. Lancet Infect Dis. 2013. [DOI] [PubMed] [Google Scholar]
- 60.Du X, Jiang X, Ye Y, Guo B, Wang W, Ding J, et al. Next generation sequencing for the investigation of an outbreak of Salmonella Schwarzengrund in Nanjing, China. Int J Biol Macromol [Internet]. 2018;107(PartA):393–6. Available from: 10.1016/j.ijbiomac.2017.09.005 [DOI] [PubMed] [Google Scholar]
- 61.Radke BR, McFall M, Radostits SM. Salmonella Muenster infection in a dairy herd. Can Vet J. 2002. [PMC free article] [PubMed] [Google Scholar]
- 62.van Cauteren D, Jourdan-da Silva N, Weill FX, King L, Brisabois A, Delmas G, et al. Outbreak of Salmonella enterica serotype Muenster infections associated with goat’s cheese, France, March 2008. Euro Surveill. 2009. [DOI] [PubMed] [Google Scholar]
- 63.Suleyman G, Tibbetts R, Perri MB, Vager D, Xin Y, Reyes K, et al. Nosocomial Outbreak of a Novel Extended-Spectrum β-Lactamase Salmonella enterica Serotype Isangi among Surgical Patients. Infect Control Hosp Epidemiol. 2016. [DOI] [PubMed] [Google Scholar]
- 64.Fonteneau L, Jourdan Da Silva N, Fabre L, Ashton P, Torpdahl M, Müller L, et al. Multinational outbreak of travel-related Salmonella Chester infections in Europe, summers 2014 and 2015. Eurosurveillance. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kagambèga A, Lienemann T, Aulu L, Traoré AS, Barro N, Siitonen A, et al. Prevalence and characterization of Salmonella enterica from the feces of cattle, poultry, swine and hedgehogs in Burkina Faso and their comparison to human Salmonella isolates. BMC Microbiol. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bonalli M, Stephan R, Käppeli U, Cernela N, Adank L, Hächler H. Salmonella enterica serotype Virchow associated with human infections in Switzerland: 2004–2009. BMC Infect Dis. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eckerle I, Zimmermann S, Kapaun A, Junghanss T. Salmonella enterica serovar virchow bacteremia presenting as typhoid-like illness in an immunocompetent patient. J Clin Microbiol. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Galanis E, Lo Fo Wong DMA, Patrick ME, Binsztein N, Cieslik A, Chalermchaikit T, et al. Web-based surveillance and global Salmonella distribution, 2000–2002. Emerg Infect Dis. 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li X, Payne JB, Santos FB, Levine JF, Anderson KE, Sheldon BW. Salmonella populations and prevalence in layer feces from commercial high-rise houses and characterization of the Salmonella isolates by serotyping, antibiotic resistance analysis, and pulsed field gel electrophoresis. Poult Sci. 2007. [DOI] [PubMed] [Google Scholar]
- 70.Pointon A, Sexton M, Dowsett P, Saputra T, Kiermeier A, Lorimer M, et al. A baseline survey of the microbiological quality of chicken portions and carcasses at retail in two Australian states (2005 to 2006). J Food Prot. 2008. [DOI] [PubMed] [Google Scholar]
- 71.Uelze L, Borowiak M, Deneke C, Szabó I, Fischer J, Tausch SH, et al. Comparative assessment of the performance and accuracy of four open-source tools for in silico serotyping of Salmonella spp. based on whole-genome short read sequencing data. Appl Environ Microbiol. 2019;(December). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou Z, Alikhan NF, Mohamed K, Fan Y, Achtman M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Williams JE, Benson ST. Survival of Salmonella typhimurium in Poultry Feed and Litter at Three Temperatures. Avian Dis. 1978. [PubMed] [Google Scholar]
- 74.Wilkinson KG, Tee E, Tomkins RB, Hepworth G, Premier R. Effect of heating and aging of poultry litter on the persistence of enteric bacteria. Poult Sci. 2011; [DOI] [PubMed] [Google Scholar]
- 75.Mollenhorst H, Van Woudenbergh CJ, Bokkers EGM, De Boer IJM. Risk factors for Salmonella enteritidis infections in laying hens. Poult Sci. 2005;84(8):1308–13. 10.1093/ps/84.8.1308 [DOI] [PubMed] [Google Scholar]
- 76.Adell E, Moset V, Zhao Y, Jiménez-Belenguer A, Cerisuelo A, Cambra-López M. Comparative performance of three sampling techniques to detect airborne Salmonella species in poultry farms. Ann Agric Environ Med. 2014. [PubMed] [Google Scholar]
- 77.Davis M, Morishita TY. Relative Ammonia Concentrations, Dust Concentrations, and Presence of Salmonella Species and Escherichia coli Inside and Outside Commercial Layer Facilities. Avian Dis. 2005. [DOI] [PubMed] [Google Scholar]
- 78.Agency A and P health. Salmonella in Livestock Production in GB. 2019;(October):147 p.-147 p.
- 79.Djeffal S, Mamache B, Elgroud R, Hireche S, Bouaziz O. Prevalence and risk factors for Salmonella spp. contamination in broiler chicken farms and slaughterhouses in the northeast of Algeria. Vet World. 2018;11(8):1102–8. 10.14202/vetworld.2018.1102-1108 [DOI] [PMC free article] [PubMed] [Google Scholar]