Abstract
Purpose of Review
This review addresses normal and pathologic functions of serum amyloid A (SAA), an enigmatic biomarker of inflammation and protein precursor of AA amyloidosis, a life-threatening complication of chronic inflammation. SAA is a small, highly evolutionarily conserved acute-phase protein whose plasma levels increase up to one thousand-fold in inflammation, infection, or after trauma. The advantage of this dramatic but transient increase is unclear, and the complex role of SAA in immune response is intensely investigated. This review summarizes recent advances in our understanding of the structure-function relationship of this intrinsically disordered protein, outlines its newly emerging beneficial roles in lipid transport and inflammation control, and discusses factors that critically influence its misfolding in AA amyloidosis.
Recent Findings
High-resolution structures of lipid-free SAA in crystals and fibrils have been determined by x-ray crystallography and electron cryo-microscopy. Low-resolution structural studies of SAA-lipid complexes, together with biochemical, cell-based, animal model, genetic, and clinical studies, have provided surprising new insights into a wide range of SAA functions. An emerging vital role of SAA is lipid encapsulation to remove cell membrane debris from sites of injury. The structural basis for this role has been proposed. The lysosomal origin of AA amyloidosis has solidified, and its molecular and cellular mechanisms have emerged.
Summary
Recent studies have revealed molecular underpinnings for understanding complex functions of this Cambrian protein in lipid transport, immune response, and amyloid formation. These findings help guide the search for much-needed targeted therapies to block the protein deposition in AA amyloidosis.
Keywords: Inflammation control and immunity; Acute-phase response; Intrinsically disordered protein; Apolipoprotein structure, dynamics, and function; Protein-lipid interactions; Systemic amyloidosis
Introduction
Serum amyloid A (SAA) is a family of ~ 12 kDa acute-phase proteins named after a disease. It is known not so much for its beneficial role in host defense but rather as a biomarker of inflammation [1•, 2] and protein precursor of amyloid A (AA) amyloidosis, a life-threatening complication of chronic inflammation [3•, 4]. Plasma levels of SAA are elevated in bacterial infections such as tuberculosis, viral infections such as hepatitis C, autoimmune disorders such as rheumatoid arthritis, autoinflammatory disorders such as familial Mediterranean fever, and metastatic cancers [1•, 2, 5, 6, 7•]. Clinical studies of viral infections in humans [5, 8, 9], including COVID-19 [10], suggest that SAA is a sensitive biomarker for early diagnostics and treatment and a predictor of the viral disease outcome. Mouse model studies report that SAA3 is a sensitive biomarker of excessive inflammation upon vaccination [11] and of macrophage infiltration of the adipose tissue in obesity [12]. Whether SAA is not just a marker but also an active participant in disease is intensely investigated, and numerous, sometimes conflicting, functions have been ascribed to this mysterious protein. The current consensus is that SAA modulates adaptive and innate immunity and mediates lipid transport during inflammation [1•, 2, 13].
Most circulating SAA binds plasma HDL, thereby mobilizing cholesterol for cell repair [14] but contributing to impaired HDL functionality in inflammation [15–17]. Elevated plasma SAA emerged as a causal risk factor for atherosclerosis [18–22, 23•]. While it is clear that chronically elevated SAA contributes to the pathogenesis in AA amyloidosis and atherosclerosis, beneficial effects of SAA are less well-understood. Importantly, following acute infection, inflammation, injury, or surgery, plasma levels of SAA increase over 24–48 h from the basal level of 1–3 μg/ml up to 1–3 mg/ml and then rapidly decline ([24, 25, 26•, 27] and references therein). The benefits for survival of this dramatic but transient increase are beginning to emerge from the research summarized below.
Overview of SAA Functions
SAA has been highly evolutionarily conserved for ~ 500 million years, suggesting that it performs a vital function in surviving injury and infection. Multiple functions have been reported for SAA depending on its isoform, expression site, lipidation status, and other factors. Of the four major SAA isoforms, acute-phase SAA1 and SAA2 (in mice and men) and SAA3 (in mice) are expressed in response to pro-inflammatory cytokines such as IL1, IL6, and TNF-α, while SAA4 is constitutively expressed [20, 24, 28, 29•, 30]. SAA is secreted mainly by hepatocytes into plasma, where it reversibly binds to HDL [29•, 31] but can also form transient SAA-only lipoproteins that are distinct from HDL [32•]. SAA can bind and activate HDL receptor SR-B1 and other cell scavenger receptors such as LOX1 and RAGE that bind modified lipoproteins; by doing so, SAA can reroute HDL transport from the reverse cholesterol transport pathway, whereby excess cellular cholesterol is removed from the body via bile, to cholesterol recycling (reviewed in [14]). SAA can also activate cell receptors involved in immune response, including TLR2 and TLR4 (reviewed in [1•, 27]). In addition to hepatic secretion, SAA is secreted locally by various cells, particularly by macrophages at the inflammation sites where it promotes cytokine production and immune cell recruitment (reviewed in [33]). SAA was reported to either promote [34] or inhibit macrophage differentiation into osteoclasts [35–37]; the latter retains macrophages for host defense but promotes bone loss in inflammation. Furthermore, SAA has been ascribed pro- or antioxidant properties [38, 39] as well as pro-inflammatory [16, 35, 40–43, 44•] or anti-inflammatory effects in various systems [45–49]. Although some discrepancies stem from bacterial contaminations and amino acid substitutions in commercial preparations of recombinant hSAA1 [50•], others reflect structural and functional differences between lipid-free and lipid-bound protein [38, 40] as well as isoform-specific and context-dependent effects. Overall, SAA has emerged as a homeostatic regulator of inflammation (reviewed in [1, 51]).
According to animal model studies, SAA induces protection against gram-negative bacteria and the lipopolysaccharide challenge [45–47, 48•]. The protective mechanisms involve SAA binding to lipopolysaccharides [46], binding and opsonizing bacterial outer-membrane proteins, and microsolubilizing the bacterial membrane [48•]. Antiviral effects of SAA are less well-explored; studies of hepatitis C suggest that SAA binds to viral glycoproteins and blocks the host cell entry via SR-B1 receptor (reviewed in [5, 6]). Another beneficial function of SAA is transport of retinol, a bioactive derivative of vitamin A that regulates innate intestinal immunity [52•, 53•, 54].
In summary, two major interconnected SAA functions have emerged: transport of lipids and lipophilic molecules and signaling in innate and adaptive immunity to control inflammation. These and other functions and their underlying genetic bases have been comprehensively reviewed elsewhere [24, 25, 26•, 27, 33]. Other excellent reviews have addressed the SAA-related pathologies in atherosclerosis [20, 21, 23•] and AA amyloidosis in animals and humans, from the cellular and molecular origins of the disease to its transmissibility, clinical challenges, and therapeutic strategies [3•, 4, 55–57]. Here, we summarize recent progress in uncovering the structural basis for the evolutionarily conserved functions of SAA and the factors critical for AA fibrillogenesis.
Functional Role of Intrinsic Structural Disorder
Multiple functions of SAA stem from its ability to bind diverse ligands. These include cell receptors involved in host defense and/or lipid metabolism (TLRs, RAGE, SR-B1, CLA-1, LOX1, P2X7, FPR2), lipids (cholesterol, zwitterionic and anionic phospholipids, lyso-phospholipids, non-esterified fatty acids (NEFA)), small lipophilic molecules (retinol), bacterial outer-membrane proteins (OmpA), basal membrane proteins (fibronectin, laminin), plasma proteins (cystatin), anions (heparan sulfate, lipopolysaccharides), and cations (Ca2+), to name a few ([26•, 58•] and references therein). These observations provoke the following question: how can a small protein of nearly 100 residues bind so many diverse ligands?
We posit that promiscuous ligand binding by SAA is facilitated by its pliable conformation [58•, 59]. Binding promiscuity is characteristic of other intrinsically disordered proteins ([60–62] and references therein). Such proteins or their domains have disordered secondary and/or tertiary structures in the absence of bound ligands, but upon binding they can fold to optimize interactions with each ligand individually. For example, in solution at near-physiologic pH and temperature, lipid-free murine SAA1 (mSAA1) is largely unfolded, but upon lipid binding, it becomes 25–50% α-helical (as measured by circular dichroism) depending on the nature and the amount of the lipid [63–67]. This structural change underlies functional differences between lipid-free and lipid-associated SAA ([32•] and references therein). Moreover, in the presence of a protein-folding osmolyte trimethylamine N-oxide, the α-helical content in mSAA1 in solution at 4 °C approaches ~ 75% (unpublished data). This maximally folded protein state probably resembles that depicted by x-ray crystallography.
X-ray Crystal Structures Reveal a Unique Protein Fold with a Concave Hydrophobic Surface
Remarkably for an intrinsically disordered protein, x-ray crystal structures of SAA have been determined to nearly 2 Å resolution by two independent teams. Four different crystal structures have been reported to date: two of human SAA1 (hSAA1) [68••] and two of murine SAA3 (mSAA3) [51, 52•]. In these crystals, the protein molecules are packed as a dimer, a dimer of dimers, a trimer, or a hexamer, forming different lattice contacts in different unit cells. Nevertheless, all structures showed a very similar monomer fold, suggesting that this fold has been evolutionally conserved [51] (Fig. 1).
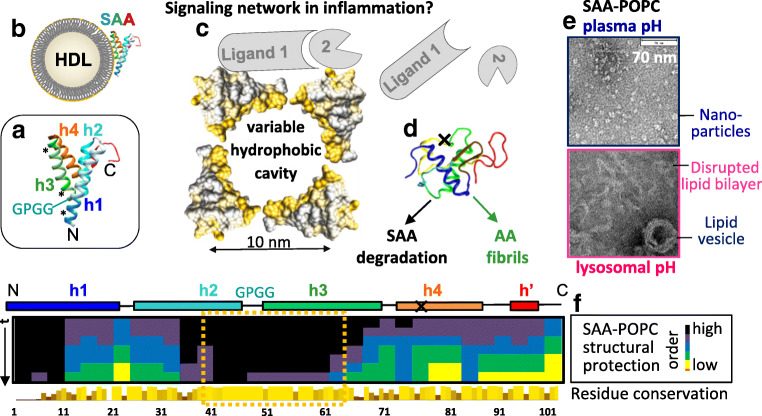
Fig. 1.
Current understanding of the structure-function relationship in SAA. (A) Ribbon diagram showing the x-ray crystal structure of hSAA1 [68••]; mSAA3 forms a very similar fold [52•]. The α-helices h1–h4 are rainbow-colored blue to red from the N- to the C-terminus. The GPGG motif in h2–h3 linker, which forms a tight interhelical turn that defines the curvature of the hydrophobic surface formed by h1 and h3 [58•], is marked; * indicates segments that are predicted to initialize hSAA1 misfolding in amyloid (residues 2–9, 53–55, and 67–70 in h1 and h3) [69]. (B) Cartoon showing SAA bound to HDL via h1 and h3. The h1–h3 segment is mostly α-helical in lipid-bound state at pH ~ 7, while the rest of the molecule lacks a stable structure [70•]. (C) Cartoon representation of an SAA-only lipoprotein. Four protein copies in a space-filling representation are shown and color-coded (yellow, hydrophobic; gray, hydrophilic). Variable hydrophobic cavity formed by helices h1 and h3 from several SAA copies can sequester diverse lipids and lipophilic molecules (e.g., retinol [53•]). Other SAA sites can bind other ligands (marked 1 and 2), potentially facilitating their interactions and signaling in inflammation [58•]. We hypothesize that such signaling via SAA-containing lipoprotein hubs may occur during acute-phase response when SAA levels are high. Upon resolution of inflammation, the SAA levels drop and these dynamic networks dissociate. (D) Lipid-free SAA, which is intrinsically disordered in solution circa pH 7, is rapidly cleared from circulation [28], yet if high SAA levels persist, it can accumulate in lysosomes and form AA deposits [3•, 71•]. N- and C-terminal truncations by the lysosomal protease cathepsin B probably contribute to this process [72, 73]; X marks a major site of the C-terminal truncation. (E) Electron micrographs of negatively stained SAA-POPC complexes. At near-neutral pH, SAA forms lipoprotein nanoparticles. However, at near-lysosomal pH, SAA oligomers disrupt lipid vesicles and cell membranes and form amyloid [71•, 74]. (F) Well-ordered protein region in SAA-containing lipoproteins is highly evolutionarily conserved. Protein structural protection in SAA-POPC nanoparticles was measured by hydrogen-deuterium exchange mass spectrometry at several time points; darker colors indicate better-ordered regions. Bar graph shows amino acid conservation throughout evolution; taller lighter bars indicate more conserved residues (for details see [70•]). Boxed region contains h2–h3 segment that is most evolutionarily conserved and is most well-ordered in lipid-bound SAA; this region contains the GPGG motif in the h2–h3 linker that defines the curvature of the concave lipid binding site formed by h1 and h3; the amphipathic character of h1 and h3 is also evolutionarily conserved [58•]. Consequently, the ability of SAA to encapsulate lipids into nanoparticles has been highly evolutionarily conserved and probably reflects its vital primordial function.
SAA monomer folds into a Y-shaped helix bundle containing α-helices h1–h4, followed by a 3/10 helix h’ and stabilized by a flexible C-terminal tail [51, 68••] (Fig. 1A). Helices h1 and h3 are predicted and observed to have a strong amphipathic character with a large, well-demarcated hydrophobic face [53•, 58•]. Similar amphipathic α-helices comprise the lipid surface-binding motif in other apolipoproteins; however, the helical packing in SAA is distinctly different.
Helix bundles in other globular proteins, including soluble lipid-free apolipoproteins apoA-I and apoE, have hydrophobic interiors and polar surfaces that confer protein solubility. These bundles readily open to expose the hydrophobic helical faces for binding lipids. In contrast, the SAA helix bundle has a hydrophilic interior partially filled with water [51] and a large hydrophobic surface approximately 1 nm × 4 nm (Fig. 1C, in yellow). This concave surface comprised the apolar faces of h1 and h3 packed at an angle of 43°; this angle defines the radius of surface curvature, r ~ 4.5 nm, which is comparable with the radius of HDL [58•]. Such a shape complementarity combined with surface hydrophobicity suggests that HDL fits neatly into the concave surface site of SAA (Fig. 1B).
Molecular Fold of SAA Has Been Evolutionarily Conserved for Function
HDL binding at the concave surface formed by h1 and h3 helps fit together several pieces of the SAA puzzle. First, it explains higher affinity of SAA for HDL (diameter d = 8–12 nm) vis-à-vis larger lipoproteins, LDL (d = 20–24 nm), and VLDL (d = 40–100 nm) ([58•] and references therein). Second, it is consistent with the demonstrated role of h1 in binding cholesterol [75] and HDL and h3 in binding retinol ([51, 52•, 68••] and references therein). Third, HDL binding at this site blocks the amyloidogenic residue segments in h1 and h3 that likely initiate SAA misfolding in amyloid [69, 76, 77] (Fig. 1A, asterisks); this explains why binding to HDL protects SAA from forming amyloid at pH ~ 7 [66, 71•, 77]. Additional indirect support for HDL binding at this site comes from the crystal structure of the SAA-retinol complex showing a retinol molecule bound in the hydrophobic cavity formed by h1 and h3 from three SAA molecules [53•]. Nevertheless, one can question the relevance of this well-ordered crystal structure, which is ~ 75% α-helical, to the functional protein conformation, which is largely unfolded in ligand-free SAA in solution and is up to 50% helical in lipid-bound SAA at 37 °C, pH ~ 7.
To explore functional conformations of SAA, hydrogen-deuterium exchange mass spectrometry was combined with circular dichroism spectroscopy and molecular dynamic simulations [70•]. Lipid-free mSAA1 in solution was compared with ~ 10 nm particles reconstituted from mSAA1 and a model lipid POPC; nanoparticles such as these are often used as models of nascent HDL [63, 64]. The results revealed that most helical structure in lipid-free and in lipid-bound SAA is confined to the h1–h3 segment (residues 1–69), while residue segment 70–104 lacks a stable structure; h1 and h3 form the lipid binding site wherein h3 is partially unstructured in solution but becomes helical upon lipid binding (Fig. 1F). Remarkably, locations of the helical segments and the interhelical linkers were similar in lipid-free SAA in solution, SAA-POPC nanoparticles, and SAA crystals, despite large differences in their α-helical content. Consequently, the crystal structures depict key aspects of functional conformational ensemble of SAA and likely represent its maximally folded state [70•].
Importantly, amino acid sequence analysis in the SAA protein family indicates that the lipid-binding site is highly evolutionarily conserved, from sea cucumber to human. In particular, the amphipathic character of h1 and h3 and the residues essential for their packing against each other are 100% conserved; this includes the GPGG motif that forms an unusually tight well-ordered turn between h2 and h3 that defines the curvature of the lipid-binding site [58•]. Importantly, the most evolutionarily conserved regions, which are located in segments from h1 and h2–h3, show the greatest structural protection in SAA-POPC complexes, implicating these regions in lipid binding (Fig. 1F). Consequently, the concave hydrophobic surface in SAA has been evolutionarily conserved to sequester lipids [70•].
SAA Acts as a Detergent and a Lipid Scavenger
Reconstituted SAA-POPC particles vary in size (8–18 nm) depending on the protein-to-lipid ratio and the preparation technique [65]; 8–18 nm SAA-lipid particles were also observed in cell-based studies [32•]. Even the most stable 8–10 nm SAA-POPC particles isolated by gel filtration formed a ladder on the native gel, suggesting different numbers of SAA copies per particle [65]. Unlike HDL, such HDL-size SAA-POPC particles contain more protein than lipid by weight, and the amount of lipid in each particle is insufficient to form a bilayer [65]. Therefore, in contrast to nascent HDL or model nanodisks, such protein-rich SAA-POPC nanoparticles must be micelle-like, with the lipid acyl chains sequestered in the hydrophobic cavity formed by several protein molecules. The size and shape of this cavity must vary depending on the number of the SAA molecules per particle and their packing. Similarly, in the crystal structures, the shape and size of the hydrophobic cavity formed by several SAA molecules vary depending on their packing [52•, 53•, 68••]. Such a variable hydrophobic cavity is expected to bind a wide array of small hydrophobic or amphipathic molecules and encapsulate them into nanoparticles (Fig. 1C).
This concept is supported by in vitro studies showing that SAA can spontaneously solubilize various liposomes and convert them into nanoparticles at pH ~ 7 [63, 66, 67] (Fig. 1E, top). This includes single- and multilamellar vesicles of zwitterionic and anionic phospholipids and their mixtures with cholesterol, oxidized lipids, lyso-phospholipids, NEFA, etc. [38, 39, 63, 65–67, 77].
Such a detergent-like activity comes in handy in vivo. One example is bactericidal activity of SAA against E. coli and S. aureus, which was observed in vitro and in vivo in a mouse model of cutaneous infection [48•]. This activity stems from the ability of SAA to solubilize bacterial cell membranes and anionic liposomes to form micelles or small vesicles at pH 5.2–6.4, which encompasses acidic conditions of the skin. The lipid-binding N-terminal region of SAA was essential for this activity [48•]. The detergent-like activity of SAA can also be important in the oxidative and hydrolytic environment at the inflammation sites. Lipid peroxides, lyso-phospholipids, and NEFA generated at these sites must be rapidly sequestered to avoid lipotoxicity. However, the levels of albumin, which normally sequesters most circulating NEFA and lyso-phospholipids, decrease in acute inflammation. Moreover, albumin’s affinity for NEFA decreases at acidic pH at the inflammation sites ([78•] and references therein), causing potential accumulation of toxic lipids. Can SAA compensate for albumin deficiency and protect the inflammation sites from lipotoxicity?
The answer is suggested by the effects of SAA on the activity of another ancient acute-phase reactant, secretory phospholipase A2 (sPLA2). Apolipoproteins such as apoA-I have long been known to stimulate various lipases by altering the physical state of lipid [79], but the levels of these proteins decrease in acute inflammation while SAA sharply increases. In vivo, SAA and sPLA2 are upregulated dramatically and concomitantly, both systemically and locally at the sites of injury [40], suggesting a potential synergy. In vitro studies support this idea and show that SAA plays dual role in augmenting the sPLA2 reaction. First, SAA solubilizes phospholipids to generate nanoparticles that provide substrates for sPLA2; second, SAA sequesters its water-insoluble products, lyso-phospholipids, and NEFA, thereby removing the rate-limiting step of lipolysis [78•]. Moreover, unlike albumin, the binding affinity of SAA for NEFA remains invariant in a broad pH range, surpassing that of albumin at acidic conditions found at the inflammation sites [78•], while the local SAA levels at these sites increase even more than its systemic levels. Together, the results suggest that SAA can help compensate for albumin’s deficiency and substantially contribute to removal of toxic lipid from the inflammation sites. Such an efficient removal of cell membrane debris is necessary for tissue healing, supporting the idea that acute-phase response serves to isolate the pathogenic species and minimize tissue damage [1•].
Hypothetical Functions of SAA-Containing Lipoproteins in Inflammation
Are lipoproteins that have SAA as their major protein found in vivo? Earlier mouse model studies detected such lipoproteins during lipopolysaccharide-induced inflammation; these HDL-size particles contained SAA as their sole protein and had higher density and, hence, higher protein content than HDL [80]. Spontaneous formation of micelle-like SAA-lipid nanoparticles was also observed in mice infected by S. aureus [48•]. Furthermore, several cell-based studies reported formation of SAA-only lipoproteins [32, 81, 82]. In some systems, this formation depended on the activity of lipid transporters such as ABCA1, the ATP-driven transporter in the plasma membrane that is necessary to generate HDL [32•, 82]. In other studies, SAA-only nanoparticles formed even in the absence of active lipid transport [81].
Since ATP-driven lipid transport is impaired in dead cells, the ability of SAA to encapsulate membrane lipids in an ATP-independent process has important implications for removing membrane debris from dead cells. We hypothesize that SAA-only lipoproteins can form in an energy-independent process at the sites of injury where SAA concentration is particularly high; they sequester membrane lipids from dead cells and incorporate them into nanoparticles. SAA-only nanoparticles are probably short-lived compared with HDL, as suggested by much faster clearance of plasma SAA [28] and much lower structural stability of free SAA and SAA-containing lipoproteins as compared with their apoA-I-containing counterparts [32•, 38, 63, 65]. Like other lipoproteins, these transient SAA-containing nanoparticles are expected to be heterogeneous in size and composition; they probably vary in the numbers of SAA copies per particle, carry diverse lipids and their degradation and oxidation products, and undergo dynamic remodeling by sPLA2 and other factors. Future in vivo studies will test these ideas.
What is the destination of the SAA-containing lipoproteins? One pathway involves cell scavenger receptors that bind such lipoproteins. Examples include LOX1, a receptor for oxidized lipoproteins; RAGE, a receptor for advanced glycation end products; and other scavenger receptors expressed on the surface of various cells, particularly macrophages. SAA can bind and activate these receptors, promoting the uptake of lipids and SAA by macrophages and other cells. This mechanism was proposed to redirect HDL during inflammation and recycle “good cholesterol” for cell repair [14]; however, it can also contribute to the pro-atherogenic lipid accumulation in the arterial macrophages. Another pro-atherogenic pathway involves enhanced retention of SAA-containing lipoproteins by arterial glycosaminoglycans (GAGs) ([16] and references therein). Ultimately, SAA undergoes lysosomal degradation; however, when SAA levels are persistently high, its lysosomal accumulation can lead to AA amyloidosis (described below).
Although molecular details of SAA interactions with cell receptors and GAGs are unknown, they probably involve flexible C-terminal region of SAA ([58•] and references therein). In particular, an acidic patch formed by h2 and h4 on the surface of hSAA1 or mSAA1 was proposed to interact with the basic apex implicated in ligand binding by SR-B1 and homologous receptors such as LOX1, RAGE, and CD36 [65], whereas basic residues from the C-terminal tail of SAA were inferred to bind GAGs [83].
One intriguing hypothesis stems from the ligand binding promiscuity of SAA and the presence of multiple SAA copies on the same lipoprotein particle [64, 65]. Each SAA copy binds lipids via the h1–h3 site, while other sites can bind other ligands (Fig. 1C). Such binding can potentially bridge several ligands on the same lipoprotein, facilitating their interactions. If so, SAA-containing lipoproteins may serve as hubs facilitating dynamic interactions in signaling networks during inflammation [58•]. The ability to act as protein hubs has been ascribed to other intrinsically disordered proteins that are also promiscuous ligand binders [60–62]. What distinguishes SAA is its ability to form multivalent lipoprotein hubs. This hypothetical scenario is in line with the proposed role of HDL as platforms for binding various proteins with diverse functions ([84] and references therein) as well as with the emerging role of SAA in controlling the onset and the resolution of inflammation [1•, 26•, 49].
SAA Misfolding in AA Amyloidosis and Its Key Effectors
AA amyloidosis results from persistently elevated SAA and affects < 5% of patients with chronic inflammation [3•, 4]. In this life-threatening disease, N- and/or C-terminally truncated SAA1 fragments, termed AA, form extracellular fibrillary deposits in kidney and other organs (liver, spleen, intestine, skin, heart), causing organ damage ([72••, 85] and references therein). AA amyloidosis used to be the major form of human amyloid disease and a major cause of death in tuberculosis patients [3•], but with improved control of infection and inflammation, this disease became relatively rare. In animals (mammals and birds), AA amyloidosis remains the major form of amyloid disease [56] that can be transmitted in a prion-like manner by a seeding mechanism [55, 56]. No animal-to-human or human-to-human transmission has been reported to date.
The mechanism of organ targeting in this and other systemic amyloidoses is unclear and probably involves local proteolytic environment and GAGs, which are ubiquitous constituents of amyloid deposits [86]. Moreover, it is unclear what protease(s) generate AA fragments in vivo and whether the proteolytic cleavage precedes or follows fibrillogenesis. The common glomerular variant of the disease has been associated with 1–76 fragment, but other C-terminal truncations and an occasional N-terminal Arg1 truncation have also been reported [3•]. Mass spectrometry analysis of renal AA deposits from several patients revealed that all fragments lacked Arg1 [85]. Fibrils from all patients had similar morphology consistent with the molecular structure of human AA fibril (fragment 2–69) determined by electron cryo-microscopy to 2.7 Å resolution [72••]. This structure, together with the structure of murine AA fragment, showed a complete conversion of the native all-α into an all-β fold, with parallel in-register twisted cross-β-sheets forming the fibril core [72••]. Fibril morphology for all AA patients was similar, suggesting a similar molecular structure [85, 87]. Importantly, the fibril structure of human AA fragment was incompatible with the presence of Arg1, suggesting that its truncation precedes fibrillogenesis. Notably, SAA cleavage by cathepsin B can account for the observed N- and C-terminally truncated AA fragments, implicating this lysosomal protease in AA amyloidosis [73]. Furthermore, most C-terminal cleavage sites were found in residue segment 64–67 [85], which is helical (protected) in lipoprotein-bound SAA but is unfolded (unprotected) in lipid-free SAA [70•] (Fig. 1D). This suggests that release from lipoproteins precedes the proteolytic cleavage of SAA and generation of AA fragments that misfold and form fibrillary deposits in vivo.
This scenario is consistent with in vitro finding that the C-terminal truncation augments SAA fibrillogenesis in vitro [76, 88]. This finding is not surprising as the amyloidogenic sequence propensity resides mainly in h1 and h3 from residue segment 1–76 ([69, 88] and references therein). Similarly, truncation of Arg1 is expected to increase the amyloid-forming propensity of the residue segment 1–7 of hSAA1, RSFFSFL, which is particularly amyloidogenic [69]. “Sticky” amyloidogenic segments such as this are usually sequestered in the hydrophobic core of globular proteins but become exposed upon proteolysis [3•]. In contrast, in lipid-free SAA, this and other amyloidogenic segments are exposed on the h1–h3 surface but are sequestered upon lipid binding [58•, 70•]. This explains why binding to plasma or model HDL protects SAA against fibrillogenesis at pH ~ 7, as shown in cell-based and in biophysical studies [66, 69, 71•]. However, like in other proteins, the effects of lipids on SAA fibrillogenesis depend on the lipid composition, protein-to-lipid ratio, solvent ionic conditions, and other factors; e.g. in HDL-size particles at near-neutral pH, POPC is protective while phosphatidylethanolamine is not [66].
A distinct feature of SAA is that at near-lysosomal pH, at which it cannot form lipoprotein nanoparticles, the role of lipids in amyloid formation drastically changes. Circa pH 4.3, free mSAA1 forms unusually stable proteolysis-resistant oligomers resembling amyloid-enhancing factors, which lyse lipid bilayers and undergo a lipid-induced α-helix to β-sheet transition culminating in fibrillogenesis [74•] (Fig. 1E, bottom). Such soluble oligomers were proposed to accumulate in lysosomes, disrupt cellular membranes, and initiate extracellular amyloid deposition. This mechanism complements cell-based studies showing that mSAA1 accumulation in lysosomes of monocytes and macrophages causes membrane disruption, lysosomal leakage, and cell death, while the pre-formed amyloid seeds the extracellular fibrils [71•, 89•]. This mechanism is also consistent with electron tomography of cell-derived AA deposits showing vesicular lipid inclusions probably originating from the amyloid-forming dead cells [90•]. Together, these findings solidify the lysosomal origin of AA amyloidosis and help establish its cellular and molecular underpinnings.
Therapeutic Targeting of AA Amyloidosis
The major therapies for AA amyloidosis target the underlying inflammatory disease and include antibacterial, anti-inflammatory, and immunosuppressive drugs; dialysis and kidney transplant are used to treat advanced renal damage. The drugs reduce SAA levels, which reduces the amyloid load and reverses the disease. However, factors other than amyloid load can also contribute to clinical symptoms [91], and the existing treatments are not always efficient [92]. For example, anti-inflammatory approaches are unsuited for patients with idiopathic AA amyloidosis without the underlying chronic inflammation [3•, 93], necessitating the development of new therapies.
Some therapeutic approaches target the protein misfolding pathway. This includes interactions with GAGs, particularly heparan sulfate, that dissociate SAA from HDL and accelerate fibrillogenesis [83, 94]. Sulfonated or sulfated GAG mimetics were proposed to block SAA-GAG interactions. Unfortunately, a sulfonated small molecule drug eprodisate failed to show efficiency in phase-3 clinical trials and was discontinued in 2016 (ClinicalTrials.gov Identifier: NCT01215747).
Other approaches propose to block amyloidogenic segments of SAA by using complementary peptides [95] or lipids. In particular, decreasing triglyceride levels in the core of HDL helps retain SAA and other apolipoproteins on the HDL surface and thereby retard fibrillogenesis [96]. If so, existing triglyceride-lowering therapies hold promise for treating AA amyloidosis, including cases without the underlying inflammation. Conversely, increased plasma triglycerides are expected to augment AA amyloidosis. This idea is consistent with clinical studies reporting a direct correlation between idiopathic AA amyloidosis and obesity, a condition wherein plasma triglycerides are elevated [93, 97].
Another proposed approach uses a cell-based system to identify nontoxic inhibitors of SAA fibrillogenesis regardless of specific mechanisms of action [98]. This approach targets the protein quality control cellular machinery and has a high screening capability, which helps select candidates for future animal model studies.
Conclusions
Major strides have been made in our understanding of the structure-function relationship in this enigmatic intrinsically disordered protein. X-ray crystal structures of hSAA1 and mSAA3, which were determined to ~ 2 Å resolution in 2014 [52•, 68••] and 2019 for SAA-retinol complex [53•], were truly eye-opening. They revealed a unique “inside-out” α-helical fold, with a large concave hydrophobic surface that provides a binding site for HDL, retinol, and various lipids and their degradation products. Lipids are encapsulated in a hydrophobic cavity which comprised several SAA molecules that self-assemble into lipoprotein nanoparticles (Fig. 1). The SAA fold has been highly evolutionarily conserved since the Cambrian period, long before the emergence of HDL, suggesting that the primordial role of SAA does not involve HDL binding. Rather, it probably involves sequestration and transport of lipids, such as cell membrane debris, to avoid lipotoxicity at the sites of injury and facilitate tissue healing [78•]. If verified in vivo, this role explains how the rapid and massive generation of SAA hours after the onset of acute injury, infection, or inflammation has benefitted the host survival throughout evolution.
Numerous other functions in homeostatic control of inflammation have been attributed to SAA, and their molecular underpinnings are being elucidated. The pro-atherogenic role of elevated SAA has been firmly established [20–22, 23•]. The lysosomal origin of AA amyloidosis has solidified, and key steps in this pathogenic pathway have been revealed, from lysosomal accumulation of SAA and its cleavage (probably by cathepsin B) and misfolding to cell membrane disruption and extracellular amyloid deposition [71•, 74] to cell-to-cell transfer [89•]. High-resolution fibril structures of human and murine AA fragments were determined by electron cryo-microscopy [72••], revealing key role of the N-terminal truncation in hSAA. These and other findings guide the search of new targeted therapies for AA amyloidosis.
In addition, numerous recent clinical studies highlight the role of SAA as a sensitive biomarker of COVID-19 infection that contributes to the severity of COVID-19 disease in diabetes and obesity [10, 99–103]. Studies such as these may help explain why diabetic and obese patients are at an increased risk of severe COVID-19 disease.
Acknowledgments
This article is dedicated to the memory of Robert Kisilevsky, MD, PhD, who made seminal contributions to our understanding of the SAA action in lipid transport and AA amyloidosis. Several concepts described in this review stem from the original research spearheaded by Drs. Shobini Jayaraman and Nicholas M. Frame in our laboratory. Special thanks are due to Dr. Jayaraman and to Emily Lewkowicz for helpful comments on the manuscript prior to publication. Continuous support by our collaborators, Drs. John R. Engen, Marcus Fändrich, and John E Straub and their teammates, is gratefully acknowledged.
Abbreviations
- SAA
Serum amyloid A
- AA
Amyloid A
- HDL
High-density lipoprotein
- apo
Apolipoprotein
- NEFA
Non-esterified fatty acid
- POPC
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- GAG
Glycosaminoglycan
Funding
This work was supported by the National Institutes of Health grants GM135158 and GM067260.
Compliance with Ethical Standards
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by the author.
Conflict of Interest
The author declares that he/she has no conflict of interest.
Footnotes
This article is part of the Topical Collection on Vascular Biology
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
- 1.Ye RD, Sun L. Emerging functions of serum amyloid A in inflammation. J Leukoc Biol. 2015;98(6):923–929. doi: 10.1189/jlb.3VMR0315-080R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y, Zhang J, Sheng H, Li H, Wang R. Acute phase reactant serum amyloid A in inflammation and other diseases. Adv Clin Chem. 2019;90:25–80. doi: 10.1016/bs.acc.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Westermark GT, Fändrich M, Westermark P. AA amyloidosis: pathogenesis and targeted therapy. Annu Rev Pathol. 2015;10:321–344. doi: 10.1146/annurev-pathol-020712-163913. [DOI] [PubMed] [Google Scholar]
- 4.Papa R, Lachmann HJ. Secondary, AA, amyloidosis. Rheum Dis Clin N Am. 2018;44(4):585–603. doi: 10.1016/j.rdc.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Perez L. Acute phase protein response to viral infection and vaccination. Arch Biochem Biophys. 2019;671:196–202. doi: 10.1016/j.abb.2019.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abouelasrar Salama S, Lavie M, De Buck M, Van Damme J, Struyf S. Cytokines and serum amyloid A in the pathogenesis of hepatitis C virus infection. Cytokine Growth Factor Rev. 2019;50:29–42. doi: 10.1016/j.cytogfr.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Lee JW, Stone ML, Porrett PM, Thomas SK, Komar CA, Li JH, Delman D, Graham K, Gladney WL, Hua X, Black TA, Chien AL, Majmundar KS, Thompson JC, Yee SS, O'Hara MH, Aggarwal C, Xin D, Shaked A, Gao M, Liu D, Borad MJ, Ramanathan RK, Carpenter EL, Ji A, de Beer MC, de Beer FC, Webb NR, Beatty GL. Hepatocytes direct the formation of a pro-metastatic niche in the liver. Nature. 2019;567(7747):249–252. doi: 10.1038/s41586-019-1004-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie N, Li Z, Zuo R, Qi S, Zhu T, Liu L, Wan L, Yuan J. Serum SAA1 and APOE are novel indicators for human cytomegalovirus infection. Sci Rep. 2017;7(1):13407. doi: 10.1038/s41598-017-13591-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Todorov I, Gospodinova M, Bocheva Y, Popcheva G. Serum amyloid A protein in the course of infectious mononucleosis. Ther Adv Infect Dis. 2019;6:2049936118811208. doi: 10.1177/2049936118811208. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Li H, Xiang X, Ren H, Xu L, Zhao L, Chen X, et al. Serum Amyloid A is a biomarker of severe coronavirus disease and poor prognosis. J Inf Secur. 2020:S0163–4453(20)30162–30166. [DOI] [PMC free article] [PubMed]
- 11.McKay PF, Cizmeci D, Aldon Y, Maertzdorf J, Weiner J, Kaufmann SH, Lewis DJ, van den Berg RA, Del Giudice G, Shattock RJ. Identification of potential biomarkers of vaccine inflammation in mice. Elife. 2019;8:e46149. doi: 10.7554/eLife.46149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanada Y, Yamamoto T, Satake R, Yamashita A, Kanai S, Kato N, van de Loo FA, Nishimura F, Scherer PE, Yanaka N. Serum amyloid A3 gene expression in adipocytes is an indicator of the interaction with macrophages. Sci Rep. 2016;6:38697. doi: 10.1038/srep38697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ather JL, Dienz O, Boyson JE, Anathy V, Amiel E, Poynter ME. Serum Amyloid A3 is required for normal lung development and survival following influenza infection. Sci Rep. 2018;8(1):16571. doi: 10.1038/s41598-018-34901-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kisilevsky R, Manley PN. Acute-phase serum amyloid A: perspectives on its physiological and pathological roles. Amyloid. 2012;19(1):5–14. doi: 10.3109/13506129.2011.654294. [DOI] [PubMed] [Google Scholar]
- 15.Vaisar T, Tang C, Babenko I, Hutchins P, Wimberger J, Suffredini AF, Heinecke JW. Inflammatory remodeling of the HDL proteome impairs cholesterol efflux capacity. J Lipid Res. 2015;56(8):1519–1530. doi: 10.1194/jlr.M059089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han CY, Tang C, Guevara ME, Wei H, Wietecha T, Shao B, Subramanian S, Omer M, Wang S, O'Brien KD, Marcovina SM, Wight TN, Vaisar T, de Beer MC, de Beer FC, Osborne WR, Elkon KB, Chait A. Serum amyloid A impairs the antiinflammatory properties of HDL. J Clin Invest. 2016;126(1):266–281. doi: 10.1172/JCI83475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zewinger S, Drechsler C, Kleber ME, Dressel A, Riffel J, Triem S, Lehmann M, Kopecky C, Säemann MD, Lepper PM, Silbernagel G, Scharnagl H, Ritsch A, Thorand B, de las Heras Gala T, Wagenpfeil S, Koenig W, Peters A, Laufs U, Wanner C, Fliser D, Speer T, März W. Serum amyloid A: high-density lipoproteins interaction and cardiovascular risk. Eur Heart J. 2015;36(43):3007–3016. doi: 10.1093/eurheartj/ehv352. [DOI] [PubMed] [Google Scholar]
- 18.Thompson JC, Jayne C, Thompson J, Wilson PG, Yoder MH, Webb N, Tannock LR. A brief elevation of serum amyloid A is sufficient to increase atherosclerosis. J Lipid Res. 2015;56(2):286–293. doi: 10.1194/jlr.M054015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McEneny J, McKavanagh P, York E, Nadeem N, Harbinson M, Stevenson M, Ball P, Lusk L, Trinick T, Young IS, McKay GJ, Donnelly PM. Serum- and HDL3-serum amyloid A and HDL3-LCAT activity are influenced by increased CVD-burden. Atherosclerosis. 2016;244:172–178. doi: 10.1016/j.atherosclerosis.2015.11.018. [DOI] [PubMed] [Google Scholar]
- 20.Getz GS, Krishack PA, Reardon CA. Serum amyloid A and atherosclerosis. Curr Opin Lipidol. 2016;27(5):531–535. doi: 10.1097/MOL.0000000000000331. [DOI] [PubMed] [Google Scholar]
- 21.Getz GS, Reardon CA, Apoproteins E. A-I, and SAA in macrophage pathobiology related to atherogenesis. Front Pharmacol. 2019;10:536. doi: 10.3389/fphar.2019.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson JC, Wilson PG, Shridas P, Ji A, de Beer M, de Beer FC, Webb NR, Tannock LR. Serum amyloid A3 is pro-atherogenic. Atherosclerosis. 2018;268:32–35. doi: 10.1016/j.atherosclerosis.2017.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shridas P, Tannock LR. Role of serum amyloid A in atherosclerosis. Curr Opin Lipidol. 2019;30(4):320–325. doi: 10.1097/MOL.0000000000000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Buck M, Gouwy M, Wang JM, Van Snick J, Opdenakker G, Struyf S, Van Damme J. Structure and expression of different serum amyloid A (SAA) variants and their concentration-dependent functions during host insults. Curr Med Chem. 2016;23(17):1725–1755. doi: 10.2174/0929867323666160418114600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun L, Ye RD. Serum amyloid A1: structure, function and gene polymorphism. Gene. 2016;583(1):48–57. doi: 10.1016/j.gene.2016.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sack GH., Jr Serum amyloid A - a review. Mol Med. 2018;24(1):46. doi: 10.1186/s10020-018-0047-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sack GH., Jr Serum amyloid a (SAA) proteins. Subcell Biochem. 2020;94:421–436. doi: 10.1007/978-3-030-41769-7_17. [DOI] [PubMed] [Google Scholar]
- 28.Kim MH, de Beer MC, Wroblewski JM, Charnigo RJ, Ji A, Webb NR, de Beer FC, van der Westhuyzen DR. Impact of individual acute phase serum amyloid A isoforms on HDL metabolism in mice. J Lipid Res. 2016;57(6):969–979. doi: 10.1194/jlr.M062174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tannock LR, De Beer MC, Ji A, Shridas P, Noffsinger VP, den Hartigh L, Chait A, De Beer FC, Webb NR. Serum amyloid A3 is a high density lipoprotein-associated acute-phase protein. J Lipid Res. 2018;59(2):339–347. doi: 10.1194/jlr.M080887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jumeau C, Awad F, Assrawi E, Cobret L, Duquesnoy P, Giurgea I, Valeyre D, Grateau G, Amselem S, Bernaudin JF, Karabina SA. Expression of SAA1, SAA2 and SAA4 genes in human primary monocytes and monocyte-derived macrophages. PLoS One. 2019;14(5):e0217005. doi: 10.1371/journal.pone.0217005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson PG, Thompson JC, Shridas P, McNamara PJ, de Beer MC, de Beer FC, Webb NR, Tannock LR. Serum amyloid A is an exchangeable apolipoprotein. Arterioscler Thromb Vasc Biol. 2018;38(8):1890–1900. doi: 10.1161/ATVBAHA.118.310979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ji A, Wang X, Noffsinger VP, Jennings D, de Beer MC, de Beer FC, Tannock LR, Webb NR. Serum amyloid A is not incorporated into HDL during HDL biogenesis. J Lipid Res. 2020;61(3):328–337. doi: 10.1194/jlr.RA119000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Buck M, Gouwy M, Wang JM, Van Snick J, Proost P, Struyf S, Van Damme J. The cytokine-serum amyloid A-chemokine network. Cytokine Growth Factor Rev. 2016;30:55–69. doi: 10.1016/j.cytogfr.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thaler R, Sturmlechner I, Spitzer S, Riester SM, Rumpler M, Zwerina J, Klaushofer K, van Wijnen AJ, Varga F. Acute-phase protein serum amyloid A3 is a novel paracrine coupling factor that controls bone homeostasis. FASEB J. 2015;29(4):1344–1359. doi: 10.1096/fj.14-265512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim J, Yang J, Park OJ, Kang SS, Yun CH, Han SH. Serum amyloid A inhibits osteoclast differentiation to maintain macrophage function. J Leukoc Biol. 2016;99(4):595–603. doi: 10.1189/jlb.3A0415-173R. [DOI] [PubMed] [Google Scholar]
- 36.Choudhary S, Goetjen A, Estus T, Jacome-Galarza CE, Aguila HL, Lorenzo J, Pilbeam C. Serum amyloid A3 secreted by preosteoclasts inhibits parathyroid hormone-stimulated cAMP signaling in murine osteoblasts. J Biol Chem. 2016;291(8):3882–3894. doi: 10.1074/jbc.M115.686576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choudhary S, Santone E, Yee SP, Lorenzo J, Adams DJ, Goetjen A, McCarthy MB, Mazzocca AD, Pilbeam C. Continuous PTH in male mice causes bone loss because it induces serum amyloid. Endocrinology. 2018;159(7):2759–2776. doi: 10.1210/en.2018-00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jayaraman S, Haupt C, Gursky O. Paradoxical effects of SAA on lipoprotein oxidation suggest a new antioxidant function for SAA. J Lipid Res. 2016;57(12):2138–2149. doi: 10.1194/jlr.M071191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sato M, Ohkawa R, Yoshimoto A, Yano K, Ichimura N, Nishimori M, Okubo S, Yatomi Y, Tozuka M. Effects of serum amyloid A on the structure and antioxidant ability of high-density lipoprotein. Biosci Rep. 2016;36(4):e00369. doi: 10.1042/BSR20160075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu S, Wang Y, Chen W, Li W, Wang A, Wong S, Bao G, Li J, Yang H, Tracey KJ, D'Angelo J, Wang H. High-density lipoprotein (HDL) counter-regulates serum amyloid A (SAA)-induced sPLA2-IIE and sPLA2-V expression in macrophages. PLoS One. 2016;11(11):e0167468. doi: 10.1371/journal.pone.0167468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shridas P, De Beer MC, Webb NR. High-density lipoprotein inhibits serum amyloid A-mediated reactive oxygen species generation and NLRP3 inflammasome activation. J Biol Chem. 2018;293(34):13257–13269. doi: 10.1074/jbc.RA118.002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dieter BP, Meek RL, Anderberg RJ, Cooney SK, Bergin JL, Zhang H, Nair V, Kretzler M, Brosius FC, Tuttle KR. Serum amyloid A and Janus kinase 2 in a mouse model of diabetic kidney disease. PLoS One. 2019;14(2):e0211555. doi: 10.1371/journal.pone.0211555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu J, Zhu H, Taheri S, Mondy W, Bonilha L, Magwood GS, Lackland D, Adams RJ, Kindy MS. Serum amyloid A-mediated inflammasome activation of microglial cells in cerebral ischemia. J Neurosci. 2019;39(47):9465–9476. doi: 10.1523/JNEUROSCI.0801-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee JY, Hall JA, Kroehling L, Wu L, Najar T, Nguyen HH, Lin WY, Yeung ST, Silva HM, Li D, Hine A, Loke P, Hudesman D, Martin JC, Kenigsberg E, Merad M, Khanna KM, Littman DR. Serum amyloid A proteins induce pathogenic Th17 cells and promote inflammatory disease. Cell. 2020;180(1):79–91. doi: 10.1016/j.cell.2019.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Linke RP, Meinel A, Chalcroft JP, Urieli-Shoval S. Serum amyloid A (SAA) treatment enhances the recovery of aggravated polymicrobial sepsis in mice, whereas blocking SAA’s invariant peptide results in early death. Amyloid. 2017;24(S1):149–150. doi: 10.1080/13506129.2017.1295950. [DOI] [PubMed] [Google Scholar]
- 46.Cheng N, Liang Y, Du X, Ye RD. Serum amyloid A promotes LPS clearance and suppresses LPS-induced inflammation and tissue injury. EMBO Rep. 2018;19(10):e45517. doi: 10.15252/embr.201745517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murdoch CC, Espenschied ST, Matty MA, Mueller O, Tobin DM, Rawls JF. Intestinal serum amyloid A suppresses systemic neutrophil activation and bactericidal activity in response to microbiota colonization. PLoS Pathog. 2019;15(3):e1007381. doi: 10.1371/journal.ppat.1007381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng H, Li H, Zhang J, Fan H, Jia L, Ma W, Ma S, Wang S, You H, Yin Z, Li X. Serum amyloid A exhibits pH dependent antibacterial action and contributes to host defense against Staphylococcus aureus cutaneous infection. J Biol Chem. 2020;295(9):2570–2581. doi: 10.1074/jbc.RA119.010626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun L, Zhou H, Zhu Z, Yan Q, Wang L, Liang Q, Ye RD. Ex vivo and in vitro effect of serum amyloid a in the induction of macrophage M2 markers and efferocytosis of apoptotic neutrophils. J Immunol. 2015;194(10):4891–4900. doi: 10.4049/jimmunol.1402164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burgess EJ, Hoyt LR, Randall MJ, Mank MM, Bivona JJ, 3rd, Eisenhauer PL, Botten JW, Ballif BA, Lam YW, Wargo MJ, Boyson JE, Ather JL, Poynter ME. Bacterial lipoproteins constitute the TLR2-stimulating activity of serum amyloid A. J Immunol. 2018;201(8):2377–2384. doi: 10.4049/jimmunol.1800503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smole U, Kratzer B, Pickl WF. Soluble pattern recognition molecules: guardians and regulators of homeostasis at airway mucosal surfaces. Eur J Immunol. 2020;50(5):624–642. doi: 10.1002/eji.201847811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Derebe MG, Zlatkov CM, Gattu S, Ruhn KA, Vaishnava S, Diehl GE, MacMillan JB, Williams NS, Hooper LV. Serum amyloid A is a retinol binding protein that transports retinol during bacterial infection. Elife. 2014;3:e03206. doi: 10.7554/eLife.03206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu Z, Bang YJ, Ruhn KA, Hooper LV. Molecular basis for retinol binding by serum amyloid A during infection. Proc Natl Acad Sci U S A. 2019;116(38):19077–19082. doi: 10.1073/pnas.1910713116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gattu S, Bang YJ, Pendse M, Dende C, Chara AL, Harris TA, Wang Y, Ruhn KA, Kuang Z, Sockanathan S, Hooper LV. Epithelial retinoic acid receptor β regulates serum amyloid A expression and vitamin A-dependent intestinal immunity. Proc Natl Acad Sci U S A. 2019;116(22):10911–10916. doi: 10.1073/pnas.1812069116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murakami T, Inoshima Y, Ishiguro N. Systemic AA amyloidosis as a prion-like disorder. Virus Res. 2015;207:76–81. doi: 10.1016/j.virusres.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 56.Gaffney PM. Amyloid A amyloidosis. Vet Pathol. 2017;54(1):5–8. doi: 10.1177/0300985816677150. [DOI] [PubMed] [Google Scholar]
- 57.Nuvolone M, Merlini G. Systemic amyloidosis: novel therapies and role of biomarkers. Nephrol Dial Transplant. 2017;32(5):770–780. doi: 10.1093/ndt/gfw305. [DOI] [PubMed] [Google Scholar]
- 58.Frame NM, Gursky O. Structure of serum amyloid A suggests a mechanism for selective lipoprotein binding and functions: SAA as a hub in macromolecular interaction networks. FEBS Lett. 2016;590(6):866–879. doi: 10.1002/1873-3468.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Colón W, Aguilera JJ, Srinivasan S. Intrinsic stability, oligomerization, and amyloidogenicity of HDL-free serum amyloid A. Adv Exp Med Biol. 2015;855:117–134. doi: 10.1007/978-3-319-17344-3_5. [DOI] [PubMed] [Google Scholar]
- 60.Uversky VN. Dancing protein clouds: the strange biology and chaotic physics of intrinsically disordered proteins. J Biol Chem. 2016;291(13):6681–6688. doi: 10.1074/jbc.R115.685859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weng J, Wang W. Dynamic multivalent interactions of intrinsically disordered proteins. Curr Opin Struct Biol. 2019;62:9–13. doi: 10.1016/j.sbi.2019.11.001. [DOI] [PubMed] [Google Scholar]
- 62.Macossay-Castillo M, Marvelli G, Guharoy M, Jain A, Kihara D, Tompa P, Wodak SJ. The balancing act of intrinsically disordered proteins: enabling functional diversity while minimizing promiscuity. J Mol Biol. 2019;431(8):1650–1670. doi: 10.1016/j.jmb.2019.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takase H, Furuchi H, Tanaka M, Yamada T, Matoba K, Iwasaki K, Kawakami T, Mukai T. Characterization of reconstituted high-density lipoprotein particles formed by lipid interactions with human serum amyloid A. Biochim Biophys Acta. 2014;1842(10):1467–1474. doi: 10.1016/j.bbalip.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 64.Jayaraman S, Haupt C, Gursky O. Thermal transitions in serum amyloid A in solution and on the lipid: implications for structure and stability of acute-phase HDL. J Lipid Res. 2015;56(8):1531–1542. doi: 10.1194/jlr.M059162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Frame NM, Jayaraman S, Gantz DL, Gursky O. Serum amyloid A self-assembles with phospholipids to form stable protein-rich nanoparticles with a distinct structure: a hypothetical function of SAA as a "molecular mop" in immune response. J Struct Biol. 2017;200(3):293–302. doi: 10.1016/j.jsb.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jayaraman S, Gantz DL, Haupt C, Fändrich M, Gursky O. Serum amyloid A sequesters diverse phospholipids and their hydrolytic products, hampering fibril formation and proteolysis in a lipid-dependent manner. Chem Commun. 2018;54(28):3532–3535. doi: 10.1039/C8CC01424H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takase H, Tanaka M, Nakamura Y, Morita SY, Yamada T, Mukai T. Effects of lipid composition on the structural properties of human serum amyloid A in reconstituted high-density lipoprotein particles. Chem Phys Lipids. 2019;221:8–14. doi: 10.1016/j.chemphyslip.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 68.Lu J, Yu Y, Zhu I, Cheng Y, Sun PD. Structural mechanism of serum amyloid A-mediated inflammatory amyloidosis. Proc Natl Acad Sci U S A. 2014;111(14):5189–5194. doi: 10.1073/pnas.1322357111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Das M, Gursky O. Amyloid-forming properties of human apolipoproteins: sequence analyses and structural insights. Adv Exp Med Biol. 2015;855:175–211. doi: 10.1007/978-3-319-17344-3_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Frame NM, Kumanan M, Wales TE, Bandara A, Fändrich M, Straub JE, Engen JR, Gursky O. Structural basis for lipid binding and function by an evolutionarily conserved protein, serum amyloid A. J Mol Biol. 2020;432(7):1978–1995. doi: 10.1016/j.jmb.2020.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Claus S, Meinhardt K, Aumüller T, Puscalau-Girtu I, Linder J, Haupt C, Walther P, Syrovets T, Simmet T, Fändrich M. Cellular mechanism of fibril formation from serum amyloid A1 protein. EMBO Rep. 2017;18(8):1352–1366. doi: 10.15252/embr.201643411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liberta F, Loerch S, Rennegarbe M, Schierhorn A, Westermark P, Westermark GT, BPC H, Grigorieff N, Fändrich M, Schmidt M. Cryo-EM fibril structures from systemic AA amyloidosis reveal the species complementarity of pathological amyloids. Nat Commun. 2019;10(1):1104. doi: 10.1038/s41467-019-09033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Röcken C, Menard R, Bühling F, Vöckler S, Raynes J, Stix B, Krüger S, Roessner A, Kähne T. Proteolysis of serum amyloid A and AA amyloid proteins by cysteine proteases: cathepsin B generates AA amyloid proteins and cathepsin L may prevent their formation. Ann Rheum Dis. 2005;64(6):808–815. doi: 10.1136/ard.2004.030429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jayaraman S, Gantz DL, Haupt C, Gursky O. Serum amyloid A forms stable oligomers that disrupt vesicles at lysosomal pH and contribute to the pathogenesis of reactive amyloidosis. Proc Natl Acad Sci U S A. 2017;114(32):E6507–E6515. doi: 10.1073/pnas.1707120114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liang JS, Schreiber BM, Salmona M, Phillip G, Gonnerman WA, de Beer FC, Sipe JD. Amino terminal region of acute phase, but not constitutive, serum amyloid A (apoSAA) specifically binds and transports cholesterol into aortic smooth muscle and HepG2 cells. J Lipid Res. 1996;37(10):2109–2116. [PubMed] [Google Scholar]
- 76.Rennegarbe M, Lenter I, Schierhorn A, Sawilla R, Haupt C. Influence of C-terminal truncation of murine serum amyloid A on fibril structure. Sci Rep. 2017;7(1):6170. doi: 10.1038/s41598-017-06419-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tanaka M, Nishimura A, Takeshita H, Takase H, Yamada T, Mukai T. Effect of lipid environment on amyloid fibril formation of human serum amyloid A. Chem Phys Lipids. 2017;202:6–12. doi: 10.1016/j.chemphyslip.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 78.Jayaraman S, Fändrich M, Gursky O. Synergy between serum amyloid A and secretory phospholipase A2. Elife. 2019;8:e46630. doi: 10.7554/eLife.46630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Massey JB, Pao Q, Van Winkle WB, Pownall HJ. Interaction of human plasma lecithin: cholesterol acyltransferase and venom phospholipase A2 with apolipoprotein A-I recombinants containing nonhydrolyzable diether phosphatidylcholines. J Biol Chem. 1985;260(21):11719–11723. [PubMed] [Google Scholar]
- 80.Cabana VG, Reardon CA, Wei B, Lukens JR, Getz GS. mSAA-only HDL formed during the acute phase response in apoA-I+/+ and apoA-I-/- mice. J Lipid Res. 1999;40(6):1090–1103. [PubMed] [Google Scholar]
- 81.Stonik JA, Remaley AT, Demosky SJ, Neufeld EB, Bocharov A, Brewer HB. Serum amyloid A promotes ABCA1-dependent and ABCA1-independent lipid efflux from cells. Biochem Biophys Res Commun. 2004;321(4):936–941. doi: 10.1016/j.bbrc.2004.07.052. [DOI] [PubMed] [Google Scholar]
- 82.Abe-Dohmae S, Kato KH, Kumon Y, Hu W, Ishigami H, Iwamoto N, Okazaki M, Wu CA, Tsujita M, Ueda K, Yokoyama S. Serum amyloid A generates high density lipoprotein with cellular lipid in an ABCA1- or ABCA7-dependent manner. J Lipid Res. 2006;47(7):1542–1550. doi: 10.1194/jlr.M600145-JLR200. [DOI] [PubMed] [Google Scholar]
- 83.Ancsin JB, Kisilevsky R. The heparin/heparan sulfate-binding site on apo-serum amyloid A. Implications for the therapeutic intervention of amyloidosis. J Biol Chem. 1999;274(11):7172–7181. doi: 10.1074/jbc.274.11.7172. [DOI] [PubMed] [Google Scholar]
- 84.Shao B, Heinecke JW. Quantifying HDL proteins by mass spectrometry: how many proteins are there and what are their functions? Expert Rev Proteomics. 2018;15(1):31–40. doi: 10.1080/14789450.2018.1402680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liberta F, Rennegarbe M, Rösler R, Bijzet J, Wiese S, Hazenberg BPC, Fändrich M. Morphological and primary structural consistency of fibrils from different AA patients (common variant) Amyloid. 2019;26(3):164–170. doi: 10.1080/13506129.2019.1628015. [DOI] [PubMed] [Google Scholar]
- 86.Kisilevsky R, Raimondi S, Bellotti V. Historical and current concepts of fibrillogenesis and in vivo amyloidogenesis: implications of amyloid tissue targeting. Front Mol Biosci. 2016;3:17. doi: 10.3389/fmolb.2016.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Annamalai K, Liberta F, Vielberg MT, Close W, Lilie H, Gührs KH, Schierhorn A, Koehler R, Schmidt A, Haupt C, Hegenbart U, Schönland S, Schmidt M, Groll M, Fändrich M. Common fibril structures imply systemically conserved protein misfolding pathways in vivo. Angew Chem Int Ed Eng. 2017;56(26):7510–7514. doi: 10.1002/anie.201701761. [DOI] [PubMed] [Google Scholar]
- 88.Tanaka M, Kawakami T, Okino N, Sasaki K, Nakanishi K, Takase H, Yamada T, Mukai T. Acceleration of amyloid fibril formation by carboxyl-terminal truncation of human serum amyloid. Arch Biochem Biophys. 2018;639:9–15. doi: 10.1016/j.abb.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 89.Claus S, Puscalau-Girtu I, Walther P, Syrovets T, Simmet T, Haupt C, Fändrich M. Cell-to-cell transfer of SAA1 protein in a cell culture model of systemic AA amyloidosis. Sci Rep. 2017;7:45683. doi: 10.1038/srep45683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kollmer M, Meinhardt K, Haupt C, Liberta F, Wulff M, Linder J, Handl L, Heinrich L, Loos C, Schmidt M, Syrovets T, Simmet T, Westermark P, Westermark GT, Horn U, Schmidt V, Walther P, Fändrich M. Electron tomography reveals the fibril structure and lipid interactions in amyloid deposits. Proc Natl Acad Sci U S A. 2016;113(20):5604–5609. doi: 10.1073/pnas.1523496113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yamagata A, Uchida T, Yamada Y, Nakanishi T, Nagai K, Imakiire T, Oshima N, Kumagai H. Rapid clinical improvement of amyloid A amyloidosis following treatment with tocilizumab despite persisting amyloid deposition: a case report. BMC Nephrol. 2017;18(1):377. doi: 10.1186/s12882-017-0799-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sayed RH, Hawkins PN, Lachmann HJ. Emerging treatments for amyloidosis. Kidney Int. 2015;87(3):516–526. doi: 10.1038/ki.2014.368. [DOI] [PubMed] [Google Scholar]
- 93.Blank N, Hegenbart U, Dietrich S, Brune M, Beimler J, Röcken C, Müller-Tidow C, Lorenz HM, Schönland SO. Obesity is a significant susceptibility factor for idiopathic AA amyloidosis. Amyloid. 2018;25(1):37–45. doi: 10.1080/13506129.2018.1429391. [DOI] [PubMed] [Google Scholar]
- 94.Noborn F, Ancsin JB, Ubhayasekera W, Kisilevsky R, Li JP. Heparan sulfate dissociates serum amyloid A (SAA) from acute-phase high-density lipoprotein, promoting SAA aggregation. J Biol Chem. 2012;287(30):25669–25677. doi: 10.1074/jbc.M112.363895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sosnowska M, Skibiszewska S, Kamińska E, Wieczerzak E, Jankowska E. Designing peptidic inhibitors of serum amyloid A aggregation process. Amino Acids. 2016;48(4):1069–1078. doi: 10.1007/s00726-015-2167-y. [DOI] [PubMed] [Google Scholar]
- 96.Jayaraman S, Sánchez-Quesada JL, Gursky O. Triglyceride increase in the core of high-density lipoproteins augments apolipoprotein dissociation from the surface: potential implications for treatment of apolipoprotein deposition diseases. Biochim Biophys Acta. 2017;1863(1):200–210. doi: 10.1016/j.bbadis.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stankovic Stojanovic K, Georgin-Lavialle S, Poitou C, Buob D, Amselem S, Grateau G, AMYLOB Study Group AA amyloidosis is an emerging cause of nephropathy in obese patients. Eur J Intern Med. 2017;39:e18–e20. doi: 10.1016/j.ejim.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 98.Puscalau-Girtu I, Scheller JS, Claus S, Fändrich M. Cell assay for the identification of amyloid inhibitors in systemic AA amyloidosis. Amyloid. 2019;26(1):24–33. doi: 10.1080/13506129.2019.1568978. [DOI] [PubMed] [Google Scholar]
- 99.Zhang Q, Wei Y, Chen M, Wan Q, Chen X. Clinical analysis of risk factors for severe COVID-19 patients with type 2 diabetes. J Diabetes Complicat. 2020;107666. 10.1016/j.jdiacomp.2020.107666 [DOI] [PMC free article] [PubMed]
- 100.Anderson MR, Geleris J, Anderson DR, Zucker J, Nobel YR, Freedberg D, et al. Body mass index and risk for intubation or death in SARS-CoV-2 infection: a retrospective cohort study. Ann Intern Med. 2020;29:M20–3214. doi: 10.7326/M20-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pranata R, Lim MA, Yonas E, Vania R, Lukito AA, Siswanto BB, et al. Body mass index and outcome in patients with COVID-19: a dose-response meta-analysis. Diabetes Metab. 2020. 10.1016/j.diabet.2020.07.005 [DOI] [PMC free article] [PubMed]
- 102.Li H, Xiang X, Ren H, Xu L, Zhao L, Chen X, Long H, Wang Q, Wu Q. Serum amyloid A is a biomarker of severe coronavirus disease and poor prognosis. J Inf Secur. 2020;80(6):646–655. doi: 10.1016/j.jinf.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Smith SM, Boppana A, Traupman JA, Unson E, Maddock DA, Chao K, et al. Impaired glucose metabolism in patients with diabetes, prediabetes, and obesity is associated with severe COVID-19. J Med Virol. 2020. 10.1002/jmv.26227. [DOI] [PMC free article] [PubMed]