Abstract
Recently, the isolation and elucidation of a series of polyhydroxyanthraquinones were reported from an organic extract of a solid phase culture of an endophytic fungus, Penicillium restrictum (strain G85). One of these compounds, ω-hydroxyemodin (1), showed promising quorum-sensing inhibition against clinical isolates of methicillin-resistant Staphylococcus aureus (MRSA) in both in vitro and in vivo models. The initial supply of 1 was 19 mg, and this amount needed to be scaled by a factor of 30 to 50 times, in order to generate material for further in vivo studies. To do so, improvements were implemented to enhance both the fermentation of the fungal culture and the isolation of this compound, with the target of generating >800 mg of study materials in a period of 13 weeks. Valuable insights, both regarding chemistry and mycology, were gained during the targeted production of 1 on the laboratory-scale. In addition, methods were modified to make the process more environmentally friendly by judicious choice of solvents, implementing procedures for solvent recycling, and minimizing the use of halogenated solvents.
Keywords: Penicillium restrictum Aspergillaceae, polyhydroxyanthraquinone, ω-hydroxyemodin, Silybum marianum Asteraceae, fungal endophyte
Introduction
Penicillium restrictum (strain G85) is a fungal endophyte that was isolated from surface-sterilized stems of the medicinal herb, milk thistle Silybum marianum (L.) Gaertn. (Asteraceae) [1]. From an organic extract of a solid phase rice culture of strain G85, a series of polyhydroxyanthraquinones were isolated (Fig. 1S, Supporting Information) [2]. Among these, ω-hydroxyemodin (1) (Fig. 1) showed promising activity, both in vitro and in vivo, as a quorum sensing inhibitor against clinical isolates of methicillin-resistant Staphylococcus aureus (MRSA) [2,3]. For example, in a murine model of MRSA, we postulated that 1 inhibited the DNA-binding capacity of a virulence regulator, AgrA, resulting in statistically significant decreases in disease severity, abscess and ulcer area, bacterial burden, and certain pro-inflammatory cytokines [3].
Fig. 1.
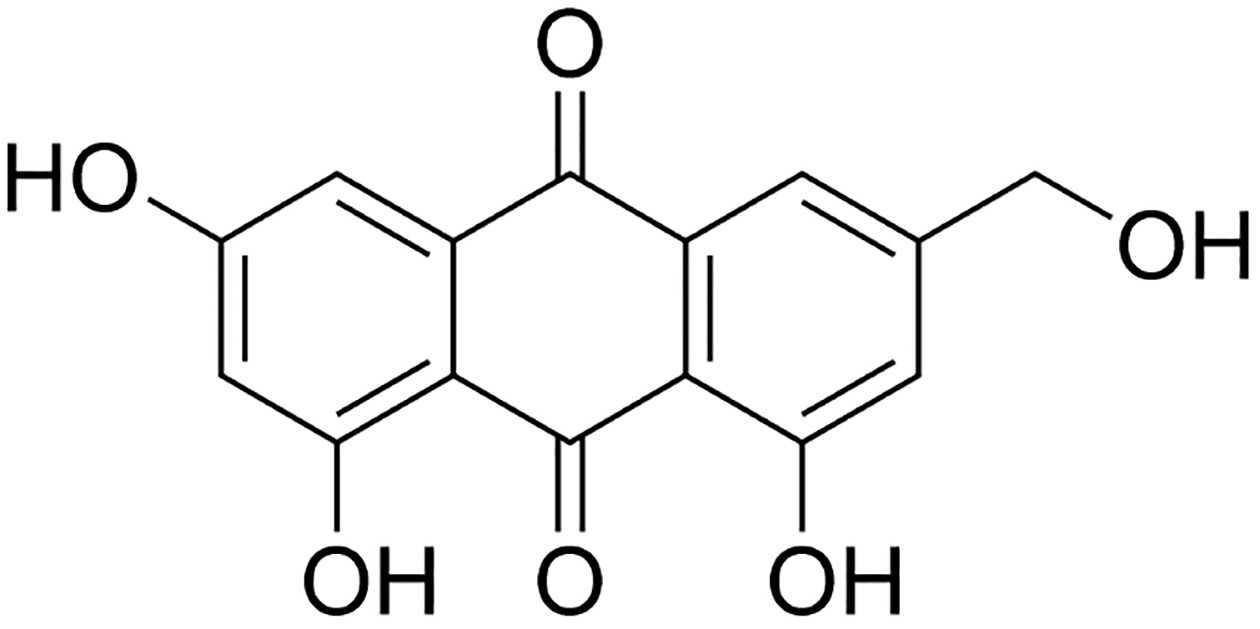
Structure of the polyhydroxyanthraquinone, ω-hydroxyemodin (1).
In the United States, the National Institute of Allergy and Infectious Diseases has called for strategic approaches to combat antibiotic-resistant strains by disarming bacteria, effectively minimizing toxins so that the body’s immune system can more easily address the infection [4]. To contribute to this, compounds with antivirulence or quorum sensing inhibition properties (also termed ‘quorum quenchers’) are needed [5], and ω-hydroxyemodin (1) has shown some promise in this regard [2,3]. With MRSA growing as a global health concern [6,7], it is important to carry out further pharmacological studies on quorum sensing inhibitors, probing the feasibility of this strategy. In the case of ω-hydroxyemodin (1), and as is likely true for many drug leads isolated from nature [8], a substantial increase in production was required for further pharmacological evaluation. While total synthesis would likely be pursued for future development, as initial preclinical work demonstrated promise, we performed fermentation studies for rapid turn around and laboratory-scale control. Moreover, since strain G85 was isolated as an endophyte, and since endophytic fungal cultures are often problematic when it comes to consistent production of secondary metabolites upon subculturing in the laboratory [9], we thought it presented an interesting test case for enhancing the supply of a promising lead on the laboratory-scale. Current synthetic approaches require emodin as a starting material for producing ω-hydroxyemodin [10], and emodin has to either be first synthesized [11,12], obtained commercially, or isolated from a natural source, such as various plant (rhubarb, aloe) and fungal sources, including lichens [2,12]. Thus, for the rapid turnaround needed for this project, it became necessary to enhance the biosynthesis and purification of 1.
Results and Discussion
In initial studies on Penicillium restrictum (strain G85), we isolated less than 20 mg of ω-hydroxyemodin (1) from one large-scale grain-based fermentation (4 × 10 g of rice/flask) [2]. Interestingly, the first report of 1 as a new compound (at that time termed citreorosein) was also from a fungal strain, P. cyclopium [13], which was more recently identified as P. chrysogenum [14,15]. In this study, various growth conditions of strain G85 were scouted for enhanced production of 1, including chemical analysis in situ from live cultures using the droplet probe instrument, the use and optimization of which has been reviewed recently [16]. In addition, we modified and optimized our extraction, separation, and isolation protocols [2,17] to more efficiently obtain >800 mg of 1. In essence, our goals were to enhance the laboratory-scale production of 1, optimizing both fermentation and isolation procedures, while working both in a more environmentally friendly manner and on a timescale of weeks to months.
Concomitant with describing the specific work accomplished in this study, an additional aim of this article was to elaborate strategies that could enhance compound production in a laboratory setting, discussing both mycological parameters and chemistry methodologies that could be refined. Solving supply issues of fermentation products have been carried out numerous times, perhaps most famously with the development of penicillin during the throws of World War II [18]. However, this study provides a modern example of a laboratory-based scale-up of solid-state fermentation cultures in an academic setting.
Media and fermentation:
A detailed explanation of how the fungal endophyte, Penicillium restrictum (strain G85), was isolated from milk thistle [1] and characterized via molecular methods [19] has been reported previously [2]. When strain G85 was first isolated and cultured on nutrient media, such as potato dextrose agar (PDA; Difco), malt extract agar (MEA; Difco), and yeast extract soy peptone dextrose agar (2% soy peptone, 2% dextrose and 1% yeast extract; YESD), it produced red guttates in both PDA and MEA media (Fig. 2a–L). The red guttates were indicative of the presence of the polyhydroxyanthraquinones (Fig. 1S, Supporting Information), including ω-hydroxyemodin (1), the target compound in this study. However, when the fungus was grown on the same three media used previously to make a seed culture for large-scale production, the red guttates were absent in all three of the media conditions (Fig. 2a–R). It has been noted previously that repeated sub-culturing of fungal strains leads to attenuation of target compounds, likely due to strain domestication [9,20–22], and the conjugation inherent in the structure of polyhydroxyanthraquinones makes this challenge visually apparent. Therefore, for the large-scale isolation of ω-hydroxyemodin, fungal strain G85 was grown on multiple media types and various pH to determine the most optimal seed culture media for scale-up purposes. Both solid and liquid broths were utilized to grow strain G85, including media used in our previous study [2].
Fig. 2.
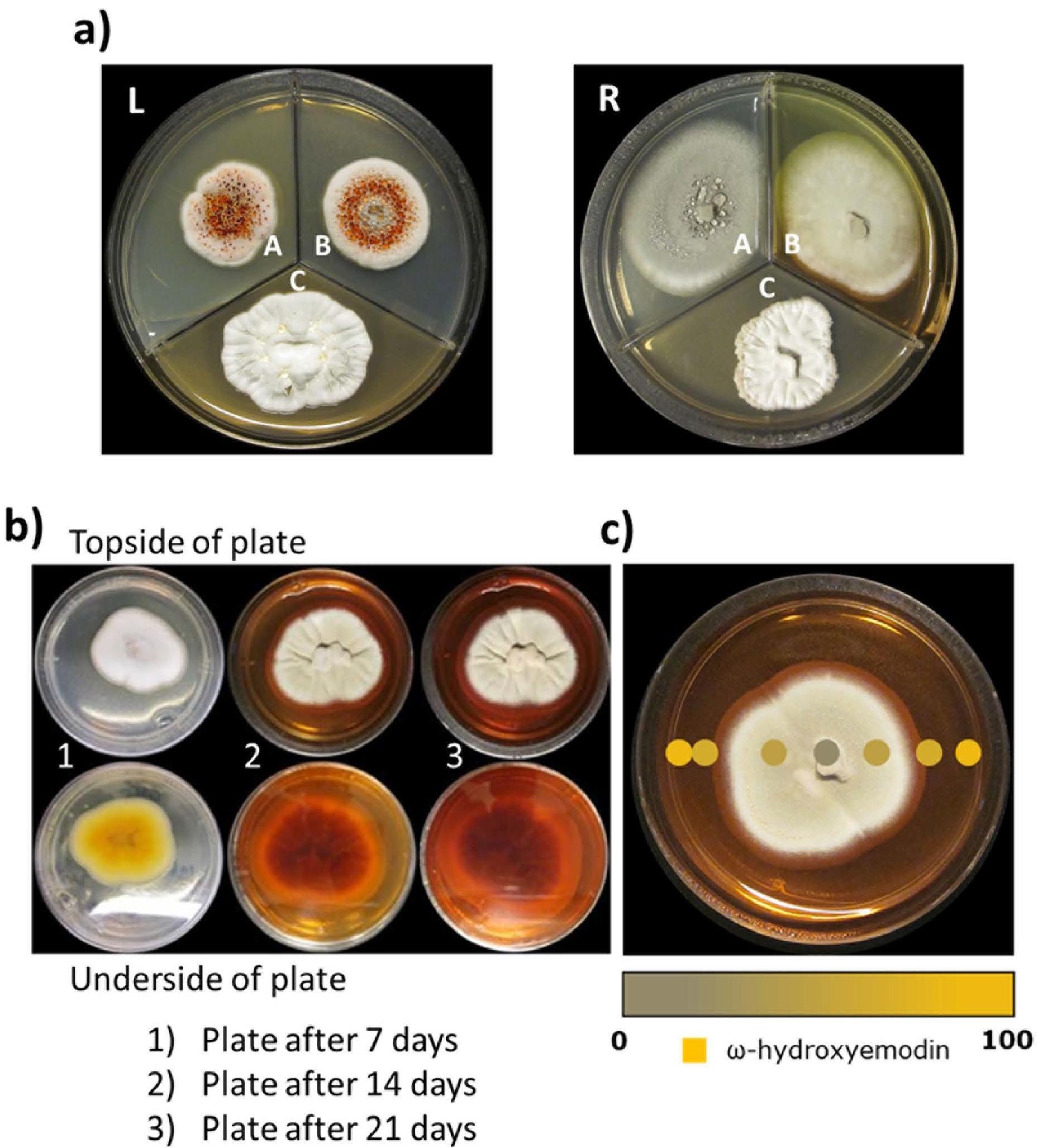
a Left:Penicillium restrictum (G85) at the time of the initial study in 2014, showing red guttates. Right: P. restrictum in 2016 using the same media conditions. Potato dextrose agar (A) Malt extract agar (B) Yeast extract soy peptone dextrose agar (C). Note how the red guttates, a key observation in the initial study [2], were no longer present after repeated transferring and culturing of P. restrictum. In our experience, this sort of ‘domestication’ of a fungus is particularly problematic with endophytic fungi; others have noted this as well [9,21,22].
Fig. 2b When Penicillium restrictum (G85) was grown on SD Agar, the fungus produced a red color that diffused into the media; this correlated well with the production of ω-hydroxyemodin, as well as other polyhydroxyanthraquinones.
Fig. 2c Surface sampling of the Penicillium restrictum strain G85 using the droplet probe [16]. A scanned image of P. restrictum on SD Agar. The intensity of the golden color of the dots indicates the relative concentration of ω-hydroxyemodin observed at that position on the plate. Via heat mapping experiments with the droplet probe, ω-hydroxyemodin (1) was shown to be present predominantly in the highly colored areas of the media.
Various growth methods were tested, with the initial aim of regenerating the original guttate-rich phenotype (Fig. 2a–L). Among the scores of media and fermentation conditions tested with strain G85 (Fig. 2S, Supporting Information), Sabouraud dextrose agar (SD Agar) turned red after 14–21 days of growth (Fig. 2b). While we could never recapitulate the original appearance of the fungal culture, repeated growth on SD Agar media gave identical results, which we found encouraging. Since 1 and other polyhydroxyanthraquinones (Fig. 1S, Supporting Information) have a strong UV-active chromophore, which was likely the cause of the plate coloration, we hypothesized that growing the culture in SD Broth for large-scale isolation could increase yield, and this could be monitored qualitatively by color. In general, SD Agar is a widely used mycological media for the isolation of a variety of molds, including both dermatophytes and common fungal strains such as Aspergillus and Penicillium spp., among other fungi. It has also been used for growing endophytic fungi [23]. In particular, we believe that the somewhat acidic pH (i.e. 5.6–5.8) conditions at room temperature (25°C) afforded by SD Agar facilitates preferential growth of fungi [24].
In situ analysis of fungal chemistry via the droplet probe:
The droplet probe has been used to monitor the production of secondary metabolites in fungal cultures [25–35], with several notable examples highlighted in a recent review [16]. One benefit of this technique for this application was the ability to generate heat maps, showing in a relative manner where secondary metabolites are concentrated. As such, a heat map was constructed with strain G85 grown on SD Agar (Fig. 2c). From this, it was apparent that the polyhydroxyanthraquinones, including 1, were exuded into the culture media (Fig. 2c). The compounds were identified by comparison of their chromatographic retention time, high-resolution mass spectrum, and tandem mass spectral data, relative to reference standards [2]. These in situ measurements had the added benefit of verifying that the red coloration, notable by visual inspection, correlated with a higher relative concentration of polyhydroxyanthraquinones.
Optimizing seed cultures for large-scale fermentation:
As the fermentation procedures were beginning to be optimized, an obvious question was whether the benefits noted with SD Agar could be leveraged for the large-scale growths. Thus, an experiment was designed where YESD broth (which is often used for seed culture propagation [36]) was replaced with Sabouraud dextrose broth (i.e., SD Broth). Strain G85 was grown on commercial rice (variety: Calrose Botan) using either SD Broth or YESD broth for the seed culture, and by visual inspection those grown with SD Broth were always more orange/red in color (Fig. 3S, Supporting Information). While the specific amounts varied, on average, the amount of 1 isolated per flask was higher when SD Broth was used for the seed culture using Calrose Botan rice compared to what was observed under the same growth conditions using YESD broth for the seed culture (Fig. 4S, Supporting Information). The deepness of the orange/red color correlated with a higher production of 1, consistent with the droplet probe analysis on SD Agar (Fig. 2c). Therefore, for all subsequent growths of strain G85, SD Broth was used for producing the most robust seed culture (Table 1 and Fig. 4S, Supporting Information).
Table 1.
Over four weeks, Penicillium restrictum (strain G85) was grown on five different solid media: Calrose Botan rice, Kokuho Rose rice, Cheerios, Oatmeal, and Calrose Botan-Indian (Udupi Sona Masoori) rice mixture. Each of these five solid media used one of two different seed cultures; Sabouraud dextrose broth (SD Broth) or YESD broth. Each condition was grown in duplicate, and their concentrations were averaged. The amount of ω-hydroxyemodin (1) per flask was determined by UPLC (see Fig. 9S, Supporting Information).
| Media Types | Quantity of 1 (μg) per flask |
|---|---|
| SD Broth-Calrose Botan rice | 1486 |
| YESD-Calrose Botan rice | 1423 |
| SD Broth-Kokuho Rose rice | 1085 |
| YESD-Kokuho Rose rice | 42 |
| SD Broth-Cheerios | 29 |
| YESD-Cheerios | 123 |
| SD Broth-Oatmeal | 39 |
| YESD-Oatmeal | 101 |
| SD Broth-1:1 Indian:Calrose Botan rice | 69 |
| YESD-1:1 Indian:Calrose Botan rice | 35 |
Optimizing grain selection for large-scale solid phase fermentations:
Solid state-fermentation was used for scaling-up production of 1 in the laboratory because it is less expensive than pure liquid broth medium and often yields higher amounts of the fungal extract [36]. Previous studies on the scaled production of fungal secondary metabolites have also successfully used solid-state fermentation [29,37].
Results of the screener scale study suggested that both Calrose Botan and Kokuho Rose varieties of rice were suitable grain-based substrates, and that using SD Broth as the seed culture produced the greatest yield of 1. Among the five different grain-based media that were evaluated, Calrose Botan rice showed the best yield (1.5 mg per 10 g of rice using SD Broth as seed culture and 1.4 mg per 10 g of rice using YESD broth as seed culture), followed by Kokuho Rose rice (1.1 mg per 10 g of rice using SD Broth as seed culture) (Table 1 and Fig. 4S, Supporting Information). For all subsequent large-scale growths, Calrose Botan rice was used with SD Broth for the seed culture.
Large-scale solid-state fermentations with Calrose Botan rice:
To permit large scale purification of the target compound, the growth conditions of strain G85 were scaled in a variety of ways, as diagramed (Fig. 3). This increase in the scale of fermentation included the use of 161 Erlenmeyer flasks (10 g of rice per 250-mL flask), 16 Fernbach flasks (100 g of rice per 2.8-L flask), two small-sized mushroom spawn bags (150 g of rice per bag), and one large-sized mushroom spawn bag (1000 g of rice). In each case, water was added at approximately double the weight of rice (e.g., 20 mL of water per 10 g of dry rice), followed by autoclaving at 121°C for 30 min.
Fig. 3.
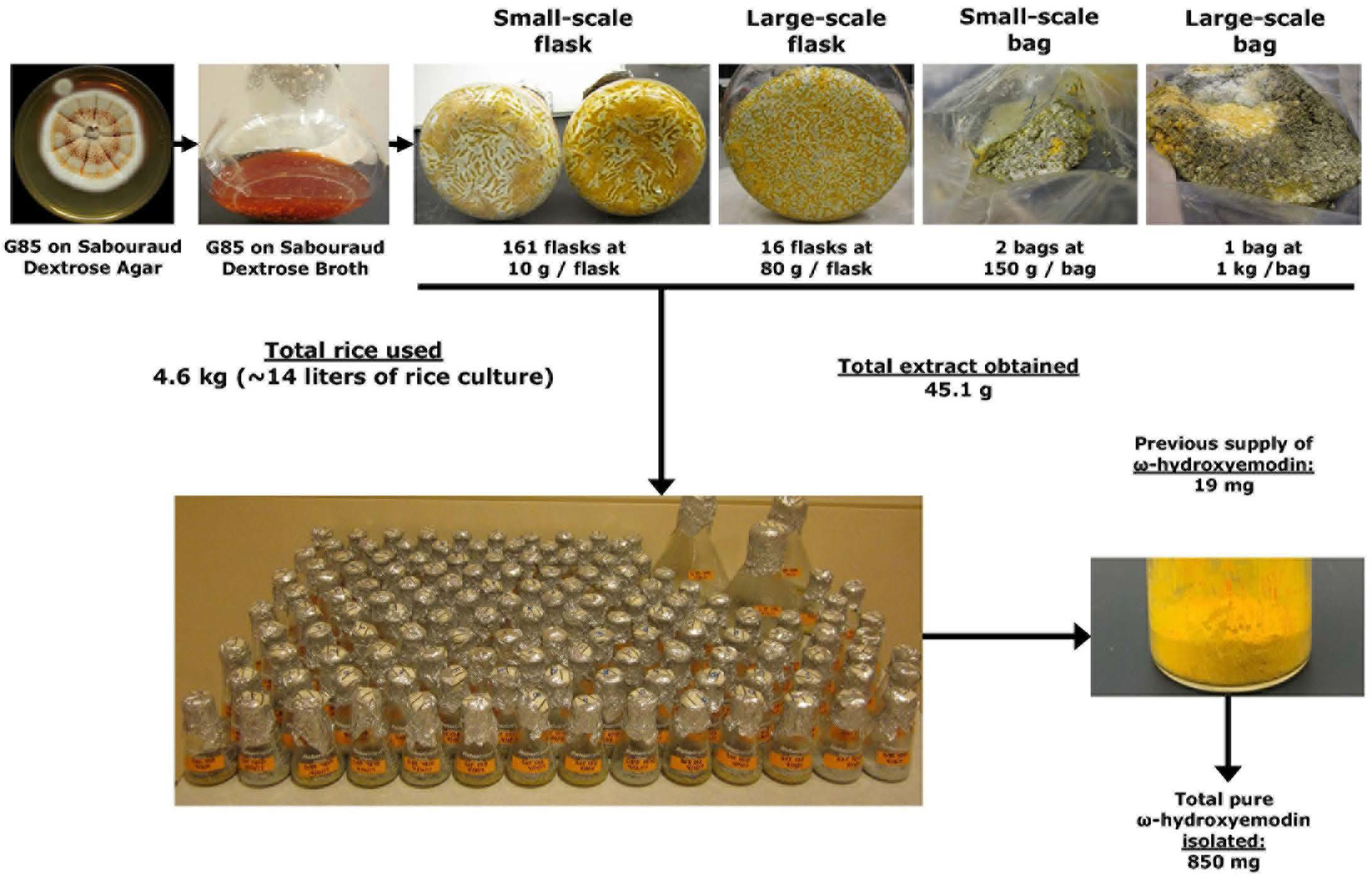
Schematic of how fermentations of Penicillium restrictum (strain G85) were modified and scaled to produce nearly a gram of ω-hydroxyemodin (1). From start to finish, it took a team of three people approximately 13 weeks to go from <20 mg to >800 mg of 1.
Modifications to extraction protocols:
The G85 fungal cultures were initially extracted using standard methods in our laboratory for extractions of fungal cultures [2,17]. An extraction of 100 g of solid-phase rice culture led to slow filtration due to clogging by fungal spores, starches, and an immiscible particle suspension-all of which led to emulsions and blockage of the filter paper. Such problems are often due to large amounts of solid material extracted along with the fungal metabolites, diluting potency, and complicating purification of fungal metabolites [37]. To overcome these issues, Celite was added to disrupt emulsions and allow for a greater surface area for filtration, resulting in filtrations that were up to ten times faster. Initial partitioning methods utilized 4:1:5 CHCl3:MeOH:H2O, however, due to the low solubility of 1 in CHCl3, as much as 50% of available 1 remained in the aqueous phase during the partitioning step (Fig. 5S-A, Supporting Information). Re-partitioning of the residual aqueous phase after a CHCl3 partition indicated that ethyl acetate was a good solvent to replace CHCl3 with for partitioning extracts (Figs. 5S-B and 5S-C, Supporting Information). Extraction of rice cultures using ethyl acetate was examined due to the higher solubility of 1 in this solvent; however, the ethyl acetate had the same aqueous miscibility issue as CHCl3, leading to a reduced penetration of the solvent into the rice media and the reduction in the recovery of 1 from the rice media. The final choice of acetone as the extraction solvent led to a higher solubility of 1, complete miscibility with water, allowing for more thorough extraction of the solid rice media, and the use of a greener (i.e. more environmentally friendly) solvent than CHCl3.
Extractions with acetone were completed within 15 min of adding this solvent to the solid rice culture (Fig. 6S, Supporting Information). We believe that this increase in speed (15 min vs. 3–12 h) was due to the ability of acetone to penetrate deeply into the water-rich rice media. Filtrations occurred quickly with the addition of Celite. Any orange/red coloration, which was indicative of the unextracted polyhydroxyanthraquinones, remaining in the residual pellet on the filter was re-extracted with acetone, for a total of two to three rounds (Figs. 6S-E and 6S-F, Supporting Information). A fourth round of acetone extraction upon the pellet yielded no further compounds of interest (Fig. 6S-G, Supporting Information). The final pellet was gray, indicating recovery of available polyhydroxyanthraquinones from the fungal culture.
Modification of partitioning methods:
Due to the more thorough extraction by acetone including very polar sugars and peptides, which had also been scaled up with enhanced production of polyhydroxyanthraquinones [38], the filtrate (90% acetone, 10% H2O) was passed through an open silica column (in vacuo) as an additional clean up step. Without this additional step, the following ethyl acetate partitioning step led to difficulties in partitioning and further purification. However, with this additional step, a layer of polar molecules remained bonded to the top of the silica (giving the silica a consistency like that of fondant, see Fig. 6S-H, Supporting Information). The clarified acetone eluent was dried under vacuum until only the H2O from the rice remained. We observed that if the acetone was thoroughly removed, the ethyl acetate partition separated very rapidly with no emulsions and could be implemented with very small volumes of solvent. The final defatting step, utilizing 1:1 ACN:MeOH and hexanes, remained unchanged from previously reported methods [2], except that the solvent volumes used were a quarter that of previous work (due to the sample being cleaner at this step). Overall, this method used less solvent, was greener than the previous methods, and virtually all the solvent used was recycled. Based on these optimizations, the increased time taken at each step during chemical processing did not increase linearly with increasing sample sizes, allowing much larger samples (e.g. those grown with hundreds of grams of rice in mushroom spawn bags, 2.8-L Fernbach flasks, or numerous small flasks) to be extracted with only slightly more effort than what was used for standard laboratory-scale studies (i.e. those using 10 g of rice in a few 250-mL Erlenmeyer flasks).
Modification of automated flash chromatography purification procedures:
The improvements to the fermentation conditions led to higher percentages of ω-hydroxyemodin (1), and this fact aided in the purification steps. For example, extracts that contained 8% or greater content of 1 produced a fine crystalline precipitate during the initial round of automated flash chromatography, specifically in the tubes that corresponded to 1. Those fractions were left overnight to maximize crystal formation, followed by rinsing with CHCl3. Initial analysis indicated the crystals were compound 1, and the sample was determined to be >98% 1 after a further clean up with CHCl3 (Figs. 7S and 8S, Supporting Information). In short, the enhanced growth conditions facilitated the isolation of 1 at an earlier stage, greatly accelerating the purification process.
Building upon this observation, each extract was evaluated for content of 1 via UPLC (Fig. 9S, Supporting Information). Those extracts that were less than 8% of 1 were processed with an additional automated flash chromatography step (Fig. 10S, Supporting Information). A rapid stepwise gradient was utilized to concentrate the polyhydroxyanthraquinones by removing the non-polar and polar compounds. Compound 1 and the other polyhydroxyanthraquinones were observed previously to elute between 4 and 10% MeOH in CHCl3. These combined fractions with greater than 8% of 1 were passed through the automated flash chromatography method (discussed above) to encourage precipitation of 1.
Modifying High Performance Liquid Chromatography:
A portion of 1 was isolated from the initial flash silica chromatography step, but that did not result in the isolation of all of the 1 present, especially in extracts/fractions with less than 8% of 1. Normal-phase HPLC methods were developed because previous studies using reverse-phase HPLC led to a substantial loss of mass on column due to the insolubility of 1 in H2O. There were still some solubility issues in CHCl3 that led to post column precipitation, but this was overcome using a make-up flow pump that was teed in directly after the column to infuse MeOH at 5 mL/min during preparative separations. This addition of MeOH prevented precipitation of 1 in the post-column flow lines and PDA detector, solving challenges that would otherwise lead to failed runs. This procedure had the added benefit of affording higher loading of the column for more efficient separations. In addition, utilization of DMSO as an injection solvent allowed for higher injection loadings (100 mg/200 μL), since 1 is approximately five times more soluble in DMSO than dioxane, which was the next best injection solvent used.
The isolates of 1 via prep-HPLC appeared clean by UPLC but demonstrated up to 30% fatty contaminants by NMR. These fatty contaminants were removed by re-suspension of 1 as a colloidal suspension in CHCl3, pelleting by centrifugation, and drawing off the supernatant. After three rounds of clean up, the fatty contaminant was removed with a small (~9%) loss of 1. For example, an 89 mg sample of 30% fat-contaminated 1 was decreased in weight to 57 mg of >98% pure 1 after this cleanup process (i.e. 9% loss of 1 during cleanup).
In summary, we increased the production of ω-hydroxyemodin (1) from < 20 mg to 850 mg within three months. We did so by utilizing a greener solvent (i.e. more environmentally friendly) for extraction (acetone rather than CHCl3), and a greener solvent for the partition step (ethyl acetate rather than CHCl3). We also designed a new method of extraction and purification that was superior to the methodology employed when 1 was isolated previously [2], taking advantage of the economies of scale. Modern instrumentation was used to evaluate the biosynthesis of secondary metabolites, specifically the droplet probe [16], so as to rapidly evaluate in situ different solid media to determine the most appropriate nutrient media for seed culture, thus optimizing fermentation conditions for laboratory-scale production of the target compound; this is the second example of the value of this tool for this purpose [29]. The benefits obtained from using the improved methods are summarized (Figs. 4, 11S, and 12S, Supporting Information). A step-by-step protocol showing these methods in use on a real sample (12 Erlenmeyer flasks of 10 g cultures) is available as Protocol 1S (Supporting Information). We believe an added benefit may be the generation of larger quantities of analogues of 1 that would otherwise be below the limit of detection during a traditional natural products investigation; studies to examine this point are ongoing.
Fig. 4.
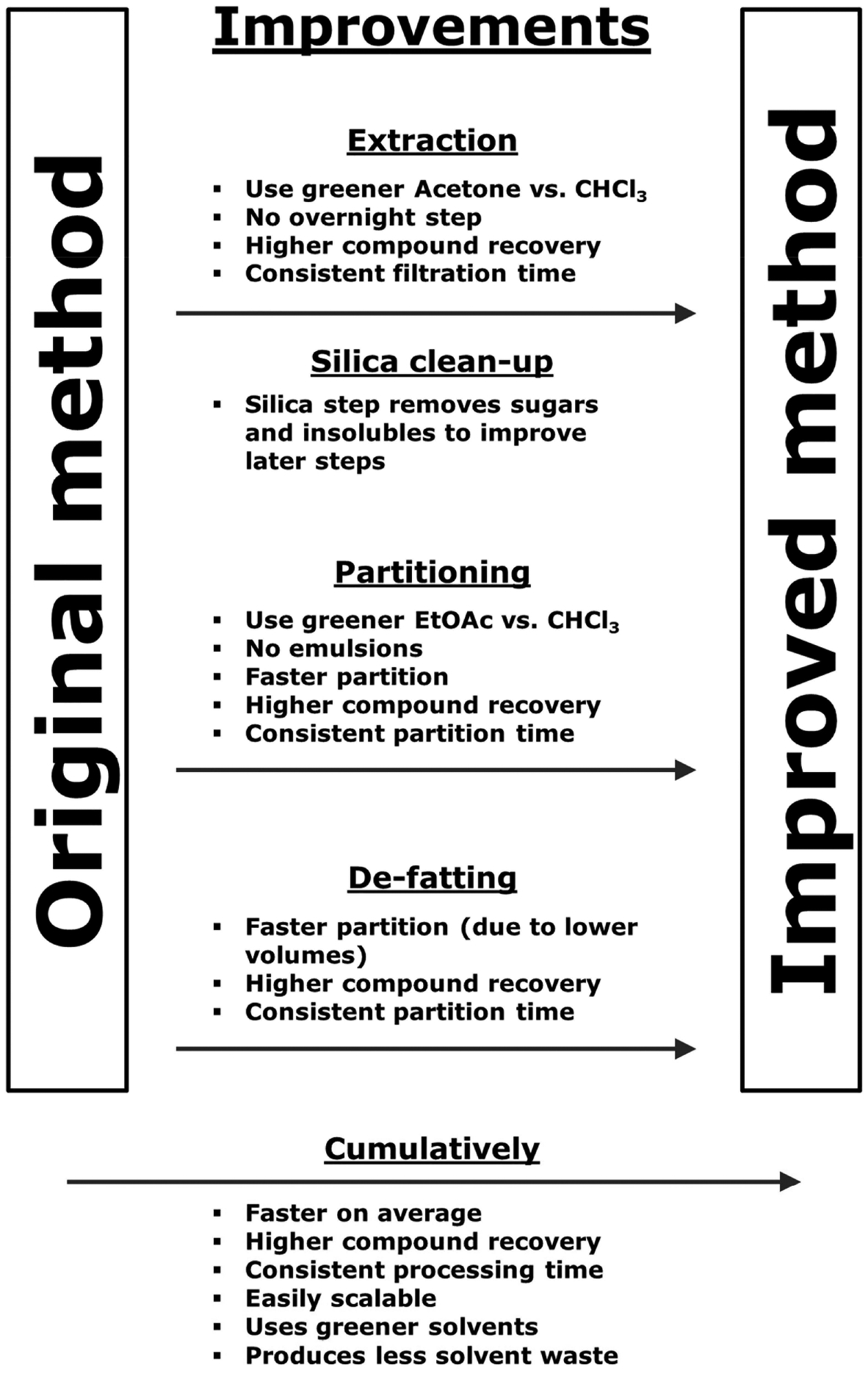
Schematic of the benefits obtained with the improved method. Benefits at each stage of processing are shown as a summary at the bottom.
Materials and Methods
Media and fermentation
All Media (solid and liquid) were prepared following the manufacturers’ instructions. For solid (agar-based) media, PDA, MEA, and Sabouraud dextrose agar (SD Agar; Difco) were all utilized. The G85 fungal strain, on PDA and MEA media was grown under three different pH conditions: 5, 8, and 11, respectively using methods outlined in earlier studies [39,40]. Further, MEA media was supplemented with L-cysteine [40] and G85 was grown in both light and dark conditions at pH 5, and 8, respectively. Strain G85 was grown on SD Agar media, which was supplemented with 20% (0.25 g/L), 40% (0.5 g/L), 60% (0.75 g/L), 80% (1 g/L), and 100% (1.25 g/L) peptone (Difco) and allowed to grow in both light and dark conditions. Finally, strain G85 was examined on SD Agar media made with tap H2O and 50, 75, and 100% deuterated H2O, both under light and dark conditions, respectively.
Liquid broths used for growing strain G85 included: potato dextrose broth (PDB; Difco), malt extract broth (MEB; Difco), 1:1 PDB:MEB, and Czapek Dox broth (CDB; Honeywell Fluka). Strain G85 was grown on PDB and MEB at pH 5, 8, or 11, (Fig. 2S, Supporting Information).
Analysis of fungal cultures in situ via the droplet probe
Monitoring the biosynthesis of secondary metabolites in agar-based fungal cultures in situ was performed using the droplet probe coupled with a Waters Acquity ultra-performance liquid chromatography (UPLC) system (Waters Corporation) to a Thermo QExactive Plus (Thermo Fisher Scientific) via procedures described previously [16,26–29] and reviewed recently [16]. This technique probes the chemistry of the cultures via a mini-extraction of about 2 μL of 1:1 MeOH:H2O, permitting the mapping of secondary metabolites in live fungal cultures [26–28,41].
Screener scale solid-state fermentations
All fungal cultures were grown by solid-state fermentation in screener scale conditions (10 g of grain) using Calrose Botan rice, Kokuho Rose rice, Cheerios, breakfast Oatmeal (old fashioned Quaker oats), or 1:1 Calrose Botan:Indian (Udupi Sona Masoori) rice, respectively in 250-mL Erlenmeyer flasks. Two different seed cultures, Sabouraud dextrose broth (SD Broth) and YESD broth, were utilized to inoculate each of the solid media (Fig. 4S, Supporting Information). The screener scales were grown to determine the most suitable grain-based substrate for scale-up.
Large scale solid-state fermentations
To prepare large-scale solid-state fermentation cultures, 50 mL of SD Broth seed cultures were initiated by cutting a piece of agar with fungal mycelium from the leading edge of a 14-day old Petri dish agar culture of strain G85 grown on SD Agar. Following incubation (7 days) at 22°C with agitation, the SD Broth seed culture was used to inoculate Calrose Botan rice. To generate a large amount of 1 rapidly, grain-based fermentation cultures were started in different sizes for scale-up, encompassing small and large sterile mushroom spawn bags, 250-mL Erlenmeyer flasks, and 2.8-L Fernbach flasks. For smaller fermentations, 10 g of Calrose Botan rice was prepared by adding 10 g of rice to a 250-mL flask with 20 mL of distilled H2O, followed by autoclaving for 30 min and inoculated with 10 mL of SD Broth seed culture. For larger fermentations, 100 g of Calrose Botan rice was autoclaved with 200 mL of distilled H2O in a Fernbach flask and inoculated with 50 mL of SD Broth seed culture; 150 g of rice was autoclaved with 300 mL of distilled H2O in small spawn bags and inoculated with 75 mL of seed culture; while 1000 g of rice were autoclaved with 2000 mL of distilled H2O in large spawn bags and inoculated with 200 mL of SD Broth seed culture. All cultures were grown for approximately 3–5 weeks before extraction. During our study, we observed that the deepest orange/red color for SD Broth cultures on Calrose Botan rice was seen at 4–5 weeks. With the deepness of orange/red color correlating to the relative concentration of 1, cultures were grown for 4–5 weeks to obtain the highest yield of the target compound.
Chemistry: Extraction and isolation of the target compound
All solvents used for HPLC and automated flash chromatography were HPLC grade. Solvents used for extractions, partitions, and open column fractionations were of ACS grade or better. All solvents were obtained from Thermo Fisher Scientific or PHARMCO-AAPER (Brookfield, CT). Filtrations during extraction steps were performed using Whatman type 1 qualitative filter paper via a Büchner funnel.
Extraction and filtration using chloroform and methanol
Solid-phase rice cultures of 100–150 g (large scale) were extracted using 400 mL of 1:1 CHCl3:MeOH. Alternatively, for solid-phase rice cultures of 10 g (small scale), 60 mL of 1:1 CHCl3:MeOH was used. All extraction mixtures were shaken overnight at room temperature, filtered on a Büchner funnel, and rinsed with a small amount of CHCl3 and MeOH. Some later samples had Celite added (10–15 g per 10 g of rice culture) to hasten filtration.
Extraction and filtration using ethyl acetate or acetone
As the fermentation and extraction methods were scaled, it became clear that the extraction methods needed to be changed to be more environmentally conscious.
Several solid-phase rice cultures totaling 500 g were extracted using 1.5 L of ethyl acetate. The extraction soaked overnight at room temperature, 150 g of Celite was added, and the sample was filtered on a Büchner funnel. The solids on the filter were resuspended in 1 L of MeOH and filtered again. The combined effluent was dried to a volume of 200 mL.
As a further improvement, solid-phase rice cultures of various sizes were mixed with Celite (15 g per 10 g of rice culture), transferred to a large mixing bowl, and acetone was added until the mixture had the consistency of wet cement (Fig. 6S-D, Supporting Information). Samples were soaked in acetone for 15 min at room temperature before being filtered using a Büchner funnel. The pellet in the funnel was transferred back into the mixing bowl, acetone added to the same consistency as before, and the sample was re-filtered using the Büchner funnel. This resuspension and filtration of the pellet was repeated two or three times until most of the orange color of the pellet was extracted out.
Extract cleanup using an open silica column
Acetone extracts were passed through a layer of silica (~500 g of silica per 100 g of rice culture) pre-wetted with acetone on a glass-fritted vacuum filter funnel (open flash chromatography), and the silica was washed with fresh acetone until little color eluted. The effluent was evaporated under a mild vacuum (~120 mbar) to recover much of the acetone for reuse.
Partitioning using chloroform and water
The 1:1 CHCl3:MeOH extract was transferred to a separatory funnel and mixed with 600 mL of CHCl3 and 2.5 L of H2O. The combined mixture was partitioned, and the CHCl3 layer was collected. The methanolic aqueous layer was repartitioned with 600 mL of CHCl3 two or three more times. The combined CHCl3 layers were dried completely, recovering the evaporated CHCl3 for recycling.
Partitioning of aqueous layers using ethyl acetate and water
Residual methanolic aqueous layers from the CHCl3 partitions were observed to contain some ω-hydroxyemodin (1) (Fig. 5S-A, Supporting Information). These methanolic aqueous layers were evaporated to remove the MeOH and then re-partitioned between ethyl acetate and H2O (250 mL of each layer per 40 g of rice culture). The aqueous layer was repartitioned with half the amount of fresh ethyl acetate two to three more times until the organic phase lost color.
Partitioning of extracts using ethyl acetate and water
Extracts from any of the extraction techniques (CHCl3:MeOH or acetone) were evaporated to remove all organic solvent (recovering much of the organic solvent for recycling) before partitioning between ethyl acetate and H2O (100–200 mL of each layer per 100 g of rice culture). The aqueous layer was repartitioned with half the initial amount of fresh ethyl acetate two to three more times (until the organic phase lost color).
Partitioning using acetonitrile and methanol versus hexanes
All CHCl3 and ethyl acetate partition products were evaporated completely (recovering the organic solvent for recycling) before partitioning between 1:1 ACN:MeOH and hexanes. Partition volumes were 50–100 mL of each phase per 100 g of rice culture. The hexanes layer was partitioned against 40–80 mL of fresh 1:1 ACN:MeOH twice more. The defatted ACN:MeOH layers were combined and evaporated completely (recovering the solvent for reuse). Hexanes layers from numerous partitions were combined and evaporated to recover the solvent for reuse.
Sample purity analysis by UPLC
Samples were analyzed using a Waters Acquity ultra-performance liquid chromatography (UPLC) system (Waters Corporation) using an Acquity BEH-shield column (2.1 × 50 mm, 1.7 μm) running a gradient elution from 10% ACN in H2O (0.1% formic acid) to 100% ACN over 4.5 minutes at a flow rate of 0.4 mL/min, a column temperature of 40°C, and observing at 288 nm. Using a reference standard of 1, a calibration curve was created for its measurement in various extracts and fractions (Fig. 9S, Supporting Information).
Automated flash silica chromatography (linear gradient method)
Samples were separated on a Teledyne ISCO CombiFlash Rf 200 using RediSep RF Gold HP Silica columns (both from Teledyne ISCO, Lincoln, NE) and chromatography monitored by UV at 288 nm and with ELSD. Defatted extracts from 2 to 4 g were separated using an 80-g gold silica RediSep column running isocratic at 100% CHCl3 for 3 column volumes, then a ramped gradient from 0 to 10 % MeOH in CHCl3 over 11 column volumes, then to 100 % MeOH over 1 column volume and washing with 100 % MeOH for 2 column volumes. ω-Hydroxyemodin (1) eluted at 6–8 % MeOH, and for isolations of extracts with > 8% w/w content of 1, crystals were observed in fractions containing it. For extracts with < 8% w/w content of 1, no crystals were observed. After allowing the crystal containing fractions to precipitate overnight, the fractions containing crystals were filtered and subsequently washed with CHCl3.
Automated flash silica chromatography (step gradient method)
Samples were separated on the CombiFlash Rf 200 using RediSep Gold Silica columns (as above). Polyhydroxyanthraquinones from extracts with less than 8% ω-hydroxyemodin content were isolated using an 80-g gold silica RediSep column running isocratic at 100% CHCl3 for 3 column volumes, isocratic at 15 % MeOH in CHCl3 for 3 column volumes and washing at 100 % MeOH for 3 column volumes. Polyhydroxyanthraquinones eluted between 4.5 and 5.5 column volumes (Fig. 10S, Supporting Information).
HPLC purifications
HPLC separations were carried out on a Varian ProStar HPLC system equipped with ProStar 210 pumps, a ProStar 701 fraction collector, a ProStar 335 diode array detector (DAD), and Galaxie Chromatography Workstation software (version 1.9.3.2, Varian Inc.). Separations were monitored by UV absorbance at 288 nm. Semi-preparative HPLC separations were carried out using a Phenomenex Luna 5 μm particle size Silica (2) column (10 × 250 mm) at a flow rate of 4.7 mL/min. Preparative HPLC separations were carried out using a Phenomenex Luna 5 μm particle size Silica (2) column (21.2 × 250 mm) at a flow rate of 21.2 mL/min. All semi-preparative and preparative separations were executed utilizing a gradient from 0:100 to 90:10 of A:B, where A = 1:9 MeOH:CHCl3 and B = CHCl3.
Chloroform cleanup
For semi-pure ω-hydroxyemodin (1), the sample was resuspended in CHCl3 (1–2 mL per 800 mg of the sample), vortexed and sonicated, then centrifuged to produce a pellet. This procedure was repeated several times while monitoring the purity of the pellet and supernatant by UPLC to determine completeness.
Final product analysis
NMR experiments were conducted in DMSO-d6 on a JEOL ECA-500 NMR spectrometer operating at 500 MHz for 1H and 125 MHz for 13C (JEOL USA, Inc., Peabody, MA, USA) (Fig. 8S, Supporting Information). HRESIMS data were obtained using a Thermo QExactive Plus mass spectrometer (ThermoFisher Scientific) paired with an electrospray ionization source.
Supplementary Material
Acknowledgements
Diana Kao was supported by the National Center for Complementary and Integrated Health, NIH via grant F31 AT009264. The mass spectrometry data were acquired in the Triad Mass Spectrometry Facility.
Footnotes
Supporting information
Structures of the polyhydroxyanthraquinones, photographs of various media studies, chromatographic analysis of samples, NMR data, schematics that outline the purification protocols, and an example extraction procedure.
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Raja HA, Kaur A, El-Elimat T, Figueroa M, Kumar R, Deep G, Agarwal R, Faeth SH, Cech NB, Oberlies NH. Phylogenetic and chemical diversity of fungal endophytes isolated from Silybum marianum (L) Gaertn. (milk thistle). Mycology 2015; 6: 8–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Figueroa M, Jarmusch AK, Raja HA, El-Elimat T, Kavanaugh JS, Horswill AR, Cooks RG, Cech NB, Oberlies NH. Polyhydroxyanthraquinones as quorum sensing Inhibitors from the guttates of Penicillium restrictum and their analysis by desorption electrospray ionization mass spectrometry. J Nat Prod 2014; 77: 1351–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daly SM, Elmore BO, Kavanaugh JS, Triplett KD, Figueroa M, Raja HA, El-Elimat T, Crosby HA, Femling JK, Cech NB, Horswill AR, Oberlies NH, Hall PR. ω-Hydroxyemodin limits Staphylococcus aureus quorum sensing-mediated pathogenesis and inflammation. Antimicrob Agents Chemother 2015; 59: 2223–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spellberg B, Bartlett JG, Gilbert DN. The future of antibiotics and resistance. New Eng J Med 2013; 368: 299–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cech NB, Horswill AR. Small-molecule quorum quenchers to prevent Staphylococcus aureus infection. Future Microbiol 2013; 8: 1511–1514 [DOI] [PubMed] [Google Scholar]
- 6.Parlet CP, Kavanaugh JS, Crosby HA, Raja HA, El-Elimat T, Todd DA, Pearce CJ, Cech NB, Oberlies NH, Horswill AR. Apicidin attenuates MRSA virulence through quorum-sensing inhibition and enhanced host defense. Cell Rep 2019; 27: 187–198. e186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parlet CP, Brown MM, Horswill AR. Commensal staphylococci influence staphylococcus aureus skin colonization and disease. Trends Microbiol 2019, DOI: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lanman BA. Addressing supply issues for natural products in the clinic. Science 2017; 358: 166–167 [DOI] [PubMed] [Google Scholar]
- 9.Sudhakar T, Dash S, Rao R, Srinivasan R, Zacharia S, Atmanand M, Subramaniam B, Nayak S. Do endophytic fungi possess pathway genes for plant secondary metabolites? Curr Sci 2013; 104: 178 [Google Scholar]
- 10.Liang JL, Cha HC, Lee SH, Son J-K, Chang HW, Eom J-E, Kwon Y, Jahng Y. A facile synthesis of emodin derivatives, emodin carbaldehyde, citreorosein, and their 10-deoxygenated derivatives and their inhibitory activities on μ-calpain. Arch Pharm Res 2012; 35: 447–454 [DOI] [PubMed] [Google Scholar]
- 11.Jacobson R, Adams R. Trihydroxy-methylanthraquinones. III. Synthesis of emodin. J Am Chem Soc 1924; 46: 1312–1316 [Google Scholar]
- 12.Teich L, Daub KS, Krügel V, Nissler L, Gebhardt R, Eger K. Synthesis and biological evaluation of new derivatives of emodin. Bioorg Med Chem 2004; 12: 5961–5971 [DOI] [PubMed] [Google Scholar]
- 13.Anslow WK, Breen J, Raistrick H. Studies in the biochemistry of micro-organisms: Emodic acid (4: 5: 7-trihydroxyanthraquinone-2-carboxylic acid) and ω-hydroxyemodin (4: 5: 7-trihydroxy-2-(hydroxymethyl)-anthraquinone), metabolic products of a strain of Penicillium cyclopium Westling. Biochem J 1940; 34: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frisvad JC, Smedsgaard J, Larsen TO, Samson RA. Mycotoxins, drugs and other extrolites produced by species in Penicillium subgenus Penicillium. Stud Mycol 2004; 49: 201–241 [Google Scholar]
- 15.Frisvad JC, Samson RA. Polyphasic taxonomy of Penicillium subgenus Penicillium. A guide to identification of food and air-borne terverticillate Penicillia and their mycotoxins. Stud Mycol 2004; 49: 1–174 [Google Scholar]
- 16.Oberlies NH, Knowles Sonja L, Amrine CSM, Kao D, Kertesz V, Raja HA. Droplet probe: Coupling chromatography to the in situ evaluation of the chemistry of nature. Nat Prod Rep 2019; 36: 944–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El-Elimat T, Raja HA, Figueroa M, Falkinham III JO, Oberlies NH. Isochromenones, isobenzofuranone, and tetrahydronaphthalenes produced by Paraphoma radicina, a fungus isolated from a freshwater habitat. Phytochem 2014; 104: 114–120 [DOI] [PubMed] [Google Scholar]
- 18.Lax E The mold in Dr. Florey’s coat: the story of the penicillin miracle. In: New York: Henry Holt; 2005 [Google Scholar]
- 19.Raja HA, Miller AN, Pearce CJ, Oberlies NH. Fungal identification using molecular tools: A primer for the natural products research community. J Nat Prod 2017; 80: 756–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El-Elimat T, Raja HA, Graf TN, Faeth SH, Cech NB, Oberlies NH. Flavonolignans from Aspergillus iizukae, a fungal endophyte of milk thistle (Silybum marianum). J Nat Prod 2014; 77: 193–199 [DOI] [PubMed] [Google Scholar]
- 21.Kusari S, Zühlke S, Spiteller M. An endophytic fungus from Camptotheca acuminata that produces camptothecin and analogues. J Nat Prod 2009; 72: 2–7 [DOI] [PubMed] [Google Scholar]
- 22.Ludwig-Müller J Plants and endophytes: Equal partners in secondary metabolite production? Biotechnol Lett 2015; 37: 1325–1334 [DOI] [PubMed] [Google Scholar]
- 23.Puri SC, Verma V, Amna T, Qazi GN, Spiteller M. An endophytic fungus from Nothapodytes foetida that produces camptothecin. J Nat Prod 2005; 68: 1717–1719 [DOI] [PubMed] [Google Scholar]
- 24.Hare JM. Sabouraud agar for fungal growth In, Laboratory Protocols in Fungal Biology: Springer; 2013: 211–216 [Google Scholar]
- 25.Sica VP, Figueroa M, Raja HA, El-Elimat T, Darveaux BA, Pearce CJ, Oberlies NH. Optimizing production and evaluating biosynthesis in situ of a herbicidal compound, mevalocidin, from Coniolariella sp. J Ind Microbiol 2016; 43: 1149–1157 [DOI] [PubMed] [Google Scholar]
- 26.Sica VP, Raja HA, El-Elimat T, Kertesz V, Van Berkel GJ, Pearce CJ, Oberlies NH. Dereplicating and spatial mapping of secondary metabolites from fungal cultures in situ. J Nat Prod 2015; 78: 1926–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sica VP, Rees ER, Raja HA, Rivera-Chávez J, Burdette JE, Pearce CJ, Oberlies NH. In situ mass spectrometry monitoring of fungal cultures led to the identification of four peptaibols with a rare threonine residue. Phytochem 2017; 143: 45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sica VP, Rees ER, Tchegnon E, Bardsley RH, Raja HA, Oberlies NH. Spatial and temporal profiling of griseofulvin production in Xylaria cubensis using mass spectrometry mapping. Front Microbiol 2016; 7: 1126–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amrine CSM, Raja HA, Darveus BA, Pearce CJ, Oberlies NH. Media studies to enhance the production of verticillins facilitated by in situ chemical analysis. J Ind Microbiol 2018; 45: 1053–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rivera-Chávez J, Raja HA, Graf TN, Burdette JE, Pearce CJ, Oberlies NH. Biosynthesis of fluorinated peptaibols using a site-directed building block incorporation approach. J Nat Prod 2017; 80: 1883–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rivera-Chávez J, Raja HA, Graf TN, Gallagher JM, Metri P, Xue D, Pearce CJ, Oberlies NH. Prealamethicin F50 and related peptaibols from Trichoderma arundinaceum: Validation of their authenticity via in situ chemical analysis. RSC Advances 2017; 7: 45733–45741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amrine CSM, Long JL, Raja HA, Kurina SJ, Burdette JE, Pearce CJ, Oberlies NH. Engineering fluorine into verticillins (Epipolythiodioxopiperazine alkaloids) via precursor-directed biosynthesis. J Nat Prod 2019; 82: 3104–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paguigan ND, Raja HA, Day CS, Oberlies NH. Acetophenone derivatives from a freshwater fungal isolate of recently described Lindgomyces madisonensis (G416). Phytochem 2016; 126: 59–65 [DOI] [PubMed] [Google Scholar]
- 34.Knowles SL, Raja HA, Wright AJ, Lee AML, Caesar LK, Cech NB, Mead ME, Steenwyk JL, Ries L, Goldman GH. Mapping the fungal battlefield: Using in situ chemistry and deletion mutants to monitor interspecific chemical interactions between fungi. Front Microbiol 2019; 10: 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knowles SL, Vu N, Todd DA, Raja HA, Rokas A, Zhang Q, Oberlies NH. Orthogonal method for double-bond placement via ozone-induced dissociation mass spectrometry (OzID-MS). J Nat Prod 2019; 82: 3421–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vandermolen KM, Raja HA, El-Elimat T, Oberlies NH. Evaluation of culture media for the production of secondary metabolites in a natural products screening program. AMB Express 2013; 3: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bills GF, Dombrowski AW, Goetz MA. The “FERMEX” method for metabolite-enriched fungal extracts In: Keller NP, Turner G eds, Fungal Secondary Metabolism: Methods and Protocols: Springer; 2012: 79–96 [DOI] [PubMed] [Google Scholar]
- 38.Kumarasamy Y Scaling-Up of Natural Products Isolation In, Natural Products Isolation: Springer; 2012: 465–472 [DOI] [PubMed] [Google Scholar]
- 39.Naga K, Suzuk K, Okada G. Studies on the distribution of alkalophilic and alkali-tolerant soil fungi II: Fungal flora in two limestone caves in Japan. Mycoscience 1998; 39: 293–298 [Google Scholar]
- 40.Raudabaugh DB, Miller AN. Nutritional capability of and substrate suitability for Pseudogymnoascus destructans, the causal agent of bat white-nose syndrome. PLoS One 2013; 8: e78300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kertesz V, Van Berkel GJ. Liquid microjunction surface sampling coupled with high-pressure liquid chromatography− electrospray ionization-mass spectrometry for analysis of drugs and metabolites in whole-body thin tissue sections. Anal Chem 2010; 82: 5917–5921 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


