Abstract
Dopamine (DA) signals in the striatum are critical for a variety of vital processes, including motivation, motor learning, and reinforcement learning. Striatal DA signals can be evoked by direct activation of inputs from midbrain DA neurons (DANs) as well as cortical and thalamic inputs to the striatum. In this study, we show that in vivo optogenetic stimulation of prelimbic (PrL) and infralimbic (IL) cortical afferents to the striatum triggers an increase in extracellular DA concentration, which coincides with elevation of striatal acetylcholine (ACh) levels. This increase is blocked by a nicotinic ACh receptor (nAChR) antagonist. Using single or dual optogenetic stimulation in brain slices from male and female mice, we compared the properties of these PrL/IL-evoked DA signals with those evoked by stimulation from midbrain DAN axonal projections. PrL/IL-evoked DA signals are undistinguishable from DAN evoked DA signals in their amplitudes and electrochemical properties. However, PrL/IL-evoked DA signals are spatially restricted and preferentially recorded in the dorsomedial striatum. PrL/IL-evoked DA signals also differ in their pharmacological properties, requiring activation of glutamate and nicotinic ACh receptors. Thus, both in vivo and in vitro results indicate that cortical evoked DA signals rely on recruitment of cholinergic interneurons, which renders DA signals less able to summate during trains of stimulation and more sensitive to both cholinergic drugs and temperature. In conclusion, cortical and midbrain inputs to the striatum evoke DA signals with unique spatial and pharmacological properties that likely shape their functional roles and behavioral relevance.
SIGNIFICANCE STATEMENT Dopamine signals in the striatum play a critical role in basal ganglia function, such as reinforcement and motor learning. Different afferents to the striatum can trigger dopamine signals, but their release properties are not well understood. Further, these input-specific dopamine signals have only been studied in separate animals. Here we show that optogenetic stimulation of cortical glutamatergic afferents to the striatum triggers dopamine signals both in vivo and in vitro. These afferents engage cholinergic interneurons, which drive dopamine release from dopamine neuron axons by activation of nicotinic acetylcholine receptors. We also show that cortically evoked dopamine signals have other unique properties, including spatial restriction and sensitivity to temperature changes than dopamine signals evoked by stimulation of midbrain dopamine neuron axons.
Keywords: DA release, dorsomedial striatum, fast-scan cyclic voltammetry, optogenetics, PFC
Introduction
The striatum receives dense innervation from midbrain DA neurons (DANs), which are the main source of DA in the striatum. DA plays a critical neuromodulatory role in regulating striatal circuitry and function (Surmeier et al., 2007; Gerfen and Surmeier, 2011; Burke et al., 2017). Disruptions in striatal DA levels are associated with many neurologic and psychiatric disorders, such as Parkinson's disease and substance abuse disorder (Gerfen, 2000; Luscher et al., 2020). DA is released from a fraction of the varicosities distributed along DAN axons, which contain specialized active zone-like sites (Sulzer et al., 2016; Liu et al., 2018; Liu and Kaeser, 2019). The axonal arborizations of DANs ramify extensively within the striatum, and a single dopamine axon can spread over a significant area (∼3% in average) of the striatum (Matsuda et al., 2009). DA released from axonal varicosities generates a rapid increase in local extracellular DA concentration. This extracellular increase leads to the activation of multiple types of DA receptors, which are localized in dendrites, somas, and presynaptic terminals. Ultimately, this impacts the network activity of the striatum and drives behavior output (Tritsch and Sabatini, 2012; Chuhma et al., 2014; Mamaligas et al., 2016; Burke et al., 2017; Shin et al., 2017; Lahiri and Bevan, 2020). This striatal DA signal is known to be triggered by two mechanisms. One mechanism involves action potential firing initiated at midbrain DAN somas, which propagates through dense axonal arborizations to reach active zone-like release sites in the striatum (Matsuda et al., 2009; Liu et al., 2018). In the other mechanism, DAN axons in the striatum are locally activated, independent of the action potential firing at DAN somas. This mode of DA release requires the activation of nAChRs expressed on DAN axons and synchronized activation of cholinergic interneurons (CINs), which is thought to give rise to local release of ACh (Cachope et al., 2012; Threlfell et al., 2012; Wang et al., 2014; Shin et al., 2015, 2017).
This local intrastriatal trigger of DA release has only recently been demonstrated and is gaining attention as it represents a newly discovered means for striatal DA modulation/transmission. However, little is known about the differential/unique biophysical and pharmacological properties of locally evoked versus DAN-evoked DA transmission. Recent in vitro studies using optogenetic stimulation and fast-scan cyclic voltammetry (FSCV) show that local DA signals can be triggered in the striatum by stimulation of glutamatergic inputs from thalamus (Threlfell et al., 2012; Kosillo et al., 2016; Johnson et al., 2017; Cover et al., 2019), motor cortex (Kosillo et al., 2016), or PFC (Mateo et al., 2017). These findings agree with previous literature showing in vivo DA signals in the striatum evoked by stimulation of PFC (Taber and Fibiger, 1993; Quiroz et al., 2016; Hill et al., 2018), hippocampus (Tritschler et al., 2018), or amygdala (Floresco et al., 1998). Furthermore, in vivo intrastriatal administration of glutamate and ACh was also shown to cause increases in extracellular DA concentration in the rat striatum (Giorguieff et al., 1976, 1977; Leviel et al., 1990; Shimizu et al., 1990). Together, this evidence supports an intrastriatal mechanism for DA signal generation that can be initiated by excitatory inputs to the striatum that require glutamate and ACh. While cortically evoked DA signals in the striatum have been reported, identifying the unique pharmacological and basic properties of these input-specific signals will allow us to selectively manipulate and target them, leading to a better understanding of their functional significance. In this study, we tackle this gap in knowledge using both in vivo microdialysis and in vitro FSCV with optogenetic stimulation. Particularly, we set up a novel approach using dual optogenetic stimulation in the same brain slices to input-specific evoke DA release and compare the properties of these DA signals in the striatum.
Materials and Methods
Animals
All animals used in the study were maintained in accordance with the guidelines of the National Institutes of Health Animal Care, and the animal research procedures were approved by the National Institute on Alcohol Abuse and Alcoholism Animal Care and Use Committee for mice and by the National Institute on Drug Abuse Intramural Research Program Animal Care and Use Committee for rats. Except for the in vivo microdialysis experiments, all experiments were conducted using male and female mice of C57BL/6 background. Heterozygote B6.SJL-Slc6a3tm1.1(cre)Bkmn/J mice (Backman et al., 2006) (The Jackson Laboratory, 006660) referred to as DATIRES-Cre+ mice and negative DATIRES-Cre− littermates considered as WT mice were used. To fluorescently label CINs, homozygote B6N.129S6(B6)-Chattm2(cre)Lowl/J mice (Rossi et al., 2011) (The Jackson Laboratory, 018957) were crossed with homozygote B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J mice (Madisen et al., 2010) (The Jackson Laboratory, 007914), and the progeny is referred to as CIN-tdTomato mice. For the microdialysis experiments, male Sprague Dawley albino rats (Charles River Laboratories) were used. Rats were housed individually for the first week after intracranial injection, until suture removal, after which the rats were housed 2 per cage. All animals were housed on a 12 h light/dark cycle (06:30 to 18:30 light) with food and water ad libitum.
Surgery and stereotaxic injection of AAV-ChR2 vectors
Injections were conducted as described previously (Adrover et al., 2014). Briefly, mice (5-6 weeks old) were anesthetized by inhalation of isoflurane-oxygen mixture and placed in a stereotaxic frame (David Kopf). Adeno-associated virus (AAV) vectors with Cre recombinase-dependent expression of Channelrhodopsin2 (ChR2) protein, AAV5-EF1a-DIO-hChR2(H134R)-EYFP (4 × 1012 IU/ml), were bilaterally injected into the VTA/SNc (AP: −3.30, ML: ±0.60, DV: −4.50) of DATIRES-Cre+ mice. The AAV vectors AAV5-CaMKII-hChR2(H134R)-EYFP (4 × 1012 IU/ml) or AAV5-Syn-ChrimsonR-tdTomato (1.7 × 1013 IU/ml), were injected in the PFC (PrL cortex, AP: 2.10, ML: ±0.35, DV: −2.30; IL cortex, AP: 1.90, ML: ±0.30, DV: −3.20) of negative littermates or DATIRES-Cre+ mice. All stereotaxic coordinates were from bregma (in mm) according to the mouse atlas by Franklin and Paxinos (2007); 300-500 nl for VTA/SNc and 100-200 nl for PFC were injected at a flow rate of 100 nl/min. Recordings were made after a minimum of 4 weeks of incubation. For mice injected in both VTA/SNc (DIO-ChR2-EYFP) and PFC (ChrimsonR-tdTomato), recordings were performed after a minimum of 8 weeks of incubation. Viral vectors were purchased from Gene Therapy Center Vector Core at University of North Carolina and Penn Vector Core at University of Pennsylvania.
Slice preparation
Mice were anesthetized and rapidly decapitated. Brains were quickly removed, mounted, and sliced using a vibratome (VT-1200S, Leica Microsystems) in an ice-cold cutting solution containing the following (in mm): 225 sucrose, 13.9 NaCl, 26.2 NaHCO3, 1 NaH2PO4, 1.25 glucose, 2.5 KCl, 0.1 CaCl2, 4.9 MgCl2, and 3 kynurenic acid. The sagittal slices (240 μm) were incubated for 20 min at 33°C in ACSF containing the following (in mm): 124 NaCl, 1 NaH2PO4, 2.5 KCl, 1.3 MgCl2, 2.5 CaCl2, 20 glucose, 26.2 NaHCO3, and 0.4 ascorbic acid, and kept in the dark at room temperature before use. Recording chamber was perfused at 2 ml/min with ACSF heated at 32°C using an inline heater (Harvard Apparatus).
FSCV and amperometry
FSCV was performed in the dorsal striatum. Carbon-fiber electrodes (CFEs) were prepared with a cylindrical carbon-fiber (7 μm diameter, ∼150 μm of exposed fiber) inserted into a glass pipette. Before use, the CFEs were conditioned with an 8-ms-long triangular voltage ramp (−0.4 to 1.2 and back to −0.4 V vs Ag/AgCl reference at 400 V/s) delivered every 15 ms. CFEs showing current >1.8 µA or <1.0 µA in response to the voltage ramp at ∼0.6 V were discarded. During the recording, the CFEs were held at −0.4 V versus Ag/AgCl, and the same triangular voltage ramp was delivered every 100 ms. DA transients were evoked by electrical or optical single pulse, or 5 pulses at 20 Hz stimulations. Using the same CFE and location, DA signals were evoked by alternating electrical and optical stimulations in some experiments. These data were combined with results from other experiments where either electrical or optical stimulation was used to evoke DA signals. For electrical stimulation, a glass pipette filled with ACSF was placed near the tip of the carbon fiber and a rectangular pulse (0.2 ms, 100 μA) was applied every 2 min. For optogenetic stimulation, a fiber-optic (200 μm diameter, 0.22 NA, ThorLabs) connected to a blue LED (470 nm, 1.8 mW of maximal output power measured at the tip of the fiber-optics, ThorLabs) was placed over the carbon fiber and light pulses (0.6 ms) were delivered every 2 min. For input-output curves, the widths of light pulse were 0.1, 0.2, 0.5, 1, 2, and 5 ms. For dual optogenetic recordings, two fiber-optics connected to a purple LED (420 nm, 3.0 mW, ThorLabs) and an orange LED (590 nm, 0.7 mW, ThorLabs), respectively, were placed over the carbon fiber. Light pulses (0.6 ms for 420 nm and 0.6-2 ms for 590 nm) were delivered in an alternating pattern every 2 min. Data were collected with a retrofit headstage (CB-7B/EC with 5 mΩ resistor) using a Multiclamp 700B amplifier (Molecular Devices) after low-pass filter at 3 kHz and digitized at 100 kHz using a DA board (NI USB-6229 BNC, National Instruments). Data acquisition and analysis were performed using a custom-written software, VIGOR, in Igor Pro (Wavemetrics) using mafPC (courtesy of MA Xu-Friedman). The current peak amplitudes of the evoked DA transients were converted to DA concentration according to the postexperimental calibration using 1-3 μm DA. Amperometric recordings were performed using the same carbon-fiber electrodes held at −0.4 V versus Ag/AgCl and a 30-s-long step to 0.6 V was applied. Single pulse or 5 pulses at 20 Hz stimulation were delivered at 20 s after switching to 0.6 V. Since we did not find major differences between PrL and IL in evoking DA transients, we combined the data obtained from two groups to represent DA transients evoked by PFC inputs (PFC-oDA).
Cell-attached recordings
Striatal CINs from CIN-TdTomato mice injected with ChR2-EYFP in the PFC were identified by fluorescence and confirmed by their characteristic spontaneous firing pattern. Cell-attached recordings were performed from CINs in the striatum using glass pipette electrodes with a resistance of ∼3-4 mΩ, filled with an internal solution containing the following (in mm): 120 cesium methanesulfonate, 20 CsCl, 10 HEPES, 0.2 EGTA, 10 sodium phosphocreatine, 4 Na2-ATP, and 0.4 Na-GTP, pH, 7.25 (290-310 mOsm), at a holding potential of 0 mV. To estimate the strength of connectivity to CINs, the lowest light intensity needed to reliably evoke action potentials in CINs was determined, ranging from 0.4 mW for “high” connectivity to 2.1 mW for “low” connectivity. The data were collected using a Multiclamp 700B amplifier after low-pass filter at 1 kHz and digitized at 5 kHz, using pClamp10 software (Molecular Devices). Spike fidelity was calculated as percentages of the number of AP evoked from five trials for both single pulse and trains stimulation.
In vivo microdialysis and optogenetic stimulation of the cortical inputs
Rats (80-90 g) received unilateral intracranial injection of AAV-CaMKIIa-hChR2(H134R)-EYFP (titer: 1012 IU/ml; Gene Therapy Center Vector Core at University of North Carolina) in the PrL and IL cortex (AP: 3.0 mm, ML: 0.5 mm, DV: −3.5 and −5 mm with respect to bregma). Two injection sites per hemisphere were used, and 300 nl of viral vector solution was delivered per site via a 105-μm-thick silica tubing injector coupled directly to a 1 μl syringe driven by an infusion pump (rate = 50 nl/min) during 10 min. The injector was left in place for an additional 10 min following each injection to allow for diffusion of the suspension. Ten weeks after viral vector injection, rats (350-400 g) underwent surgery for probe implantation according to previously published procedures (Quiroz et al., 2016). Briefly, a modified microdialysis probe with an embedded light-guiding optic fiber was implanted into the NAc shell (AP: 1.2 mm, ML: 0.5 mm, DV: −8.0 mm with respect to bregma). The probe was fixed to the skull with a stainless-steel screw and glass-ionomer dental cement. All surgical procedures were performed under anesthesia with 3 ml/kg of Equithesin (4.44 × g of chloralhydrate, 0.972 × g of Na pentobarbital, 2.124 × g of MgSO4, 44.4 ml of propylene glycol, 12 ml of ethanol, and distilled H2O up to 100 ml of final solution; National Institute on Drug Abuse Pharmacy). To build the microdialysis probe with embedded optic fiber, the tip of an optic fiber (105 μm diameter core, 0.22 NA) was sculpted into a conical shape using a Flaming-Brown pipette puller fitted with a custom platinum heating filament (Sutter Instruments) to allow for a larger area of stimulation. The conical optic fiber tip was embedded inside the microdialysis probe and implanted. Microdialysis experiments were performed in freely moving rats 24 h after probe implantation. Optical fiber was coupled to a 473 nm solid-state laser module and light stimulation was driven by a Grass S88 stimulator. Light stimulation was delivered for 20 min as trains of light pulses (2 ms pulse duration; at 100 Hz for 160 ms; trains repeats once per second; intensity = 5-8 mW at probe tip). ACSF containing (in mm) 144 NaCl, 4.8 KCl, 1.7 CaCl2, and 1.2 MgCl2 was pumped through the optogenetic-microdialysis probe (rate = 1.25 µl/min). After a washout period of 90 min, dialysate samples were collected at 20 min intervals. After 80 min of baseline sampling, optogenetic stimulation was applied for 20 min, and samples were collected for 80 additional minutes after the end of the stimulation. Samples were split and analyzed separately for glutamate, DA, and ACh content. ACh and glutamate contents were measured by HPLC coupled to an ACh oxidase and glutamate oxidase enzyme reactors, respectively, and electrochemical detection (Eicom). DA was measured by HPLC coupled with a coulometric detector (5200a Coulochem III, ESA). At the end of the microdialysis experiment, animals were deeply anesthetized with Equithesin and perfused transcardially with 0.1 m PBS, followed by 4% formaldehyde in 0.1 m PBS, pH 7.4. Brains were postfixed in the same fixative for 2 h and immersed in 20% sucrose/0.1 m PBS, pH 7.4, solution for 48 h at 4°C. Forty-micron-thick coronal sections were cut in a Leica Microsystems CM3050S cryostat at −20°C, collected in PBS, and stored in antifreeze-buffered solution (20% ethylene glycol, 10% glycerol, and 10% sucrose in PBS) at −80°C until processing. Sections were then evaluated for localization of implanted probes and ChR2-EYFP expression. Wide-field images were acquired with a Typhoon laser scanner (GE Healthcare). Confocal fluorescence microscopy images were acquired with an Examiner Z1 microscope (Carl Zeiss) fitted with a confocal laser module (LSM-710, Carl Zeiss).
Drugs
Dihydro-β-erythroidine hydrobromide (DHβE) was purchased from Tocris. Kynurenic acid (sodium salt), NBQX, and 3-((R)-2-carboxypiperazin-4-yl)-propyl-1-phosphonic acid (CPP) were purchased from Abcam. All other chemicals were purchased from Sigma Millipore.
Statistical analysis
Statistical analysis was performed with Prism (GraphPad). One-sample t test, two-tailed paired t test, repeated-measures one-way ANOVA with or without mixed-effects models, or two-way ANOVA was used as specified. Tukey's or Dunnett's multiple comparisons test was used for post hoc analysis as specified. The number of experiments, n, was expressed as the number of slices or cells/the number of mice or number of rats for the in vivo microdialysis experiments.
Results
In vivo stimulation of PrL/IL inputs increases striatal ACh and DA concentration, and the increase in DA requires nAChR activation
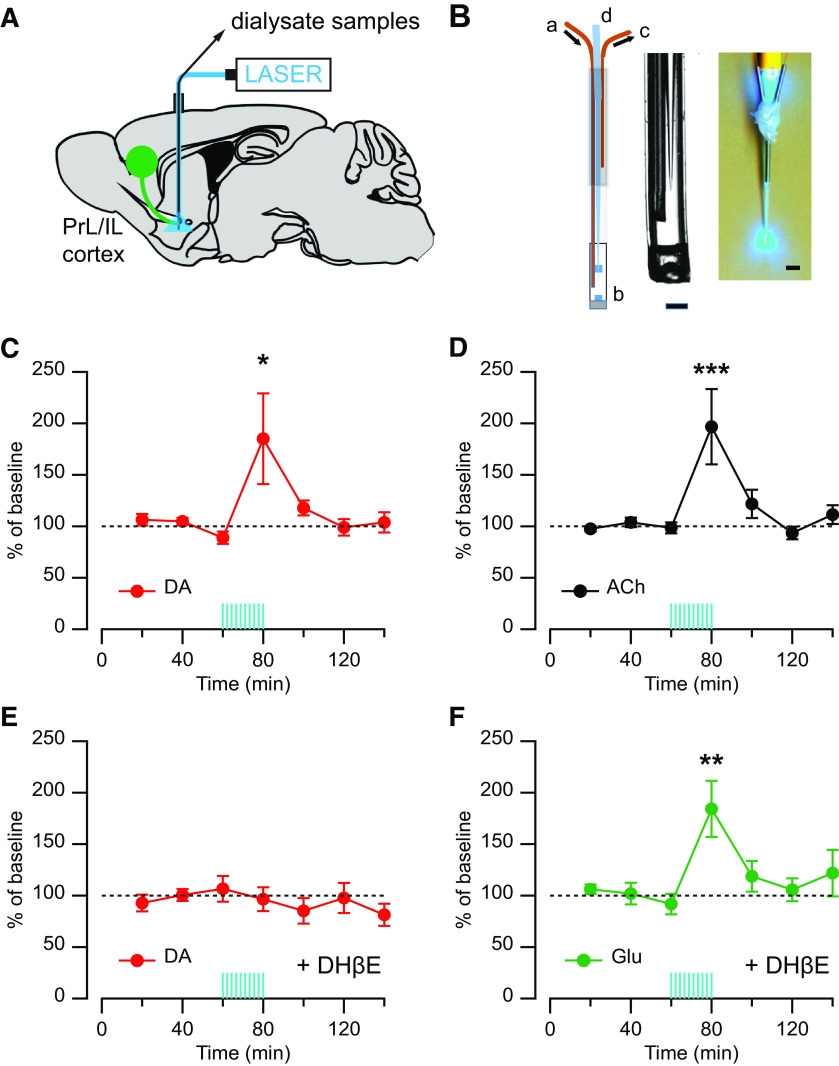
Published work in vivo shows that optogenetic stimulation of cortical inputs to the striatum can evoke DA release (Quiroz et al., 2016). However, this study provides no evidence that activation of these cortical inputs recruits striatal CINs or that the cortically evoked DA release in vivo requires activation of striatal nAChRs. To assess these matters, we used in vivo microdialysis combined with optogenetic stimulation, as previously described (Quiroz et al., 2016), and measured extracellular levels of ACh and DA in response to optogenetic stimulation of cortical inputs. A modified microdialysis probe with an embedded light-guiding optic fiber was implanted into the striatum of rats expressing ChR2-EYFP in the PrL/IL cortex (Fig. 1A,B). On the experiment day, dialysate samples were collected at baseline for 80 min before stimulation started. Optogenetic stimulation of cortical axons within the striatum was delivered around the microdialysis probe as trains of light pulses (16 pulses at 100 Hz every 1 s for 20 min). Samples were collected for an additional 80 min after stimulation and were split and analyzed separately for DA and ACh content or DA and glutamate. This long train of in vivo optogenetic stimulation of PrL/IL cortical axon fibers within the striatum produced an increase in extracellular concentration of DA in the striatal dialysate (185.1 ± 44.0% of baseline, n = 7; F = 5.27, p = 0.0002, repeated-measures one-way ANOVA; p = 0.0002 for 60 min vs 80 min; Dunnett's multiple comparisons test; Fig. 1C), which coincided with an increase of extracellular ACh concentration (196.7 ± 36.6% of baseline, n = 7; F = 3.00, p = 0.0002, repeated-measures one-way ANOVA; p = 0.004 for 60 min vs 80 min; Dunnett's multiple comparisons test; Fig. 1D). These in vivo microdialysis findings support the hypothesis that cortical stimulation recruits CINs. In addition, when the in vivo optogenetic stimulation was performed in constant perfusion of the nAChR antagonist DHβE (10 μm) delivered locally via reverse dialysis, the same stimulation protocol did not cause any change in extracellular DA concentration in the striatal dialysates (96.5 ± 11.6% of baseline, n = 9; F = 0.64, p = 0.70, repeated-measures one-way ANOVA; Fig. 1E), indicating that activation of nAChRs in the striatum is required for the cortically evoked DA release in vivo. As a control, we measured glutamate concentration in these same dialysates, which showed increased glutamate levels and confirmed the delivery of cortical stimulation in the presence of the nAChR antagonist (184.3 ± 27.3% of baseline, n = 9; F(6,45) = 4.01, p = 0.003, repeated-measures one-way ANOVA with mixed-effects model; p = 0.0006 for 60 min vs 80 min; Dunnett's multiple comparisons test; Fig. 1F).
Figure 1.
Effect of the nAChR antagonist DHβE on the extracellular DA level evoked by local optogenetic stimulation of PFC fibers in vivo. A, Diagram showing the injection of ChR2-EYFP in the PrL and IL cortices (green circle) and projection to the striatum with the microdialysis-optogenetics probe in a sagittal brain view. B, Left, Optogenetic-microdialysis probe schematics: a, liquid inlet; b, dialysis membrane; c, liquid outlet; d, sculpted optic fiber. Middle, Probe tip detail. Scale bar, 0.2 mm. Right, Picture of the optogenetic-microdialysis probe showing light spread pattern at the probe tip. Scale bar, 1 mm. C, D, Time course of extracellular concentrations of (C) DA (red) and (D) ACh (black) in the NAc. E, F, Time course of extracellular concentrations of (E) DA (red) and (F) glutamate (Glu, green) in the NAc with constant perfusion (reverse dialysis) of the nAChR antagonist DHβE (10 μm). Time 0-60 represents the values of samples before stimulation. Blue vertical lines indicate the period of optogenetic stimulation (20 min). Results are expressed as mean ± SEM of percentage of the average of three values before stimulation. *p < 0.05, **p < 0.01, ***p < 0.001.
Stimulation of PrL/IL inputs to striatum is sufficient to evoke local DA signals in brain slices
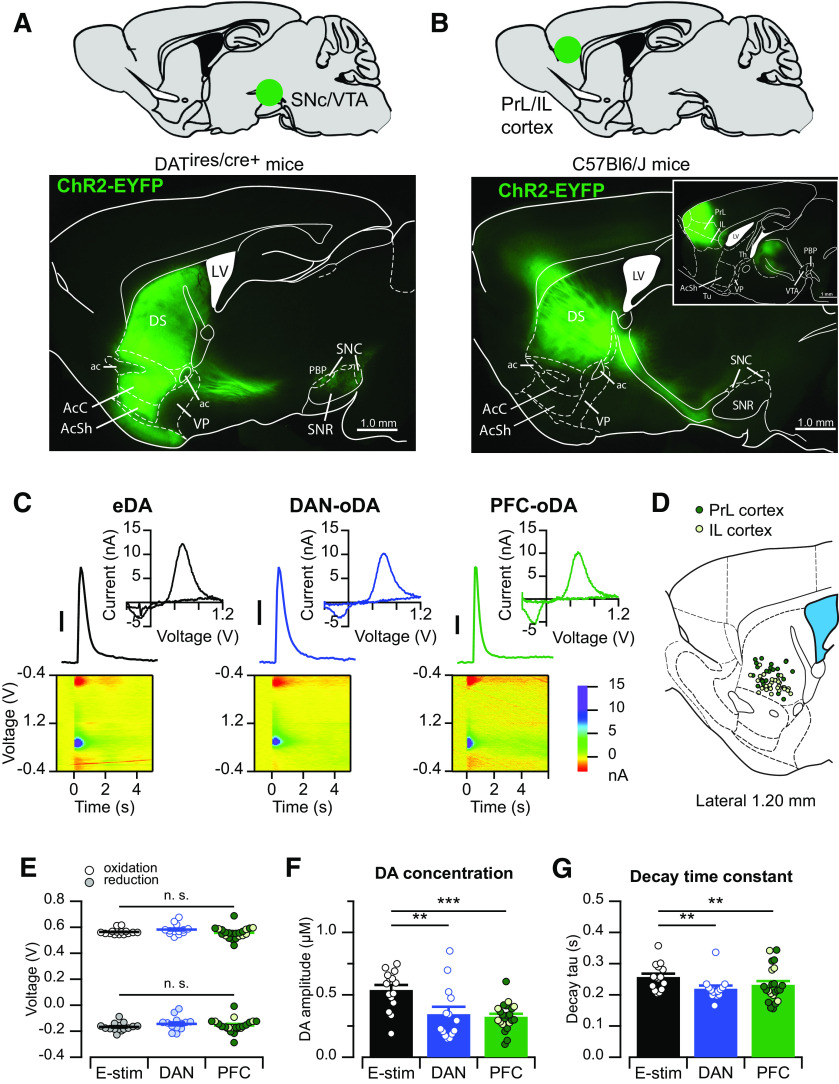
In order to precisely access the specific inputs and apply pharmacological agents, we used transgenic mice with combination of optogenetic stimulation. DA signals were recorded in the striatum using FSCV in mouse brain slice preparations. Release of DA from axonal projections within the striatum was evoked by electrical stimulation and compared with DA signals evoked by input-specific optogenetic stimulation. To achieve input-specific stimulation, ChR2-EYFP was expressed in either midbrain DANs and their axonal projections to the striatum using Cre-dependent viral vector injection in DATIRES-Cre+ mice (Fig. 2A) or ChR2-EYFP in cortical neurons of the PrL/IL cortex and their projections to the striatum using viral injection in Cre-negative littermate mice (Fig. 2B).
Figure 2.
Striatal DA signals evoked by midbrain and cortical inputs. A, Example of the fluorescence pattern (bottom) observed in a sagittal brain slice from a DATIRES-Cre+ mouse injected with DIO-ChR2-EYFP in the midbrain (top). B, Example of the fluorescence pattern (bottom) observed in a sagittal brain slice from a C57Bl6/J (DATIRES-Cre–) mouse injected with ChR2-EYFP in the PrL or IL cortex (top). Inset, A more medial slice with the site of injection. C, Representative DA transients, current–voltage (CV) plots, and color voltammograms when evoked by electrical stimulation (eDA, left), optogenetic stimulation of DAN fibers (DAN-oDA, middle), or optogenetic stimulation of PFC inputs (PFC-oDA, right). All three CV plots show the electrochemical profile of DA oxidation. Scale bar, 100 nM. D, A sagittal brain section modified from the Mouse Brain Atlas (Franklin and Paxinos, 2007) showing the FSCV recording sites from mice injected with ChR2-EYFP in the PrL (filled) and IL (empty) cortex. E, The oxidation (top) and reduction (bottom) voltages from the CV plots of eDA (left), DAN-oDA (middle), and PFC-oDA (right) were plotted as average with SEM. Open and filled circles represent individual values. F, DA peak concentrations and G, decay time constants of eDA (left), DAN-oDA (middle), and PFC-oDA (right) were plotted as average with SEM. Open circles represent individual values. For PFC-oDAs, the same color code was applied as in D for PrL and IL according to injection location. E–G, Data points include experiments where electrical and optogenetic stimulation was delivered alternatingly in the same slice or in different slices (see Materials and Methods). **p < 0.01, ***p < 0.001. LV, Lateral ventricle; DS, dorsal striatum; ac, anterior commissure; AcC, accumbens core; AcSh, accumbens shell; VP, ventral pallidum; PBP, parabrachial pigmented nucleus; SNR, substantia nigra reticulata; Th, thalamus; Tu, olfactory tubercle. n.s., not significant.
As expected, single brief pulses of electrical stimulation delivered via an electrode placed within the striatal tissue reliably evoked DA (electrically evoked DA transient [eDA]) signals in striatal brain slices (Fig. 2C). Also, direct optogenetic stimulation of midbrain DAN axonal projections within the striatum also evoked DA signals (referred here as DAN-oDA) on delivery of a single brief pulse of blue light (0.6 ms), in agreement with previous published studies (Adrover et al., 2014; Melchior et al., 2015). Last, similar to the in vivo findings in Figure 1, selective optogenetic stimulation of axon projections from PrL/IL cortex into the striatum was also sufficient to trigger DA signals (referred as PFC-oDA) in the striatum of in slice preparation (Fig. 2C). These PFC-oDA signals were also reliably evoked by brief single pulses of blue light (0.6 ms) in the ventral portion of the dorsomedial striatum in sagittal brain slices (Fig. 2D). The viral expression around injection site was often large enough to include both PrL and IL regions of the cortex. For this reason, we combined data from the two regions. It is worth noting that these brain sections mainly contain the axon projections from PrL/IL neurons, but not their cell bodies, which are localized to more medial sagittal sections of the brain (Fig. 2B, inset).
The current–voltage plots of DA signals evoked via the three different types of stimulation (electrical, optogenetic stimulation of DANs, and optogenetic stimulation of PrL/IL) were indistinguishable and displayed current peaks at the expected oxidation (0.56 ± 0.01 V for eDA, n = 15/n = 8; 0.58 ± 0.01 V for DAN-oDA, n = 14/n = 6; 0.56 ± 0.01 V for PFC-oDA, n = 22/n = 13; F = 0.49, p = 0.62, repeated-measures one-way ANOVA with mixed-effects model; Fig. 2E) and reduction voltages (−0.17 ± 0.01 V for eDA, −0.14 ± 0.01 V for DAN-oDA, −0.16 ± 0.02 V for PFC-oDA; F = 0.76, p = 0.49, repeated-measures one-way ANOVA with mixed-effects model; Fig. 2E) for DA. The peak amplitude of eDA was significantly higher than DAN-oDA or PFC-oDA (556 ± 42 nm for eDA; 367 ± 58 nm for DAN-oDA; 325 ± 23 nm for PFC-oDA; F(2,12) = 13.430.76, p > 0.05, repeated-measures one-way ANOVA with mixed-effects model; p = 0.01 for eDA vs DAN-oDA, p = 0.0005 for eDA vs PFC-oDA, and p = 0.99 for DAN-oDA vs PFC-oDA; Tukey's multiple comparisons test; Fig. 2F), and the decay time constants (tau) were similarly longer with eDA (0.25 ± 0.01 s for eDA, 0.22 ± 0.01 s for DAN-oDA, 0.23 ± 0.01 s for PFC-oDA; F(2,12) = 13.43, p = 0.0009, repeated-measures one-way ANOVA with mixed-effects model; p = 0.004 for eDA vs DAN-oDA, p = 0.009 for eDA vs PFC-oDA, and p = 0.83 for DAN-oDA vs PFC-oDA; Tukey's multiple comparisons test; Fig. 2G). Thus, we were able to reliably evoke steady DA signals by selective optogenetic stimulation of axon projections from PrL/IL cortex in the ventral portion of the dorsomedial striatum. The DA signals recorded using FSCV showed indistinguishable chemical and temporal characteristics, even when evoked by different inputs.
Unique physical and pharmacological properties across input-specific DA signals
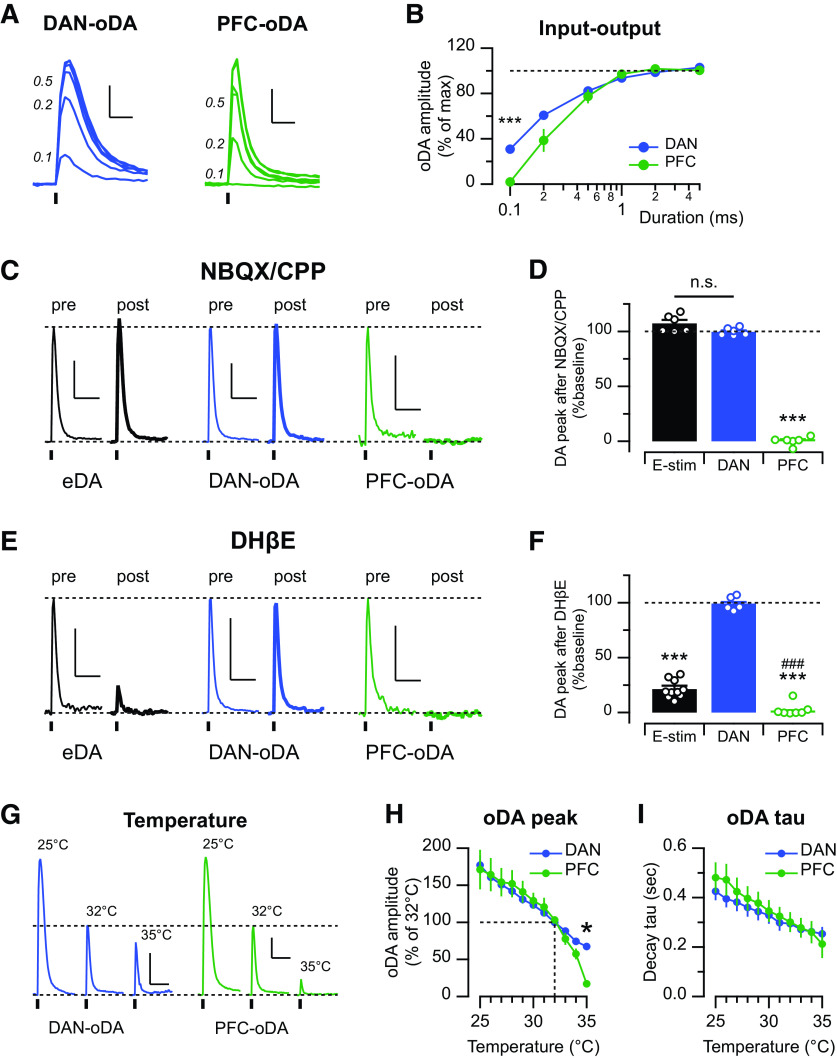
We first explored the threshold for triggering the input-specific optogenetically evoked DA signals. The threshold was determined by constructing input-output curves and varying the light pulse durations from 0.1 to 5 ms in brain slices from mice expressing ChR2 in midbrain DANs or the PrL/IL region (Fig. 3A,B). The relative amplitudes of DA transients were overlapping for pulses of 0.5-5 ms duration, and both DAN-oDA and PFC-oDA signals showed maximal amplitude with 1-5 ms light pulse durations. However, very short light pulses (0.1 ms) evoked measurable DA signals only in slices in which ChR2 was expressed in midbrain DAN projections to the striatum, indicating a lower threshold for evoking DA signals with direct stimulation of DA neuron axons than with stimulation of cortical inputs (duration × input, F(5,90) = 11.58, p < 0.0001, repeated-measures two-way ANOVA; post hoc for the duration of 0.1 ms: 33 ± 4% for DAN-oDA, n = 12/n = 8, and 1.9 ± 0.7% for PFC-oDA, n = 8/n = 5, p < 0.0001; Fig. 3B). A higher threshold for the cortically evoked DA signals could reflect technical differences, such as opsin expression levels, or other biological factors, such as the density of innervation of cortical versus DAN axonal projections, difference in the release probability of the DA varicosities that respond to direct stimulation of DAN axons from those responding to cortical inputs, and/or the indirect and polysynaptic nature of the cortical-evoked DA signals.
Figure 3.
DAN-oDA and PFC-oDA show different physical and pharmacological properties. A, Representative DAN-oDA (left) and PFC-oDA (right) transients evoked with different light pulse duration (in ms). Scale bar, 100 nM, 0.5 s. B, oDA amplitudes normalized to their maximum response were averaged and plotted as a function of the stimulus duration. C, Representative traces of eDA, DAN-oDA, and PFC-oDA transients before and after bath application of the glutamate receptor antagonists, NBQX and CPP (both 5 μm). Dotted line (top) indicates the amplitude of DA transients before the drugs. Scale bar, 200 nM, 2 s. D, Averages with SEM of DA amplitude after NBQX and CPP were plotted. Open circles represent individual value. E, Representative traces of eDA, DAN-oDA, and PFC-oDA transients before and after bath application of the β2-contatining nAChR antagonist, DHβE (1 μm). Scale bar, 200 nM, 2 s. F, Averages with SEM of DA amplitude after DHβE were plotted. Open circles represent individual value. ***Versus DAN-oDA. ###Versus eDA. G, Representative traces of DAN-oDA (left) and PFC-oDA (right) transients at 25°C, 32°C, and 35°C. Dotted line (top) indicates the oDA amplitude at 32°C. Scale bar, 200 nM, 2 s. H, Average oDA peak amplitudes normalized to 32°C. I, Average oDA decay time constants were plotted as a function of temperature. *p < 0.05, ***p < 0.001, ###p < 0.001, n.s., not significant.
To determine whether PrL/IL activation elicits striatal DA release and engages the CIN-dependent mechanism as shown in in vivo microdialysis experiment (Fig. 1), we then studied the basic pharmacological characteristic of DA signals evoked by stimulation of PFC inputs and compared them with those evoked by electrical and DAN input stimulation. In striatal brain slices, PFC-oDA signals were completely abolished by a mixture of AMPA and NMDA receptor antagonists (NBQX/CPP), whereas eDA and DAN-oDA signals were unaffected (amplitude after the antagonists = 0.3 ± 1.6% of baseline for PFC-oDA, n = 6/n = 5; 107.3 ± 3.0% for eDA, n = 6/n = 4; 99.8 ± 1.4% for DAN-oDA, n = 6/n = 4; F(2,3) = 694.8, p < 0.0001, repeated-measures one-way ANOVA with mixed-effects model; p = 0.14 for eDA vs DAN-oDA, p values < 0.0001 for PFC-oDA vs eDA or PFC-oDA vs DAN-oDA; Tukey's multiple comparisons test; Fig. 3C,D). These findings indicate a requirement for ionotropic glutamate transmission for PrL/IL inputs to evoke local DA signals, which is in agreement with previous reports on the requirement of glutamate transmission for DA signals evoked by motor cortex and thalamic inputs to the striatum (Threlfell et al., 2012; Kosillo et al., 2016; Cover et al., 2019). Further, similar to our in vivo data (Fig. 1D), DHβE also abolished PFC-oDA signals, and significantly depressed eDA signals, while having no effect on DAN-oDA signals (1.9 ± 1.4% of baseline for PFC-oDA, n = 11/n = 7; 21 ± 3% for eDA, n = 10/n = 5; 99 ± 3% for DAN-oDA, n = 5/n = 2; F(2,7) = 395.2, p < 0.0001, repeated-measures one-way ANOVA with mixed-effects model; p < 0.0001 for eDA vs DAN-oDA, p = 0.0006 for eDA vs PFC-oDA, and p < 0.0001 for DAN-oDA vs PFC-oDA; Tukey's multiple comparisons test; Fig. 3E,F). Together, the pharmacology results support the hypothesis that PFC-oDA signals are mediated by activation of glutamate synapses from PrL/IL cortex to striatal CINs, which excites them to fire and trigger ACh release, thereby driving DA release via activation of nAChRs on DAN axon fibers within the striatum.
ACh is rapidly cleared by enzymatic degradation by AChE (Quinn, 1987). The extremely dense presence of AChE in the striatum (Zhou et al., 2001) assures the termination of cholinergic action. We also have previously shown the influence of the AChE on the DA transmission in the striatum (Shin et al., 2015, 2017). As AChE activity shows temperature dependence (Vidal et al., 1987), we then tested the temperature dependence of the input-specific DA transients by varying the temperature of the bath solution. The amplitude of both DAN-oDA and PFC-oDA signals increased linearly as the temperature was lowered from the standard 32°C to 25°C, and both showed a tight negative regression with similar slopes significantly different from zero (−10.8 ± 0.6% per °C, n = 8/n = 3, for DAN-oDA, and −10.8 ± 2.0% per °C, n = 6/n = 3, for PFC-oDA; p values < 0.0001; Fig. 3G,H). The temperature dependence of the DA signal amplitude likely reflects the change in rate of DA transporter activity with temperature. DA transporter activity was reported to have a temperature coefficient Q10 ranging from 1.39 to 2.95 (Zhu and Hexum, 1992), from which we estimate a Q10 of ∼2.3 for the DA transporter for 24°C–37°C range. Our estimate predicts that large changes in the rate of DA reuptake by the transporter would be around the tested and physiological temperatures. Indeed, the decay of the DA transients, which is a parameter that reflects the clearance of extracellular DA by the transporter, was also affected as the temperature was lowered. The decay time constant of both DAN-oDA and PFC-oDA increased linearly with the decrease of the temperature (DAN-oDA: −0.017 ± 0.001 s/°C; PFC-oDA: −0.025 ± 0.001 s/°C; Fig. 3I), which corresponded with the linear increase in amplitude. However, raising the temperature from 33°C to 35°C caused a dramatic and selective drop in the amplitude of the PFC-oDA signals, compared with DAN-oDA signals (Fig. 3G,H). The slope for PFC-oDA signals increased threefold from −11% to −31% per °C.
Vidal et al. (1987) have reported on a high dependence with temperature of the activity of AChE from rat brain, which reaches maximal rate at ∼35°C. Our in vivo and in vitro pharmacology experiments showed that PFC-oDA signals require activation of nAChR. Therefore, we speculate that the observed steep temperature dependence of the signals is in large part mediated by rate increase in AChE activity. Furthermore, at physiological temperatures, the PFC-evoked DA signals may be very localized near ACh release sites, presumably overlapping with nAChR expression in DAN axons.
Activity dependent summation of the midbrain and cortical DA signals
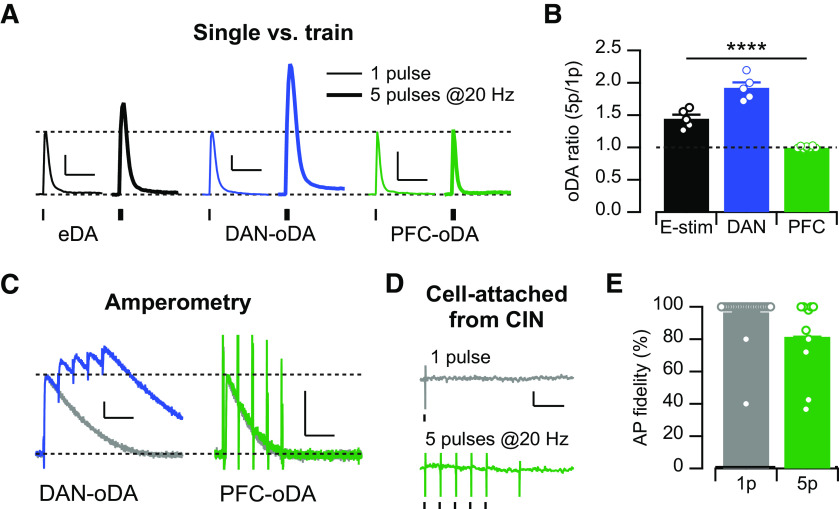
Trains of stimulation pulses have been shown to produce sublinear summation in the amplitude of DA signals evoked by electrical and optogenetic stimulation of DAN axon projections in the striatum in vitro (Zhang and Sulzer, 2004; Threlfell et al., 2010; Melchior et al., 2015; Shin et al., 2017). We then tested the degree of summation of the DA signals in response to short trains of stimulation pulses (5 pulses at 20 Hz) and compared it with the transients evoked by a single pulse (Fig. 4A,B). This pattern of train stimulation was chosen based on burst firing patterns recorded in vivo for DANs during behavior (Schultz et al., 1993; Hyland et al., 2002). Indeed, we found that trains of 5 pulses at 20 Hz evoke DA transients 50% larger than those evoked by single pulse stimulation for eDA signals and almost double for DAN-oDA (5p/1p: 1.44 ± 0.06, n = 5/n = 3, for eDA; 1.92 ± 0.19, n = 5/n = 2, for DAN-oDA; 1.00 ± 0.02, n = 10/n = 7; F(2,2) = 110.6, p = 0.009, repeated-measures one-way ANOVA with mixed-effects model; p = 0.04 for eDA vs DAN-oDA, p = 0.03 for eDA vs PFC-oDA, and p = 0.008 for DAN-oDA vs PFC-oDA; Tukey's multiple comparisons test; Fig. 4A,B). On the contrary, PFC-oDA transients evoked by this train stimulation had similar amplitudes compared with those evoked by a single pulse, indicating no summation of DA signals evoked by PrL/IL inputs. In order to measure DA signals with better temporal resolution, we performed amperometric recordings and again compared DA signals evoked by single pulse and trains (Fig. 4C). In line with the FSCV results, amperometric recordings showed that DAN-oDA signals display sublinear summation, whereas PFC-oDA signals are indistinguishable between single pulse or trains of 5 pulses at 20 Hz. The lack of summation in the PFC-oDA signals in response to trains resembles the findings obtained with DA signals evoked by synchronized activation of CINs (Threlfell et al., 2012; Shin et al., 2017) and further supports the idea that the PFC-evoked local DA signals are mediated by synchronized action of CINs in response to stimulation of PrL/IL inputs. The large transient current deflection in response to each pulse of stimulation (n = 5/n = 4; Fig. 4C, right) is suggestive of the action potential firing evoked in CINs and other striatal neurons by the optogenetic stimulation of PrL/IL inputs. The fact that every pulse of the train stimulation triggers a current deflection reflects the ability of PrL/IL inputs to evoke action potentials firing in downstream neurons and supports the idea that the lack of summation in DA signals is not likely because of the failure of action potential firing by CINs. To directly test the contribution of failure of action potential firing by CINs to the lack of summation in PFC-oDA signals, we conducted cell-attached recordings from CINs and measured action potential fidelity in response to a single pulse and trains of 5 pulses at 20 Hz (single pulse: 96.5 ± 2.7%, n = 23/n = 4, t = 1.28, p = 0.21; trains of 5: 81.5 ± 7.7%, n = 10/n = 4, t = 2.42, p = 0.04; both with one-sample t test to 100% value; Fig. 4D,E). Although there was a small but significant percentage of failure, the totality of the data indicates that the lack of summation is not likely caused by action potential failure, but instead can be attributed to other downstream mechanisms, such as desensitization of nAChRs (Threlfell et al., 2012; Shin et al., 2017).
Figure 4.
PFC-oDA shows no summation by train stimulations. A, Representative DA traces of eDA, DAN-oDA, and PFC-oDA transients evoked by single pulse (1p, thin traces) or train of 5 pulses at 20 Hz (5p, thick traces). Scale bar, 200 nM, 2 s. B, Averages with SEM of the DA amplitude ratio (5p/1p) for eDA, DAN-oDA, and PFC-oDA transients were plotted. Open circles represent individual values. C, Representative amperometric traces for DAN-oDA (left) and PFC-oDA (right) transients evoked by single pulse (gray traces) or train of 5 pulses at 20 Hz (color traces). PFC-oDA amperometric transients evoked by 50 at 20 Hz were indistinguishable from 1p stimulation, except the large deflection at the stimulation time for 5p pulses. Scale bars: 200 pA, 100 ms. D, Representative cell-attached recordings from CINs with single (top, gray) or train of 5 pulses at 20 Hz (bottom, green). Scale bars: 20 pA, 100 ms. E, Averages with SEM of the action potential fidelity were plotted for the single pulse and train stimulation. Open circles represent individual values. ****p < 0.0001.
Input-specific DA signals are evoked by dual optogenetic stimulation in the same brain slice
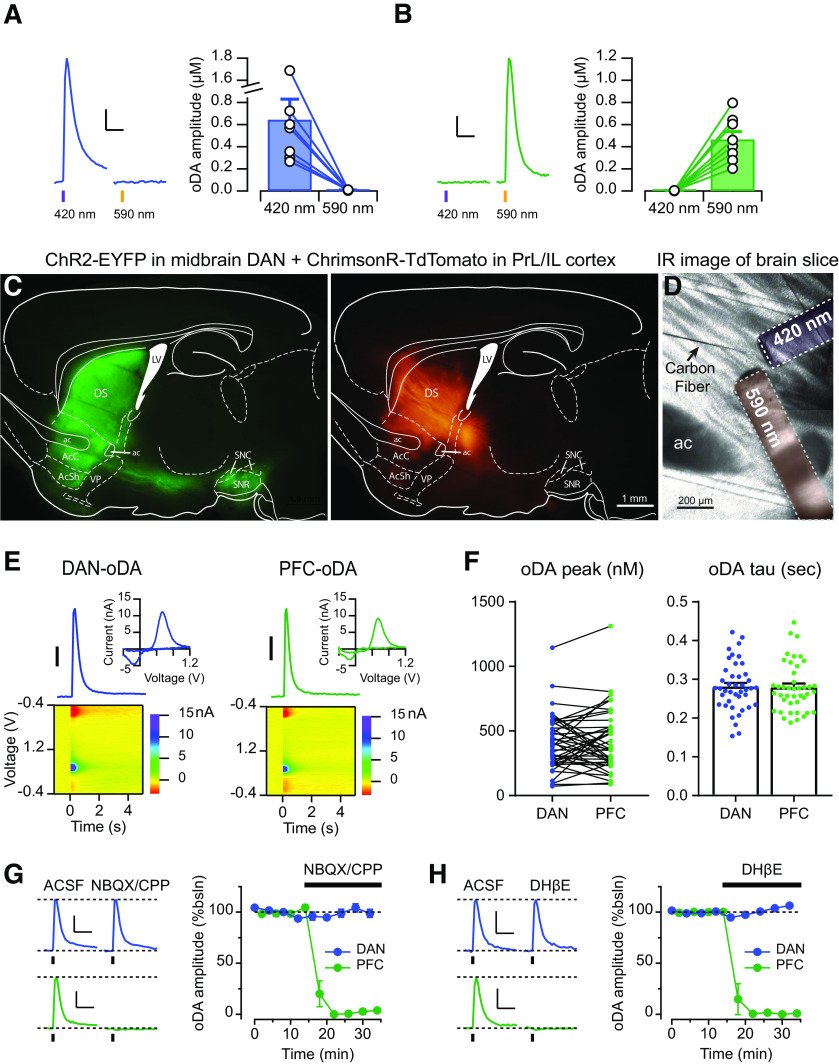
Thus far, DA signals evoked by the different inputs were recorded in brain slices from different mice that expressed ChR2 in either midbrain DANs or in PrL/IL cortex. In order to strictly test the segregation/distinction of these two pathways, it was important to examine these two inputs and compare the input-specific DA signals in the slice preparation from the same mice. We took advantage of opsins activated by different light wavelengths, specifically, ChR2 and the red-shifted opsin ChrimsonR (Klapoetke et al., 2014). We first set up the conditions for selective stimulation of each opsin without cross-activation. In brain slices expressing only ChR2 in midbrain DANs, pulses of purple light (420 nm) evoked reliable DA signal, whereas similar and longer pulses of orange light (590 nm) did not trigger any detectable signals (Fig. 5A). Conversely, in slices expressing only ChrimsonR in the PrL/IL cortex, brief orange light pulses evoked DA signal, but not purple light (Fig. 5B). Thus, under these experimental conditions, ChR2 and ChrimsonR can be used in combination to selectively stimulate two different inputs by delivering pulses of two different light wavelengths without any detectable cross-activation.
Figure 5.
Dual-opsin expression to evoke cortical and midbrain DA signals in the same brain slices. A, B, Left, Representative traces of DA signals evoked by either 420 or 590 nm light pulses from mice expressing (A) only ChR2 in midbrain DANs or (B) only ChrimsonR in PrL/IL cortex. Right, oDA amplitudes were plotted as pairs for each wavelength. Scale bars: 200 nM for A 100 nM for B; 1s C, Example of the fluorescence patterns with filter set for yellow signal (left) or red signal (right) from a sagittal brain slice of a DATIRES-Cre+ mice injected with ChrimsonR-TdTomato in the PFC and DIO-ChR2-EYFP in the midbrain. D, Configuration to the FSCV DA recording using a carbon fiber and two fiber-optics delivering 420 and 590 nm, respectively. E, Representative DA transients, CV plots, and color voltammograms of DAN-oDA and PFC-oDA. Scale bar: 100 nM F, Left, Amplitudes were plotted as pairs for DAN-oDA and PFC-oDA recorded from the same slices. Right, Averages with SEM of decay time constant were plotted for DAN-oDA and PFC-oDA. Dots represent individual values. G, Left, Representative traces of DAN-oDA (top) and PFC-oDA (bottom) before and after the application of glutamate receptor antagonists NBQX and CPP. Right, Averages with SEM of DAN-oDA and PFC-oDA were plotted as a function of time as NBQX/CPP was applied. H, Left, Representative traces of DAN-oDA (top) and PFC-oDA (bottom) before and after the application of nAChR antagonists DHβE. Right, Averages with SEM of DAN-oDA and PFC-oDA were plotted as a function of time as DHβE was applied. Scale bars for G–H: 100 nM; 1 s.
Next, in the same DATIRES-Cre+ mice, ChR2-YFP was expressed in midbrain DANs with a Cre-dependent expression vector, and ChrimsonR-TdTomato was expressed in PrL/IL cortex. Figure 5C shows examples of the fluorescent expression pattern of ChR2-YFP (green) and ChrimsonR (red) in the same sagittal brain slice from this double-injected mouse. Figure 5D shows the experimental arrangement for the dual-input recordings with the placement of the carbon fiber and the two fiber-optics for delivering the purple and orange light pulses that were used to activate ChR2 and ChrimsonR, respectively. In these slices with dual-opsin expression, brief pulses of either purple or orange light evoked DA signals with indistinguishable current–voltage plots, showing the characteristic peaks for DA oxidation and reduction (Fig. 5E). The DA concentration transients had comparable peak amplitudes with means of 407 ± 32 nm for DAN-oDA and 420 ± 36 nm for PFC-oDA (n = 43/n = 13, t = 0.47, df = 42, p = 0.64, paired t test; Fig. 5F) and overlapping time courses as measured at 10 Hz sample rate of FSCV, in agreement with the results shown in Figure 2. The decay time constant of the DA transients was similar (0.28 ± 0.01 s for DAN-oDA and 0.28 ± 0.01 s for PFC-oDA, n = 43/n = 13, t = 0.41, df = 41, p = 0.68 from paired t test; Fig. 5F).
Then, we tested the pharmacological properties of DAN-oDA and PFC-oDA by delivering alternating purple and orange light pulses and recording from a single carbon fiber. Application of the glutamate receptor antagonists NBQX/CPP (99.2 ± 3.1% of the baseline for DAN-oDA and 1.6 ± 1.1% for PFC-oDA, n = 5/n = 3; t = 25.3, df = 4, p < 0.0001, two-tailed paired t test; Fig. 5G) or the nAChR antagonist DHβE (101.9 ± 0.6% of the baseline for DAN-oDA and 0.8 ± 0.8% for PFC-oDA, n = 5/n = 3; t = 79.13, df = 4, p < 0.0001, two-tailed paired t test; Fig. 5H) completely abolished PFC-oDA signals while leaving DAN-oDA signals in the same location intact. These results confirm the findings from Figure 3, which use antagonists and single-input optogenetic stimulation to evoke either DAN-oDA or PFC-oDA in separate slices/mice. These results also validate the experimental approach by showing input specificity with no apparent crossover in the optogenetic stimulation of ChR2 and ChrimsonR with the described wavelengths. More importantly, these pharmacological findings highlight the different mechanisms and the unique nature of the input-specific DA signals evoked by PFC and midbrain inputs.
Cortically evoked signals are spatially restricted
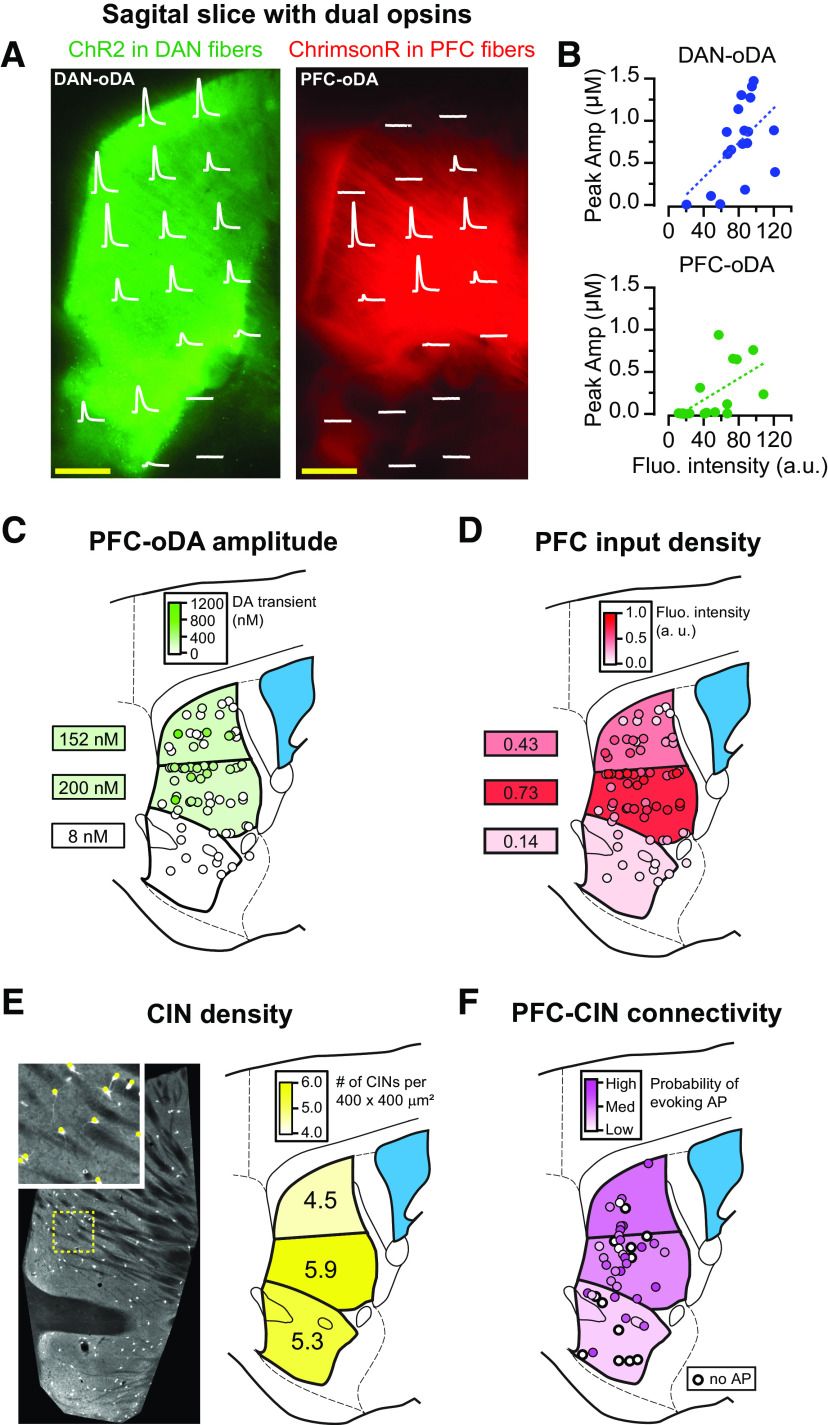
As shown in Figure 2D, DA signals evoked by selective stimulation of PrL/IL inputs were preferentially detected between the NAc core and dorsal striatum. We then set out to investigate in more detail the spatial distribution of the input-specific DA signals using the dual-opsin expression approach by sampling the amplitude of DA signals across 18 different striatal areas of the sagittal brain slice while evoking responses with purple and orange light. While DAN-oDA signals were detected throughout the whole striatum, PFC-oDA signals were spatially restricted (Fig. 6A,B). Both DAN-oDA and PFC-oDA signals showed significant correlations with the fluorescent intensity of the projections (p = 0.03 and r2 = 0.28 for DAN-oDA; p = 0.01, r2 = 0.37 for PFC-oDA; Fig. 6B). However, the slope of the linear regression was larger for DAN-oDA signals than PFC-oDA signals (10 vs 6 nm/AU, respectively; Fig. 6B). Thus, the intensity of the DAN-labeled fibers was a good predictor of the magnitude of DAN-oDA signals. On the other hand, the presence of cortical axon fluorescence was not always a predictor of PFC-oDA signals, likely because of the larger density of passing cortical axons that are not synaptic terminal run through the striatum.
Figure 6.
Cortically evoked DA signals are spatially restricted. A, The locations of carbon fiber and two fiber-optics were adjusted to measure DAN-oDA (left) and PFC-oDA (right) from each location where the corresponding oDAs were superimposed on the fluorescence patterns. Scale bars, 500 µm. B, Peak amplitudes of DAN-oDA (top) and PFC-oDA (bottom) were plotted as a function of florescence intensity at each location shown in A. Dotted lines indicate linear regression between the oDA amplitudes and the fluorescence intensities. C, PFC-oDAs were measured from 74 locations (between ∼1.0 and 1.2 mm in mediolateral coordinate) of mice injected with ChR2-EYFP in PrL/IL cortex and color-coded according to their peak DA concentrations. The average PFC-oDA amplitudes were calculated and color-coded for the three subregions. D, The PFC input fluorescence intensities were color-coded for the same 74 locations in C. The averages from the three subregions were calculated and color-coded. E, Left, Fluorescence image of striatal CINs labeled with td-Tomato. Inset, Examples of identified CINs (red) from the area with the dotted yellow line. Right, The average numbers of CINs per 400 × 400 µm2 (area as shown in the inset) were calculated and shown for the three subregions. F, Using cell-attached patch recording, action potentials evoked by PFC stimulation were observed from a total of 46 CINs. For each CIN, a minimum light intensity to evoke action potentials was determined to score the connectivity. White circles with a thicker line represent CINs which did not show any evoked action potentials even with the maximum light intensity.
Averaging the area responses in 5 brain slices from 3 mice, we found that the mean amplitude of the PFC-oDA signals was largest in the ventral part of the dorsal striatum, which again maps in gross terms with the location of the brightest intensity of ChR2-EYFP-labeled projections from PrL/IL cortex (Fig. 6C,D). However, the correlation between fluorescence intensity and amplitude of PFC-oDA signals is not always strong; for example, the more intensely labeled caudal areas near the globus pallidus show almost no PFC-oDA signal (Fig. 6C,D). The evidence of small or no PFC-oDA signals in the more caudal striatum could possibly reflect greater fiber density rather than innervation of the caudal portion of the striatum by the axonal projections from PrL/IL cortex.
The number of CINs is <1% of the total striatal neurons, and they are sparsely distributed throughout the striatum (Burke et al., 2017). We then quantified the density of CINs from the same area to further determine whether the distribution of CINs contribute to the spatial restriction of PFC-oDA signals. For this purpose, we took advantage of CIN-tdTomato mouse line, which fluorescently labels CINs throughout the striatal slices (Fig. 6E). The quantification analysis of cell numbers in 400 × 400 µm2 showed that the density of CINs was also highest in the middle part of the dorsomedial striatum (6 slices from 3 mice), suggesting that an uneven distribution of CINs could also contribute to the spatial restriction of the local DA signals evoked by PrL/IL cortical inputs to the ventral portion of the dorsomedial striatum. To further test whether the spatial profile of PFC-oDA signals is in part determined by the strength of the innervation of CINs by PrL/IL cortical neurons, we performed cell-attached recordings from the same CINs of CIN-TdTomato mice and measured the ability of PFC optogenetic stimulation to evoke action potentials in CINs throughout the striatum (46 CINs from 3 mice). We found that the connectivity followed a similar pattern to the PFC-oDA profile, and the probability that CINs will fire an action potential in response to PFC stimulation was higher in the central part of the striatum than in the NAc and dorsal regions of the dorsal striatum (Fig. 6F).
In conclusion, this study offers a series of in vitro and in vivo evidence that stimulation of cortical inputs from the PrL/IL cortex can evoke DA release in the striatum via a local mechanism that recruits CINs and requires activation of nAChRs. Since these cortically evoked DA signals are spatially restricted and have different properties, we speculate that they are engaged in distinctive striatal functions from those assigned for DA signals evoked by midbrain DANs.
Discussion
This study provides strong evidence that DA signals in the striatum can occur both in vivo and in vitro in response to stimulation of PrL/IL cortical inputs. These PrL/IL-evoked DA signals have distinctive pharmacological and physiological properties, compared with the DA signals evoked by DAN inputs. The in vitro experiments also introduce a dual optogenetic stimulation approach with which we can activate two different inputs to the striatum in the same brain slice, without apparent cross-activation. This study further reveals the spatial localization of the DA signals evoked by PFC inputs.
We showed that the PrL/IL-evoked DA signals require activation of glutamate and nicotinic ACh receptors, in line of a series of elegant published work. Using in vitro radioactive assays and in vivo microdialysis, it was first shown that locally administered ACh and glutamate can trigger DA release in the striatum (Giorguieff et al., 1976, 1977). Giorguieff et al. (1976) showed that an nAChR antagonist blocked ACh-evoked DA signals, and speculated that “… the release of DA from dopaminergic terminals can be regulated by cholinergic presynaptic receptors exhibiting nicotinic characteristics.” To their credit, their conclusions agree with our interpretations of the findings from the current study as well as other recent optogenetic studies where it was shown that selective stimulation of CINs is sufficient to evoke DA signals (Cachope et al., 2012; Threlfell et al., 2012; Wang et al., 2014; Shin et al., 2017). The involvement of CINs in the glutamate-dependent DA signals was also suggested by early work by Taber and Fibiger (1994) who showed that electrical stimulation of PFC can increase levels of ACh in the striatum and further supported by more recent optogenetic studies (Threlfell et al., 2012; Kosillo et al., 2016; Johnson et al., 2017; Mateo et al., 2017; Cover et al., 2019).
Here, our experiments confirmed the requirement for ionotropic glutamate receptor activation in the PFC-evoked DA signals as previously shown (Mateo et al., 2017). We also show that nAChRs are required for the PrL/IL-evoked DA signals in vitro, similar to other findings when stimulating inputs from other cortical (Kosillo et al., 2016) and thalamic areas (Threlfell et al., 2012; Kosillo et al., 2016; Cover et al., 2019) and indirectly in vivo (Mateo et al., 2017). Our results from the in vivo microdialysis experiments reproduce the original findings from Quiroz et al. (2016) showing that optogenetic stimulation of PFC inputs increases striatal levels of DA in vivo (and glutamate, as expected; Fig. 1). More importantly, we showed that stimulation of PrL/IL inputs to the striatum also elevates striatal levels of ACh in vivo and that this cortically evoked DA signals are blocked when a nAChR antagonist is perfused (Fig. 1C,D). Thus, these findings support that idea PrL/IL inputs form excitatory synapses on striatal CINs and can activate then to evoke ACh release (Fig. 1C). In agreement with this conclusion, recent work showed that optogenetic stimulation of either M1 motor cortex or parafascicular nucleus of the thalamus can induce release of ACh that was detected using exogenous G-protein-coupled inward rectifying K-current expressed in medium spiny neurons (Mamaligas et al., 2019). Together, our findings from both in vitro and in vivo strongly support the hypothesis of a local mechanism for evoking DA release in the striatum, in addition to the more conventional mechanism based on midbrain DAN firing. This local mechanism engages striatal CINs, the targets of inputs from PFC, which have an essential role in evoking DA release from DAN fibers.
DA signals evoked by PrL/IL inputs and those evoked by DAN fibers stimulation share common properties, such as overlapping electrochemical profiles of the voltammetric currents and similar concentration range, time course, and decay time constant of the signals (Figs. 2, 5E,F). However, there are also several differences between the input-specific DA signals. First, DAN-evoked DA signals do not require activation of either ionotropic glutamate receptors or nAChRs (Figs. 3C–F, 5G,H), confirming the direct nature of this mechanism that is triggered by ChR2-evoked action potentials in DAN axon fibers. Second, PrL/IL-evoked DA signals display different temperature sensitivity and a higher threshold compared with DAN-oDA, requiring a longer duration of light stimulation as determined in the input-output relationship (Figs. 3, 4). This finding may reflect the polysynaptic nature of the cortically evoked signals and the requirement of synchronized activation of CINs to trigger DA release by this local mechanism (Threlfell et al., 2012; Kosillo et al., 2016; Liu et al., 2018). Third, the amplitude of DAN-evoked DA signals increases as the number of stimulation pulses increases, indicating some summation in the DA signals evoked by a train of stimulation pulses (Fig. 4A,B). In contrast, the amplitude of DA signals in response to a train of pulses of cortical input stimulation is similar to the amplitudes obtained in response to a single pulse of stimulation, indicating no summation under this condition for the local mechanism (Fig. 4A,B). ChR2 displays use-dependent inactivation when stimulated at high frequency (Hass and Glickfeld, 2016). However, the lack of summation by the train stimulation is unlikely because of this use-dependent inactivation of ChR2 since 20 Hz train stimulation was still able to evoke action potentials in CINs recorded in cell-attached mode (Fig. 4D,E) and amperometry recording showed large deflections in response to train stimulation (Fig. 4C). Anecdotally, these current deflections disappeared when the ionotropic glutamate receptor antagonists were applied, also supporting the idea that firing in CINs and other downstream neurons occurs in response to 20 Hz train stimulation. The lack of summation was also reported in DA signals evoked by direct optogenetic stimulation of CINs in the dorsal striatum (Threlfell et al., 2012; Shin et al., 2017), which was suggested to be because of nAChR desensitization. We speculate then that the lack of summation we report here by 20 Hz trains in PrL/IL-evoked DA signals, which also require nAChRs activation, is also because of desensitization.
To directly study and compare the input-specific DA signals in the same animal, we established dual optogenetic stimulation using a red-shifted opsin, ChrimsonR, in combination with the blue-light activated ChR2. Two wavelengths for excitation were chosen based on the excitation spectrum of each opsin (Klapoetke et al., 2014), 590 nm for ChrimsonR and 420 nm for ChR2, which showed no cross-activation (Fig. 5A,B). This dual optogenetic approach was further validated using pharmacology in animals expressing ChR2 in midbrain DANs and ChrimsonR in PrL/IL neurons. We found that PrL/IL-evoked DA signals were completely blocked by antagonists of either ionotropic glutamate receptors or nAChRs, without affecting DAN-oDA signals in the same slice (Fig. 5G,H). Therefore, we determined that the two inputs can be activated independently using this dual optogenetic approach in brain slices from the same animal.
Interestingly, using this dual optogenetic approach, an intriguing spatial pattern was revealed for the cortically evoked PrL/IL DA signals. While DAN-oDA signals were recorded throughout the striatum, PrL/IL oDA signals were spatially restricted to a “hot-spot” region around the boundary between the NAc core and dorsal medial striatum (Fig. 6). This hot-spot area for the PrL/IL oDA signals corresponds to the same area where we found the highest density of CINs in our analysis. Thus, CIN density could contribute in part to the generation of this hot-spot area for the PrL/IL-evoked DA signals. Further, we also found that this area has the highest intensity of fluorescently labeled PrL/IL fibers, as well as the highest degree of synaptic connections to CINs, which is also in agreement with reported patterns of cortical connectivity (Voorn et al., 2004). Our initial interest was to study the PFC projections to the accumbens. However, since PrL/IL-evoked DA signals were largest and more reliable in the ventral DS (Fig. 6), recordings were performed in this area. Together, these findings suggest that PrL/IL inputs and the synaptic innervation to CINs are preferentially localized to the dorsomedial subregion, giving rise to the largest cortically evoked DA signals.
From the mechanism described here for this PrL/IL-evoked DA signals, we propose that any input to the striatum that strongly activates CINs (cortical, thalamic, or any other brain region) can in theory evoke local DA signals in the striatum. We also speculate, based on ours and other groups' works, that these glutamate-driven DA signals can similarly happen in other striatal subregions. The selective localization of PrL/IL-evoked DA signals suggests the existence of a “topographic map” for the local DA signals where inputs from different cortical subregions can trigger local DA signals at the striatal subregion which they innervate. Spatial segregation of input-specific DA signals in the striatum is a new concept introduced here that is rarely considered in the field. This concept may become helpful in understanding how DA signals in the striatum can be involved in so many diverse processes ranging from reinforcement learning and motivation to motor output and action selection.
Footnotes
This work was supported by the Intramural Programs of National Institute on Alcohol Abuse and Alcoholism, National Institute of Neurological Disorders and Stroke (ZIA-AA000421), and National Institute on Drug Abuse. We thank Roland Bock (National Institute on Alcohol Abuse and Alcoholism, National Institutes of Health) for development of the VIGOR acquisition and analysis software; Drs. Karl Deisseroth (Stanford University) and Ed Boyden for providing the channelrhodopsin-2 and ChrimsonR constructs, respectively; and members of the A.A.V. laboratory for valuable comments on the manuscript.
The authors declare no competing financial interests.
References
- Adrover MF, Shin JH, Alvarez VA (2014) Glutamate and dopamine transmission from midbrain dopamine neurons share similar release properties but are differentially affected by cocaine. J Neurosci 34:3183–3192. 10.1523/JNEUROSCI.4958-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backman CM, Malik N, Zhang Y, Shan L, Grinberg A, Hoffer BJ, Westphal H, Tomac AC (2006) Characterization of a mouse strain expressing Cre recombinase from the 3′ untranslated region of the dopamine transporter locus. Genesis 44:383–390. 10.1002/dvg.20228 [DOI] [PubMed] [Google Scholar]
- Burke DA, Rotstein HG, Alvarez VA (2017) Striatal local circuitry: a new framework for lateral inhibition. Neuron 96:267–284. 10.1016/j.neuron.2017.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cachope R, Mateo Y, Mathur BN, Irving J, Wang HL, Morales M, Lovinger DM, Cheer JF (2012) Selective activation of cholinergic interneurons enhances accumbal phasic dopamine release: setting the tone for reward processing. Cell Rep 2:33–41. 10.1016/j.celrep.2012.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuhma N, Mingote S, Moore H, Rayport S (2014) Dopamine neurons control striatal cholinergic neurons via regionally heterogeneous dopamine and glutamate signaling. Neuron 81:901–912. 10.1016/j.neuron.2013.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cover KK, Gyawali U, Kerkhoff WG, Patton MH, Mu C, White MG, Marquardt AE, Roberts BM, Cheer JF, Mathur BN (2019) Activation of the rostral intralaminar thalamus drives reinforcement through striatal dopamine release. Cell Rep 26:1389–1398.e3. 10.1016/j.celrep.2019.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floresco SB, Yang CR, Phillips AG, Blaha CD (1998) Basolateral amygdala stimulation evokes glutamate receptor-dependent dopamine efflux in the nucleus accumbens of the anaesthetized rat. Eur J Neurosci 10:1241–1251. 10.1046/j.1460-9568.1998.00133.x [DOI] [PubMed] [Google Scholar]
- Franklin KB, Paxinos G (2007) The mouse brain in stereotaxic coordinates, Ed 3 Amsterdam: Elsevier. [Google Scholar]
- Gerfen CR. (2000) Dopamine-mediated gene regulation in models of Parkinson's disease. Ann Neurol 47:S42–S50; discussion S50-S52. [PubMed] [Google Scholar]
- Gerfen CR, Surmeier DJ (2011) Modulation of striatal projection systems by dopamine. Annu Rev Neurosci 34:441–466. 10.1146/annurev-neuro-061010-113641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorguieff MF, Le Floc'h ML, Westfall TC, Glowinski J, Besson MJ (1976) Nicotinic effect of acetylcholine on the release of newly synthesized (3H)dopamine in rat striatal slices and cat caudate nucleus. Brain Res 106:117–131. 10.1016/0006-8993(76)90077-9 [DOI] [PubMed] [Google Scholar]
- Giorguieff MF, Kemel ML, Glowinski J (1977) Presynaptic effect of L-glutamic acid on the release of dopamine in rat striatal slices. Neurosci Lett 6:73–77. 10.1016/0304-3940(77)90068-4 [DOI] [PubMed] [Google Scholar]
- Hass CA, Glickfeld LL (2016) High-fidelity optical excitation of cortico-cortical projections at physiological frequencies. J Neurophysiol 116:2056–2066. 10.1152/jn.00456.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DF, Parent KL, Atcherley CW, Cowen SL, Heien ML (2018) Differential release of dopamine in the nucleus accumbens evoked by low-versus high-frequency medial prefrontal cortex stimulation. Brain Stimul 11:426–434. 10.1016/j.brs.2017.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyland BI, Reynolds JN, Hay J, Perk CG, Miller R (2002) Firing modes of midbrain dopamine cells in the freely moving rat. Neuroscience 114:475–492. 10.1016/s0306-4522(02)00267-1 [DOI] [PubMed] [Google Scholar]
- Johnson KA, Mateo Y, Lovinger DM (2017) Metabotropic glutamate receptor 2 inhibits thalamically-driven glutamate and dopamine release in the dorsal striatum. Neuropharmacology 117:114–123. 10.1016/j.neuropharm.2017.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapoetke NC, Murata Y, Kim SS, Pulver SR, Birdsey-Benson A, Cho YK, Morimoto TK, Chuong AS, Carpenter EJ, Tian Z, Wang J, Xie Y, Yan Z, Zhang Y, Chow BY, Surek B, Melkonian M, Jayaraman V, Constantine-Paton M, Wong GK, et al. (2014) Independent optical excitation of distinct neural populations. Nat Methods 11:338–346. 10.1038/nmeth.2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosillo P, Zhang YF, Threlfell S, Cragg SJ (2016) Cortical control of striatal dopamine transmission via striatal cholinergic interneurons. Cereb Cortex 26:4160–4169. 10.1093/cercor/bhw252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri AK, Bevan MD (2020) Dopaminergic transmission rapidly and persistently enhances excitability of D1 receptor-expressing striatal projection neurons. Neuron 106:277–290.e6. 10.1016/j.neuron.2020.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leviel V, Gobert A, Guibert B (1990) The glutamate-mediated release of dopamine in the rat striatum: further characterization of the dual excitatory-inhibitory function. Neuroscience 39:305–312. 10.1016/0306-4522(90)90269-a [DOI] [PubMed] [Google Scholar]
- Liu C, Kaeser PS (2019) Mechanisms and regulation of dopamine release. Curr Opin Neurobiol 57:46–53. 10.1016/j.conb.2019.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Kershberg L, Wang J, Schneeberger S, Kaeser PS (2018) Dopamine secretion is mediated by sparse active zone-like release sites. Cell 172:706–718.e15. 10.1016/j.cell.2018.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscher C, Robbins TW, Everitt BJ (2020) The transition to compulsion in addiction. Nat Rev Neurosci 21:247–263. 10.1038/s41583-020-0289-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H (2010) A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13:133–140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamaligas AA, Cai Y, Ford CP (2016) Nicotinic and opioid receptor regulation of striatal dopamine D2-receptor mediated transmission. Sci Rep 6:37834. 10.1038/srep37834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamaligas AA, Barcomb K, Ford CP (2019) Cholinergic transmission at muscarinic synapses in the striatum is driven equally by cortical and thalamic inputs. Cell Rep 28:1003–1014.e3. 10.1016/j.celrep.2019.06.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo Y, Johnson KA, Covey DP, Atwood BK, Wang HL, Zhang S, Gildish I, Cachope R, Bellocchio L, Guzman M, Morales M, Cheer JF, Lovinger DM (2017) Endocannabinoid actions on cortical terminals orchestrate local modulation of dopamine release in the nucleus accumbens. Neuron 96:1112–1126.e5. 10.1016/j.neuron.2017.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda W, Furuta T, Nakamura KC, Hioki H, Fujiyama F, Arai R, Kaneko T (2009) Single nigrostriatal dopaminergic neurons form widely spread and highly dense axonal arborizations in the neostriatum. J Neurosci 29:444–453. 10.1523/JNEUROSCI.4029-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior JR, Ferris MJ, Stuber GD, Riddle DR, Jones SR (2015) Optogenetic versus electrical stimulation of dopamine terminals in the nucleus accumbens reveals local modulation of presynaptic release. J Neurochem 134:833–844. 10.1111/jnc.13177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn DM. (1987) Acetylcholinesterase: enzyme structure, reaction dynamics, and virtual transition states. Chem Rev 87:955–979. 10.1021/cr00081a005 [DOI] [Google Scholar]
- Quiroz C, Orrú M, Rea W, Ciudad-Roberts A, Yepes G, Britt JP, Ferré S (2016) Local control of extracellular dopamine levels in the medial nucleus accumbens by a glutamatergic projection from the infralimbic cortex. J Neurosci 36:851–859. 10.1523/JNEUROSCI.2850-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi J, Balthasar N, Olson D, Scott M, Berglund E, Lee CE, Choi MJ, Lauzon D, Lowell BB, Elmquist JK (2011) Melanocortin-4 receptors expressed by cholinergic neurons regulate energy balance and glucose homeostasis. Cell Metab 13:195–204. 10.1016/j.cmet.2011.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz W, Apicella P, Ljungberg T (1993) Responses of monkey dopamine neurons to reward and conditioned stimuli during successive steps of learning a delayed response task. J Neurosci 13:900–913. 10.1523/JNEUROSCI.13-03-00900.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu N, Duan SM, Hori T, Oomura Y (1990) Glutamate modulates dopamine release in the striatum as measured by brain microdialysis. Brain Res Bull 25:99–102. 10.1016/0361-9230(90)90258-2 [DOI] [PubMed] [Google Scholar]
- Shin JH, Adrover MF, Alvarez VA (2017) Distinctive modulation of dopamine release in the nucleus accumbens shell mediated by dopamine and acetylcholine receptors. J Neurosci 37:11166–11180. 10.1523/JNEUROSCI.0596-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JH, Adrover MF, Wess J, Alvarez VA (2015) Muscarinic regulation of dopamine and glutamate transmission in the nucleus accumbens. Proc Natl Acad Sci USA 112:8124–8129. 10.1073/pnas.1508846112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulzer D, Cragg SJ, Rice ME (2016) Striatal dopamine neurotransmission: regulation of release and uptake. Basal Ganglia 6:123–148. 10.1016/j.baga.2016.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surmeier DJ, Ding J, Day M, Wang Z, Shen W (2007) D1 and D2 dopamine-receptor modulation of striatal glutamatergic signaling in striatal medium spiny neurons. Trends Neurosci 30:228–235. 10.1016/j.tins.2007.03.008 [DOI] [PubMed] [Google Scholar]
- Taber MT, Fibiger HC (1993) Electrical stimulation of the medial prefrontal cortex increases dopamine release in the striatum. Neuropsychopharmacology 9:271–275. 10.1038/npp.1993.63 [DOI] [PubMed] [Google Scholar]
- Taber MT, Fibiger HC (1994) Cortical regulation of acetylcholine release in rat striatum. Brain Res 639:354–356. 10.1016/0006-8993(94)91754-x [DOI] [PubMed] [Google Scholar]
- Threlfell S, Clements MA, Khodai T, Pienaar IS, Exley R, Wess J, Cragg SJ (2010) Striatal muscarinic receptors promote activity dependence of dopamine transmission via distinct receptor subtypes on cholinergic interneurons in ventral versus dorsal striatum. J Neurosci 30:3398–3408. 10.1523/JNEUROSCI.5620-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threlfell S, Lalic T, Platt NJ, Jennings KA, Deisseroth K, Cragg SJ (2012) Striatal dopamine release is triggered by synchronized activity in cholinergic interneurons. Neuron 75:58–64. 10.1016/j.neuron.2012.04.038 [DOI] [PubMed] [Google Scholar]
- Tritsch NX, Sabatini BL (2012) Dopaminergic modulation of synaptic transmission in cortex and striatum. Neuron 76:33–50. 10.1016/j.neuron.2012.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritschler L, Kheirbek MA, Dantec YL, Mendez-David I, Guilloux JP, Faye C, Doan J, Pham TH, Hen R, David DJ, Gardier AM (2018) Optogenetic activation of granule cells in the dorsal dentate gyrus enhances dopaminergic neurotransmission in the nucleus accumbens. Neurosci Res 134:56–60. 10.1016/j.neures.2017.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal CJ, Chai MS, Plummer DT (1987) The effect of temperature on the activity of acetylcholinesterase preparations from rat brain. Neurochem Int 11:135–141. 10.1016/0197-0186(87)90001-5 [DOI] [PubMed] [Google Scholar]
- Voorn P, Vanderschuren LJ, Groenewegen HJ, Robbins TW, Pennartz CM (2004) Putting a spin on the dorsal-ventral divide of the striatum. Trends Neurosci 27:468–474. 10.1016/j.tins.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Wang L, Shang S, Kang X, Teng S, Zhu F, Liu B, Wu Q, Li M, Liu W, Xu H, Zhou L, Jiao R, Dou H, Zuo P, Zhang X, Zheng L, Wang S, Wang C, Zhou Z (2014) Modulation of dopamine release in the striatum by physiologically relevant levels of nicotine. Nat Commun 5:3925. 10.1038/ncomms4925 [DOI] [PubMed] [Google Scholar]
- Zhang H, Sulzer D (2004) Frequency-dependent modulation of dopamine release by nicotine. Nat Neurosci 7:581–582. 10.1038/nn1243 [DOI] [PubMed] [Google Scholar]
- Zhou FM, Liang Y, Dani JA (2001) Endogenous nicotinic cholinergic activity regulates dopamine release in the striatum. Nat Neurosci 4:1224–1229. 10.1038/nn769 [DOI] [PubMed] [Google Scholar]
- Zhu J, Hexum TD (1992) Characterization of cocaine-sensitive dopamine uptake in PC12 cells. Neurochem Int 21:521–526. 10.1016/0197-0186(92)90083-4 [DOI] [PubMed] [Google Scholar]